Abstract
Weedy rice is a valuable germplasm resource characterized by its high tolerance to both abiotic and biotic stresses. Abscisic acid (ABA) serves as a regulatory signal in plant cells as part of their adaptive response to stress. However, a global understanding of the response of weedy rice to ABA remains to be elucidated. In the present study, the sensitivity to ABA of weedy rice (WR04-6) was compared with that of temperate japonica Shennong9816 (SN9816) in terms of seed germination and post-germination growth via the application of exogenous ABA and diniconazole, an inhibitor of ABA catabolism. Physiological analysis and a transcriptomic comparison allowed elucidation of the molecular and physiological mechanisms associated with continuous ABA and diniconazole treatment. WR04-6 was found to display higher ABA sensitivity than SN9816, resulting in the rapid promotion of antioxidant enzyme activity. Comparative transcriptomic analyses indicated that the number of differentially expressed genes (DEGs) in WR04-6 seedlings treated with 2 μM ABA or 10 μM diniconazole was greater than that in SN9816 seedlings. Genes involved in stress defense, hormone signal transduction, and glycolytic and citrate cycle pathways were highly expressed in WR04-6 in response to ABA and diniconazole. These findings provide new insight into key processes mediating the ABA response between weedy and cultivated rice.
Subject terms: Gene expression analysis, Gene expression profiling, Developmental biology, Physiology, Plant sciences
Introduction
Weedy rice (Oryza. sativa f. spontanea) is taxonomically classified as the same species as cultivated rice (Oryza. sativa L.)1 and is among the most persistent and problematic weeds found in paddy fields worldwide. The phenotypic characteristics of weedy rice are highly variable and appear to be intermediate between wild rice and cultivated rice, retaining the characteristics of both the wild relatives (black hull, long awn, red pericarp, and seed shattering) and the developed crop mimicries (plant height and heading time), enhancing their adaptation and competitiveness in the agroecosystem2. Weedy rice is considered a valuable germplasm resource because of its enhanced tolerance to deep sowing3, higher nitrogen use efficiency4, greater photosynthetic activity5, and resistance to blast disease6 compared with cultivated rice. Furthermore, weedy rice displays significant versatility in its adaption to a wide range of adverse conditions. It has many useful stress-related genes that respond to biotic and abiotic stresses7. There are weedy rice genotypes that tolerate short-term low temperature8,9, high salinity, and drought stress10 at the seedling stage owing to excellent stress adaptation mechanisms. Crop domestication and improvement have dramatically reduced genetic variability by 50–60% in comparison with ancestral wild species11, therefore, weedy rice provides the possibility to widen the gene pool of cultivated rice and breed cultivars with improved yield, quality, and stress tolerance.
To adapt and survive in adverse conditions, plants have evolved and established mechanisms that are plastic at multiple levels, including molecular, physiological, and developmental levels, which permit them to cope with unfavorable environments12,13. Abscisic acid (ABA) is generally considered a stress hormone, and its biosynthesis plays a role in rapid metabolic adjustment in response to stress14. The expression of stress-inducible genes in plants is primarily regulated by both ABA-dependent and ABA-independent pathways15. When plants encounter stress conditions, endogenous ABA rapidly accumulates. ABA receptors (PYR/RCARs) sense ABA and bind to subclass A protein type 2C phosphatases (PP2Cs), resulting in the activation of SNF-related protein kinases (SnRK2s)16. Activated SnRK2s are able to phosphorylate ABRE binding factor (ABF) transcription factors, which in turn promote the expression of ABA-dependent genes17. The ABRE-binding proteins/AREB-binding factors (AREB/ABFs) transcription factors belong to an ABA-responsive gene family. Of these, AREB1/ABF2, AREB2/ABF4, and ABF3 are induced by dehydration, high salinity, or ABA treatment during the vegetative stage18, and overexpression of AREB/ABFs in transgenic Arabidopsis displays enhanced drought tolerance via an ABA-dependent pathway19. Additional signaling factors associated with the principal ABA signaling pathway have also been identified. Stress-inducible SlMYB102 and GhMYB73 belonging to the R2R3 MYB family enhance salt tolerance in transgenic tomatoes and Arabidopsis, respectively20,21. Dehydration-responsive element-binding (DREB) transcription factors play a crucial role in plant growth, development, and stress responses. They function as components of both ABA-dependent and ABA-independent pathways in response to abiotic stress22. In rice, DREB1A, DREB1B, and DREB1C regulate plant cold tolerance by recognizing the GCC box of the target gene23. DREB2A and DREB2B become highly induced by dehydration and salinity, and overexpression of soybean GmDREB2 in Arabidopsis enhances tolerance to salinity24.
ABA catabolism is an alternative method of regulating ABA levels. ABA is primarily catabolized to form 8ʹ-hydroxy ABA through hydroxylation by ABA 8ʹ-hydroxylase and final rearrangement to phaseic acid (PA)25. ABA 8′-hydroxylase is presumed to be a rate-limiting enzyme in the ABA catabolic pathway. Studies have shown that ABA catabolism under stress and recovery is principally regulated at the transcriptional level of the ABA 8ʹ-hydroxylase gene. In Arabidopsis, there are four genes (AtCYP707A1-4) that encode ABA 8ʹ-hydroxylase. The expression of the four CYP707As is induced by dehydration stress and subsequent rehydration26. Of the three ABA 8ʹ-hydroxylase genes (OsABA8ox1-3) in rice, the expression of OsABA8ox1 is induced by cold stress27. Overall, ABA 8ʹ-hydroxylase plays an important role in controlling ABA homeostasis when experiencing abiotic stress.
The regulation of ABA in abiotic stress in plants has been extensively studied, while ABA-associated regulatory networks in response to the accumulation of ABA in weedy rice remain elusive. In the present study, the sensitivity of weedy and cultivated rice to ABA during seed germination and post-germination growth was compared following the administration of exogenous ABA and diniconazole, an inhibitor of ABA catabolism. Physiological analysis was combined with a comparative transcriptomic investigation to elucidate the molecular and physiological mechanisms associated with continuous ABA or diniconazole treatment. The present study aimed to identify differentially expressed genes and pathways regulating the ABA response and provide deeper insights into the regulatory networks of ABA in weedy rice that could provide information for further crop improvements.
Results
Variation in ABA sensitivity during seed germination
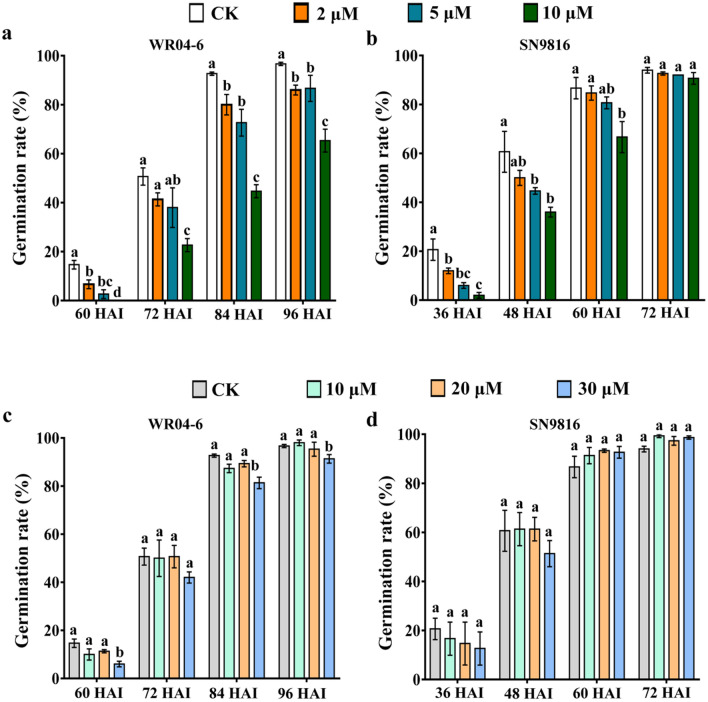
The effects of ABA on the germination of WR04-6 and SN9816 seeds after exposure to ABA and its catabolic inhibitor diniconazole were examined. Homogeneity of variance test (p > 0.05), frequency test (Skewness and Kurtosis values were less than 1), and single-sample K–S test (Z and P values were more than 0.05) showed that the phenotypic data were in line with normal distribution, and the results of ANOVA indicated the significant difference on germination rate of WR04-6 and SN9816 after ABA and diniconazole treatment. The WR04-6 seeds began to germinate 60 h after imbibition (HAI) and reached a 100% germination rate at 96 HAI under normal conditions. The proportion of germinated seeds decreased significantly when seeds were treated with ABA in a concentration-dependent manner (Fig. 1a). At 10 μM ABA, the seed germination rate was only 65% at 96 HAI. Conversely, SN9816 seeds were capable of germination at all ABA concentrations, although slightly delayed, indicating that the SN9816 seeds were insensitive to ABA relative to WR04-6 seeds (Fig. 1b).
