Abstract
Intelectins are ancient carbohydrate binding proteins, spanning chordate evolution and implicated in multiple human diseases. Previous GWAS have linked SNPs in ITLN1 (also known as omentin) with susceptibility to Crohn's disease (CD); however, analysis of possible functional significance of SNPs at this locus is lacking. Using the Ensembl database, pairwise linkage disequilibrium (LD) analyses indicated that several disease-associated SNPs at the ITLN1 locus, including SNPs in CD244 and Ly9, were in LD. The alleles comprising the risk haplotype are the major alleles in European (67%), but minor alleles in African superpopulations. Neither ITLN1 mRNA nor protein abundance in intestinal tissue, which we confirm as goblet-cell derived, was altered in the CD samples overall nor when samples were analyzed according to genotype. Moreover, the missense variant V109D does not influence ITLN1 glycan binding to the glycan β-D-galactofuranose or protein–protein oligomerization. Taken together, our data are an important step in defining the role(s) of the CD-risk haplotype by determining that risk is unlikely to be due to changes in ITLN1 carbohydrate recognition, protein oligomerization, or expression levels in intestinal mucosa. Our findings suggest that the relationship between the genomic data and disease arises from changes in CD244 or Ly9 biology, differences in ITLN1 expression in other tissues, or an alteration in ITLN1 interaction with other proteins.
Subject terms: Gastroenterology, Molecular medicine, Lectins
Introduction
Intelectins are ancient proteins, spanning chordate evolution and have been implicated in various human disease states, including inflammatory bowel disease (IBD), obesity, non-insulin dependent diabetes mellitus, and asthma1–3. Intelectins are multimeric, secreted proteins that exhibit calcium-dependent carbohydrate binding, and each monomer is comprised of a fibrinogen-related domain and C-terminal glycan-binding domain4–6. Both preservation across evolution and abundant expression at mucosal surfaces suggest important function of these lectins in innate immunity4. Mammalian intelectins can be broadly grouped into ITLN1-like and ITLN2-like proteins based on distinctive N-terminal primary sequence and amino acid length (~ 313 versus ~ 325)4,7. The first mammalian intelectin identified (Itln1) was found to be expressed in Paneth cells of mice8. Some strains of mice have an expanded intelectin locus of up to six Itln1-like genes but lack clear ITLN2-like orthologs, whereas humans encode a single ITLN1 gene and a single ITLN2 gene7,9,10. Human ITLN1 is expressed in the small and large intestine, as well as in extra-intestinal tissues, including visceral adipose where the alternative name “omentin” is sometimes used6,8,11–13. Orthologs of human ITLN1 are induced during intestinal parasitemia and related Th2-type immune responses, including asthma9,14–18.
Human ITLN1 was found to recognize β-D-galactofuranose, a galactose isomer incorporated into the glycans of microorganisms, including protozoa, fungi, and bacteria, but not mammalian cells6,19,20. Within β-D-galactofuranose, ITLN1 binds to the exocyclic vicinal 1,2-diol, a chemical moiety present on other microbial carbohydrate-containing structures, such as Kdo2 (Di[3-deoxy-D-manno-octulosonic acid]) of lipopolysaccharide6,20–22. This carbohydrate specificity suggests microbial pattern recognition binding for ITLN1, in its otherwise elusive role in innate immunity6,19,20.
Genome-wide association studies (GWAS) have identified several single nucleotide polymorphisms (SNPs) present at the ITLN1 locus as risk alleles for Crohn’s disease, a main subtype of IBD, which is characterized by dysbiosis, epithelial barrier dysfunction, and immune function perturbations; processes consistent with an innate immune dysfunction1,23–28. Herein, we further characterize the ITLN1 locus in the context of IBD-associated SNPs, examine the intestinal expression and glycan binding of ITLN1 associated with risk and non-risk alleles, and localize ITLN1 protein expression to goblet cells of the human small and large intestine.
Results
The intelectin-1 locus and Crohn’s disease
To gain insight into the potential influence of IBD-associated allelic variants on expression and protein function, we genotyped surgical specimens from a previously reported cohort of individuals requiring surgical resection for Crohn’s disease or ulcerative colitis (n = 134)29. Accordingly, we found by direct sequence analysis of PCR products that the GWAS-identified SNP, rs2274910 (T/C), located in intron 3 of ITLN1, was associated with rs2274907 (A/T), which encodes a missense variant in exon 4, where valine is substituted with aspartic acid at amino acid position 109 (V109D)1,30. Utilizing the 1000 Genomes Project Phase 3 of the Ensembl database we found rs2274910 and rs2274907 are in linkage disequilibrium (LD) across human populations, consistent with findings of association in our U.S. cohort (Supplemental Table 1)31,32. We sequenced the entire coding region of ITLN1 cDNA in all specimens of this cohort, exploring the potential for additional exon variants in IBD subpopulations. We found rs2274908 (C/T: H89H/Q) exclusively as a synonymous (silent) variant associated with rs227490730. No additional missense variants were identified.
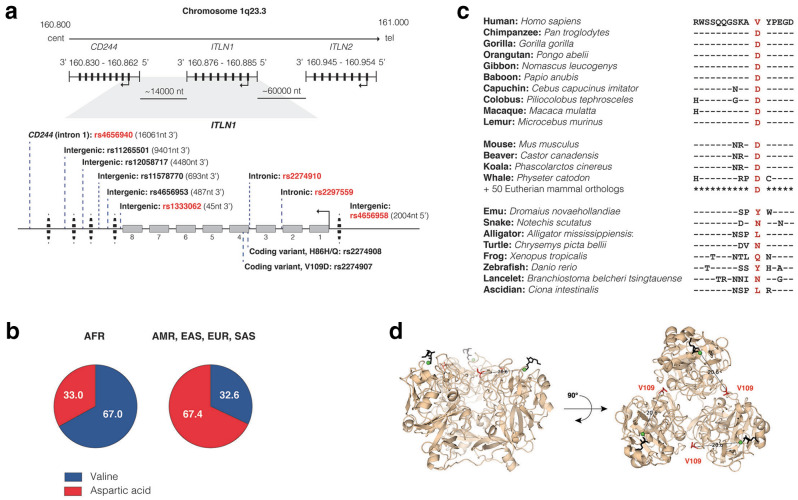
To display allelic variants associated with disease, we constructed a locus map of ITLN1 (Fig. 1A) annotated with published SNPs as outlined by the National Human Genome Research Institute—European Bioinformatics Institute (NHGRI-EBI) Catalog and NCBI-LitVar database, including variants within ITLN1 and its flanking regions intergenic to CD244 (3’) and ITLN2 (5’)33,34. ITLN1 and CD244 (rs1333062 and rs4656940, respectively) have been annotated together as risk loci for IBD, due in part to their close proximity (~ 14,000 nt)23,24. The SNP rs133062 is located in the 3’-flanking region of ITLN1, 46 nt intergenic to exon 8, where rs4656940 is located within intron 1 of CD244. The odds ratio (OR) for Crohn’s disease of rs4656940 is approximately equivalent (1.15; 95% confidence interval [CI] = 1.09–1.21) to that of rs2274910 (1.14; 95% CI = NR) of ITLN11,23. Several annotated SNPs in CD244 (rs4656942 and rs11265498) and in the adjacent gene LY9 (rs540254 and rs560681), but not any in neighboring SLAMF7, are in LD with GWAS-identified rs2274910 and rs4656940 (Supplemental Table 2)35. In addition, rs4656958 located in the 5' flanking region of ITLN1 (2004 nt 5’ to exon 1) has also been identified by GWAS as a risk allele for IBD (OR = 1.06; 95% CI = 1.03–1.09)25–27. Thus, GWAS-identified IBD-associated SNPs are located within ITLN1 and its flanking regions, and these SNPs are in LD with SNPs in CD244 and Ly9, which are genes located 3’ to ITLN1. Data in the GTEx browser revealed that quantitative trait loci (QTL) for expression (eQTL) in blood for ITLN1 and CD244 are both affected by several disease risk SNPs, including rs1333062, rs2274907 and rs2297559 (Supplemental Table 3)36. In addition, QTL for splicing (sQTL) of USF1 was similarly affected by these SNPs, as was sQTL for Ly9 by rs465940. No significant eQTLs or sQTLs were identified in small intestine or colon tissue36. Although other genes located at 1q23.3 may be responsible for the disease association, we chose to investigate ITLN1 as the strongest candidate based on our prior knowledge of the function of intelectins.
