Abstract
Atypical Teratoid Rhabdoid Tumor (AT/RT) is a rare pediatric central nervous system cancer often characterized by deletion or mutation of SMARCB1, a tumor suppressor gene. In this study, we found that SMARCB1 regulates Human Endogenous Retrovirus K (HERV-K, subtype HML-2) expression. HML-2 is a repetitive element scattered throughout the human genome, encoding several intact viral proteins that have been associated with stem cell maintenance and tumorigenesis. We found HML-2 env expression in both the intracellular and extracellular compartments in all AT/RT cell lines (n = 4) and in 95% of AT/RT patient tissues (n = 37) evaluated. SMARCB1 knock-down in neural stem cells (NSCs) led to an upregulation of HML-2 transcription. We found that SMARCB1 binds adjacent to the HML-2 promoter, repressing its transcription via chromatin immunoprecipitation; restoration of SMARCB1 expression in AT/RT cell lines significantly downregulated HML-2 expression. Further, targeted downregulation of HML-2 transcription via CRISPR-dCas9 coupled with suppressor proteins led to cellular dispersion, decreased proliferation, and cell death in vitro. HML-2 knock-down with shRNA, siRNA, and CRISPR-dCas9 significantly decreased Ras expression as measured by qRT-PCR, suggesting that HML-2 modulates MAPK/ERK signaling in AT/RT cells. Overexpression of NRAS was sufficient to restore cellular proliferation, and MYC, a transcription factor downstream of NRAS, was bound to the HERV-K LTR significantly more in the absence of SMARCB1 expression in AT/RT cells. We show a mechanism by which these undifferentiated tumors remain pluripotent, and we demonstrate that their formation is aided by aberrant HML-2 activation, which is dependent on SMARCB1 and its interaction with MYC.
Subject terms: Cancer, Paediatric cancer, Genetics, Epigenomics
Introduction
Atypical teratoid rhabdoid tumor (AT/RT) is a rare embryonal central nervous system (CNS) cancer diagnosed most often on the basis of biallelic loss of SMARCB1 (SWI/SNF Related, Matrix Associated, Actin Dependent Regulator Of Chromatin, subfamily B, Member 1), a master chromatin regulator which is essential during development1. Mortality remains high in patients with AT/RT due to limited treatment options such as tumor resection, chemotherapy, and in a select population, radiation therapy2. AT/RTs are comprised of undifferentiated cancer cells characterized by expression of epithelial, mesenchymal, and neuroectodermal markers and loss of SMARCB1 expression1. This cancer is most common in children under 3 years of age and is largely due to a failure of proper development, neuronal cell migration, and cell differentiation3,4.
SMARCB1 is one of the core proteins in the SWItch/Sucrose Non Fermentable (SWI/SNF) chromatin remodeling complex, which contributes to conformational changes in the nucleosome, altering DNA-histone binding, and resulting in transcription factor access to gene promoters4. When SMARCB1 is absent, there is residual function of SMARCA2/4, the ATPase subunit of the chromatin remodeling complex; however regulation of enhancers which are key for development and differentiation are lost4. The activity of SMARCA2/4 is present at a barely detectable level and occupies super enhancers, areas with high transcription factor density, resulting in the maintenance of a stem-cell identity4. SMARCB1 loss of expression has different effects depending on the timing of the inactivation. In a study using conditional knock out mice, the early loss of SMARCB1 expression in neural crest cells caused the development of rhabdoid tumors5. A loss of both NF2 (neurofibromatosis 2) and SMARCB1 at a later stage of development in the Schwann cell lineage, led to the development of schwannomas5. In addition, mutations in SMARCB1 have been connected to brain midline defects such as glial aberrations leading to agenesis of the corpus callosum in the conditional SMARCB1 knockout mouse6. The defects observed in the SMARCB1 conditional knockout mouse are largely due to ineffective neuronal migration and neuronal maturation6.
SMARCB1 is a transcriptional repressor of integrated Human Immunodeficiency Virus (HIV) long terminal repeat (LTR) promoter7. When SMARCB1 expression is depleted, HIV transcription increases; further, the reappearance of SMARCB1 expression results in decreased viral transcription and epigenetic silencing7. Long terminal repeats are a consequence of retroviral integration and also flank Human Endogenous Retroviruses (HERVs) which comprise about 8% of the human genome and were acquired throughout human evolution8,9. Each integrated retrovirus has an LTR, which acts as the promoter for the HERV; LTR activity is regulated by methylation of CpGs in the LTR10–12 and by many transcription factors that can bind to its sequence such as C-MYC13. Currently it is not known how SMARCB1-mediated chromatin remodeling around the LTR regulates endogenous retroviral activity. In the human genome, transcription factor binding, CpG methylation, and chromatin remodeling all work in conjunction to alter expression of endogenous retroviruses14. There are only a few chromatin remodeling proteins which have been linked to endogenous retroviral suppression in humans such as SETDB1 and ATRX15,16.
When chromatin is tightly closed, it is difficult for transcription factors to access DNA; further, targeted binding of transcription factors can prompt chromatin remodeling resulting in an accessible promoter17. The SWI/SNF complex, including SMARCB1, does not bind to DNA based on sequence18; rather, the N-terminus of SMARCB1 interacts with acidic transcription sites and the SWI/SNF complex binding is facilitated by ultrastructural interaction and nucleosome positioning17,19. SMARCB1 binds C-MYC protein under non-pathogenic conditions; when SMARCB1 is absent, there is more C-MYC freely available to bind transcription factor sites13. C-MYC has not been definitively shown to bind an HERV-K LTR; however, many HERV-K LTRs in the human genome harbor multiple C-MYC binding sites20.
HERVs originated from exogenous viruses that were incorporated into the genome of the host germline and thereby maintained in subsequent generations21. The most recent additions to the genome are LTR5_Hs (Hs for human specific) and HERV-K (subtype HML-2)22,23; these gene features are relevant to disease phenotype because they often encode one or more intact viral proteins24. Normally, HML-2 expression is tightly controlled both temporally and spatially in stem cells and during development25. Further, HML-2 RNA and proteins are not highly expressed in differentiated cells or tissue; if expressed, they induce cytotoxicity in non-dividing cells such as neurons26.
Expression of HML-2 envelope (env) confers stem cell like features in dividing cells if not silenced27. Improper regulation of HML-2 protein expression in dividing cells may result in uncontrolled growth that leads to cancer28. Further, it has been shown that ras is downregulated in concordance with HML-2 downregulation in vivo, suggesting that its expression may be regulated by HML-229,30.
Herein, we sought to understand the role of SMARCB1 in mediating endogenous retrovirus expression in the undifferentiated embryonal cancer AT/RT. Further, we evaluated the membrane for vesicle production and neurite outgrowth, which we expected to be abnormal given the undifferentiated and neural crest-like cells that comprise these tumors31–33. We hypothesize that loss of SMARCB1 chromatin remodeling coupled with enhanced C-MYC binding to the HML-2 LTR results in constitutive expression of HML-2 elements, allowing cells to maintain a stem-cell like identity and persist as this developmental tumor.
Results
Detection of HERV-K (HML-2) in AT/RT tumors
AT/RT tissues derived from 35 of 37 (95%) patients demonstrated clusters of cells strongly expressing HML-2 env protein as measured by a monoclonal env antibody immunohistochemistry on a tissue microarray (Fig. 1, Supplemental Table S1, Supplemental Fig. S1). No env expression was observed in the normal cerebellum or in non-AT/RT brain samples (Fig. 1C, Supplemental Table S1, Supplemental Fig. S1). All ten of the AT/RT tissues originating from recurring tumors expressed HML-2 env (Supplemental Table S1). Placenta was stained as a positive control34 due to its mixed composition of mesenchymal and progenitor cells (Supplemental Fig. S1).
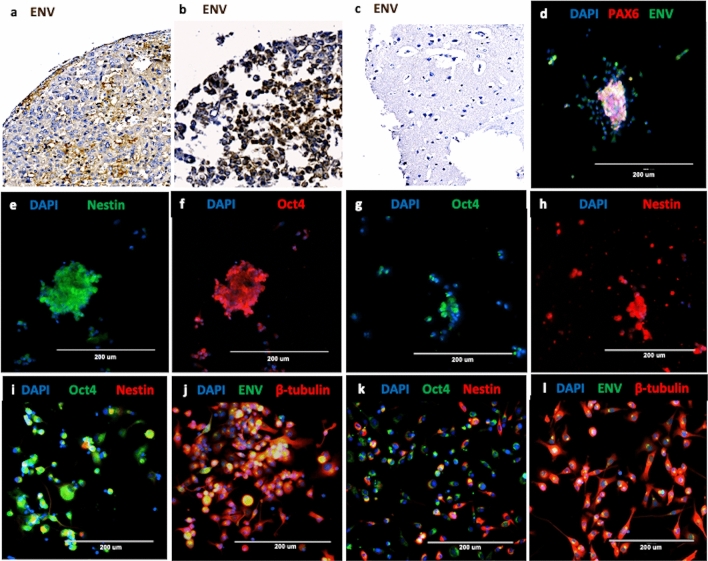
Figure 1.