Figure 1.
Germination rate (%) of WR04-6 and SN9816 seeds for different concentrations of ABA or diniconazole. (a) Germination rate (%) of WR04-6 seeds after ABA treatment. (b) Germination rate (%) of SN9816 seeds after ABA treatment. (c) Germination rate (%) of WR04-6 seeds after diniconazole treatment. (d) Germination rate (%) of SN9816 seeds after diniconazole treatment. Different letters above the bars indicate significant differences in the germination rate for different concentrations of ABA or diniconazole (one-way ANOVA. p < 0.05). Error bars represent SEs (n = 3).
Diniconazole treatment increased the ABA content in seeds by inhibiting ABA catabolism28. Compared with untreated WR04-6 seeds, low concentrations of inhibitor (10 μM or 20 μM) had no significant effect on seed germination, but a relatively lower germination rate was observed with 30 μM diniconazole, suggesting that catabolism of ABA plays a role in seed germination (Fig. 1c). In contrast, there was no significant effect of treatment with diniconazole on the germination of SN9816 seeds (Fig. 1d).
Effects of ABA on post-germination growth
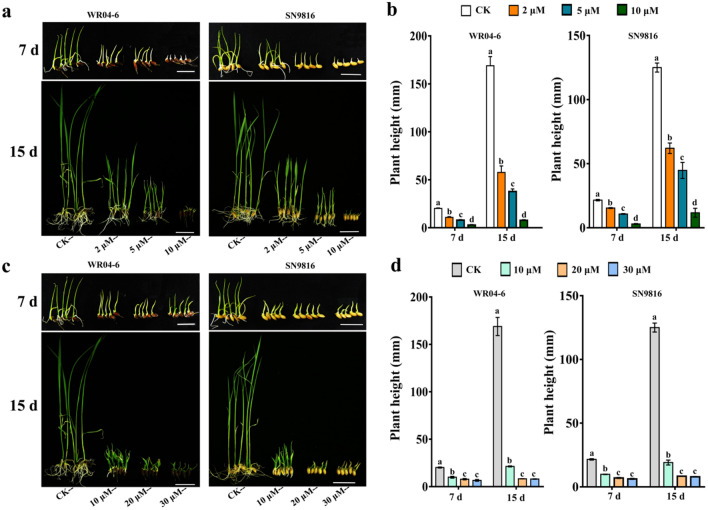
To further measure the inhibition of post-germination growth when seedlings were continuously exposed to ABA or diniconazole, the development of seedlings at 7 days and 15 days was investigated (Fig. 2). The height of WR04-6 plants grown hydroponically without treatment was approximately 169 mm, which was significantly higher than that of SN9816 after 15 days (125 mm, t < 0.05) (Fig. 2a, b). ABA increasingly retarded both WR04-6 and SN9816 plant development at increasing concentrations. Similarly, the height of WR04-6 and SN9816 plants decreased significantly with increasing diniconazole concentrations at 7 days and 15 days (Fig. 2c, d). Plant growth was completely inhibited in both WR04-6 and SN9816 plants when diniconazole concentrations in the medium increased to 20 μM and 30 μM. It should be noted that across different ABA and diniconazole concentrations, the degree of reduction in plant height for WR04-6 was much greater than that for SN9816 at 15 days.
Figure 2.
The growth and development of WR04-6 and SN9816 seedlings were retarded when they were continuously treated with ABA or diniconazole. (a) Growth of WR04-6 and SN9816 seedlings after 7 d and 15 d of continuous treatment with ABA. Bar = 2 cm. (b) Comparison of plant height (mm) of WR04-6 and SN9816 seedlings after treatment with different concentrations of ABA for 7 days and 15 days. (c) Growth of WR04-6 and SN9816 seedlings with continuous diniconazole treatment for 7 d and 15 d. Bar = 2 cm. (d) Comparison of plant height (mm) of WR04-6 and SN9816 seedlings after treatment with different concentrations of diniconazole for 7 and 15 days. Error bars represent SEs (n = 3). Different letters over the bars indicate treatments that are significantly different (p < 0.05).
Evaluation of oxidative stress and antioxidative enzyme activity
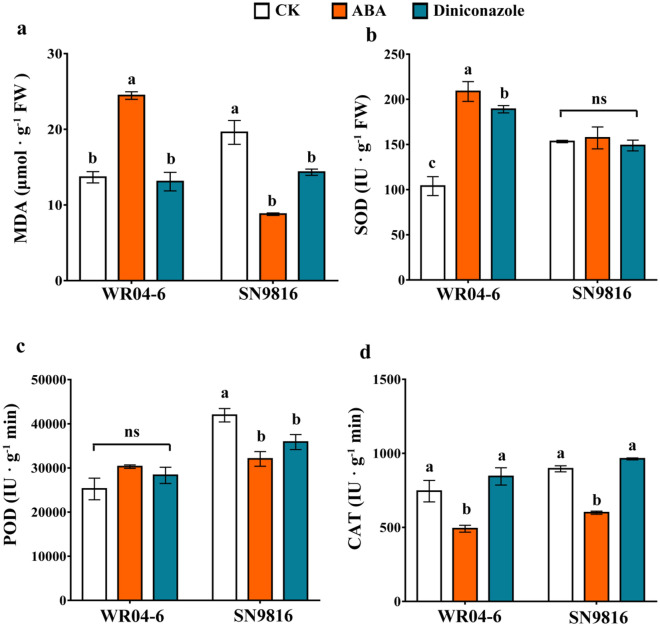
In preliminary experiments to determine the effective ABA and diniconazole concentrations for plant growth, seedlings were treated with 2 μM ABA and 10 μM diniconazole for the analysis of physiological indices. A significant accumulation of malondiadehyde (MDA), 1.79-fold greater than that of the control group, was observed in WR04-6 seedlings following ABA treatment (Fig. 3a), while diniconazole treatment did not induce MDA accumulation. The MDA concentration in unstressed SN9816 seedlings was higher than that in the WR04-6 control group. Both ABA and diniconazole resulted in a considerable decrease in the MDA concentration in SN9816 seedlings, which was 0.45- and 0.73-fold lower than that in the unstressed seedlings.
Figure 3.
Evaluation of MDA concentration and antioxidant enzyme activity in WR04-6 and SN9816 seedlings after treatment with 2 μM ABA or 10 μM diniconazole. (a) MDA content. (b) SOD activity. (c) POD activity. (d) CAT activity. Error bars represent SEs (n = 3). Different letters indicate significant differences between treatments. ns indicates no difference (p < 0.05).
The activity of the antioxidative enzymes SOD, POD, and CAT was evaluated after treatment with ABA and diniconazole. WR04-6 seedlings exposed to ABA and diniconazole experienced significantly increased SOD activity (Fig. 3b). The increased SOD activity resulting from ABA treatment was significantly greater than that after diniconazole treatment. However, there was no difference in SOD activity in the control compared with SN9816 seedlings treated with ABA or diniconazole. Similarly, POD activity in WR04-6 seedlings treated with ABA or diniconazole did not change significantly (Fig. 3c), while a significant decline was observed in SN9816 seedlings treated with ABA or diniconazole compared with the control. CAT activity in both varieties displayed similar trends when plants were exposed to stimulated stress conditions, decreasing considerably when treated with ABA but not changing when treated with diniconazole (Fig. 3d).
Transcriptome sequencing and de novo assembly
To elucidate the possible molecular mechanisms of the difference in response to exogenous ABA or diniconazole in WR04-6 and SN9816 plants, eighteen cDNA libraries (three replicates per treatment: WR_CK, WR_A, WR_D, SN_CK, SN_A, and SN_D) were constructed, and RNA-Seq libraries were sequenced (Table S1). After the removal of low-quality reads, an average of 53,584,168 high-quality clean reads were obtained from each sample, accounting for 96.79% of the 53,697,638 raw reads. The mean GC content was 54.62%. The proportions of Q20 and Q30 quality scores were 97.88% and 94.02%, respectively, indicating that all libraries were of high quality. Clean reads of each sample were sequentially aligned with the designated reference, and the alignment efficiency ranged from 96.42%to 97.39%.