Figure 1.
ITLN1 polymorphisms. (a) The physical arrangement of ITLN1 relative to centromere/telomere orientation and flanking genes on chromosome 1q23.3 is diagramed, along with single nucleotide polymorphisms associated with human disease, including IBD: rs465694023, rs1126550158, rs1205871759, rs1157877060, rs4656953 61, rs133306224, rs227490730, rs227490830, rs22749101, rs229755925, and rs465695825–27. Red text highlights GWAS-identified SNPs. (b) 1000 Genomes Project Phase 3 super population allele frequency (percentages) of rs2274907 (A/T, V109D): AFR, African; AMR, American, EAS, East Asian; EUR, European; SAS, South Asian31,32. (c) 3D structure of human trimeric ITLN1 bound to β-D-galactofuranose (black). The distance between V109 residue (red) and coordinating calcium (green) are indicated on each monomer (PDB: 4WMY). (d) Amino acid sequence adjacent to coding variant rs2274907 (V109D) in orthologs of human ITLN1 (“—” identical residue present ITLN1).
Further review of ITLN1 rs2274910 (C/T) revealed a distinction in the frequency of the GWAS-identified risk allele (C) between African (28%) and non-African (67%) superpopulations (SP)31,32. This prompted us to further explore allele frequency in the context of different SP groups. The study populations for most IBD-focused GWAS are comprised primarily of non-African SP, including investigations identifying ITLN1 as a risk locus1,23,25–27,37. Using the Ensembl database, we performed pairwise LD analyses of disease-associated ITLN1 variants (Fig. 1a, Table 1) in African and European SP using rs2274907 (A/T: V109D) as a focus point simply due to it being a missense variant. In addition to being in complete LD (D’ = 1.0, r2 = 1.0) with rs2274910, rs2274907 (V109D) was found also to be in LD with GWAS-identified alleles across the ITLN1 locus, where risk alleles are the major alleles in European SP and minor alleles in African SP (Table 1, Fig. 1b)31,32,35.
Table 1.
European and African superpopulation haplotype analysis at the ITLN1 locus.
| rs4656940 (A/G) |
rs11265501 (G/A) |
rs12058717 (C/T) |
rs11578770 (C/T) |
rs4656953 (C/T) |
rs1333062 (T/G) |
rs2274907 (A/T) |
rs2274908 (G/A) |
rs2274910 (T/C) |
rs2297559 (G/A) |
rs4656958 (A/G) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Location: Chromosome 1q23.3; GRCh38:CM000663.2 (Forward strand) |
(CD244) 160,860,478 |
3’ Intergenic 160,867,138 |
3’ Intergenic 160,872,059 |
3’ Intergenic 160,875,846 |
3’ Intergenic 160,876,052 |
3’ Intergenic 160,876,494 |
ITLN1 Exon 4 160,882,036 |
ITLN1 Exon 4 160,882,104 |
ITLN1 Intron 3 160,882,256 |
ITLN1 Intron 2 160,884,736 |
5’ Intergenic 160,887,174 |
| Regional feature | – | – | – | Putative Regulatory Region | Putative Regulatory Region | Putative Regulatory Region | Coding (V109D) | Coding (H86H/Q) | – | – | Putative Regulatory Region |
|
Distance (nucleotides) to rs2274907 (V109D) [Allele correlated with rs2274907 (T)] |
−21,558 [A] |
−14,898 [G] |
−9977 [C] |
−6119 [C] |
−5984 [T] |
−5542 [G] |
0 |
+ 68 [A] |
+ 220 [C] |
+ 2700 [A] |
+ 5138 [G] |
|
European LD: rs2274907 D’, r2 [AF] |
0.83–0.95, 0.28–0.46 [G, 19.0] |
0.84–0.95, 0.33–0.47 [A, 20.0] |
Abs [T, 0.0] |
1.00, 0.17–0.36 [T, 10.0] |
1.00, 1.00 [T, 68.0] |
1.00, 1.00 [G, 68.0] |
[T, 68.0] |
1.00, 1.00 [A, 68.0] |
1.00, 1.00 [C, 68.0] |
0.97–1.00, 0.87–0.98 [A, 68.0] |
1.00, 0.97–1.00 [G, 68.0] |
|
African LD: rs2274907 D’, r2 [AF] |
0.41–0.83, 0.05–0.27 [G, 40.0] |
0.89–1.00, 0.65–0.78 [A, 63.0] |
0.71–1.00, 0.11–0.17 [T, 32.0] |
Abs [T, 0.0] |
0.97–1.00, 0.62–0.81 [T, 26.0] |
0.92–1.00, 0.78–0.96 [G, 32.0] |
[T, 33.0] |
1.00, 0.87–1.00 [A, 31.0] |
1.00, 0.68–0.87 [A, 28.0] |
0.50–0.91, 0.09–0.36 [A, 55.0] |
0.43–0.81, 0.07–0.31 [G, 54.0] |
The allele genotype frequencies in European SP: VV (7.6%), VD (49.3%), and DD (43.1%) contrasts sharply with that in the African SP VV (45.7%), VD (43.1%), and DD (11.2%) (Supplemental Fig. 1). In our small cohort of surgical specimens, allele frequencies were not significantly different for those diagnosed as either Crohn’s disease or ulcerative colitis as compared to respective controls, but there was a suggestion of skewing in specimens diagnosed as ileal compared to colonic Crohn’s disease, worthy of follow-up analysis in a larger cohort (Supplemental Table 1). Our findings clarify that rs2274907 (T, D109), which is in complete LD with GWAS-identified rs2274910 risk allele (C), is the major allele in non-African SP; in Crohn’s disease multiple GWAS report a further skewing of this high frequency of the risk allele observed in the studied populations1,23–27.