Immunostaining and Immunohistochemistry of patient-derived AT/RT cells and tissues. (a,b) Two resected patient AT/RT demonstrate strong immunostaining with HML-2 Envelope (Env) monoclonal antibody (brown). (c) Cerebrum from a normal brain is negative when stained with the same antibody. (d–l) AT/RT cell lines express markers of multiple stages of differentiation; representative images are shown. (d) CHLA 02, a patient derived AT/RT cell line, expresses Pax6 (red), a marker for neuroectoderm, merged with immunostaining for HML-2 Env (green). (e): This cell line also expresses Nestin (green), a marker of neural stem cells. (f) The same cell line expresses Oct4 (red), a pluripotency marker. (g,h) CHLA 04 cells, another AT/RT line, express Oct4 (green) and Nestin (red). (i) CHLA 05 cells also express Oct4 (green) and Nestin (red). (j) The same cell line also expresses βIII tubulin (red), a marker for neurons and HML-2 envelope (green). (k) CHLA 06 cells also express Nestin (red) and Oct4 (green). (l) The same cell line also expresses βIII tubulin (red) and HML-2 Env (green). Nuclei are stained with DAPI (blue). For IHC images, slides were scanned with a OptraScan Digital Pathology Scanner and processed in Adobe Photoshop for incorporation into the figure. For immunofluorescent images, an EVOS fluorescence microscope (AMG) was used. Images were acquired with the native software installed on the EVOS microscope and processed in Microsoft PowerPoint (https://www.thermofisher.com/us/en/home/technical-resources/software-downloads/evos-fl-cell-imaging-system.html).
Characterization of AT/RT cell lines
Four AT/RT cell lines CHLA 02, CHLA 04, CHLA 05, and CHLA 06, were used in this study. All four cell lines lacked SMARCB1 expression35,36 and expressed markers indicative of pluripotent cells, including: HML-2 env and OCT4 (a marker for stem cells), Pax6 (a neuroectodermal marker), NES (a neural stem cell marker, nestin), and ΒIII-tubulin (a neuronal marker) (Fig. 1d–l). Immunocytochemical characterization revealed that the AT/RT cells are a mixed population at various stages of differentiation but they all express HML-2 env in the plasma membrane (Fig. 1). In previous publications, identification of subgroups of AT/RT tumors and tumor cell lines were described with regard to genetic and epigenetic expression and specific therapeutic treatment responses36,37. CHLA 02, CHLA 04, and CHLA 05 cell lines were all classified as group 1 neurogenic tumors while CHLA 06 cell line was classified as group 2 due to differences in methylation patterns and distinct clinical and genotypic features36,37. With an updated analysis of AT/RT subgroups, group 1 neurogenic tumors have been classified as ATRT-SHH, sonic hedgehog, and group 2 tumors fall into either an ATRT-MYC, MYC proto-oncogene, or a ATRT-TYR, tyrosine, subtype37. Even though one of the four cell lines was classified as belonging to a different group, all four cell lines expressed HML-2 env suggesting env expression may be a broad marker for AT/RT tumors.
Cellular and extracellular vesicles express of HML-2 env
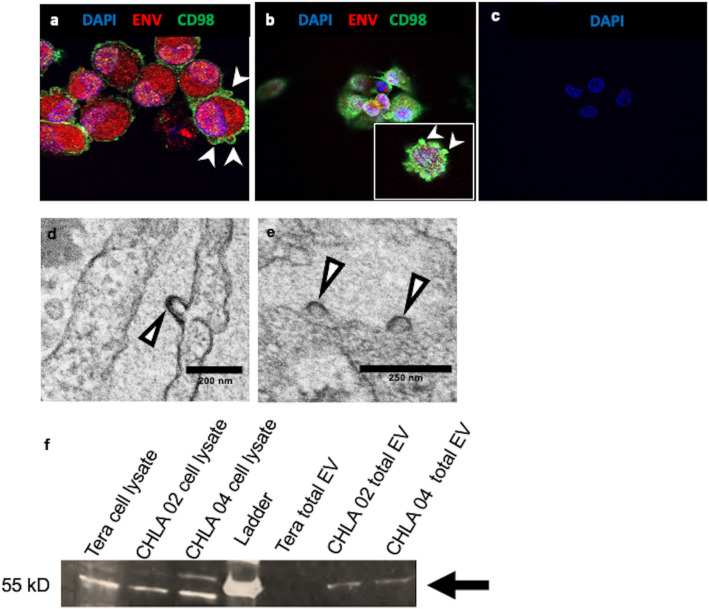
HML-2 env was identified in the cytoplasm and the plasma membrane by confocal microscopy in the CHLA 02 and CHLA 04 lines (Fig. 2a–c). Env was found to colocalize with CD98, a putative receptor for HML-2 env35. We further evaluated the CHLA 02 cells by electron microscopy and found no evidence of typical viral budding in approximately 100 cells. However, membrane-like structures resembling exocytosis of vesicles were noted (Fig. 2d,e). These appear to correspond to vesicles of similar morphology seen on confocal imaging to contain HML-2 env protein (Fig. 2). We confirmed that the atypical membrane structures observed by EM and confocal imaging contain HML-2 env protein by immunoblotting of enriched extracellular vesicles from cell free media via Nanotrap particles (Fig. 2f).
Figure 2.
Extracellular Vesicles are released from CHLA 02 and CHLA 04 cells and contain HML-2 Envelope (Env). (a,b) AT/RT cells release extracellular vesicles contain HML-2 Env. (a) Immunostaining CHLA 02 cells with HML-2 env polyclonal antibody (surface unit) (red) and CD98 (green), a cell-surface marker, shows Env is expressed in extracellular vesicles enclosed by the plasma membrane. Cell nuclei are stained with DAPI (blue); (b) CHLA 04 cells similarly immunostained; magnification is at 63X. (c) CHLA 02 cells incubated with secondary antibody only as a control for nonspecific immunostaining. (d,e) Electron microscopy shows extracellular vesicles forming on plasma membrane of CHLA 02 AT/RT cells (arrow heads). (f) Cell lysates from Tera, CHLA 02, and CHLA 04 cells express HML-2 env (left three lanes) while only purified extracellular vesicles from CHLA 02 and CHLA 04 express HML-2 env (three right lanes) on immunoblot (arrow). A fluorescent Zeiss confocal microscope (LSM510) was used to acquire images processed with Zeiss LSM 5 Image Browser and Microsoft PowerPoint for figure creation (https://www.embl.de/eamnet/html/downloads.html). The gel image has been cropped to focus on the bands of interest, the full gel image can be found in the supplementary material. Gel Image was taken with a FluorChem E system (https://www.proteinsimple.com/fluorchem.html) and images were processed with ImageJ and Microsoft PowerPoint for relative densitometry quantification and display. Electron microscopy images were taken with a JEOL 1200 EXII Transmission Electron Microscope (AMT digital camera system).
Regulation of HML-2 repetitive elements by SMARCB1
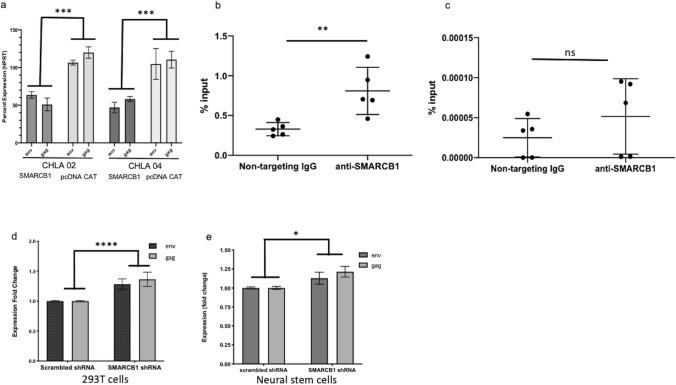
Overexpression of SMARCB1 in CHLA 02 and CHLA 04 cells led to significantly reduced transcription of HML-2 genes, suggesting SMARCB1 regulates HML-2 transcription (Fig. 3a). We observed increased binding of SMARCB1 to the HML-2 LTR compared to the housekeeping gene Hypoxanthine–guanine phosphoribosyltransferase (HPRT) (Fig. 3b,c). HML-2 transcription increased following knockdown of SMARCB1 in both 293 T cells (Fig. 3d) and in NSCs (Fig. 3e).
Figure 3.
SMARCB1 regulates HERV-K (HML-2) env expression. (a) Restored SMARCB1 expression in CHLA 02 and CHLA 04 AT/RT cell lines results in downregulation of HML-2 transcription measured at 48 h by qRT-PCR. (b,c) SMARCB1 binds the HML-2 LTR significantly more than the promoter of control gene, HPRT. (b) SMARCB1 transfected 293 T cells show a significantly greater proportion of HML-2 LTR bound to SMARCB1 following immunoprecipitation compared to control, non-targeting IgG. (c) SMARCB1 transfected 293 T cells show no significant difference between non-targeting IgG bound to genomic HPRT and SMARCB1 bound to genomic HPRT. Percent input is a normalized value with input set to 100% (e.g. % input = 100*2^(input Ct − immunoprecipitated chromatin Ct). Ct is cycle threshold. (d,e) SMARCB1 knockdown results in increased transcription of HML-2 transcripts as measured with qRT-PCR. (d) HML-2 transcripts in 293 T cells transfected with scrambled shRNA control compared to shRNA targeting SMARCB1 at 24 h as measured by qRT-PCR. (e) HML-2 transcripts in neural stem cells transfected with scrambled shRNA control are significantly higher compared to transcripts in cells transfected with shRNA targeting SMARCB1 at 48 h. (qRT-PCR) Data was entered into Prism v9 for graph creation. [Error bars represent SEM. Statistics in Supplemental Table S6].