Differential transcriptome response to exogenous treatments
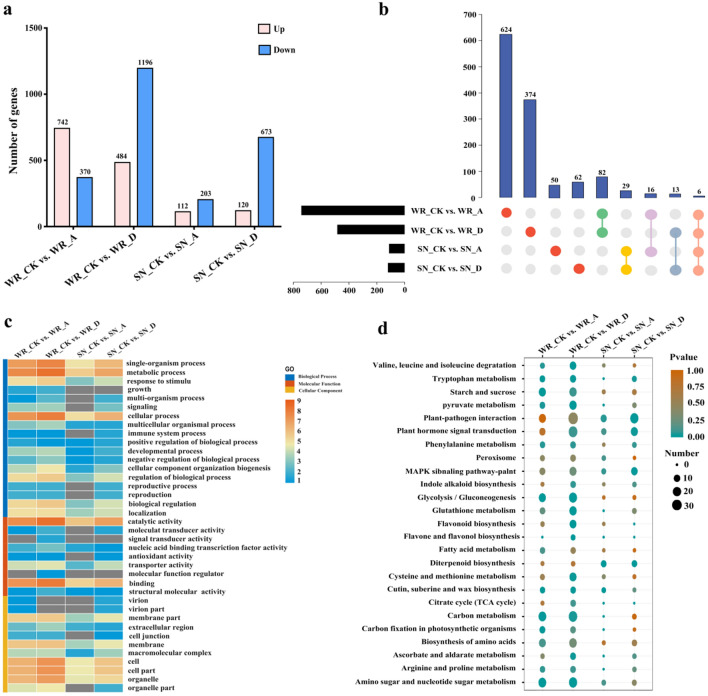
WR04-6 and SN9816 seedlings displayed a significantly different transcriptional response to different treatments. A total of 1112 genes were differentially expressed in the WR_CK versus WR_A group, of which 742 were up-regulated and 370 were down-regulated (Fig. 4a; Table S2). A total of 1680 genes were differentially expressed in the WR_CK versus WR_D group, of which 484 were significantly up-regulated and 1196 were down-regulated (Fig. 4a; Table S3). A considerably smaller number of genes in SN9816 displayed altered expression levels compared with WR04-6 when exposed to ABA and diniconazole. Only 112 up-regulated and 203 down-regulated genes were identified in the comparison between SN_CK and SN_A in SN9816 (Table S4), and 120 up-regulated and 673 down-regulated genes were identified for the comparison SN_CK versus SN_D (Table S5).
Figure 4.
Plot of differentially expressed genes (DEGs) from different comparisons of samples from WR04-6 and SN9816 plants after ABA and diniconazole treatments. (a) Numbers of DEGs resulting from pairwise comparison of samples between WR04-6 and SN9816 after ABA and diniconazole treatments. A in WR_A or SN_A indicates ABA treatment, D in WR_D or SN_D indicates diniconazole treatment, and CK in WR_CK or SN_CK indicates the control group without treatment. (b) Congruence of the up-regulation of DEGs from pairwise comparisons of samples after treatment with ABA or diniconazole in WR04-6 and SN9816 plants. Red represents DEGs uniquely expressed in each treatment for WR04-6 and SN9816 plants; green represents DEGs uniquely expressed in WR_A and WR_D; yellow represents DEGs uniquely expressed in SN_A and SN_D; purple represents DEGs uniquely expressed in WR_A and SN_A; Blue-grey represents DEGs uniquely expressed in WR_D and SN_D; and pink represents DEGs uniquely expressed after both ABA and diniconazole treatments in the two varieties. (c) GO enrichment analysis of the differentially expressed genes presented as a heat map. (d) KEGG enrichment analysis of differentially expressed genes presented as a bubble chart.
To better understand the difference in the transcriptional response of WR04-6 and SN9816 seedlings to exogenous treatment, the congruence of genes from pairwise comparisons of samples after ABA or diniconazole treatment was explored by screening up-regulated DEGs (Fig. 4b). A total of 624, 374, 50, and 62 DEGs were identified that were uniquely expressed in the WR_CK versus WR_A, WR_CK versus WR_D, SN_CK versus SN_A, and SN_CK versus SN_D groups, respectively. There were 82, 29, 16, and 13 common DEGs that were shared across the comparisons WR_A versus WR_D, SN_A versus SN_D, WR_A versus SN_A, and WR_D versus SN_D, respectively. However, only six genes were commonly up-regulated in the two varieties in response to ABA or diniconazole treatment.
DEG annotation and functional categorization
GO enrichment analysis was performed on the DEGs obtained from the four pairwise transcriptome comparisons. GO annotation allocated all DEGs into three categories: biological process, molecular function, or cellular process. A total of 36 terms were significantly enriched in both the WR_CK versus WR_A and WR_CK versus WR_A comparisons, while 24 and 35 terms were significantly enriched in the SN_CK versus SN_A and SN_CK versus SN_D comparisons, respectively (Fig. 4c). The enriched terms varied greatly in WR_CK versus WR_A compared with SN_CK versus SN_A. The enriched terms were grouped into biological processes (signaling, growth, reproduction, reproductive process, immune system process, and multi organism), molecular functions (organelle part, cell junction, virion, and virion part), and cellular processes (antioxidant activity and molecular transducer activity), suggesting that the WR04-6 and SN9816 seedlings had different sensitivities to ABA. However, WR04-6 and SN9816 exhibited similar GO terms when exposed to diniconazole (Fig. S1).
KEGG pathway enrichment analysis was conducted to determine the changes in metabolic pathways following treatment with ABA and diniconazole. The DEGs were enriched in 103 and 105 KEGG pathway terms in WR04-6 plants after ABA and diniconazole treatments, respectively. However, a total of 58 and 78 pathways were found to be enriched in SN9816 after ABA and diniconazole treatment, respectively. The top 25 enrichment pathways for each group of DEGs are shown in Fig. 4d. DEGs common to both varieties were enriched in pathways related to plant-pathogen interaction (KO04626), plant hormone signal transduction (KO04075), and MAPK signaling pathways (KO04016). Furthermore, several genes involved in carbon metabolism (KO01200), glycolysis/gluconeogenesis (KO00010), starch and sucrose metabolism (KO00500), biosynthesis of amino acids (KO01230), glutathione metabolism (KO00480), arginine and proline metabolism (KO00330), and peroxisomes (KO04146) were expressed at higher levels in WR04-6 plants than in SN9816 plants.
Effect of exogenous ABA and diniconazole on ROS homeostasis
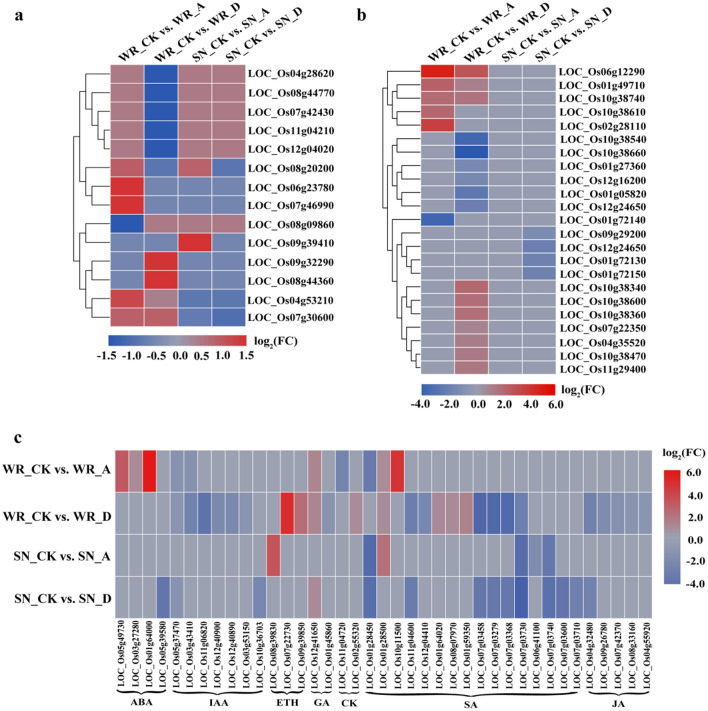
Reactive oxygen species (ROS) serve as signals of cellular homeostasis, particularly in the cells of the immune system. Several genes related to peroxidase had higher expression in WR04-6 than in SN9816 under ABA and diniconazole treatments. In the WR_CK versus WR_A group, five genes encoding peroxidase were identified, of which four were up-regulated and only one was down-regulated (Fig. 5a). There were eight genes related to peroxidase in the WR_CK versus WR_D group, of which only three (LOC_Os04g53210, LOC_Os09g32290, and LOC_Os08g44360) were up-regulated. A smaller number of genes with altered expression levels were identified in SN9816 than in WR04-6 when exposed to ABA and diniconazole. One down-regulated and two up-regulated genes were identified in a comparison of SN_CK and SN_A in SN9816. In the SN_CK versus SN_D group, only LOC_Os07g30600 was enriched and significantly down-regulated.
Figure 5.
DEGs relevant to peroxisome, glutathione metabolism, and hormone signaling after treatment with ABA or diniconazole. (a) Change in transcript expression levels associated with peroxisomes in WR04-6 and SN9816 plants after ABA and diniconazole treatment. (b) Expression patterns of genes involved in the glutathione metabolism pathway in the four pairwise transcriptome comparisons. (c) DEGs involved in hormone signaling in response to ABA and diniconazole treatment in WR04-6 and SN9816 plants. Gene expression levels were transformed by log2(FPKM) values. Red and blue represent up-regulation and down-regulation, respectively. A in WR_A or SN_A indicates ABA treatment, D in WR_D or SN_D indicates diniconazole treatment, and CK in WR_CK or SN_CK indicates the control group without treatment.