Intestinal intelectin-1 expression, glycan-binding, and oligomerization is unaltered by missense variant rs2274907 (A/T, V109D)
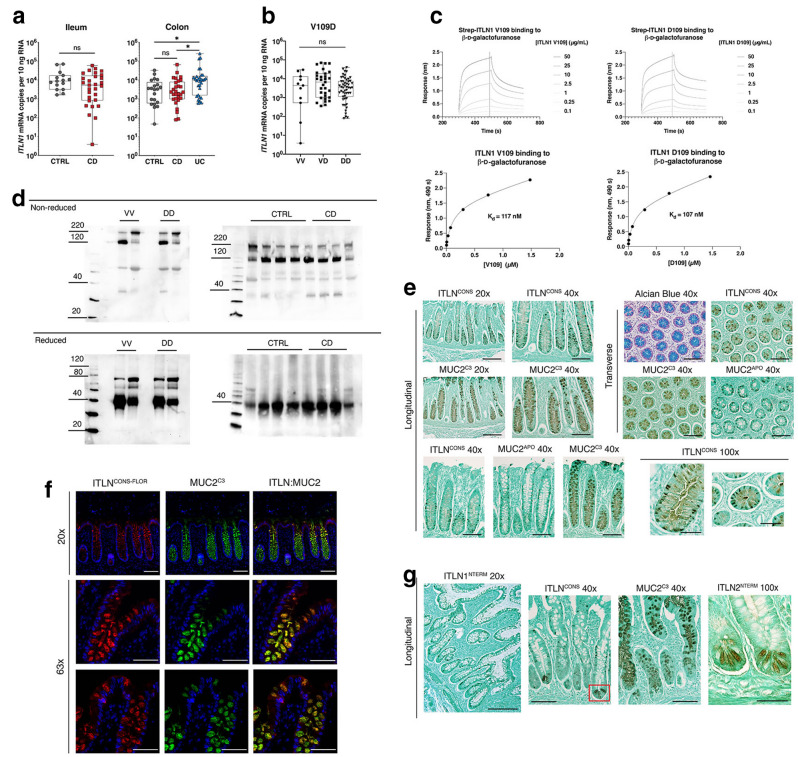
Di Narzo, et al. examined blood and intestinal eQTLs from a therapy-resistant Crohn's disease cohort38. Their analysis indicated eQTLs for ITLN1 in intestine (but not blood) are linked to GWAS-identified risk alleles rs4656958 and rs2274910, and eQTLs for CD244 in blood (but not intestine) are linked to the risk alleles rs4656958, rs4656940 and rs2274910. In both cases, a statistically significant decrease in expression is associated with the GWAS-identified SNPs. To better clarify expression patterns of ITLN1 within intestinal tissue, we generated an ITLN1-specific quantitative real-time PCR assay to enumerate the absolute mRNA transcript levels. ITLN1 mRNA was abundant in each anatomic segment across both small and large intestine (Supplemental Fig. 2a). Similarly, high levels of ITLN1 gene expression were observed in surgical specimens of both ileum and colon (Fig. 2a). Crohn’s disease diagnosis did not alter transcript levels in either small or large intestine (Fig. 2a). Of note, ITLN1 expression in the colon was elevated (p < 0.05) in specimens from individuals diagnosed with ulcerative colitis (Fig. 2a). To determine whether ITLN1 expression is modified by risk-associated haplotype, we stratified the data by genotype. In ileum, colon, and combined sample sets, the risk associated haplotype (queried by rs2274907 genotype) did not modify ITLN1 expression (Fig. 2b and Supplemental Fig. 2b). Together, these findings suggest that the mucosal transcript level of ITLN1 is unaltered in Crohn’s disease. Moreover, the risk-associated haplotype does not modify mucosal ITLN1 expression (Table 1).
Figure 2.
ITLN1 mRNA and protein expression in the small intestine and colon. (a) ITLN1 mRNA expression in surgical specimens of ileum and colon according to diagnosis. The mRNA copy number per 10 ng total RNA was determined with quantitative real-time RT-PCR using external cDNA standards (see methods). (b) ITLN1 mRNA expression by V109D genotype (excluding ulcerative colitis samples). Box plots present the median and first and third quartiles, whiskers present the range. Statistical analysis of ITLN1 mRNA expression was performed using either a Kruskal–Wallis test with Dunn’s multiple comparisons or two-tailed Mann–Whitney U test; * p < 0.05. (c) Representative real-time bio-layer interferometry responses of recombinant ITLN1 V109 and D109 binding to immobilized β-D-galactofuranose (upper). Responses at 490 s were plotted against ITLN1concentration and fit to a single site binding equation to determine Kd (lower). Data is representative of three separate experiments. (d) SDS-PAGE (4–20% gradient) immunoblots of human ileum by V109D genotype (left, n = 2 per genotype) and disease state (right). (e) Immunohistochemistry of human colon using the following antibodies: ITLN conserved (ITLNCONS), ITLN1-specific (ITLN1NTERM), MUC2 mature (MUC2C3), deglycosylated MUC2 (MUC2APO, PH1900). Tissue counterstained by DAPI (blue). (f) Immunofluorescence of human colon using ITLNCONS-FLOR (anti-chicken; Alexa Flour 647) and MUC2C3 (anti-rabbit; Alexa Flour 488). (g) Immunohistochemistry of human ileum; ITLN2-specific (ITLN2NTERM) antibody. Red box indicates Paneth cells identified with the ITLNCONS antibody. CTRL = control (non-Crohn’s disease and non-ulcerative colitis), CD = Crohn’s disease, UC = ulcerative colitis. Images in (e–g) are representative of multiple (> 6 fields) from 9 specimens. Light microscopy scale bars: 20x (200 µm), 40x (100 µm), and 100x (50 µm); fluorescence microscopy scale bars: 20x (100 µm) and 63x (50 µm).
The distinguishing chemical properties of valine and aspartic acid provided impetus to further explore the potential of missense variant V109D to modify ITLN1 oligomeric structure or function. Using PyMOL to visualize the published crystal structure of human ITLN1 (PBD: 4WMY), residue 109 was found to be located at the protein surface; however, it is not located at the sites annotated for glycan binding and/or calcium coordination (Fig. 1c)20,39. Bio-layer interferometry analysis found that recombinant V109-ITLN1 reversibly bound to immobilized β-D-galactofuranose with kinetics and affinity that mirror recombinant D109-ITLN1 (117 nM versus 107 nM, respectively) (Fig. 2c). Without finding discernable influence on glycan binding, we next tested if the V109D change altered ITLN1 oligomerization state and stability. For other secreted lectins (e.g., ficolins and mannose-binding lectin), oligomerization is important for avidity in binding cell-surface glycans as well as subsequent effector functions40–42. Previous studies report that human ITLN1 forms stable homotrimers, consisting of ~ 35 kDa monomers, connected through intermolecular disulfides (C31 and C48)6,11,43. Analysis of human ileal lysates confirmed that ITLN1 forms a stable homotrimer at approximately 110 kDa; however, a consistent ITLN1 band near the ~ 220 kDa marker was also identified using a gradient SDS-PAGE (4–20%). This suggests that human ITLN1 forms higher molecular weight oligomers (likely dimers of homotrimers), which are stable under denaturing (but not reducing) conditions (Fig. 2d). These findings agree with those reported for an amphibian ITLN1 ortholog, where dimers of homotrimers were shown to mediate bacterial agglutination40. We also performed immunoblots of ileal specimens isolated from V109 and D109 homozygote individuals to determine the potential influence on oligomer formation. Neither VV or DD genotype affected the formation of homotrimers or hexamers (Fig. 2d). These data suggest that that it is unlikely that the role of V109D variation in Crohn’s disease pathogenesis results from alterations in ITLN1 glycan binding or oligomerization.
Alternatively, we reasoned that D109 might influence protein stability, and that such differences would be observed as changes in protein abundance. In human ileal samples, we observed that under reducing conditions ITLN1 was present primarily as a monomer at ~ 40 kDa, with the exception of a minor fraction migrating at ~ 70 kDa (Fig. 2d). This minor band may represent a covalent non-reducible dimer but further study is needed to fully characterize this entity. Human ITLN1 possesses a single glycosylation site at N163, which slightly increases the molecular weight from that predicted (33 kDa) by the deduced amino acid sequence43. Ileal samples from individuals diagnosed with Crohn’s disease demonstrated equivalent relative proportions of protein oligomers and no additional degradation products compared to control specimens (Fig. 2d). Thus, ITLN1 was found to be an abundant protein in the small intestine, where expression was comparable in specimens from control and Crohn’s disease biopsies, consistent with mRNA expression data (Fig. 2a, d).