RNA Sequencing Analysis of AT/RT Cell Lines
RNA expression profiles determined by next generation sequencing supported the immunohistochemical findings (Fig. 1), indicating that each tumor originated from cells at different stages of development/differentiation. We found HML-2 repetitive elements within highly expressed genes in these cell lines. Two groups of highly expressed genes containing either sequences of HML-2 internal coding sequences (HML-2 int) or LTR5_Hs were found by DAVID functional clustering gene ontology analysis38. The first group contained genes related to Krüppel associated box (KRAB) and zinc finger proteins both of which play roles in transcriptional regulation (Supplemental Table S2). The genes in the second group were affiliated with the neuroblastoma breakpoint family (NBPF) (Supplemental Table S2); genes in this family, such as putative tumor suppressor NBPF1, which is highly expressed in the brain, have repetitive structures containing both intragenic and intergenic sequences39.
Proteomic analysis of these cell lines confirmed the high expression observed by RNA sequencing analysis and the immunohistochemistry (IHC). Two groups of highly expressed proteins were found after analysis with DAVID of the CHLA 02 and CHLA 04 AT/RT cells by liquid chromatography mass spectrometry (LCMS). Both cell lines expressed proteins associated with the cellular immune response to viral proteins, in addition to neural stem cell marker (nestin), neuronal marker (βIII-tubulin), and glial cells (vimentin), which clustered as the first group (Supplemental Table S3). One of the proteins highly expressed in CHLA 02 and CHLA 04 AT/RT cells is adenovirus early region 1B associated protein 5, a protein known to be activated during adenoviral infection, which has been suggested as a marker for undifferentiated embryonic stem cells40. An enrichment of proteins related to cellular adhesion, such as genes affiliated with cadherin cell–cell binding, cell–cell junction, and cell–cell adhesion was detected in the second group (Supplemental Table S3).
Identification of activated HML-2 loci due to SMARCB1 loss
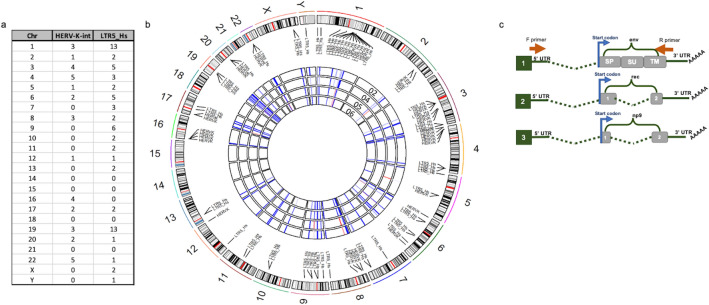
Initial RNA sequencing analysis from all four AT/RT cell lines revealed the expression of several loci from both LTR5_Hs and HML-2 int (Fig. 4a–c). However, while at least 36 loci expressed transcripts of HML-2 internal sequences, a minority of loci encoded potential proteins (Supplemental Table S4). HML-2 elements were actively expressed from all chromosomes except for 14, 15, 18, and 21 (Fig. 4a,b). The most actively transcribed HML-2 sequences were from chromosomes 1 and 19 each of which had 13 LTR5_Hs transcripts and 3 HML-2 int (Fig. 4a). The expressed transcripts aligning to elements encoding at least one full-length HML-2 gene are listed in Supplemental Table S4. Of these, only the locus on Chr19q11 encodes full length and potentially functional env protein (highlighted in Supplemental Table S4). Using a second analysis method, the TEtranscripts pipeline41,42 (Supplemental Fig. S3), transcripts were detected from all chromosomes except 21 (Supplemental Fig. S3a). Three loci identified by the TEtranscripts pipeline were capable of producing full length env protein: Chr7p22.1a, Chr7p22.1b, and Chr19q11 (Supplemental Fig. S3b, highlighted in Supplemental Table S4). Transcripts originating from both Chr19q11, Chr7p22.1a, and Chr7p22.1b were confirmed with RT-PCR and Sanger sequencing (Supplemental Table S5). The loci on chromosome 7 are a pair of duplications24 whose sequences are nearly identical, and both contain an intact env gene. Both methods suggested env expression originated from Chr19q11, but with the TEtranscripts pipeline which uses expectation maximization algorithm also suggested the Chr7 loci.
Figure 4.
RNA sequencing of AT/RT cell lines. (a,b) HERV-K (HML-2) internal coding genes and LTR5_Hs are expressed from most chromosomes. (a) Table shows quantity of loci expressed on each chromosome (Chr). (b) Graphical depiction of HERV-K (HML-2) internal coding genes in (HERV-K-int) and LTR5_Hs in (long terminal repeat 5 human specific) expression in four AT/RT cell lines. The outer ring is comprised of the chromosomes from the human genome with the list of both “HERV-K-int” (internal coding sequence) and LTR5_Hs loci and black lines connecting them to their approximate location on the chromosome. The four rings in the center correspond to the CHLA 02, 04, 05, and 06 cell lines with the 02 cells being the outermost circle and the 06 cells the innermost circle. The segments of the central circles represent the chromosomes; blue and red lines correspond to the level of expression of either the LTR5_Hs or HERV-K (HML-2) labeled at that location (red denotes higher expression, blue denotes less). (c) Scheme for PCR amplification and Sanger sequencing validation of HML-2 transcripts. Shows a representation of primer position relative to potential env, rec, and np9 transcripts which are transcribed in the AT/RT cells. The forward primer used for amplifying env (1), rec (2), and np 9 (3) transcripts is positioned in the 5’ UTR while the reverse primer is positioned in the 3’ UTR. Because both primers are in the UTRs, env, rec and np9 transcripts can all be amplified with the same primer set. The image in (b) was generated with the RCircos package version 1.2.1 (https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-14-244).
RNA from AT/RT cells was reverse transcribed and then amplified by PCR to confirm the RNA sequencing data (Fig. 4c, Supplemental Table S5). There are two types of HERV-K (HML-2) present in the human genome. Type 1 proviruses possess a 292 bp deletion in the env gene whereas type 2 proviruses have an intact env14. Type 1 proviruses express np9, a spliced transcript containing part of the env preceding the deletion, and a sequence 3’ of the deletion14. Type 2 proviruses can express env as well as rec, another gene spliced from the beginning of the env transcript and an abbreviated 3’ sequence of env. The primers used to amplify env were designed to amplify full-length transcript and its smaller spliced products rec, or np9, (Fig. 4c). Rec and np9 sequences were derived from multiple loci including Chr1p31.1, 1q22, 3q12.3, 3q21.2, 7p22.1a, 7p22.1b, 10q24.1, 11q23.3, 19q11, 19q13.12, and 22q11.21 (Supplemental Table S5). After aligning the Sanger sequences to the human genome and consensus env transcript, we compared the data to the expression profile obtained with RNA sequencing (Supplemental Table S4). Only three HML-2 loci which could make a functional full length env protein were identified, namely, Chr19q11, Chr7p22.1a, and Chr7p22.1b.
Effect of down regulation of HML-2 expression on AT/RT cells
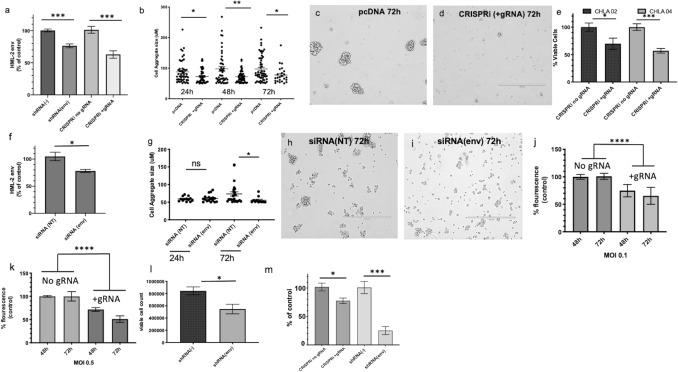
HML-2 transcription was downregulated 48 h post transfection with a construct containing shRNA targeting HERV-K env or CRISPRi and gRNA targeting the HML-2 LTR and led to a significant decrease in HERV-K env transcription (Fig. 5a) and in the size of cellular aggregates (Fig. 5a–d). Due to the importance of cellular adhesion for pluripotency43–45, we also analyzed cell viability following CRISPRi transfections and found a significant decrease at 96 h post transfection in CHLA 02 and CHLA 04 (Fig. 5e). Downregulation of HML-2 env with siRNA (Table 1) resulted in a significant decrease in env transcripts (Fig. 5f) and protein expression (Supplemental Fig. S2) accompanied by a decrease in cellular aggregate size at 72 h post transfection (Fig. 5g–i).
Figure 5.
Effect of the Downregulation of HML-2 on AT/RT. (a–d) A decrease in HML-2 expression leads to reduced cellular aggregates in CHLA 02 AT/RT cells. (a) HML-2 env transcript levels in CHLA 02 cells 48 h post transfection with either shRNA targeting env or CRISPRi targeting HML-2 LTR. (b) CHLA 02 cells transfected with CRISPRi + gRNA have reduced aggregate size following reduced expression of HML-2 mRNA at 24 h, 48 h, and 72 h post transfection. (c) CHLA 02 cells at 72 h post transfection with either pcDNA or (d) with CRISPRi + gRNA. (e–i) Downregulation of HML-2 results in AT/RT cell death and reduced cell aggregates. (e) CHLA 02 and CHLA 04 cells transfected with CRISPRi with or without gRNA targeting HML-2 LTR results in different proportions of viable cells at 96 h post transfection. (f) Percent of control (siRNA non-targeting) env transcription following transfection with siRNA targeting HML-2 env in CHLA 02 at 24 h. (g) CHLA 02 cell aggregates were reduced in size after 72 h following siRNA transfection targeting HML-2 env. (h) CHLA 02 at 72 h post transfection with both non-targeting siRNA and (i) cells treated with HML-2 env siRNA. (j–m) Lentiviral vectors with CRISPRi + gRNA targeting HML-2 and shRNA targeting HML-2 env result in decreased AT/RT viability and a reduction in HML-2 env protein expression. (j,k) CHLA 02 cytotoxicity at multiple time points post-transduction with lentivirus expressing either a CRISPRi construct with CRISPRi + gRNA targeting HERV-K (HML-2) LTR5_Hs or with CRISPRi without gRNA. (l) Percent viable cells 48 h post-transfection shRNA targeting HML-2 env. (m) env protein post-transfection as a percent of control transfection (either with CRISPRi and no gRNA or shRNA (−)). Graphs generated with Prism v9. [Error bars represent SEM. Statistics in Supplemental Table S6].