Furthermore, the induction of glutathione metabolic genes was prominent in WR04-6 under ABA and diniconazole treatments. Seven genes encoding glutathione S-transferases (GSTs) were identified in WR_CK versus WR_A, of which five were up-regulated and two were down-regulated. There were more DEGs in the WR_CK versus WR_D group than in the WR_CK versus WR_A group, and a total of 16 genes were identified, of which 10 were up-regulated and six were down-regulated. However, no gene was differentially expressed in SN9816 after ABA treatment, and four genes were differentially expressed in the SN_CK versus SN_D group and were significantly down-regulated.
Response of phytohormone-related genes to exogenous treatment
Genes related to phytohormone signaling were also differentially expressed in response to ABA and diniconazole. A total of 40 DEGs involved in seven hormones, including abscisic acid (ABA), auxin (IAA), ethylene (ETH), gibberellic acid (GA), cytokinin (CK), salicylic acid (SA), and jasmonic acid (JA), were identified from four pairwise transcriptome comparisons (Fig. 5c, Table S6). A total of 10 DEGs were identified in the WR_CK versus WR_A group, and genes related to ABA-, GA-, and SA-responsive genes were dramatically up-regulated, while two (LOC_Os05g37470 and LOC_Os03g43410) IAA-responsive genes and one (LOC_Os11g04720) CK-responsive gene were down-regulated. There were more DEGs in the WR_CK versus WR_D group than in the WR_CK versus WR_A group. IAA and JA displayed a negative response to diniconazole, as all DEGs related to IAA- and JA-responsive genes were significantly down-regulated. Of the ten DEGs related to the SA signaling pathway, four (LOC_Os01g28500, LOC_Os01g64020, LOC_Os08g07970, and LOC_Os01g59350) were significantly up-regulated, and six (LOC_Os11g04600, LOC_Os12g04410, LOC_Os07g03458, LOC_Os07g03279, LOC_Os07g03368, and LOC_Os07g03730) were down-regulated. In addition, ETH and GA displayed a positive regulatory effect due to exogenous treatment. SLRL1 (LOC_Os01g45860), orthologous to SLR1 and a negative regulator of the GA signaling pathway, was significantly down-regulated in the WR_CK versus WR_D group. Expression levels of DEGs in SN9816 were considerably lower than those in WR04-6 after treatment with ABA and diniconazole. Only one ETH-responsive gene (LOC_Os08g39830) was identified in the SN_CK versus SN_A group, and that was up-regulated. The expression of SA-responsive genes was lower in response to ABA treatment, with four genes (LOC_Os01g28450, LOC_Os07g03730, LOC_Os06g41100, and LOC_Os07g03740) significantly down-regulated and one gene, LOC_Os01g28500, up-regulated. A total of 14 DEGs were identified in the SN_CK versus SN_D group, of which one GA-responsive gene (LOC_Os12g41650) was up-regulated while the other genes involved in the ABA, IAA, SA, and JA signaling pathways were down-regulated.
Cluster analysis of defense-related genes in exogenous treatments
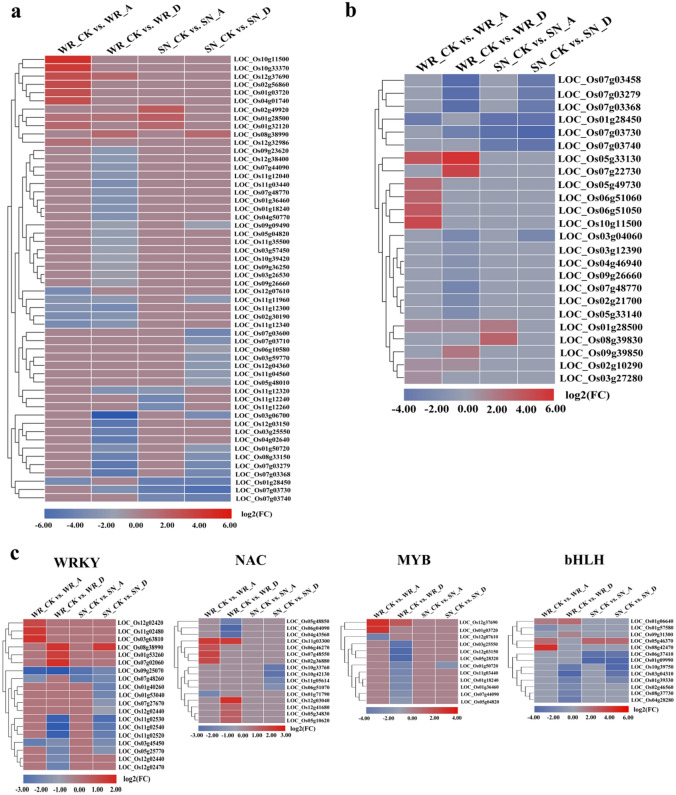
An analysis of DEGs indicated the presence of a milieu of defense-related genes that participated in the ABA and diniconazole response. In the WR_CK versus WR_A group, 15 genes were identified that participate in the interaction between plants and pathogens, of which nine were up-regulated and six were down-regulated. There were 35 genes involved in the plant-pathogen interaction pathway in the WR_CK versus WR_D group, of which only three (LOC_Os01g28500, LOC_Os08g38990, and LOC_Os12g37690) were up-regulated (Fig. 6a, b, Table S7). A total of nine and 20 genes were identified that were involved in the plant-pathogen interaction pathway in SN9816 following ABA or diniconazole treatment, respectively. Three genes (LOC_Os01g28500, LOC_Os01g32120, and LOC_Os02g49920) were found to be up-regulated in SN_CK versus SN_A, while one (LOC_Os08g38990) was identified in SN_CK versus SN_D.
Figure 6.
Expression profiles of defense-related genes in WR04-6 and SN9816 plants after exogenous treatments. Comparative analysis of pathogen interactions (a), MAPK signaling pathway transduction (b), and the expression of transcription factor related genes (c) in WR04-6 and SN9816 plants after exogenous treatment. Gene expression levels were transformed by log2(FPKM) values. Red and blue represent up-regulation and down-regulation, respectively. A in WR_A or SN_A indicates ABA treatment, D in WR_D or SN_D indicates diniconazole treatment, and CK in WR_CK or SN_CK indicates the control group without treatment.
The mitogen-activated protein kinase (MAPK) signaling pathway plays a positive role in plants exposed to adverse conditions. The pathogen-related genes LOC_Os01g28500, LOC_Os07g48770, LOC_Os07g03730, LOC_Os09g26660 (OsrbohB), LOC_Os01g28450, LOC_Os07g03740, and LOC_Os10g11500 were also involved in the MAPK signaling pathway. LOC_Os05g33130, LOC_Os05g49730, LOC_Os06g51050, and LOC_Os06g51060 were uniquely expressed in the WR_CK versus WR_A group (Fig. 6b).
Transcription factors (TFs) also play an important role in the response of plants to stress. There were 56 (41 up-regulated, 15 down-regulated), 111 (39 up-regulated, 72 down-regulated), 20 (6 up-regulated, 14 down-regulated), and 50 (7 up-regulated, 43 down-regulated) TFs identified in the WR_CK versus WR_A, WR_CK versus WR_D, SN_CK versus SN_A, and SN_CK versus SN_D groups, respectively (Table S8). The up-regulated TFs in the present study were members of a variety of families, with the majority representing WRKY, NAC, MYB, and bHLH families in response to ABA or diniconazole treatment. The genes LOC_Os11g02480, LOC_Os12g02420, and LOC_Os03g63810, belonging to the WRKY family, were up-regulated in the WR_CK versus WR_A group. Twelve WRKY family genes were identified in the WR_CK versus WR_D group, of which LOC_Os08g38990, LOC_Os07g02060, and LOC_Os01g53260 were up-regulated. Only one gene (LOC_Os08g38990) was positively regulated in SN9816 in response to diniconazole treatment (Fig. 6c, Table S8). Furthermore, genes belonging to the NAC, MYB, and bHLH TF families were positively regulated in WR04-6 after ABA or diniconazole treatment. Considerably fewer NAC, MYB, and bHLH TFs were identified in the SN_CK versus SN_A and SN_CK versus SN_D groups, and the majority were down-regulated.