To further investigate the possible significance of V109D variation in Crohn’s disease, we performed a sequence analysis (NCBI BLAST) of the neighboring amino acid region across various ITLN1-like orthologs. In all Eutherian mammals queried (n = 63), aspartic acid (D) was reported as the amino acid at the corresponding position (Fig. 1d). Even amongst primates, valine at position 109 is unique to Homo sapiens. Nevertheless, other chordates were found to encode hydrophobic amino acids like valine (e.g., leucine) at this position, including ancient ascidians (Fig. 1d). While aspartic acid marks the risk-allele associated with Crohn's disease, the V109D variation is not a “mutation”, despite its location in a conserved domain of the protein and differences in chemical properties of these residues. Furthermore, V109, or the genetic variants in LD with this allele, could be considered “protective” in the context of Crohn’s disease.
Human intelectin-1 is a goblet cell product
In mice, where mammalian intelectins were first discovered, Itln1 is expressed in Paneth cells8. In contrast, a recent single-cell RNA sequence analysis investigation identified ITLN1 mRNA in goblet cells of the ileum and colon44. We sought to localize ITLN1 protein in this these tissues to confirm and extend this cell-type expression pattern in humans. Both ITLN1 and ITLN2 are expressed in the human small intestine and cannot be distinguished using most commercially available antibodies; therefore, we generated paralog-specific antibodies to clarify the cellular origin of ITLN1. Colon biopsies of non-IBD subjects, preserved in fixatives to retain mucus (either Carnoy’s fixative or HistoChoice™), were incubated with either antisera targeting a domain highly conserved in intelectins (ITLNCONS) or targeting the N-terminal specific domain (ITLN1NTERM). Both ITLNCONS and ITLN1NTERM immunoreactivity were localized to colonic goblet cells (Fig. 2e, Supplemental Fig. 2c). To confirm goblet cell localization, samples were stained with a mucin-2 (MUC2) antibody generated against a peptide sequence located within the C-terminal domain (adjacent to the autocatalytic sequence) of the mature, glycosylated MUC2 protein (MUC2C3); this antibody targets goblet cell granules and secreted protein45. While MUC2C3 stained goblet cells most strongly in their secretory granule, the cellular staining pattern of ITLN1 was different. Specifically, punctate staining of ITLN1 adjacent to the granule and relatively weak staining within the granule was a feature of all ITLN1-positive samples tested (n = 6 colon, n = 3 ileum). To further characterize the staining pattern of ITLN1 within goblet cells, we used another MUC2 antibody that targets the tandem repeat region of the apo-MUC2 protein (PH1900, MUC2APO). This antibody identifies non-O-glycosylated MUC2, which resides within the endoplasmic reticulum region of goblet cells46,47. MUC2APO staining of the cytoplasmic region in goblet cells mimicked the staining pattern for ITLN1NTERM (Fig. 2e, Supplemental Fig. 2C). At higher magnification, ITLN1NTERM staining was identified in both cytoplasmic and granule compartments, and readily apparent in both longitudinally and transversely orientated tissue sections (Fig. 2e). To better localize ITLN1 within goblet cells, we generated a chicken anti-ITLN antibody (conserved, ITLNCONS-FLOR) for dual labeling with the rabbit-derived MUC2C3 reagent in fluorescence microscopy. As observed by conventional microscopy, ITLN1 and MUC2 largely colocalized within colonic goblet cells, where ITLN1 was also found occasionally in the cytosolic compartments adjacent to nuclear (DAPI) staining (Fig. 2f). Immunohistochemistry of human ileum using the ITLN1NTERM antibody also identified goblet cells, where the aforementioned cytosolic and granular staining pattern was also evident (Fig. 2g). It is worth noting that as a feature distinct from findings in mice, human Paneth cells remained unstained with the ITLN1NTERM antibody (Supplemental Fig. 2D). Although staining of human ileum with the ITLNCONS antibody identified Paneth cells in addition to goblet cells, paralog-specific antibodies determined Paneth cell staining to be human ITLN2 (Fig. 2g, and Nonnecke, et al.: unpublished observations). Together, our results demonstrate that human ITLN1 is expressed in goblet cells, both in the ileum and colon.
Discussion
Various GWAS investigations have identified SNPs at the ITLN1 locus as risk alleles for Crohn’s disease1,23–27. We observed that these SNPs are in LD, forming a risk haplotype that extends into the adjacent CD244 and Ly9 genes. Given the abundant expression of ITLN1 in intestinal tissues and the likely role of this lectin in innate immunity, we focused our investigation on ITLN1 in the intestine.
The first GWAS-identified SNP at this locus (rs2274910) is located in intron 3 of ITLN11. We determined that this variant was associated with an adjacent SNP in exon 4 (rs2274907), which encodes a valine to aspartic acid amino change at residue 109. Although valine is the most common allele in African SP, aspartic acid is both the most common and the disease-associated allele in European SP—the primary population studied in IBD GWAS1,23–27. Our results clarify that V109D is in LD with other GWAS-identified Crohn's disease-associated risk SNPs, resulting in a risk haplotype. Our data suggest that mRNA transcript and protein abundance of mucosal ITLN1, now confirmed to be goblet-cell derived, is unaltered in the Crohn’s disease samples overall, as well as in samples analyzed according to rs2274907 (A/T, V109D) genotype in both disease and non-disease specimens. We clarify that ITLN1 D109 is the ancestral amino acid in mammalian orthologs, supporting that D109 is not a mutation. Furthermore, our data indicate that any role(s) of V109D in Crohn’s disease pathogenesis is unlikely due to changes in ITLN1 carbohydrate recognition or protein oligomerization.
Although multiple GWAS have identified ITLN1 SNPs in Crohn’s disease, the calculated OR(s) noted above are modest compared to variants of nucleotide-binding oligomerization domain-containing protein 2 (NOD2/CARD15): rs2066847 (frameshift variant L1007fsX1008: OR = 3.99; CI = NR) and interleukin 23 receptor (IL23R): rs11465804 (intron variant T/G: OR = 1.38—2.56)1,48–50. The OR for Crohn’s disease risk associated with the susceptible haplotype should be viewed from the perspective that the risk alleles are also the major alleles in European SP. Unlike NOD2/CARD15 (rs2066847) and IL23R (rs11465804), which are rare variants (1% and 3% globally), ITLN1 rs2274910 (T/C), and other alleles forming the risk haplotype are common, where T and C alleles of rs2274910 are present at 43% and 57%, respectively, across Phase 3 Individuals. Similarly, the Crohn’s disease risk allele of autophagy-related 16-like 1 (ATG16L1) rs2241880 (A/G) is common (60:40%) and associated with a modest OR (1.45; CI = 1.27—1.64)31,32.
Further investigation is required to determine the mechanism by which genetic variation at this locus affects risk for Crohn's disease. In this study, we tested relevant possibilities, and our data support that it is unlikely to be due to ITLN1 expression in intestinal goblet cells. It is possible that expression of ITLN1 in another tissue could underlie disease risk. Moreover, expression of other genes at this locus, notably either CD244 or Ly9, which include SNPs in LD with the risk haplotype might explain disease risk33–35. Based on available QTL data, expression of USF1 or CD244 in whole blood would also be reasonable candidates for future investigation36.