Table 1.
Sequences of siRNA, shRNA, and gRNAs targeting HML-2 expression.
| Reagent | Nucleotide sequence | Alignment to consensus gene or consensus LTR5_Hs |
|---|---|---|
| siRNA ENV 1 | CUGACGCAGUUAGCUACAAUU | 166–184 |
| siRNA ENV 2 | GAUUCACUUAUCACAUGGUUU | 572–590 |
| gRNA sequence 1 | GATAGGGAAAAACCGCCTTAAGGG | 563–585 |
| gRNA sequence 2 | AAAGCAGTATTGCTGCCCGCAGG | 600–620 |
| gRNA sequence 3 | TCCTGCCTGTCCCTGGGCAATGG | 538–560 |
| gRNA sequence 4 | AGTAGATGGAGCATACAATCGGG | 498–520 |
| shRNA_469 | GGGTATCGTTATCCTCCTATT | 469–489 |
| shRNA_866 | CAGCTGTTGATAGCGACTTAA | 866–886 |
| shRNA_1620 | CATGAGCTTAGAACATCGTTT | 1620–1640 |
A significant and time-dependent increase in cytotoxicity occurred at 48 h and 72 h in CHLA 02 cells post transduction with HML-2 targeted CRISPRi lentivirus at a MOI of 0.1 and 0.5 (Fig. 5j,k). Two days following nucleofection, significantly fewer viable cells were observed as measured with propidium iodide staining in the shRNA (env) transfected cells (Fig. 5l). Targeting HML-2 env with shRNA or CRISPRi + gRNA resulted in decreased cell proliferation at 48 h and a significant decrease in env protein (Fig. 5m, Supplemental Fig. S2).
HML-2 expression induces cell proliferation through the NRAS pathway
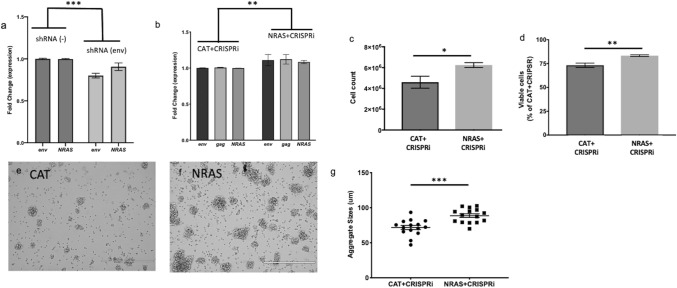
In previous studies of HML-2 downregulation in tumors, a concurrent decrease in N-Ras protein has been observed suggesting possible regulation of Ras genes by HML-229,30,46. When CHLA 02 cells were treated with shRNA to HML-2 env, there was a significant decrease in N-Ras expression alongside HML-2 expression in the CHLA 02 and CHLA 04 cells (Fig. 6a). Forty-eight hours after CHLA 04 transfection with both the CRISPRi construct (+ gRNA targeting HML-2) and with an NRAS plasmid, higher transcription of HML-2 and NRAS were found by qRT-PCR relative to cells transfected with a pcDNA CAT which served as a control (Fig. 6b). This result indicates that NRAS overexpression is sufficient to restore HML-2 transcription. At 5 days post transfection, we observed a significant difference between both the viability and cell number of the NRAS and CAT transfected cells (Fig. 6c,d). Additionally, we observed the cell aggregates were significantly larger in the cells transfected with both CRISPRi and NRAS (Fig. 6e–g), demonstrating a restoration of the cell proliferation of the AT/RT cells.
Figure 6.
NRAS expression is downregulated post HML-2 downregulation, and its overexpression is sufficient to restore cellular proliferation to AT/RT cells. (a) Downregulation of HML-2 transcription with shRNA results in a decrease in NRAS expression. Transcription level of HML-2 env and NRAS measured with qPCR 48 h post transfection with shRNA targeting HML-2 env in CHLA 02. (b–g) Co-transfection with NRAS overexpression plasmid can overcome the effects of HML-2 downregulation. (b) Expression of env, gag, and NRAS transcripts 48 h post transfection in CHLA 04 cells with both CRISPRi + gRNA and NRAS plasmids. (c) Cell number in CHLA 04 AT/RT cells transfected with CRISPRi + gRNA and either a CAT plasmid or an NRAS construct and 5 days post-transfection. (d) Viable cell percentage 5 days post transfection with CRISPRi and either CAT or NRAS plasmids. Image of cells transfected with CRISPRi plus HML-2 gRNA and either CAT (e) or NRAS (f). (g) Quantification of cell aggregate size 5 days post-transfection with CRISPRi + gRNA and either CAT or NRAS plasmids. Graphs generated with Prism v9. [Error bars represent SEM. Statistics in Supplemental Table S6].
The role of C-MYC in HERV-K transcription activation
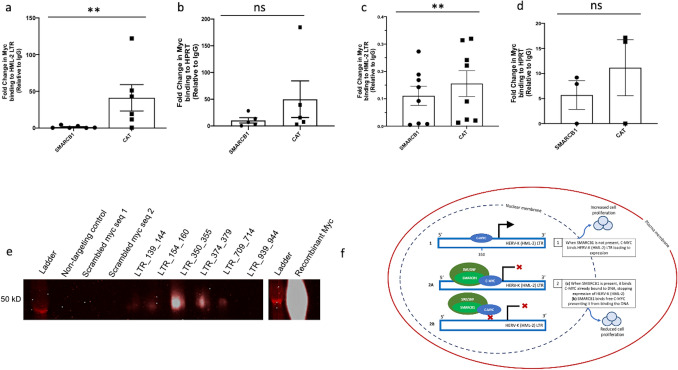
Many transcription factor binding sites (TFBS) have been reported in the LTR of HERV-K (HML-2) loci; however, not all suggested TFBS have been confirmed13,14. In the LTR of Chr7p22.1a and Chr7p221b, both highly expressed loci in AT/RT, there are multiple potential C-MYC binding sites present47. Using chromatin immunoprecipitation of crosslinked DNA in 293 T cells, we observed significantly fewer HML-2 LTR5_Hs sequences bound to C-MYC protein when SMARCB1 was overexpressed (Fig. 7a,b). In the CHLA 02 AT/RT cells, the chromatin immunoprecipitation of C-MYC revealed fewer HERV-K LTR sequences bound to C-MYC in the absence of SMARCB1 expression (Fig. 7c,d). There was no significant difference between the SMARCB1 transfected and the control (CAT) transfected cells when comparing C-MYC bound to a housekeeping gene, HPRT, in both the 293 T and CHLA 02 cells (Fig. 7b,d). We tested binding sites predicted via bioinformatics by PROMO47 and other manually identified sites, using a pulldown using biotinylated HERV-K (HML-2) LTR sequences combined with purified C-MYC protein (Supplemental methods). C-MYC protein bound to multiple tested HERV-K sequences and was absent in the non-targeting control as well as the scrambled C-MYC sequences tested (Fig. 7e).
Figure 7.
C-MYC binding to the HERV-K (HML-2) LTR. (a–d) C-MYC binds HML-2 LTR significantly less when SMARCB1 is expressed. Panel a: Relative quantification of HML-2 LTR bound to C-MYC transcription factor binding protein immunoprecipitated from either 293 T cells transfected with a SMARCB1 plasmid or a control (CAT) plasmid. (b) Relative quantification of housekeeping gene HPRT bound to C-MYC post-immunoprecipitation from 293 T cells transfected with either SMARCB1 or a control (CAT) plasmid. (c) Relative quantification of C-MYC bound to the HML-2 LTR post immunoprecipitation in CHLA 02 cells transfected with either SMARCB1 or with CAT plasmid. (d) Relative quantification of CMYC bound to housekeeping gene HPRT post immunoprecipitation in CHLA 02 cells transfected with either SMARCB1 or with CAT plasmid. Percent input is a normalized value with input set to 100% (e.g. % input = 100*2^(input Ct − immunoprecipitated chromatin Ct). Ct is cycle threshold. (e) C-MYC binds the HML-2 LTR in vitro. Biotinylated nucleotides with a non-targeting sequence, a scrambled canonical C-MYC binding sequence, or varying sequences from the Chr7p22.1a or Chr7p22.1b HML-2 LTR were incubated with recombinant C-MYC protein, washed, and run on an immunoblot to detect specific binding between C-MYC and different sequences. There is a white line between the the final LTR_939_944 and another ladder depicting that a portion of the gel was deleted to simplify the figure. The full gel image can be found in the supplemental information. (f) Contains a diagram of the mechanism by which HML-2 expression is regulated in the absence or presence of SMARCB1 protein. (f1) shows how HML-2 is actively expressed when C-MYC is bound to the LTR (promoter) when SMARCB1 protein is not expressed, while (f2a,b) depict the SMARCB1 mediated inhibition of C-MYC activation of HML-2 transcription. Graphs generated with Prism v9. (f) Generated in Microsoft PowerPoint. [Error bars represent SEM. Statistics in Supplemental Table S6].