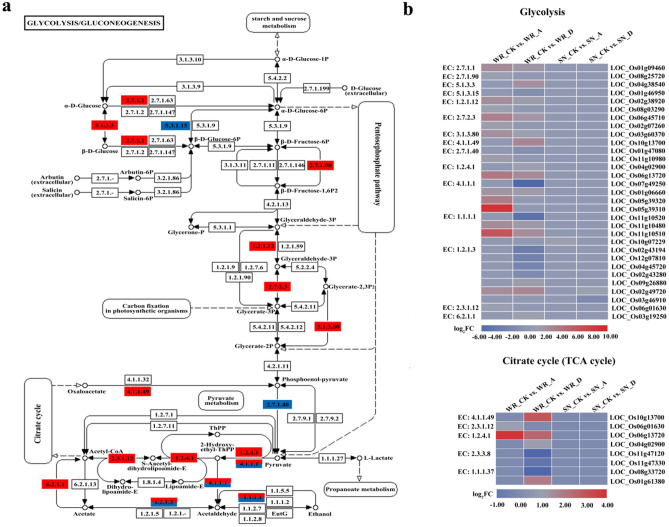
DEGs involved in glycolysis and the citrate (TCA) cycle
The distribution of DEGs revealed that glycolysis/gluconeogenesis and the citrate cycle (TCA cycle) were strongly affected in WR04-6 plants following treatment with 2 μM ABA and 10 μM diniconazole (Fig. 7a). In the present study, 31 DEGs that participate in the glycolysis pathway were identified in the WR_CK versus WR_A and WR_CK versus WR_D groups, of which 21 encoding enzymes important to glycolysis were significantly up-regulated (Fig. 7b). These were hexokinase (EC: 2.7.1.1), diphosphate-dependent phosphofructokinase (EC: 2.7.1.90), aldose 1-epimerase (EC: 5.1.3.3), glyceraldehyde 3-phosphate dehydrogenase (EC: 1.2.1.12), phosphoglycerate kinase (EC: 2.7.2.3), 2,3-bisphosphoglycerate 3-phosphatase (EC: 3.1.3.80), phosphoenolpyruvate carboxykinase (EC: 4.1.1.49), pyruvate kinase (EC: 2.7.1.40), pyruvate dehydrogenase E1 (EC: 1.2.4.1), pyruvate decarboxylase (EC: 4.1.1.1), alcohol dehydrogenase 1/7 (EC: 1.1.1.1), aldehyde dehydrogenase (EC: 1.2.1.3), pyruvate dehydrogenase E2 (EC: 2.3.1.12), and acetyl-CoA synthetase (EC: 6.2.1.1). Only three DEGs (two up-regulated and one down-regulated) were identified in the SN_CK versus SN_A and SN_CK versus SN_D groups. The up-regulated genes encoded alcohol dehydrogenase 1/7 (EC: 1.1.1.1) and aldehyde dehydrogenase (EC: 1.2.1.3).
Figure 7.
DEGs relevant to glycolytic metabolism and the citrate cycle after treatment with ABA or diniconazole. (a) Change in transcript expression levels associated with glycolysis and citrate cycle pathways in WR04-6 and SN9816 plants after ABA and diniconazole treatment. (b) Expression patterns of genes involved in glycolysis and citrate cycle pathways in the four pairwise transcriptome comparisons.
A total of eight DEGs (five up-regulated and three down-regulated) were identified from the WR_CK versus WR_A and WR_CK versus WR_D groups that participated in the TCA cycle (Fig. 7b). These genes primarily encode key enzymes, including phosphoenolpyruvate carboxykinase (EC: 4.1.1.49), pyruvate dehydrogenase E2 (EC: 2.3.1.12), pyruvate dehydrogenase E1 (EC: 1.2.4.1), ATP citrate (pro-S)-lyase (EC: 2.3.3.8), and malate dehydrogenase (EC: 1.1.1.37). However, no genes from the SN_CK versus SN_A and SN_CK versus SN_D groups participated in the TCA cycle.
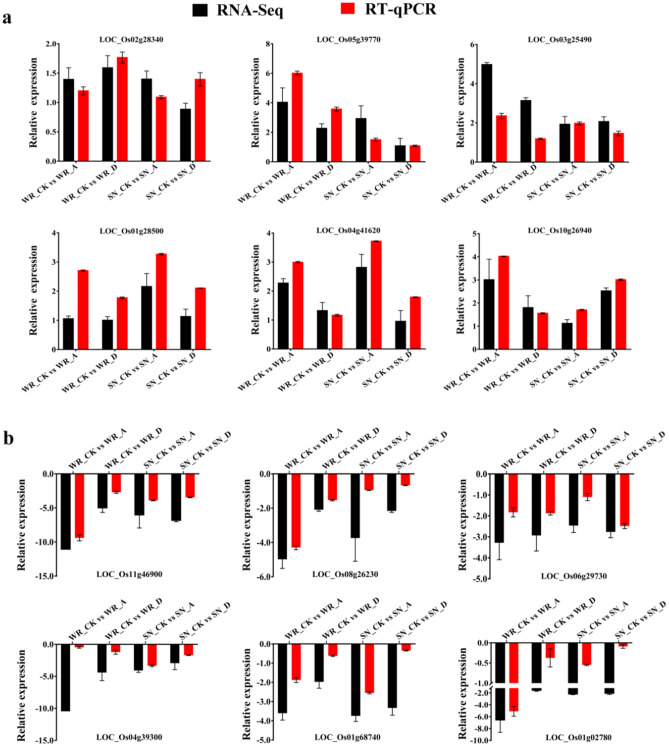
Validation of RNA-Seq data by RT-qPCR
To further validate the RNA-Seq expression profile data, twelve genes were selected from representative transcripts commonly expressed in each treatment for RT-qPCR assay. The Pearson correlation coefficients (PCCs) between the expression profiles determined by RNA-Seq and by RT-qPCR were analyzed by a Gaussian distribution yielding 95% confidence for a PCC = 0.890. The results indicate that the expression profiles of the selected genes were largely consistent with data derived from the RNA-Seq results, suggesting that they were reliable and repeatable (Fig. 8).
Figure 8.
Comparison of relative expression level of twelve genes by RNA-Seq and RT-qPCR. (a) up-regulated genes; (b) down-regulated genes. Black and red bas represent relative expression level from FPKM and RT-qPCR, respectively. Error bars represent SEs (n = 3).
Discussion
ABA is generally known to inhibit seed germination and post-germination growth29. The current analysis revealed that ABA delayed seed germination of WR04-6 and SN9816 plants in an ABA concentration-dependent manner. This is similar to Arabidopsis, where a statistical correlation was confirmed between ABA concentration and the rate of seed germination30. However, a slight delay in SN9816 seed germination was observed for each concentration of ABA (Fig. 1a). ABA represses germination through metabolism and signaling31. Diniconazole has been shown to inhibit ABA 8′-hydroxylase activity, resulting in a twofold increase in endogenous ABA concentration in potatoes32. As expected, high concentrations of diniconazole slightly delayed seed germination in WR04-6 but had no effect on the germination of SN9816 at the same concentration (Fig. 1b). Furthermore, continuous ABA or diniconazole treatment significantly inhibited the post-germination growth of both WR04-6 and SN9816 seeds, although ABA and diniconazole more significantly inhibited plant height in WR04-6 (Fig. 2). Similar results have been observed in Arabidopsis33, which demonstrated that the ABA 8′-hydroxylase gene inhibited seedling development by influencing ABA metabolism. These results indicate that ABA plays a larger role in WR04-6 seed germination and post-germination growth than it does in SN9816 plants.
Damage to membranes and H2O2 accumulation are considered the principal modes of damage that comprise the oxidative stress process34. MDA is the product of membrane lipid peroxidation and is indicative of compromised membrane integrity and structure. ABA significantly increased the content of MDA in WR04-6, while the opposite was observed in SN9816 (Fig. 3a). Plants enhance stress tolerance by regulating antioxidant activity. SOD is considered a critical enzyme for the regulation of O2- and H2O2 levels35. In the present study, SOD activity increased significantly following exogenous ABA or diniconazole treatment in WR04-6 plants (Fig. 3b). Similar results have previously been observed in tomato plants after ABA treatment36, demonstrating up-regulation of SOD activity. However, only a slight effect was observed on SOD activity in SN9816 plants after ABA or diniconazole treatment. POD and CAT break down H2O2, producing water and molecular oxygen37. POD activity in WR04-6 changed slightly after ABA or diniconazole application, but CAT activity decreased significantly after the application of ABA. Surprisingly, POD and CAT activity in SN9816 plants decreased significantly after ABA or diniconazole treatment (Fig. 3c, d). In agreement with this result, several antioxidant enzyme-related genes were differentially expressed, including GSTs, SODs, and APXs (Fig. 5a, b). In this study, LOC_Os01g49710, LOC_Os06g12290, and LOC_Os10g38740, which are involved in glutathione metabolism, were up-regulated in WR04-6 under both ABA and diniconazole applications. LOC_Os10g38470, LOC_Os11g29400, LOC_Os07g22350, LOC_Os10g38600, LOC_Os10g38340, and LOC_Os10g38360 were uniquely up-regulated in the WR_CK versus WR_D group, and these genes may play an important role in maintaining ROS homeostasis in WR04-6 plants.