Methods
Human specimens and ethics statement
All methods were carried out in accordance with relevant guidelines and regulations. Surgical specimens were obtained at The Cleveland Clinic Foundation (Cleveland, OH) with informed consent from all patients, and deidentified under protocols approved by the Institutional Review Board at The Cleveland Clinic Foundation as previously described29. These specimens were collected from subjects undergoing surgery for Crohn’s disease (CD), ulcerative colitis (UC), or non-CD/UC-related (control, CTRL) surgical indications (i.e., colon cancer, bowel obstruction, and familial adenomatous polyposis). Race was recorded for these specimens based on medical record reporting. Diagnoses were based on standard criteria using clinical, radiological, endoscopic and histopathological findings29. Exclusion criteria included the diagnoses of backwash ileitis, indeterminate colitis, concurrent CMV or C. difficile infection. Samples were immediately either snap frozen in liquid nitrogen (for RNA, DNA, and protein analysis) or placed into tissue fixative for histology. The mucosa, utilized for mRNA and protein expression, was dissected away from the intestinal/colonic serosa and associated adipose tissue. The overview of regional mRNA expression (Suppl. Figure 1) used commercially-obtained RNAs of human tissue (Biochain, Newark, CA).
Databases
Single nucleotide polymorphisms (SNPs) referenced to human ITLN1 were identified using the NHGRI-EBI Catalog of human genome-wide association studies (https://www.ebi.ac.uk/gwas/home) and NCBI-LitVar (https://www.ncbi.nlm.nih.gov/CBBresearch/Lu/Demo/LitVar/#!?query =) databases33,34. Further investigation of identified SNPs, including descriptions of putative function and population frequency, was procured from Ensembl and the 1000 Genomes Project (Phase 3)31,32. Allele (i.e., nucleotide) associations and linkage disequilibrium were studied using the NCBI-LDlink (LDpair) database (https://ldlink.nci.nih.gov/?tab=help#LDpair)35. QTL analysis was queried at The Genotype-Tissue Expression (GTEx) Portal (https://www.gtexportal.org/home/)36. Missense variant rs2274907 (V109D) was applied to the referenced crystal structure of human ITLN1 (PBD ID: 4WMQ) using PyMOL software (The PyMOL Molecular Graphics System, Version 1.8.2.1 Schrödinger, LLC., https://pymol.org/2/) to orientate location.
Genotyping
Genomic DNA was isolated and purified using the QIAamp DNA minikit (Qiagen, Germantown, MD) according to the manufacturer’s protocol. Isolated DNA was quantified by ultraviolet absorbance spectroscopy (260 nm) using a NanoDrop spectrophotometer (Thermo Scientific/NanoDrop Products, Wilmington, DE). Exon variants in each specimen of our IBD cohort were explored by Sanger sequencing of cDNA corresponding to the entire coding region of ITLN1 amplified using specific primers (2 s: 5’- GGAGCGTTTTTGGAGAAAGCTGCA-3’ and 1147a: 5’- ATTAACATTCTAGCTACTGGGT-3’). The sequenced PCR products were aligned to the NCBI-ITLN1 reference (NM_017625.3), identifying rs2274907 (A/T: V109D) and rs2274908 (C/T: H89H/Q) in some specimens. Association of single nucleotide polymorphisms rs2274910 (T/C) and rs2274907 (A/T) was identified by PCR amplification from genomic DNA across adjacent regions in multiple samples, including both homozygous, as well as heterozygous (T/C and A/T) genotypes using specific oligonucleotide primers (1062 s: 5’-ACAGGAGCCTTTAGGCCATGT-3’, 1431a: 5’-AGTGGCACCAACCTTGTAGTC-3’). The PCR reactions were initiated by denaturation of the DNA template at 95 °C for 10 min followed by 45 cycles consisting of 95 °C for 15 s, a -1 °C per two-cycle ‘touchdown’ annealing temperature for 5 s (i.e., 65 °C to 58 °C), and 10 s at 72 °C. Primers were designed using MacVector Software (version 16, MacVector, Apex, NC), and synthesized by Invitrogen Life Technologies (Invitrogen, Carlsbad, CA).
RNA isolation
The general procedures for RNA isolation and synthesis of cDNA were previously described by our group51,52. For the current study, RNA was freshly isolated for all specimens from frozen tissue29. Briefly, frozen tissue was dispersed using Brinkmann Polytron homogenizer in guanidinium thiocyanate (GTC) buffer (4–5 M GTC, 0.5% sodium-N-lauryl sarcosine, 10 mM EDTA, 20 mM Tris–HCl 0.1 M 2-mercaptoethanol). Homogenates were layered above 5.7 M CsCl/EDTA in diethyl pyrocarbonate (DEPC) H2O in polyallomer tubes (11 × 34 mm; Beckman, Indianapolis, IN). Using a TLS-55 swinging bucket rotor (Beckman, Indianapolis, IN) samples were centrifuged for 3 h at 55,000 rpm in a TL-100 ultracentrifuge (Beckman). Following centrifugation, excess GTC and CsCl were removed by aspiration and the RNA pellet was solubilized with RNA sample buffer (0.5% N-lauroylsarcosine, 5 mM EDTA, 5 mM tris, and 5% 2-mercaptoethanol). The RNA-buffer solution was transferred to tubes containing an equal volume of 50:50 phenol–chloroform (Calbiochem/EMD Millipore, San Diego, CA), vortexed, and centrifuged for 3 min at 13,000 rpm. The resulting supernatant was washed with 100% chloroform (Sigma-Aldrich, St. Louis, MO), centrifuged, and added to tubes containing 100% ETOH and 0.3 M sodium acetate. Following overnight incubation at -80 °C, RNA was precipitated by centrifugation for 20 min at 10,000 rpm and washed once with 80% ETOH. The resulting RNA pellet was re-suspended in DEPC H2O, and concentration was determined by ultraviolet absorbance spectroscopy (260 nm)51,52.
cDNA synthesis
RNA (2–5 μg) was precipitated under centrifugation, washed once with 80% ETOH and re-suspended in DEPC H2O. cDNA synthesis was performed according to the procedures outlined by the SuperScript III First-Strand cDNA Synthesis System (Invitrogen, Thermo Fischer Scientific, Waltham, MA). The cDNA product was purified using a column QIAquick PCR Purification Kit (Qiagen, Germantown, MD).
Quantitative real time PCR (qRT-PCR)
Primers specific for human ITLN1 (211 s: 5’-CTTCGTCTCCATCTCTGCCC-3’, 297a: 5’-CTGGTAGATAACACCATTCTC-3’) and ACTB (128 s: 5’- TGATGGTGGGCATGGGTCAG-3’, 220a: 5’- CGTGCTCGATGGGGTACTTCAG-3’) were designed using MacVector version 16 Software. Real-time PCR was performed on cDNA template corresponding to 10 ng of RNA using a Roche Diagnostics Lightcycler 2.0 (Roche, Indianapolis, IN) as previously described51,52. Briefly, each reaction included 4 mM MgCl2, 0.5 μM forward and reverse primers, and 1 × LightCycler FastStart SYBR Green I mix (Roche, Mannheim, Germany). A control without cDNA template was included in the procedural assay. The PCR conditions were: initial denaturation at 95 °C for 10 min, followed by 45 cycles with each cycle consisting of denaturation, 95 °C for 15 s; annealing at 60 °C for 5 s; and extension at 72 °C for 10 s. Following the cycle runs, samples were denatured to establish the melting temperature(s) of the PCR product. The sample melt temperatures were compared to that of the internal standard to confirm template specificity. Absolute quantification of target gene within each sample was obtained by inclusion of an external cloned cDNA standard of known concentration, where crossing point (i.e., cycle number) is matched to a standard curve. Data was plotted in Prism version 9.0.0 (GraphPad).