Discussion
We report here that the absence of SMARCB1 expression in AT/RT leads to aberrant HML-2 env expression in cells which are meant to differentiate along a neuronal lineage. Our study establishes that aberrant regulation of HML-2 expression at the level of chromatin remodeling is sufficient to drive cellular proliferation. In our study of AT/RT, HML-2 activates cell proliferation through the Ras pathway, and the consequences of a targeted decrease in HML-2 expression can be reversed by an overexpression of NRAS. These results suggest that improper chromatin remodeling during early neural crest migration and neuronal differentiation provide a unique opportunity for HML-2 env expression to be retained via C-MYC binding and thereby maintain an undifferentiated phenotype. We show that HML-2 env downregulation through transcriptional repression reduces functional env protein, resulting in reduced proliferation and cytotoxicity.
It has been suggested that some HERVs are selectively maintained in the human genome due to their highly regulated expression patterns during development25,48. In cells where HML-2 has been dysregulated during key developmental stages, development cannot proceed25. Using tissue arrays we found that 95% of tested AT/RT samples expressed HML-2 env. We confirmed expression of a pluripotency gene, OCT4, and HML-2 env genes in AT/RT cell lines by immunostaining. The cells also expressed neuronal stem cell and neuronal markers suggesting these cells are partially differentiated along a neuronal lineage. Consistent with the cellular heterogeneity and multiple stages of differentiation in AT/RT, HML-2 Env was not uniformly expressed in the tumors. Our results support previous findings in pluripotent stem cells, embryonic stem cells, embryonic carcinoma cells, and many other cancers10,25,29,30,46,49–67.
We observed HML-2 Env expression in two AT/RT cell lines in extracellular vesicles by immunocytochemistry (ICC), Immunoblot, and with electron microscopy. The extracellular vesicles containing HML-2 Env could serve as a continuous signal to adjacent cells to maintain pluripotency thereby supporting tumor survival68. Further, the expression of proteins related to viral nucleoproteins, nucleocapsids, virions, cell–cell junctions, cadherin cell–cell binding, and cellular adhesion likely contribute to tumor survival and growth. The enrichment of cellular interaction and attachment proteins reflects the morphology of the cells in vitro and affirms the importance of plasma membrane contact between adjacent cells44. Cadherins are calcium dependent transmembrane proteins which are vital for cell adhesion45, and E-cadherin expression promotes adhesion between adjacent cells43,69,70. Cell–cell communication is key in a tumor to adapt to microenvironmental shifts. In AT/RT, extracellular vesicles with HML-2 env combined with the expression of cellular adhesion proteins may enable the tumor to efficiently adapt to its microenvironment.
RNA sequencing showed that there were several active loci from which HML-2 env transcripts were produced. This was confirmed by cloning and sequencing the transcripts. All four cell lines expressed transcripts originating from three loci capable of encoding a full length Env protein on Chr19q11, Chr7p22.1a, or Chr7p22.1b. Targeting HML-2 env with shRNA has been previously shown to lead to reduced cellular proliferation and decreased metastatic potential of breast cancer in vitro through the inhibition of tumor associated genes like Ras, p-RSK, and p-ERK29. To further investigate the role of HML-2 env in AT/RT tumorigenesis, we modulated its expression via siRNA and shRNA and found a significant increase in cytotoxicity and decrease in cellular proliferation. After transfecting CHLA 02 with shRNA targeting HML-2 env, we observed a decrease in NRAS expression concurrent with the decrease in proliferation. An overexpression of NRAS was sufficient to rescue AT/RT cells from the loss of HML-2 env expression. Further, C-MYC binding to the HERV-K LTR is significantly increased in the absence of SMARCB1 resulting in high HERV-K expression. These findings suggest that HML-2 env plays a role in cellular viability and proliferation.
Our study establishes a new connection between chromatin remodeling and the regulation of endogenous retroviral elements in disease. We demonstrate that the regulation of HML-2 env expression early in development or differentiation is critical to maintenance of pluripotent stem cell identity in AT/RT. When the SWI/SNF chromatin remodeling complex loses a core subunit, SMARCB1, during development, the aberrant activation of human endogenous retroviral genes can contribute to the maintenance of stem cell features in cells which were meant to differentiate during CNS development. In addition to aberrant HML-2 transcription in the absence of SMARCB1, the transcription factor binding protein C-MYC binds the HML-2 LTR more frequently and upregulates its transcription. This study introduces a new mechanism of endogenous virus-mediated tumorigenesis. Identification of HML-2 expression as a marker of AT/RT following the loss of SMARCB1 is a primary step in establishing the role of endogenous retroelements in this central nervous system tumor development. Based on the findings in our study, HML-2 env expression may have a role as a marker for diagnosis of AT/RT in conjunction with SMARCB1. Further studies are needed to determine if HML-2 expression may be of prognostic significance and if it may be a therapeutic target.
Methods
Cell culture
CHLA AT/RT cell lines and 293 T cells were purchased from ATCC and maintained as recommended in DMEM:F12 with 20 ng/mL human recombinant basic EGF, 20 ng/mL human recombinant basic FGF, and B-27 supplement to a final concentration of 2%. iPSCs, NSCs, and neurons were cultured as documented in51. Cell line information from ATCC is as follows: CHLA 02 (CRL-3020), CHLA 04 (CRL-3036), CHLA 05 (CRL-3037), and CHLA 06 (CRL-3038).
Immunofluorescence
The AT/RT cell lines CHLA 02, CHLA 04, CHLA 05, and CHLA 06 all grow in suspension; therefore, the cells were attached to the bottom of the plate with Matrigel for staining and grew overnight at 37 °C. Next, they were fixed with 4% paraformaldehyde (PFA) for 10 min. The cells were washed with 1X phosphate buffered saline (PBS) 2 × for 5 min each and then permeated with PBS with 0.05% triton X 100 (PBST) for 10 min. Cells were incubated with a blocking solution of 5% donkey or goat serum in PBS for 1 h and then washed with PBS 3 × for 5 min each and primary antibodies diluted in blocking solution were applied to the samples. After an overnight incubation at 4 °C, the cells were washed 3 × for 10 min each with PBS and then incubated with the appropriate secondary antibodies at 1:400 dilution for 1 h. Following the secondary antibody, the cells were treated with 4′,6-diamidino-2-phenylindole (DAPI) at 1:10,000 and washed 2 × with PBS before imaging with a fluorescent Zeiss confocal microscope (LSM510) in Fig. 2, images were acquired with Zeiss LSM 5 Image Browser and processed in Microsoft PowerPoint for figure creation. For Fig. 1 immunoflourescence images, an EVOS fluorescence microscope (AMG) was used. Images were acquired with the native software installed on the EVOS microscope and processed in Microsoft PowerPoint. Primary antibodies: AntiCD98 antibody ab108300; anti-Tubulin β3 801213 (Bio-legend), Anti-Oct4 antibody AB3209 (Millipore sigma), Anti-nestin Mab 5326 (Millipore), Anti-Pax6 NBP1 51622 (Novus biologicals). Secondary antibodies: Goat Anti-Rabbit IgG (Alexa Fluor 488, ab150077), Goat Anti-Mouse IgG (Alexa Fluor 594 ab150116), Goat-anti Rabbit IgG (Alexa Fluor 594, ab150080), Goat anti-mouse IgG (Alexa Fluor 488, ab150113) used at 1:500 dilution. Validation of the polyclonal antibody (PAb) is shown in Supplemental Fig. S3. The peptides used to make HERV-K env polyclonal antibody were as follows:
QRKAPPRRRRHRNRC (HERV-K env amino acid position: 8–21), CSDLTESLDKHKHKK (env amino acid position: 294–307), and CSKRKGGNVGKSKRD (env amino acid position: 680–693).
Electron microscopy (EM)
CHLA 02 cells were pelleted at 2,000 RPM for 5 min and 3 million cells were resuspended in 700 ul of media. 4% room temperature glutaraldehyde was made in a 1% cacodylate buffer. 700 ul of glutaraldehyde solution was added to cell/media mixture and gently mixed in Eppendorf tube. One drop of 22% albumin was added to tube and the sample was incubated at room temperature for 30 min. Following room temperature incubation, sample was stored at 4 °C until tubes are ready to be further processed for.
EM. When ready for silver enhancement, cells were washed with deionized water thoroughly (5 × 5 min). Silver enhancement was then performed (under safety light) and cells were washed in water (at least 5 times over 10 min). The sample was washed in 0.1 M phosphate buffer, then with 0.2% OsO4 in 0.1 M phosphate buffer for an additional 30 min. Next, dehydration was performed in ETOH and sample was embedded in resin. Samples were then cut with an ultramicrotome into ultrathin sections and are placed on a copper metal grid.