Hormonal regulation plays an important role and could be an important factor that defines sensitivity and tolerance in plant systems38. A complex relationship in stress-responsive genes that depends on the transduction of phytohormone signaling likely exists in the present study. Exposure of WR04-6 seedlings to ABA significantly increased the expression of ABA-related genes, and genes related to SA signal transduction were additionally induced. Subsequently, IAA and CK signal transduction was negatively regulated by ABA, further suppressing cell enlargement and plant growth. Distinct from ABA treatment, the accumulation of endogenous ABA induced by diniconazole promoted the expression of EIL- and ERF-related genes. As JAZs are located upstream of ERFs, overexpression of ERF feedback suppresses JAZs, thus inhibiting the JA signal transduction pathway (Fig. 5c). A similar hormone regulatory network was established in wheat roots after ABA treatment15, indicating that phytohormone signaling transduction pathways are critical in stress responses.
SA can increase the activity of superoxide dismutase-producing enzymes, leading to increased H2O2 and causing a series of hypersensitive responses in plants that ultimately enhance resistance to plant disease39. Sixteen SA-related genes were identified in this study, with genes LOC_Os01g28500 LOC_Os01g28450, and LOC_Os07g03730 identified in both varieties. LOC_Os01g64020, LOC_Os08g07970, and LOC_Os01g59350 (OsbZIP64) had unique expression profiles in the WR_CK versus WR_D group, and these genes may play an important role in plant disease resistance in WR04-6 plants.
The WRKY, MYB, NAC, and bHLH families of TFs play an important role in plant immunity by inducing the expression of downstream defense-related genes. Previous studies have shown that drought, low temperature, and bacterial infection induce the overexpression of WRKY family TFs39–41. In the present study, LOC_Os11g02480 (OsWRKY46a), LOC_Os12g02420 (OsWRKY46b), and LOC_Os03g63810 (OsWRKY80), which belong to the WRKY family, were uniquely up-regulated in the WR_CK versus WR_A group (Fig. 6c). In addition, LOC_Os07g02060 (OsWRKY29) and LOC_Os01g53260 (OsWRKY23) were uniquely up-regulated in the WR_CK versus WR_D group. It has been reported that LOC_Os03g63810 (OsWRKY80) functions as a positive transcriptional regulator against Rhizoctonia solani by directly binding to the promoter of OsWRKY442. These results are consistent with WRKY TFs being involved in plant defense responses. NAC TFs appear to be widespread in plants, and the expression of multiple NAC genes is induced by abiotic and biotic stress43. Many studies have used NAC TFs to improve stress tolerance in plants by genetic engineering44. In the present study, 17 NAC TFs were identified in the two plant varieties, and the genes LOC_Os11g03300 (OsNAC10), LOC_Os07g48550 (–), LOC_Os06g46270 (OMTN4), LOC_Os02g36880 (OMTN1), LOC_Os05g34830 (OsNAC52), LOC_Os12g03040 (ONAC131), LOC_Os12g41680 (OMTN3), and LOC_Os05g10620 (–) were uniquely expressed in the WR_CK versus WR_A and WR_CK versus WR_D groups. The rice pathogen-responsive gene LOC_Os12g03040 (ONAC131), plays important roles in the disease resistance response through the regulated expression of other defense- and signaling-related genes45. Overexpression of LOC_Os11g03300 (OsNAC10) and LOC_Os05g34830 (OsNAC52) can improve tolerance to drought and low temperature, respectively46,47. These data suggest that NAC TFs possess potential utility in improving stress tolerance in crops. Additionally, the novel genes LOC_Os12g37690 and LOC_Os01g03720 belonging to the MYB family of TFs, and LOC_Os08g42470, LOC_Os01g06640, and LOC_Os09g31300 genes belonging to the bHLH family were expressed in WR04-6 after application of ABA or diniconazole.
Changes in carbohydrate content are of particular importance and affect the activity of plant physiological processes, such as photosynthesis, respiration, and metabolism48. Previous studies have shown that the carbohydrate metabolic pathway is the most sensitive in plants to stress conditions40. DEGs identified by transcriptome sequencing were mostly related to glycolysis, starch and sucrose metabolism, the TCA cycle, and pyruvate metabolic pathways. These observations are consistent with our results in which multiple DEGs related to carbohydrate metabolism were mapped to glycolysis and TCA cycle pathways. Glycolysis is a key respiratory pathway that provides ATP, reducing agents, and metabolites for plant growth and development49. When experiencing abiotic stress, glycolysis is often involved in the regulation of adaptability of plants to the environment50. In the present study, both ABA and diniconazole treatment in WR04-6 plants caused enrichment in the glycolysis pathway, with 16 and 22 DEGs enriched after ABA and diniconazole treatment, respectively. However, only one gene (LOC_Os10g07229) that participates in glycolysis was induced in SN9816 after ABA treatment, and only two genes (LOC_Os02g49720 and LOC_Os03g46910) were induced after diniconazole treatment. The TCA cycle is the principal means of aerobic decomposition of glucose during respiration, which gradually oxidizes and decomposes the glycolytic product pyruvate into CO2 and H2O2, generating NADH, FADH2, and ATP that support stress tolerance51. RNA-Seq data demonstrated that three genes were enriched after ABA treatment and seven after the application of diniconazole. Similar results have been previously observed in Dendrobium officinale52 and Poa pratensis53 exposed to abiotic stress conditions. These results indicate that WR04-6 can respond to ABA by actively regulating glycolysis and TCA cycle pathway-related genes and enzyme activities. Taken together, weedy rice (WR04-6) and cultivated rice (SN9816) showed different transcriptional characteristics under ABA or diniconazole treatment. Several differentially expressed genes had unique expression profiles in the WR_CK versus WR_A and WR_CK versus WR_D groups, these genes may serve as candidates to be used for further crop improvement (Table S9).
Conclusion
This study revealed different physiological and molecular mechanisms in response to ABA and diniconazole between weedy rice and cultivated rice with contrasting ABA sensitivity. The transcriptomic analysis uncovered global and weedy-specific reactions of WR04-6 in response to ABA and diniconazole, such as the enhancement of antioxidative actions, stress defense, hormone signaling transduction, and glycolytic and citrate cycle activities. The candidate genes screened in this study will provide novel material for genetic and functional studies of stress in cultivated rice, which can be used for further crop improvement and breeding of resistant varieties.
Materials and methods
Materials
WR04-6, a representative weedy rice accession and temperate japonica Shennong9816 (SN9816) as a control rice sample were obtained from the Rice Research Institute of Shenyang Agricultural University. Both of them were grown in germplasm resource field at Rice Research Institute of Shenyang Agricultural University, Shenyang, China. The samples were collected from the germplasm resource field with the permission of Rice Research Institute of Shenyang Agricultural University. Seeds were sown on April 15, 2019, with seedlings transplanted to their final locations on May 26, 2019. Plants were spaced 30.0 × 13.4 cm apart. Fertilizer and water management followed the local standard management. Seeds were harvested 35 days after anthesis and stored at 4 °C until use for experimentation. Experimental research and field studies on plants must comply with relevant institutional, national, and international guidelines and legislation.
Seed germination and coleoptile sheath length after treatment with ABA and diniconazole
For germination assays, seeds were presoaked in a thermostatic incubator at 50 °C for 3 days to prevent any possible dormancy. Fifty sterilized WR04-6 and SN9816 seeds were sown in distilled water supplemented with different concentrations of ABA (0, 2, 5, or 10 μM) or the ABA catabolism inhibitor diniconazole (0, 10, 20, or 30 μM). Seeds were considered germinated when radicles had emerged from the seed coats. The percentage (%) germination was calculated every 12 h after initiation. All experiments were performed independently three times. After germination, 30 germinated seeds with uniform growth potential for each treatment were transferred to 1/2 × Murashige and Skoog (MS) medium (pH = 5.8) supplemented with ABA or diniconazole appropriate for the experimental grouping and grown at 30 °C within a 16 h light/8 h dark cycle. The coleoptile sheath length was measured and photographed on days 7 and 15 to determine the growth performance for each treatment.
Analysis of malondialdehyde (MDA) and antioxidant enzyme activity
Following exposure to continuous ABA and diniconazole treatment, seedlings treated with 2 μM ABA and 10 μM diniconazole were selected for analysis of physiological indices. Approximately 0.9 g of fresh leaves of each treatment were collected and ground into powder in liquid nitrogen and homogenized in an ice bath with 5 mL of 50 mmol/L sodium phosphate buffer (pH = 7.8) containing 5 mmol/L ethylenediaminetetraacetic acid (EDTA) and 1% polyvinylpyrrolidone. The homogenate was centrifuged at 12,000 rpm for 20 min at 4 °C, and the supernatant was analyzed for MDA concentration and catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD) activity.
MDA concentration was measured using the thiobarbituric acid (TBA) method, as described in a previous study54 with slight modifications. A 2.5 mL mixture of 0.5% trichloroacetic acid (TCA) and 0.5% TBA was added to 1 mL of leaf supernatant, and the sample was heated for 15 min and then cooled quickly in an ice bath. The mixture was then centrifuged at 3000 rpm for 30 min, and the absorbance of the supernatant was measured at 532 nm, from which nonspecific absorption at 600 nm was subtracted. The results were expressed as μmol g−1 FW.