Rabbit anti-ITLN1 polyclonal antibodies
Analysis of the pre-ITLN1 deduced protein sequence using the SignalP4.0 algorithm indicated signal cleavage would occur between G16 and W1753. The adjacent 19 amino acids of ITLN1 (WSTDEANTYFKEWTCSSSP) comprised a completely unique sequence compared to the corresponding region of the human ITLN2 sequence (AAASSLEMLSREFETCAFSFS). Adjacent to this discriminating 19-amino acid sequence (W17 to P35) of ITLN1 is a domain beginning at amino acid S36 that has very high sequence identity to ITLN2 as well as other intelectin orthologs. BLASTp search of the non-redundant protein database with the 19-amino acid W17 to P35 peptide sequence yielded numerous primate ITLN1 orthologs with highly similar sequence (> 80%), but no other mammalian proteins. This 19-amino acid peptide was synthetically prepared, purified and characterized to confirm proper mass, as previously described54. The peptide was coupled to ovalbumin (OVA, Thermo Fischer Scientific, Waltham, MA) at a ratio of 20 molecules peptide per molecule OVA using two strategies. First, one equivalent volume of 0.2% glutaraldehyde was added to the 20:1 peptide/ovalbumin mixture in PBS; the mixture was then stirred for 1 h at room temperature. 1 M glycine was added to a final concentration of 200 mM and stirred for an additional 1 h. Second, the peptide was reacted with maleimide-activated-ovalbumin (Imject™ Maleimide-Activated Ovalbumin, Thermo Fischer Scientific, Waltham, MA) according to the suppliers’ protocol. Briefly, the peptide was mixed with the activated ovalbumin at a 20:1 molar ratio in PBS and allowed to react at room temperature for 2 h. Each of the two peptide/OVA conjugates were dialyzed against PBS for 24 h at 4 °C, changing dialysis buffer every 6–8 h. The protein concentration was adjusted to 750 µg/ml with PBS and then equal portions of the two peptide/OVA conjugates were mixed to then use as antigen. In a separate set of reactions, the peptide was coupled to keyhole limpet hemocyanin (KLH, Thermo Fischer Scientific, Waltham, MA) at a ratio of 200 molecules peptide per molecule KLH using the identical protocols (glutaraldehyde and maleimide-activated-KLH (Imject™ Maleimide-Activated KLH, Thermo Fischer Scientific, Waltham, MA) and equal portions of the resulting two peptide/KLH conjugates were mixed to use as antigen. Rabbits were immunized by Antibodies Inc. (Davis, CA) with the ITLN1 peptide/OVA antigen on days 1, 14, 21, and then boosted by immunization with ITLN1 peptide/KLH antigen on days 35 and 49 to produce the N-terminal polyclonal antisera (termed ITLN1NTERM). Preimmunization sera was collected and used as control antisera. All methods for these procedures were in accordance with relevant guidelines and regulations, and approved by the U.S. Department of Health and Human Services Public Health Service Animal Welfare Assurance Committee (Assurance ID D16-00576). Rabbit polyclonal antiserum generated by immunization with human ITLN1-derived synthetic peptides was described and characterized previously (termed ITLNCONS)11,55.
Chicken anti-ITLN1 IgY
The ITLN1 protein sequence was analyzed using the online algorithm for design of peptide-directed antibodies provided by the NHLBI (https://hpcwebapps.cit.nih.gov/AbDesigner/)56. A 28 aa peptide (CTVGDRWSSQQGSKADYPEGDGNWANYN) was identified as an attractive antigen candidate. This peptide spanned the V108D polymorphic site, and was highly similar in sequence to human ITLN2 and C57BL/6 mouse Itln1 (27/28 identical residues to both of these orthologs). This peptide was synthetically prepared, purified and characterized54. In two separate reactions, the peptide was coupled to KLH, using the two chemical coupling strategies (glutaraldehyde and maleimide) and protocols, as described above. Each reaction product was dialyzed against PBS and protein concentration adjusted to 750 µg/ml. Equal portions of the two peptide/KLH conjugates were mixed to use as antigen. Leghorn chickens were given a primary inoculation with the KLH conjugate in complete Freund's adjuvant, followed by two booster inoculations at 3 and 6 weeks (with incomplete Freund's adjuvant) by Aves Labs, Inc. (Davis, CA). An enriched IgY fraction from egg yolk of immunized chickens (termed ITLNCONS-FLOR) was obtained by organic extraction de-lipidation, high-salt precipitation and then extensive dialysis against PBS.
Bio-layer interferometry
Dissociation constants for recombinant ITLN1 with biotin-β-D-galactofuranose were determined using a bio-layer interferometry OctetRed instrument (ForteBio, Fremont, CA). Biotin-β-D-galactofuranose was diluted to 5 μM in PBS, pH 7.5, and loaded (120 s) onto Streptavidin biosensors (Forte Bio). The sensor was washed in PBS for 60 s, and then a baseline (60 s) established in (20 mM HEPES, pH 7.4, 150 mM sodium chloride, 10 mM calcium chloride, 0.1% Tween-20, 0.1% BSA). Recombinant ITLN1 was diluted to 50, 25, 10, 2.5, 1, 0.25, and 0.1 µg/mL in (20 mM HEPES, pH 7.4, 150 mM sodium chloride, 10 mM calcium chloride, 0.1% Tween-20, 0.1% BSA) and allowed to associate with the biotin-β-D-galactofuranose coated biosensor (200 s). Dissociation (200 s) was monitored by dipping biosensor in (20 mM HEPES, pH 7.4, 150 mM sodium chloride, 10 mM calcium chloride, 0.1% Tween-20, 0.1% BSA). Biosensors were regenerated between samples by triple-washing biosensors in 10 mM glycine pH 2.5 (5 s) followed by neutralization (5 s) in (20 mM HEPES, pH 7.4, 150 mM sodium chloride, 10 mM calcium chloride, 0.1% Tween-20, 0.1% BSA). The shake rate was set to 1000 rpm and binding assays were performed at room temperature. Data was plotted in Graphpad Prism software (version 9.0.0,Graphpad Software, San Diego, CA). The response at equilibrium (490 s) was plotted against concentration of ITLN1 and curves fit to a one-site total non-linear regression equation to determine equilibrium dissociation constants.
Immunoblotting
Ileal non-CD and CD specimens, genotyped for ITLN1 rs2274907 (V109D), were homogenized in assay buffer (150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) containing a cocktail (1:100) of protease inhibitors (Protease Inhibitor Cocktail III: 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF, 1 mM), aprotinin (600 nM), bestatin (50 µM), E-64 (15 µM), leupeptin (20 µM),pepstatin (10 µM); Calbiochem/EMD Millipore, Burlington, MA). Protein concentration was determined by bicinchoninic acid (BCA) assay (Peirce, ThermoFischer Scientific). Following heating at 95 °C for 30 min (with or without 2.5% v/v 2-mercaptoethanol for reducing conditions), protein (25–50 μg/lane) was loaded to SDS–polyacrylamide gels (10% and 4–20% acrylamide) and electrophoresed for 40–50 min at 200 V, prior to wet transfer (Towbin Buffer: 25 mM Tris, 192 mM glycine, 20% v/v methanol, pH 8.3) to nitrocellulose membranes (0.2 μM pore, Bio-Rad Laboratories, Hercules, CA) at 350 mA for 60–80 min. Estimated molecular sizes of ITLN1 monomers and higher order oligomers were deduced using the MagicMark™ XP Western Protein Standard (ThermoFisher Scientific). Membranes were blocked in PBS-T containing 5% w/v skim milk for 30 min, and incubated with primary antibodies at 4 °C. Following overnight incubation, membranes were washed with PBS-T for 1 h, probed with donkey anti-rabbit IgG horseradish peroxidase (HRP) linked secondary antibody (GE Healthcare Amersham, Pittsburgh, PA) for 3 h at room temperature, rinsed in PBS-T, and visualized using Femto and ECL West chemiluminescent substrates (ThermoFisher Scientific). Chemiluminescent signal was detected using a Biospectrum AC Imaging System (UVP, Upland, CA).