Transfections
The CHLA cell lines were transfected using the P3 nucleofector kit (Lonza, catalog no. V4XP-3012) and the CA -137 program at 1 ug of plasmid per million cells. After transfection, the cells were re-suspended in at least 4 mL of media. Sequences for shRNA, siRNA, and gRNAs are included in Table 1. The 293 T cells were transfected with Lipofectamine 3000 following manufacturer’s recommendations using 5 ug of plasmid DNA per million cells. SMARCB1 lentiviral plasmid was obtained from Addgene. The pcDNA CAT plasmid contained a bacterial acetylcholine transferase. The CAT construct was used to control for effects of transfection as a plasmid of similar size to the SMARCB1 plasmid71. pDONR223_SMARCB1_WT was a gift from Jesse Boehm & William Hahn & David Root (Addgene plasmid # 81791; http://n2t.net/addgene:81791; RRID:Addgene_81791)72. pHAGE-NRAS was a gift from Gordon Mills & Kenneth Scott (Addgene plasmid # 116767; http://n2t.net/addgene:116767; RRID:Addgene_116767). A CRISPRi construct was used, comprised of a plasmid with a CRISPR-dCas9 (dead Cas9), four fused Sin3 repressive interacting domains (SID), along with four gRNAs targeting HML-2 LTR5_Hs (SID4X) (Table 1).
SID is a chromatin remodeling protein73,74 that prevents transcriptional machinery from accessing the DNA resulting in decreased expression of the targeted gene. As a control, cells were transfected with pcDNA and the transfection related toxicity was normalized accordingly in the CRISPRi transfected cells.
Immunoblotting
Immunoblotting was performed as done previously26 with the exception of the transfer which was performed with the iBlot 2 (ThermoFisher, IB24001). In addition, a new polyclonal antibody to detect HML-2 SU (surface unit) Env was used (additional information and antibody validation can be found in Supplemental Fig. S2 and Supplemental methods.) All images were quantified with ImageJ. The HML-2 Env antibody used in Supplemental Fig. S2 targets the transmembrane portion of the protein. The transmembrane antibody was made using the immunogen CSKRKGGNVGKSKRD, used at a concentration of 1:1000, and was first mentioned in a previous manuscript75.
Immunohistochemistry
Immunohistochemistry detected cells with Human endogenous retroviral Env expression primary antibody (Austral Biologicals, HERM 1855) at a 1:500 concentration within formalin fixed paraffin embedded tissue samples. Further detail can be found in Supplemental methods.
Enrichment of extracellular vesicles using nanotrap particles
Nanotrap particles, NT82 particles (#CN2010) and NT80 particles (#CN1030) (Ceres Nanosciences, Inc.) have been previously shown to be effective in the enrichment of extracellular vesicles from cell culture supernatant and patient biofluids76,77. Equal volumes of these two particles were combined with 1X PBS without calcium or magnesium to create a 30% slurry. For capture of extracellular vesicles (EVs) from cell culture supernatant, 30 µL of the 30% slurry was added to 1 mL of cell-free supernatant and rotated at 4 °C overnight. The following day, nanoparticles were pelleted at 10,000×g for 10 min at room temperature.
The resulting pellets were washed once with 1X PBS without calcium and magnesium and resuspended in 20 µL of Tris–Glycine SDS sample buffer with 10% 2-Mercaptoethanol. Samples were heated at 95 °C for 15 min with vortexing and loaded onto a 4–20% Tris/Glycine gel (Invitrogen). Gels were run at 100 V and transferred at 20 V for 7 min using iBlot 2 Gel Transfer Device. Membranes were blocked in 5% milk in PBST for 2 h at 4 °C. then incubated at 4 °C overnight in PBS-T with HML-2 envelope polyclonal antibody against the transmembrane protein (see Immunoblot section in Supplementary methods).
RT-PCR, PCR, and sequencing of env transcripts
RNA was isolated from iPSC, NSC, neuronal, and AT/RT cell lines with a Qiagen RNeasy kit (Qiagen, 74104) and reverse transcribed with the SuperScript First-Strand Synthesis RT-PCR kit (ThermoFisher Scientific, 11904018). Polymerase chain reactions were used to amplify HML-2 envelope transcripts with primers which target the full length env transcript (Table 1). Q5 high fidelity 2X master mix was used for the PCR with the following cycling conditions: 98 °C for 90 s, [98 °C 10 s, 55 °C 20 s, 72 °C 4 min] repeat 34 times, final extension at 72 °C for 6 min, 4 °C forever. The PCR primers were used to amplify the full length env gene as following: forward primer 5’- cccactagacatttgaagttctaca-3’, and reverse primer 5’- ggagtctcctatgtctacttcttt-3’. One primer aligned to the 3’ LTR of the HML-2 element and the other primer was positioned 5’ (upstream) of the start of the env gene. For the AT/RT cells, we cloned products into Topo-TA vectors (ThermoFisher Scientific, K203001) and sent them for Sanger sequencing to determine which env loci were actively transcribed in the samples. For the iPSCs, NSCs, and neurons, PCR products were run in a 2% agarose gel with GelStar Nucleic Acid Gel stain (Lonza, 50535) and the gel was imaged using Flourchem Protein Simple imager.
Alamar Blue viability assay
To distinguish the effect of HML-2 downregulation on AT/RT viability the Alamar Blue cell viability reagent (Thermofisher, DAL1100) was used. The fluorescence of the cell media resulting from the reduction of the Resazurin to resorufin indicated the percent of viable cells. Viability was calculated by comparing the fluorescence of each treatment concentration divided by the viability of the control treatment/ transfection.
Each experiment was performed with at least three technical replicates and three biological replicates.
Plates were read on the FlexStation 3 using Softmax Pro with the ‘blue fluorescence’ at 590 nm viability assay setting.
Lentiviral production
Lipofectamine 3000 (Invitrogen L3000008) kit was used to transfect 293 T cells with either a lentiviral construct containing four repressive Sin3 domains, dead Cas9, and four gRNAs designed for HERVK LTR5_Hs or one without gRNA. The aforementioned CRISPRi construct was incorporated into a lentiviral vector, both with and without gRNA targeting the LTR5_Hs, and AT/RT cells were transduced with the lentivirus at MOIs of 0.1 and 0.5. Manufacturer’s guidelines were followed for the transfection. Further information can be found in the Supplementary methods.
RNA sequencing
Libraries were both prepared and sequenced at NYGC (www.nygenome.org). For library preparation, the Illumina TruSeq Stranded Total RNA protocol was used (Illumina, San Diego, CA). Per sequencing, each library was paired-end sequenced (125 bp) for a target depth of 40 million reads (HiSeq 2500 Illumina); providing for a pair of .fastq files per library post CASAVA deplexing accessible at NCBI as GSE124210.
For further detail regarding analysis, please see Supplemental methods.
Chromatin immunoprecipitation (ChIP) and qRT-PCR
Five million cells were collected for each ChIP experiment from either pcDNA CAT transfected cells or cells transfected with SMARCB1. Cells were fixed with 1% PFA in fresh media and crosslinking was stopped by the addition of 1.25 M glycine to a final concentration of 0.125 M. Cell pellets were then snap frozen for storage at − 80 °C until ChIP was performed. Specific buffers and additional information about the procedure can be found in the Supplemental methods. To obtain the ratio of specific sequences pulled down during ChIP, semi-quantitative PCR was performed using primers that spanned the HML-2 transcription start site (TSS): LTR Forward (5'-GTT TGT CTG CTG ACC CTC TC-3') and Reverse (5’-AGC CTC TGA GTT CCC TTA GT-3’); qPCR was also performed using primers for an unrelated genomic region (hypoxanthine phosphoribosyltransferase 1; HPRT1), Forward (5’-GCT GAC CTG CTG GAT TAC AT-3’) and Reverse (5’-GGT TTG CAG AGA TTC AAA GAA-3’). Results are shown as percent of input chromatin, calculated using the formula % input = 100*2(Ct[input] − Ct[IP]). Antibodies used for ChIP: Go-ChIP grade purified Anti-RNA polymerase II antibody, 904,004, Biolegend. Mouse Mab IgG XP (R) isotype control antibody, Cell Signaling, 3900S. Ini1 antibody (A-5): sc-166165 (Ini1 is another name for SMARCB1), Santa Cruz.
Mass spectrometry
In-gel digestion of PAGE fractionated cell lysates
The entire lane was excised from the PAGE gel, sectioned into 16 approximately equivalent sections, transferred to 1.5 mL microfuge tubes and subjected to a modified in-gel digestion protocol78. Briefly, excised bands were destained by adding 0.5 mL 100 mM NH4HCO3:CH3OH (50:50, v/v) and incubated in a thermomixer (800 RPM, 37 °C) for 60 min. Solvent decanted and discarded, process repeated as necessary. Gel-immobilized proteins were reduced by adding 0.5 mL 100 mM NH4HCO3, 5 mM DTT, incubated in a thermomixer (800 RPM, 50 °C) for 30 min, and allowed to cool to RT. Reduced proteins were carboxamidomethylated by buffer exchange into 0.5 mL 100 mM NH4HCO3, 12.5 mM IAM and incubated in the dark for 30 min at RT. Gel bands were washed extensively by sequential buffer exchange using 0.5 mL aliquots of 100 mM NH4HCO3, 100 mM NH4HCO3:CH3CN (50:50, v/v; 0.5 mL), and neat CH3CN (0.25 mL). In each instance, the gel bands were incubated in a thermomixer (800 RPM, 37 °C) and the solvent decanted and discarded.
Gel bands were rehydrated using 25 uL 100 mM NH4HCO3 containing sequencing grade trypsin (Promega Corp.) at 10 ng/uL (0.4 pmol/uL). After 30 min, additional 100 mM NH4HCO3 was added to completely cover the gel bands (approx. 200 uL total vol.). Samples were incubated in a thermomixer (800 RPM, 37 °C) for 18 h.
Peptides were recovered by decanting the supernatant to a new 1.5 mL microfuge tube and extracting the gel bands with an equivalent volume of H2O:CH3CN:CF3CO2H (20:80:0.25; v/v/v) and sonicating for 30 min.