SOD activity was measured using a nitroblue tetrazolium (NBT) assay55. The 3 mL reaction mixture contained enzyme extract, 130 mM methionine, 0.75 mM NBT, 0.1 mM EDTA, 20 μM riboflavin and 50 mM phosphate buffer (pH 7.8). Tubes were reacted at 4000 lx for 20 min, after which absorbance at 560 nm was measured using a spectrophotometer (Hitachi, U-5100). POD activity was measured in accordance with the method of Zhang et al.56. The reaction mixture contained 0.1 mL of enzyme extract, 0.3 mL of 0.6% H2O2, and 2.6 mL of 0.3% guaiacol. The oxidation of guaiacol by H2O2 was quantified at 420 nm as a measure of POD activity. CAT activity was measured in accordance with the method described by Kraus et al.57. The reaction mixture comprised 0.2 of mL distilled water, 2.8 mL of 60 mM H2O2, and 0.2 mL of enzyme extract. The decrease in absorbance at 240 nm due to H2O2 over 3 min was recorded. The results are expressed as IU·g−1 FW and IU·g−1 min for POD and CAT, respectively.
RNA isolation, cDNA library construction, and RNA sequencing
Seedlings treated with 2 μM ABA and 10 μM diniconazole were selected for further RNA-Seq analysis. For each treatment, leaf tissue from fifteen plants (three biological replicates with five plants per replicate) with the same growth status was collected 15 days after treatment with hormones. Total RNA was extracted from each sample using TRIzol reagent (Thermo Fisher Scientific, MA, USA) in accordance with the manufacturer’s protocol. RNA concentration was assessed in a Nanodrop 8000 spectrophotometer (Thermo Fisher Scientific, MA, USA), and integrity was verified by agarose gel electrophoresis. After total RNA was extracted, mRNA was enriched by oligo (dT) beads, and then the enriched mRNA was fragmented into short fragments by fragmentation buffer and reverse transcribed into cDNA with random primers. Second-strand cDNA was synthesized by DNA polymerase I, RNase H, dNTPs and buffer. Then, the cDNA fragments were purified with a QiaQuick PCR extraction kit (Qiagen, Venlo, The Netherlands), end repaired, poly(A) added, and ligated to Illumina sequencing adapters. The ligation products were size selected by agarose gel electrophoresis and PCR amplified. The cDNA libraries were sequenced using an Illumina HiSeq 2500 sequencing platform (Illumina Crop., San Diego, CA, USA) at Gene De novo Biotechnology Co., Ltd. (Guangzhou, China).
Normalization and analysis of differentially expressed genes (DEGs)
Raw data were processed through a quality check using the FastQC protocol (https://github.com/OpenGene/fastp), and clean reads were obtained by removing adapter sequences, low-quality sequences (bases with Q-value ≤ 20) and reads with poly-N. The reference genome of Oryza sativa ssp. japonica (Os-Nipponbare-Reference-IRGSP-1.0) was obtained from The Rice Annotation Project Database (RAP-DB) (https://rapdb.dna.affrc.go.jp/index.html). Cleaned short reads were aligned to all exon sequences using HISAT258. Gene expression was calculated with the RSEM tool59 using default parameters. Relative expression levels were normalized in terms of fragments per kilobase of exon per million reads (FPKM). DEGs were identified using DESeq260, and genes with |Log2 (Fold Change, FC)|> 1 and a false discovery rate < 0.05 were considered to be significant DEGs. The DEGs underwent gene ontology (GO) enrichment (available online: http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (available online: http://kobas.cbi.pku.edu.cn/) pathway analysis. TBtools software was used to determine the differential expression and overlap of transcripts between comparison sets for the treatment and control groups (WR04-6 and SN9816, respectively).
Quantitative real-time PCR validation
Commonly expressed genes were quantified in samples from each treatment using RT-qPCR. Gene-specific primers were designed using Primer Premier 5 software, as detailed in Table S10. First-strand cDNA was synthesized using a PrimeScript RT regent kit combined with gDNA Eraser (TaKaRa, Dalian, China) in accordance with the manufacturer’s instructions. TB Green Premix Ex Taq II (Tli RNaseH Plus, RR820A) (TaKaRa, Dalian, China) was used to perform RT-qPCR in an Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher Scientific, USA). The reaction conditions were as follows: 95 °C for 30 s, then 40 cycles of 95 °C for 5 s and 60 °C for 34 s. The expression of transcripts was normalized to the reference gene OsActin using the 2−ΔΔCt method61. Three biological replicates were quantified by RT-qPCR analysis.
Statistical analysis
Treatment differences were statistically evaluated via one-way analysis of variance (ANOVA), followed by individual comparisons with Duncan’s multiple range tests, using SPSS software v19.0. Student’s t-tests (t < 0.05) were performed to compare expression between samples. The results are presented as the means ± standard error (SE) of three biological replicates. Pearson correlation analysis was performed to evaluate the correlation between RNA-Seq and RT-qPCR (p < 0.05).
Data availability statement
Raw sequence data were deposited in the NCBI Short Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA723912.
Supplementary Information
Author contributions
D.M. and H.L. conceived the project; H.L., Y.H. and F.Z. conducted gene expression and antioxidative enzyme activity measurements; F.X., and M.Z. analyzed and interpreted the data; H.L. drafted the manuscript; and D.M., F.X., and M.Z. critically revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Liaoning Revitalization Talents Program (Grant No. XLYC1808003).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92504-5.
References
- 1.Kanapeckas KL, et al. Contrasting patterns of variation in weedy traits and unique crop features in divergent populations of US weedy rice (Oryza sativa sp.) in Arkansas and California. Pest Manag. Sci. 2018;74:1404–1415. doi: 10.1002/ps.4820. [DOI] [PubMed] [Google Scholar]
- 2.Cao Q, et al. Genetic diversity and origin of weedy rice (Oryza sativa f. spontanea) populations found in north-eastern China revealed by simple sequence repeat (SSR) markers. Ann. Bot. 2006;98:1241–1252. doi: 10.1093/aob/mcl210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bevilacqua BN, et al. Analysis of stress-responsive gene expression in cultivated and weedy rice differing in cold stress tolerance. Plos One. 2015 doi: 10.1371/journal.pone.0132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgos NR, Norman RJ, Gealy DR, Black H. Competitive N uptake between rice and weedy rice. Field Crop Res. 2006;99:96–105. doi: 10.1016/j.fcr.2006.03.009. [DOI] [Google Scholar]
- 5.Dai L, Song X, He B, Valverde BE, Qiang S. Enhanced photosynthesis endows seedling growth vigour contributing to the competitive dominance of weedy rice over cultivated rice. Pest Manag. Sci. 2017;73:1410–1420. doi: 10.1002/ps.4471. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, et al. QTL Analysis for resistance to blast disease in U.S. weedy rice. Mol. Plant Microbe Interact. 2015;28:834–844. doi: 10.1094/mpmi-12-14-0386-r. [DOI] [PubMed] [Google Scholar]
- 7.He Q, Kim KW, Park YJ. Population genomics identifies the origin and signatures of selection of Korean weedy rice. Plant Biotechnol. J. 2017;15:357–366. doi: 10.1111/pbi.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oh CS, Choi YH, Lee SJ, Yoon DB, Moon HP. Mapping of quantitative trait loci for cold tolerance in weedy rice. Breeding Sci. 2004;54:373–380. doi: 10.1270/jsbbs.54.373. [DOI] [Google Scholar]
- 9.Guan S, et al. Transcriptomics profiling in response to cold stress in cultivated rice and weedy rice. Gene. 2019;685:96–105. doi: 10.1016/j.gene.2018.10.066. [DOI] [PubMed] [Google Scholar]
- 10.Tang L, et al. Utilization of weedy rice for development of japonica hybrid rice (Oryza sativa L.) Plant Sci. 2011;180:733–740. doi: 10.1016/j.plantsci.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Subudhi PK, et al. A chromosome segment substitution library of weedy rice for genetic dissection of complex agronomic and domestication traits. Plos One. 2015 doi: 10.1371/journal.pone.0130650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin J, et al. Ethylene insensitive3-like2 (OsEIL2) confers stress sensitivity by regulating OsBURP16, the β subunit of polygalacturonase (PG1β-like) subfamily gene in rice. Plant Sci. 2020 doi: 10.1016/j.plantsci.2019.110353. [DOI] [PubMed] [Google Scholar]