Immunohistochemistry (IHC)
Surgical tissue of human ileum and colon, fixed in Carnoy's fixative or HistoChoice™ Tissue Fixative (Sigma Aldrich, St. Louis, MO), were paraffin embedded and sectioned (4–5 μM) and mounted on X-tra™ positive-charged slides (Leica Biosystems, Buffalo Grove, IL). Sections were deparaffinized, rehydrated in gradient ethanol, H2O, and PBS and blocked with 5% donkey serum (in 1 × PBS) for 30 min prior to overnight incubation at 4 °C with antibody51. Following incubation, slides were washed, and probed with Vectastain ABC kit goat anti-rabbit/goat secondary antibodies (Vector Laboratories, Burlingame, CA), activated with 3,3-diaminobenzidine peroxidase (Vector Laboratories, Burlingame, CA), and counterstained in 0.4% light green (Sigma Aldrich, St. Louis, MO), 0.2% glacial acetic acid for 5 min prior to dehydration and mounting51. Samples were visualized using an Olympus BX51 microscope (Center Valley, PA) and images were processed using Adobe Creative Suite software (version 25, Adobe Inc., San Jose, CA).
Fluorescence IHC
Specimen slides were prepared as before. Following overnight incubation with primary antibodies, samples were incubated with two secondary antibodies (goat anti-chicken IgY Alexa Flour Plus 647 [Thermo Fischer Scientific, Waltham, MA] and goat anti-rabbit IgG Alexa Flour 488 [Abcam, Cambridge MA]), rinsed, and then stained with DAPI using the TrueVIEW Autofluorescence Quenching Kit (Vector Laboratories, Burlingame, CA) as outlined by the manufacturer. Images were captured using a Leica SP8 STED 3 × microscope (Leica Microsystems Inc., Buffalo Grove, IL). Acquired Z-stacks were further processed to generate figure images using Fiji ImageJ software (version 1.0, https://imagej.net/Fiji)57.
Statistical analysis
Statistical analysis was performed using Graphpad Prism software (version 9.0.0,Graphpad Software, San Diego, CA). Specific details are outlined in figure and table legends.
Supplementary Information
Acknowledgements
We thank Dr. Ingrid Brust-Mascher and the Health Sciences District Advanced Imaging Facility at UC Davis for assistance with confocal microscopy. Research was supported by National Institutes of Health (U01AI125926 [Mucosal Immunology Study Team], R37AI32738, U01CA231079), and the MIT Center for Microbiome Informatics and Therapeutics.
Author contributions
E.B.N., B.L., and C.L.B. designed research; E.B.N., P.A.C., A.E.D., F.A. and W.Y. performed research; C.A.D.L.M., W.Y., W.L., B.S., and M.E.V.J. contributed new reagents/analytic tools; E.B.N., P.A.C., A.E.D., M.A.U., C.A.D.L.M., B.S., M.E.V.J., L.L.K., E.J.H., B.L., and C.L.B. analyzed data; and E.B.N. and C.L.B. wrote the paper. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Eric B. Nonnecke, Email: ebnonnecke@ucdavis.edu
Charles L. Bevins, Email: clbevins@ucdavis.edu
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92198-9.
References
- 1.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Souza Batista, C. M. et al. Omentin plasma levels and gene expression are decreased in obesity. Diabetes56, 1655–1661, doi:10.2337/db06-1506 (2007). [DOI] [PubMed]
- 3.Pemberton AD, Rose-Zerilli MJ, Holloway JW, Gray RD, Holgate ST. A single-nucleotide polymorphism in intelectin 1 is associated with increased asthma risk. J Allergy Clin Immunol. 2008;122:1033–1034. doi: 10.1016/j.jaci.2008.08.037. [DOI] [PubMed] [Google Scholar]
- 4.Yan J, et al. Comparative genomic and phylogenetic analyses of the intelectin gene family: implications for their origin and evolution. Dev Comput Immunol. 2013;41:189–199. doi: 10.1016/j.dci.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 5.Lee JK, Baum LG, Moremen K, Pierce M. The X-lectins: a new family with homology to the Xenopus laevis oocyte lectin XL-35. Glycoconj J. 2004;21:443–450. doi: 10.1007/s10719-004-5534-6. [DOI] [PubMed] [Google Scholar]
- 6.Tsuji S, et al. Human intelectin is a novel soluble lectin that recognizes galactofuranose in carbohydrate chains of bacterial cell wall. J Biol Chem. 2001;276:23456–23463. doi: 10.1074/jbc.M103162200. [DOI] [PubMed] [Google Scholar]
- 7.Lee JK, et al. Human homologs of the Xenopus oocyte cortical granule lectin XL35. Glycobiology. 2001;11:65–73. doi: 10.1093/glycob/11.1.65. [DOI] [PubMed] [Google Scholar]
- 8.Komiya T, Tanigawa Y, Hirohashi S. Cloning of the novel gene intelectin, which is expressed in intestinal paneth cells in mice. Biochem Biophys Res Commun. 1998;251:759–762. doi: 10.1006/bbrc.1998.9513. [DOI] [PubMed] [Google Scholar]
- 9.Pemberton AD, et al. Innate BALB/c enteric epithelial responses to Trichinella spiralis: inducible expression of a novel goblet cell lectin, intelectin-2, and its natural deletion in C57BL/10 mice. J Immunol. 2004;173:1894–1901. doi: 10.4049/jimmunol.173.3.1894. [DOI] [PubMed] [Google Scholar]
- 10.Lu ZH, et al. Strain-specific copy number variation in the intelectin locus on the 129 mouse chromosome 1. BMC Genomics. 2011;12:110. doi: 10.1186/1471-2164-12-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki YA, Shin K, Lönnerdal B. Molecular cloning and functional expression of a human intestinal lactoferrin receptor. Biochemistry. 2001;40:15771–15779. doi: 10.1021/bi0155899. [DOI] [PubMed] [Google Scholar]
- 12.Yang R, et al. Cloning of omentin, a new adipocytokine from omental fat tissue in humans. Diabetes. 2003;52:1. doi: 10.2337/diabetes.52.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Schäffler A, et al. Genomic structure of human omentin, a new adipocytokine expressed in omental adipose tissue. Biochim Biophys Acta. 2005;1732:96–102. doi: 10.1016/j.bbaexp.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 14.French AT, et al. The expression of intelectin in sheep goblet cells and upregulation by interleukin-4. Vet Immunol Immunopathol. 2007;120:41–46. doi: 10.1016/j.vetimm.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 15.Voehringer D, et al. Nippostrongylus brasiliensis: identification of intelectin-1 and -2 as Stat6-dependent genes expressed in lung and intestine during infection. Exp Parasitol. 2007;116:458–466. doi: 10.1016/j.exppara.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.French AT, et al. Up-regulation of intelectin in sheep after infection with Teladorsagia circumcincta. Int J Parasitol. 2008;38:467–475. doi: 10.1016/j.ijpara.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Kerr SC, et al. Intelectin-1 is a prominent protein constituent of pathologic mucus associated with eosinophilic airway inflammation in asthma. Am J Respir Crit Care Med. 2014;189:1005–1007. doi: 10.1164/rccm.201312-2220LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe T, et al. Expression of intelectin-1 in bronchial epithelial cells of asthma is correlated with T-helper 2 (Type-2) related parameters and its function. Allergy Asthma Clin Immunol. 2017;13:35. doi: 10.1186/s13223-017-0207-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuji S, et al. Capture of heat-killed Mycobacterium bovis bacillus Calmette-Guérin by intelectin-1 deposited on cell surfaces. Glycobiology. 2009;19:518–526. doi: 10.1093/glycob/cwp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wesener DA, et al. Recognition of microbial glycans by human intelectin-1. Nat Struct Mol Biol. 2015;22:603–610. doi: 10.1038/nsmb.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMahon CM, et al. Stereoelectronic effects impact glycan recognition. J Am Chem Soc. 2020;142:2386–2395. doi: 10.1021/jacs.9b11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rose NL, et al. Expression, purification, and characterization of a galactofuranosyltransferase involved in Mycobacterium tuberculosis arabinogalactan biosynthesis. J Am Chem Soc. 2006;128:6721–6729. doi: 10.1021/ja058254d. [DOI] [PubMed] [Google Scholar]