Extracts were combined with original supernatants and lyophilized to dryness using a vacuum centrifuge.
All samples were desalted by solid phase extraction using TARGA C18 microspin columns according to manufacturer’s instructions (The Nest Group). Desalted samples were resolubilized in 10 μL 1% CF3CO2H transferred to an autosampler vial.
Liquid chromatography-mass spectrometry
Nanoflow LC–MS/MS was performed using an UltiMate 3000 RSLCnano UHPLC directly coupled to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, Inc., San Jose, CA) similarly to previous studies35. An integrated autosampler was used to load samples at 5 μL/min onto a nanoViper trap column (75 mm × 20 mm, Acclaim PepMap 100 C18 resin, 3 μm particle size, 100 Å pore size, PN: 164535). Reversed phase HPLC was performed using an EasySpray C18 column (75 mm ID × 750 mm; Acclaim PepMap 100 C18 resin, 2 μm particle size, 100 Å pore size, PN: ES805). Mobile phases were Buffer A: H2O: (CH3)2SO: HCO2H (95:5:0.1; v/v/v) and Buffer B: CH3CN: (CH3)2SO: HCO2H (95:5:0.1; v/v/v). After 5 min of isocratic flow (200 nL/min, 2%B) a linear gradient from 2 to 35%B was developed over 66 min, 35–80% B over 4 min, and isocratic flow at 80%B for 10 min. LC–MS/MS experiments were performed using data dependent acquisition with dynamic exclusion enabled (exclusion width = + /− 10 ppm, repeat count = 2, repeat duration = 15 s, exclusion duration 22.5 s). MS1 scans (scan range 400–1600) were acquired in the Orbitrap mass analyzer (resolution 120,000 at m/z 400), AGC Target = 5.0 × 105, max. injection time = 50 ms).
Charge state exclusion enabled (1 + , ≥ 5 +). Based on relative intensity (threshold = 1.0 × 104), up to 20 ions from each survey scan were sequentially isolated and fragmented by HCD (quadrupole isolation width = 0.7 Da, normalized collision energy = 35%, AGC Target = 1.0 × 104).
Raw MS files were processed using Proteome Discoverer 2.2 software. MS/MS spectra were searched against a custom database constructed from the SwissProt portion of the UniProt database (release 2018_7). The database included all HUMAN sequences in SwissProt (curated) and was augmented with common reagent protein sequences (ASPN_PSEFR, CLOS_CLOHI, CTRA_BOVIN, CTRB_BOVIN, CTRC_BOVIN, GST26_SCHJA, LYSC_PSEAE, PNGF_ELIMR, SAV_STRAV, SPA_STAA8, SPA_STAAU, SPG1_STRSG, SPG2_STRSG, and TRYP_PIG).
Processing workflow parameters were: enzyme = trypsin, maximum missed cleavages = 2, fixed modifications = carbamidomethyl-Cys (C), variable modifications = N-acetyl (Protein), pyro-Glu (Q), and oxidation (M), precursor ion mass tolerance = 10 ppm, product ion mass tolerance = 0.6 Da, mass values = monoisotopic. Consensus workflow parameters: use only high confidence peptide, apply strict parsimony, 1% FDR, and 2 peptide IDs minimum.
Statistics
Prism ‘analyze’ tool used for all t-tests and ANOVA calculations. For all biological assays, at least 3 technical replicates and 3 biological replicates were measured and used for calculation of significance unless otherwise noted. P < 0.05 was considered significant for all statistical tests performed and generally * denotes P < 0.05, ** denotes P < 0.01, *** denotes P < 0.001, and **** denotes P < 0.0001. See statistics table for further information (Supplemental Table S6).
Supplementary Information
Acknowledgements
This study was funded by intramural funds provided to the National Institute of Neurological Disorders and Stroke and the National Cancer Institute at the NIH. The authors would like to thank Sadhana Jackson, M.D. Ph.D. and Ulisses Santamaria, B.A. for their careful review of the manuscript. The authors would also like to thank the NINDS Light Imaging Facility.
Author contributions
Conceptualization, T.D.O. and A.N.; Methodology, T.D.O., B.L.D., C.D., K.R.J., M. G. M., J.K., M.S., T.W., L.J.H., M.H.L., K.S., W.L., J.S. and A.N.; Investigation, T.D.O., B.L.D., C.D., A.A., K.R.J., M.G.M., J.K., S.J.A., B.A.O., M.S., L.J.H., S.F., and M.H.L.; Formal Analysis: T.D.O., B.L.D., C.D., J.S.R.; K.R.J., M.G.M., J.K., L.J.H., and A.N.; Writing—Original Draft, T.D.O., B.L.D., and A.N.; Writing—Review & Editing, T.D.O., B.L.D., J.S.R., Z.Z., and A.N.; Supervision, T.D.O. and A.N.
Funding
Open Access funding provided by the National Institutes of Health (NIH). This work was funded in part by the Intramural Program of the NCI and NINDS, NIH.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92223-x.
References
- 1.Nemes K, Fruhwald MC. Emerging therapeutic targets for the treatment of malignant rhabdoid tumors. Expert Opin. Ther. Targets. 2018;22(4):365–379. doi: 10.1080/14728222.2018.1451839. [DOI] [PubMed] [Google Scholar]
- 2.Rorke LB, Packer RJ, Biegel JA. Central nervous system atypical teratoid/rhabdoid tumors of infancy and childhood: Definition of an entity. J. Neurosurg. 1996;85(1):56–65. doi: 10.3171/jns.1996.85.1.0056. [DOI] [PubMed] [Google Scholar]
- 3.Biswas A, et al. Atypical teratoid/rhabdoid tumors: Challenges and search for solutions. Cancer Manag. Res. 2016;8:115–125. doi: 10.2147/CMAR.S83472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang XF, et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat. Genet. 2017;49(2):289–295. doi: 10.1038/ng.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vitte J, et al. Timing of Smarcb1 and Nf2 inactivation determines schwannoma versus rhabdoid tumor development. Nat. Commun. 2017;8(1):300. doi: 10.1038/s41467-017-00346-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filatova A, et al. Mutations in SMARCB1 and in other Coffin-Siris syndrome genes lead to various brain midline defects. Nat. Commun. 2019;10(1):2966. doi: 10.1038/s41467-019-10849-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boese A, et al. Integrase interactor 1 (Ini1/hSNF5) is a repressor of basal human immunodeficiency virus type 1 promoter activity. J. Gen. Virol. 2009;90:25032512. doi: 10.1099/vir.0.013656-0. [DOI] [PubMed] [Google Scholar]
- 8.Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 2012;10(6):395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 9.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 10.Buzdin A, et al. At least 50% of human-specific HERV-K (HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription. J. Virol. 2006;80(21):10752–10762. doi: 10.1128/JVI.00871-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovalskaya E, et al. Functional human endogenous retroviral LTR transcription start sites are located between the R and U5 regions. Virology. 2006;346(2):373–378. doi: 10.1016/j.virol.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Lavie L, et al. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2) J. Virol. 2005;79(2):876–883. doi: 10.1128/JVI.79.2.876-883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manghera M, Douville RN. Endogenous retrovirus-K promoter: A landing strip for inflammatory transcription factors? Retrovirology. 2013;10:1–11. doi: 10.1186/1742-4690-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Montojo M, et al. Human endogenous retrovirus-K (HML-2): A comprehensive review. Crit. Rev. Microbiol. 2018;44(6):715–738. doi: 10.1080/1040841X.2018.1501345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, et al. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells (vol 28, pg 2041, 2014) Genes Dev. 2015;29(1):108–108. doi: 10.1101/gad.244848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoelper D, et al. Structural and mechanistic insights into ATRX-dependent and -independent functions of the histone chaperone DAXX. Nat. Commun. 2017;8:1–13. doi: 10.1038/s41467-017-01206-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreira ME, et al. Mechanism of transcription factor recruitment by acidic activators. J. Biol. Chem. 2005;280(23):21779–21784. doi: 10.1074/jbc.M502627200. [DOI] [PubMed] [Google Scholar]
- 18.Neely KE, et al. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 2002;22(6):1615–1625. doi: 10.1128/MCB.22.6.1615-1625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han Y, et al. Cryo-EM structure of SWI/SNF complex bound to a nucleosome. Nature. 2020;579(7799):452–455. doi: 10.1038/s41586-020-2087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sammak S, et al. The structure of INI1/hSNF5 RPT1 and its interactions with the c-MYC:MAX heterodimer provide insights into the interplay between MYC and the SWI/SNF chromatin remodeling complex. FEBS J. 2018;285(22):4165–4180. doi: 10.1111/febs.14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boeke JD, Stoye JP. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor; 1997. [PubMed] [Google Scholar]
- 22.Barbulescu M, et al. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 1999;9(16):861–868. doi: 10.1016/S0960-9822(99)80390-X. [DOI] [PubMed] [Google Scholar]
- 23.Okahara G, et al. Expression analyses of human endogenous retroviruses (HERVs): Tissue specific and developmental stage-dependent expression of HERVs. Genomics. 2004;84(6):982990. doi: 10.1016/j.ygeno.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian RP, et al. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011;8:1–22. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grow EJ, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522(7555):221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 2015;7(307):307ra153. doi: 10.1126/scitranslmed.aac8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang T, et al. Regulation of stem cell function and neuronal differentiation by HERV-K via mTOR pathway. Proc. Natl. Acad. Sci. U S A. 2020;117(30):17842–17853. doi: 10.1073/pnas.2002427117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen T, et al. The viral oncogene Np9 acts as a critical molecular switch for co-activating betacatenin, ERK, Akt and Notch1 and promoting the growth of human leukemia stem/progenitor cells. Leukemia. 2013;27(7):1469–1478. doi: 10.1038/leu.2013.8. [DOI] [PubMed] [Google Scholar]