- 13.Liu K, et al. Young seedlings adapt to stress by retaining starch and retarding growth through ABA-Dependent and -independent pathways in Arabidopsis. Biochem. Biophys. Res. Commun. 2019;515:699–705. doi: 10.1016/j.bbrc.2019.06.023. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida T, Christmann A, Yamaguchi-Shinozaki K, Grill E, Fernie AR. Revisiting the basal role of ABA roles outside of stress. Trends Plant Sci. 2019;24:625–635. doi: 10.1016/j.tplants.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Li C, et al. Transcriptome analysis of osmotic-responsive genes in ABA-dependent and -independent pathways in wheat (Triticum aestivum L.) roots. Peer J. 2019 doi: 10.7717/peerj.6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- 17.Kim JA, et al. Transcriptome analysis of ABA/JA-Dual responsive genes in rice shoot and root. Curr. Genomics. 2017 doi: 10.2174/1389202918666170228134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida T, et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- 19.Kang J, Choi H, Im M, Kim SY. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 2002;14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, et al. Over-expression of an R2R3 MYB Gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis. Plant Sci. 2019;286:28–36. doi: 10.1016/j.plantsci.2019.05.021. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Chen L, Shi Q, Ren Z. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato. Plant Sci. 2020 doi: 10.1016/j.plantsci.2019.110356. [DOI] [PubMed] [Google Scholar]
- 22.Je J, Chen H, Song C, Lim CO. Arabidopsis DREB2C modulates ABA biosynthesis during germination. Biochem. Biophys. Res. Commun. 2014;452:91–98. doi: 10.1016/j.bbrc.2014.08.052. [DOI] [PubMed] [Google Scholar]
- 23.Donde R, et al. Computational characterization of structural and functional roles of DREB1A, DREB1B and DREB1C in enhancing cold tolerance in rice plant. Amino Acids. 2019;51:839–853. doi: 10.1007/s00726-019-02727-0. [DOI] [PubMed] [Google Scholar]
- 24.Chen M, et al. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 2007;353:299–305. doi: 10.1016/j.bbrc.2006.12.027. [DOI] [PubMed] [Google Scholar]
- 25.Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 26.Umezawa T, et al. CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 2006;46:171–182. doi: 10.1111/j.1365-313X.2006.02683.x. [DOI] [PubMed] [Google Scholar]
- 27.Ji X, et al. Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiol. 2011;156:647–662. doi: 10.1104/pp.111.176164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu G, Ye N, Zhang J. Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol. 2009;50:644–651. doi: 10.1093/pcp/pcp022. [DOI] [PubMed] [Google Scholar]
- 29.Gu L, et al. A chloroplast-localized S1 domain-containing protein SRRP1 plays a role in Arabidopsis seedling growth in the presence of ABA. J. Plant Physiol. 2015;189:34–41. doi: 10.1016/j.jplph.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Utsugi S, Ashikawa I, Nakamura S, Shibasaka M. TaABI5, a wheat homolog of Arabidopsis thaliana ABA insensitive 5, controls seed germination. J. Plant Res. 2020;133:245–256. doi: 10.1007/s10265-020-01166-3. [DOI] [PubMed] [Google Scholar]
- 31.Carrillo-Barral N, Matilla AJ, Rodríguez-Gacio MC, Iglesias-Fernández R. Nitrate affects sensu–stricto germination of after-ripened Sisymbrium officinale seeds by modifying expression of SoNCED5, SoCYP707A2 and SoGA3ox2 genes. Plant Sci. 2014;217–218:99–108. doi: 10.1016/j.plantsci.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Suttle JC, Abrams SR, Stefano-Beltrán DL, Huckle LL. Chemical inhibition of potato ABA-8′-hydroxylase activity alters in vitro and in vivo ABA metabolism and endogenous ABA levels but does not affect potato microtuber dormancy duration. J. Exp. Bot. 2012;63:5717–5725. doi: 10.1093/jxb/ers146. [DOI] [PubMed] [Google Scholar]
- 33.Zhu G, Liu Y, Ye N, Liu R, Zhang J. Involvement of the abscisic acid catabolic gene CYP707A2 in the glucose-induced delay in seed germination and post-germination growth of Arabidopsis. Physiol. Plant. 2011;143:375–384. doi: 10.1111/j.1399-3054.2011.01510.x. [DOI] [PubMed] [Google Scholar]
- 34.Bayat L, et al. Effects of growth under different light spectra on the subsequent high light tolerance in rose plants. AoB Plants. 2018 doi: 10.1093/aobpla/ply052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Mou W, Li D, Luo Z, Mao L, Ying T. Transcriptomic analysis reveals possible influences of ABA on secondary metabolism of pigments, flavonoids and antioxidants in tomato fruit during ripening. Plos One. 2015 doi: 10.1371/journal.pone.0129598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F, et al. Senescence-specific change in ROS scavenging enzyme activities and regulation of various SOD isozymes to ROS levels in psf mutant rice leaves. Plant Physiol. Biochem. 2016;109:248–261. doi: 10.1016/j.plaphy.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Tiwari S, Lata C, Chauhan PS, Prasad V, Prasad MA. Functional genomic perspective on drought signalling and its crosstalk with phytohormone-mediated signalling pathways in plants. Curr. Genomics. 2017 doi: 10.2174/1389202918666170605083319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu L, Yang D, Tang D, Li S, Chen Z. Transcriptome analysis of different rice cultivars provides novel insights into the rice response to bacterial leaf streak infection. Funct. Integr. Genomics. 2020;20:681–693. doi: 10.1007/s10142-020-00744-x. [DOI] [PubMed] [Google Scholar]
- 40.Zhao Y, Wei X, Long Y, Ji X. Transcriptional analysis reveals sodium nitroprusside affects alfalfa in response to PEG-induced osmotic stress at germination stage. Protoplasma. 2020;257:1345–1358. doi: 10.1007/s00709-020-01508-x. [DOI] [PubMed] [Google Scholar]
- 41.Jia Y, et al. Transcriptome sequencing and iTRAQ of different rice cultivars provide insight into molecular mechanisms of cold-tolerance response in japonica rice. Rice. 2020 doi: 10.1186/s12284-020-00401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng X, et al. OsWRKY80-OsWRKY4 Module as a positive regulatory circuit in rice resistance against Rhizoctonia solani. Rice. 2016 doi: 10.1186/s12284-016-0137-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao H, Wang H, Tang X. NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 2015 doi: 10.3389/fpls.2015.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thirumalaikumar VP, et al. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018;16:354–366. doi: 10.1111/pbi.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun L, et al. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol. Biol. 2012;81:41–56. doi: 10.1007/s11103-012-9981-3. [DOI] [PubMed] [Google Scholar]
- 46.Hu H, et al. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 2008;67:169–181. doi: 10.1007/s11103-008-9309-5. [DOI] [PubMed] [Google Scholar]
- 47.Jeong JS, et al. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010;153:185–197. doi: 10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kerepesi I, Galiba G. Changes in carbohydrate content are of particular importance that affect plant physiological activities, such as photosynthesis, respiration, and metabolic processes. Crop Sci. 2014;40:482–487. doi: 10.2135/cropsci2000.402482x. [DOI] [Google Scholar]
- 49.Plaxton WC. The organization and regulation of plant glycolysis. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:185–214. doi: 10.1146/annurev.arplant.47.1.185. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, Zhang Y, Du Y, Chen S, Tang H. Dynamic metabonomic responses of tobacco (Nicotiana tabacum) plants to salt stress. J. Proteome Res. 2011;10:1904–1914. doi: 10.1021/pr101140n. [DOI] [PubMed] [Google Scholar]
- 51.Wang L, et al. Metabolomic and proteomic profiles reveal the dynamics of primary metabolism during seed development of lotus (Nelumbo nucifera) Front. Plant Sci. 2016 doi: 10.3389/fpls.2016.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu ZG, et al. Insights from the cold transcriptome and metabolome of dendrobium officinale: global reprogramming of metabolic and gene regulation networks during cold acclimation. Front. Plant Sci. 2016 doi: 10.3389/fpls.2016.01653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong W, Ma X, Jiang H, Zhao C, Ma H. Physiological and transcriptome analysis of Poa pratensis var. anceps cv. Qinghai in response to cold stress. BMC Plant Biol. 2020 doi: 10.1186/s12870-020-02559-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Draper HH, Hadley M. Malondialdehyde determination as index of lipid peroxidation. Methods Enzymol. 1990;186:421–431. doi: 10.1016/0076-6879(90)86135-I. [DOI] [PubMed] [Google Scholar]
- 55.Giannopolitis CN, Ries SK. Superoxide dismutases I. occurrence in higher plants. Plant Physiol. 2007;59:309–314. doi: 10.1104/pp.59.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H, Jiang Y, He Z, Ma M. Cadmium accumulation and oxidative burst in garlic (Allium sativum) J. Plant Physiol. 2005;162:977–984. doi: 10.1016/j.jplph.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 57.Kraus TE, Fletcher RAF. Paclobutrazol protects wheat seedlings from heat and paraquat injury is detoxification of active oxygen involved. Plant Cell Physiol. 1994;35:45–52. [Google Scholar]
- 58.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love M, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2004;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data were deposited in the NCBI Short Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA723912.