- 23.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellinghaus D, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Lange KM, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumgart DC, Sandborn WJ. Crohn's disease. Lancet. 2012;380:1590–1605. doi: 10.1016/s0140-6736(12)60026-9. [DOI] [PubMed] [Google Scholar]
- 29.Wehkamp J, et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci USA. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schäffler A, et al. Frequency and significance of the novel single nucleotide missense polymorphism Val109Asp in the human gene encoding omentin in Caucasian patients with type 2 diabetes mellitus or chronic inflammatory bowel diseases. Cardiovasc Diabetol. 2007;6:3. doi: 10.1186/1475-2840-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yates AD, et al. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–d688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buniello A, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–d1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allot A, et al. LitVar: a semantic search engine for linking genomic variant data in PubMed and PMC. Nucleic Acids Res. 2018;46:W530–w536. doi: 10.1093/nar/gky355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The Genotype-Tissue Expression (GTEx) project. Nat Genet45, 580–585, doi:10.1038/ng.2653 (2013). [DOI] [PMC free article] [PubMed]
- 37.Brant SR, et al. Genome-wide association study identifies African-specific susceptibility loci in African Americans with inflammatory bowel disease. Gastroenterology. 2017;152:206–217. doi: 10.1053/j.gastro.2016.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Narzo AF, et al. Blood and intestine eQTLs from an anti-TNF-resistant Crohn's disease cohort inform IBD genetic association loci. Clin Transl Gastroenterol. 2016;7:e177. doi: 10.1038/ctg.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, et al. iCn3D, a web-based 3D viewer for sharing 1D/2D/3D representations of biomolecular structures. Bioinformatics. 2020;36:131–135. doi: 10.1093/bioinformatics/btz502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wangkanont K, Wesener DA, Vidani JA, Kiessling LL, Forest KT. Structures of xenopus embryonic epidermal lectin reveal a conserved mechanism of microbial glycan recognition. J Biol Chem. 2016;291:5596–5610. doi: 10.1074/jbc.M115.709212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garlatti V, et al. Structural insights into the innate immune recognition specificities of L- and H-ficolins. Embo j. 2007;26:623–633. doi: 10.1038/sj.emboj.7601500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wesener DA, Dugan A, Kiessling LL. Recognition of microbial glycans by soluble human lectins. Curr Opin Struct Biol. 2017;44:168–178. doi: 10.1016/j.sbi.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuji S, et al. Differential structure and activity between human and mouse intelectin-1: human intelectin-1 is a disulfide-linked trimer, whereas mouse homologue is a monomer. Glycobiology. 2007;17:1045–1051. doi: 10.1093/glycob/cwm075. [DOI] [PubMed] [Google Scholar]
- 44.Wang, Y. et al. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J Exp Med217, doi:10.1084/jem.20191130 (2020). [DOI] [PMC free article] [PubMed]
- 45.Johansson ME, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asker N, Axelsson MA, Olofsson SO, Hansson GC. Dimerization of the human MUC2 mucin in the endoplasmic reticulum is followed by a N-glycosylation-dependent transfer of the mono- and dimers to the Golgi apparatus. J Biol Chem. 1998;273:18857–18863. doi: 10.1074/jbc.273.30.18857. [DOI] [PubMed] [Google Scholar]
- 47.Axelsson MA, Asker N, Hansson GC. O-glycosylated MUC2 monomer and dimer from LS 174T cells are water-soluble, whereas larger MUC2 species formed early during biosynthesis are insoluble and contain nonreducible intermolecular bonds. J Biol Chem. 1998;273:18864–18870. doi: 10.1074/jbc.273.30.18864. [DOI] [PubMed] [Google Scholar]
- 48.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raelson JV, et al. Genome-wide association study for Crohn's disease in the Quebec Founder Population identifies multiple validated disease loci. Proc Natl Acad Sci U S A. 2007;104:14747–14752. doi: 10.1073/pnas.0706645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGovern DP, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum Mol Genet. 2010;19:3468–3476. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wehkamp J, et al. Paneth cell antimicrobial peptides: topographical distribution and quantification in human gastrointestinal tissues. FEBS Lett. 2006;580:5344–5350. doi: 10.1016/j.febslet.2006.08.083. [DOI] [PubMed] [Google Scholar]
- 52.Castillo PA, et al. An experimental approach to rigorously assess paneth cell α-defensin (Defa) mRNA expression in C57BL/6 mice. Sci Rep. 2019;9:13115. doi: 10.1038/s41598-019-49471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petersen, T. N., Brunak, S., von Heijne, G. & Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods8, 785-786, doi:10.1038/nmeth.1701 (2011). [DOI] [PubMed]
- 54.Szyk A, et al. Crystal structures of human alpha-defensins HNP4, HD5, and HD6. Protein Sci. 2006;15:2749–2760. doi: 10.1110/ps.062336606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez V, Kelleher SL, Lonnerdal B. Lactoferrin receptor mediates apo- but not holo-lactoferrin internalization via clathrin-mediated endocytosis in trophoblasts. Biochem J. 2008;411:271–278. doi: 10.1042/bj20070393. [DOI] [PubMed] [Google Scholar]
- 56.Pisitkun T, Hoffert JD, Saeed F, Knepper MA. NHLBI-AbDesigner: an online tool for design of peptide-directed antibodies. Am J Physiol Cell Physiol. 2012;302:C154–164. doi: 10.1152/ajpcell.00325.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu X, et al. Regulation of gene expression in autoimmune disease loci and the genetic basis of proliferation in CD4+ effector memory T cells. PLoS Genet. 2014;10:e1004404. doi: 10.1371/journal.pgen.1004404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jablonski KA, et al. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 2010;59:2672–2681. doi: 10.2337/db10-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suhre K, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen BH, et al. Peripheral blood transcriptomic signatures of fasting glucose and insulin concentrations. Diabetes. 2016;65:3794–3804. doi: 10.2337/db16-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.