- 29.Zhou FL, et al. Activation of HERV-K Env protein is essential for tumorigenesis and metastasis of breast cancer cells. Oncotarget. 2016;7(51):84093–84117. doi: 10.18632/oncotarget.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, et al. Downregulation of human endogenous retrovirus Type K (HERV-K) Viral env RNA in pancreatic cancer cells decreases cell proliferation and tumor growth. Clin. Cancer Res. 2017;23(19):5892–5911. doi: 10.1158/1078-0432.CCR-17-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang G, et al. Human endogenous retroviral K element encodes fusogenic activity in melanoma cells. J. Carcinog. 2013;12:5. doi: 10.4103/1477-3163.109032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motegi A, et al. ALK receptor tyrosine kinase promotes cell growth and neurite outgrowth. J. Cell. Sci. 2004;117(Pt 15):3319–3329. doi: 10.1242/jcs.01183. [DOI] [PubMed] [Google Scholar]
- 33.Jiang MR, Stanke J, Lahti JM. The connections between neural crest development and neuroblastoma. Cancer Dev. 2011;94:77–127. doi: 10.1016/B978-0-12-380916-2.00004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kämmerer U, et al. Human endogenous retrovirus K (HERV-K) is expressed in villous and extravillous cytotrophoblast cells of the human placenta. J. Reprod. Immunol. 2011;91(1–2):1–8. doi: 10.1016/j.jri.2011.06.102. [DOI] [PubMed] [Google Scholar]
- 35.Xu J, et al. Pediatric brain tumor cell lines. J. Cell Biochem. 2015;116(2):218–224. doi: 10.1002/jcb.24976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torchia J, et al. Integrated (epi)-genomic analyses identify subgroup-specific therapeutic targets in CNS rhabdoid tumors. Cancer Cell. 2016;30(6):891–908. doi: 10.1016/j.ccell.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho B, Johann PD, Grabovska Y, Andrianteranagna MJDD, et al. Molecular subgrouping of atypical teratoid/rhabdoid tumors: A reinvestigation and current consensus. Neurol. Oncol. 2020;22(5):613–624. doi: 10.1093/neuonc/noz235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 39.Andries V, et al. NBPF1, a tumor suppressor candidate in neuroblastoma, exerts growth inhibitory effects by inducing a G1 cell cycle arrest. BMC Cancer. 2015;15:1–25. doi: 10.1186/s12885-015-1408-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi HS, et al. Identification and characterization of adenovirus early region 1B-associated protein 5 as a surface marker on undifferentiated human embryonic stem cells. Stem Cells Dev. 2011;20(4):609–620. doi: 10.1089/scd.2010.0265. [DOI] [PubMed] [Google Scholar]
- 41.Jin Y, Hammell M. Analysis of RNA-Seq data using TEtranscripts. Methods Mol. Biol. 2018;1751:153–167. doi: 10.1007/978-1-4939-7710-9_11. [DOI] [PubMed] [Google Scholar]
- 42.Jin Y, et al. TEtranscripts: A package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics. 2015;31(22):3593–3599. doi: 10.1093/bioinformatics/btv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat. Rev. Cancer. 2004;4(2):118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 44.Farahani E, et al. Cell adhesion molecules and their relation to (cancer) cell stemness. Carcinogenesis. 2014;35(4):747–759. doi: 10.1093/carcin/bgu045. [DOI] [PubMed] [Google Scholar]
- 45.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 2005;6(8):622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 46.Zhou FL, et al. Chimeric antigen receptor T cells targeting HERV-K inhibit breast cancer and its metastasis through downregulation of Ras. Oncoimmunology. 2015;4(11):e1047582. doi: 10.1080/2162402X.2015.1047582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Messeguer X, et al. PROMO: Detection of known transcription regulatory elements using speciestailored searches. Bioinformatics. 2002;18(2):333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 48.Goke J, et al. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015;16(2):135141. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 49.Agoni L, Guha C, Lenz J. Detection of human endogenous retrovirus K (HERV-K) transcripts in human prostate cancer cell lines. Front. Oncol. 2013;3:180. doi: 10.3389/fonc.2013.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burmeister T, et al. Insertional polymorphisms of endogenous HERV-K113 and HERV-K115 retroviruses in breast cancer patients and age-matched controls. AIDS Res. Hum. Retroviruses. 2004;20(11):1223–1229. doi: 10.1089/aid.2004.20.1223. [DOI] [PubMed] [Google Scholar]
- 51.Buscher K, et al. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005;65(10):4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 52.Contreras-Galindo R, et al. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J. Virol. 2008;82(19):9329–9336. doi: 10.1128/JVI.00646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flockerzi A, et al. Expression patterns of transcribed human endogenous retrovirus HERVK(HML-2) loci in human tissues and the need for a HERV Transcriptome Project. Bmc Genom. 2008;9:1–17. doi: 10.1186/1471-2164-9-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goering W, et al. Human endogenous retrovirus HERV-K(HML-2) activity in prostate cancer is dominated by a few loci. Prostate. 2015;75(16):1958–1971. doi: 10.1002/pros.23095. [DOI] [PubMed] [Google Scholar]
- 55.Golan M, et al. Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia. 2008;10(6):521–U3. doi: 10.1593/neo.07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krishnamurthy J, et al. Genetic engineering of T cells to target HERV-K, an ancient retrovirus on melanoma. Clin. Cancer Res. 2015;21(14):3241–3251. doi: 10.1158/1078-0432.CCR-14-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lemaitre C, et al. A human endogenous retrovirus-derived gene that can contribute to oncogenesis by activating the ERK pathway and inducing migration and invasion. PLoS Pathog. 2017;13(6):e1006451. doi: 10.1371/journal.ppat.1006451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmitt K, et al. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML2) loci in melanoma. Genome Biol. Evol. 2013;5(2):307–328. doi: 10.1093/gbe/evt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Serafino A, et al. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp. Cell Res. 2009;315(5):849–862. doi: 10.1016/j.yexcr.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 60.Stengel S, et al. Regulation of human endogenous retrovirus-k expression in melanomas by CpG methylation. Genes Chromosom. Cancer. 2010;49(5):401–411. doi: 10.1002/gcc.20751. [DOI] [PubMed] [Google Scholar]
- 61.Wang-Johanning F, et al. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22(10):1528–1535. doi: 10.1038/sj.onc.1206241. [DOI] [PubMed] [Google Scholar]
- 62.Wang-Johanning F, et al. Expression of human endogenous retrovirus K envelope transcripts in human breast cancer. Clin. Cancer Res. 2001;7(6):1553–1560. [PubMed] [Google Scholar]
- 63.Wang-Johanning F, et al. Human endogenous retrovirus type K antibodies and mRNA as serum biomarkers of early-stage breast cancer. Int. J. Cancer. 2014;134(3):587–595. doi: 10.1002/ijc.28389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang-Johanning F, et al. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int. J. Cancer. 2007;120(1):81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- 65.Wang-Johanning F, et al. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008;68(14):5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang-Johanning F, et al. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. Jnci-J. Natl. Cancer Inst. 2012;104(3):189–210. doi: 10.1093/jnci/djr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wildschutte JH, et al. The distribution of insertionally polymorphic endogenous retroviruses in breast cancer patients and cancer-free controls. Retrovirology. 2014;11:1–13. doi: 10.1186/s12977-014-0062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maacha S, et al. Extracellular vesicles-mediated intercellular communication: Roles in the tumor microenvironment and anti-cancer drug resistance. Mol. Cancer. 2019;18(1):55. doi: 10.1186/s12943-019-0965-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohgushi M, Sasai Y. Lonely death dance of human pluripotent stem cells: ROCKing between metastable cell states. Trends Cell Biol. 2011;21(5):274–282. doi: 10.1016/j.tcb.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 70.Zohn IE, et al. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell. 2006;125(5):957–969. doi: 10.1016/j.cell.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 71.Lesueur LL, Mir LM, Andre FM. Overcoming the specific toxicity of large plasmids electrotransfer in primary cells in vitro. Mol. Ther. Nucleic Acids. 2016;5:e291. doi: 10.1038/mtna.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim E, et al. Systematic functional interrogation of rare cancer variants identifies oncogenic alleles. Cancer Discov. 2016;6(7):714–26. doi: 10.1158/2159-8290.CD-16-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wotton D, et al. The Smad transcriptional corepressor TGIF recruits mSin3. Cell Growth Differ. 2001;12(9):457–63. [PubMed] [Google Scholar]
- 75.Tyagi R, et al. Inhibition of human endogenous retrovirus-K by antiretroviral drugs. Retrovirology. 2017;14(1):21. doi: 10.1186/s12977-017-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.DeMarino C, et al. Purification of high yield extracellular vesicle preparations away from virus. Jove-J. Vis. Exp. 2019;151:e59876. doi: 10.3791/59876. [DOI] [PubMed] [Google Scholar]
- 77.Jaworski E, et al. The use of nanotrap particles technology in capturing HIV-1 virions and viral proteins from infected cells. PLoS ONE. 2014;9(5):e96778. doi: 10.1371/journal.pone.0096778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsudaira PT. A Practical Guide to Protein and Peptide Purification for Microsequencing. 2. Academic Press; 1993. p. 184. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.