Abstract
COPD is a clinically heterogeneous syndrome characterized by injury to airways, airspaces, and lung vasculature and usually caused by tobacco smoke and/or air pollution exposure. COPD is also independently associated with nonpulmonary comorbidities (eg, cardiovascular disease) and malignancies (eg, GI, bladder), suggesting a role for systemic injury. Since not all those with exposure develop COPD, there has been a search for plasma and lung biomarkers that confer increased cross-sectional and longitudinal risk. This search typically focuses on clinically relevant COPD outcomes such as FEV1, FEV1 decline, CT measurements of emphysema, or exacerbation frequency. The rapid advances in omics technology and the molecular phenotyping of COPD cohorts now permit large-scale evaluation of genetic, transcriptomic, proteomic, and metabolic biomarkers. This review focuses on protein biomarkers associated with clinically relevant COPD outcomes. The prototypic COPD protein biomarker is alpha-1 antitrypsin; however, this biomarker only accounts for 1% to 5% of COPD. This article reviews and summarizes the evidence for other validated biomarkers for each COPD outcome, and discusses their advantages, weaknesses, and required regulatory steps to move the biomarker from the bench into clinic. Although we highlight the emergence of many novel biomarkers (eg, fibrinogen, soluble receptor for advanced glycation, surfactant protein D, club cell secretory protein), there is increasing evidence that individual biomarkers only explain a fraction of the increased COPD risk and that multiple biomarker panels are needed to completely explain clinical variation and risk in individuals and populations.
Key Words: biomarker network, COPD, multiplex platforms, protein biomarker
Abbreviations: AAT, alpha-1 antitrypsin; COPDGene, COPD Genetic Epidemiology; ECLIPSE, Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points; FDA, Food and Drug Administration; MESA-Lung study, Multi-Ethnic Study of Atherosclerosis; SPIROMICS, SubPopulations and InteRmediate Outcome Measures In COPD Study; sRAGE, soluble receptor for advanced glycation
The Need for Biomarkers in COPD
A typical patient with COPD has had decades of cigarette smoke exposure and does not manifest signs of disease until the latter stages of life. Furthermore, traditional measures of disease progression (eg, changes in postbronchodilator spirometry or CT emphysema) occur over many years. Unfortunately, there are currently no Food and Drug Administration (FDA)-approved medications that reduce disease progression endpoints or mortality in clinical trials; therefore, there is an unmet need for surrogate endpoints (ie, biomarkers) that can be used to identify subjects at high risk of progression or that could serve as targets for particular subphenotypes or COPD.1,2 This contrasts with COPD exacerbator phenotype, which has intermittent and short-term manifestations and multiple approaches which have been shown to reduce exacerbations.3,4
Definition of COPD Biomarkers
The FDA-NIH Biomarker Working Group’s BEST (Biomarkers, EndpointS and Tools)5 describes a biomarker as a “characteristic that is measured as an indicator of normal biological processes, pathogenic processes, or responses to an exposure or intervention, including therapeutic interventions. Molecular, histologic, radiographic, or physiologic characteristics are types of biomarkers. A biomarker is not an assessment of how an individual feels, functions, or survives.”
Potentially useful COPD biomarkers might include those that could differentiate between the following: (1) healthy individuals vs individuals with COPD (diagnostic biomarkers), (2) active/progressing COPD vs stable COPD (disease activity biomarkers), and (3) treatment responders vs nonresponders (response to treatment biomarkers).
Molecular biomarkers come in a large variety of types and they serve mostly as diagnostic and disease activity markers. Next to proteins, other COPD biomarkers include the following: (1) genetic biomarkers which are linked to candidate loci and single nucleotide polymorphisms in COPD risk genes (eg, SERPINA1 identifies individuals with alpha-1 deficiency-related COPD, chromosome 15q25: CHRNA3/CHRNA5/IREB2 associated with smoking behavior and COPD, independent of smoking; chromosome 19q13 linked to increased susceptibility to smoking, or PTPN6 linked to early onset familial emphysema)6, 7, 8; (2) epigenomic biomarkers to identify the methylation status of risk genes (eg, HDAC6, PTEN, Nrf2)9, 10, 11 frequently associated with the clinical response to corticosteroids in individuals with COPD; (3) transcriptomic biomarkers to identify profiles of differentially expressed genes (eg, ASAH1, CEBPD, FOXP1, TCF7)9, 10, 11 that are associated with low FEV1 and/or FEV1/FVC ratio; (4) proteomic biomarkers to identify directional changes of relevant systemic/lung-specific proteins (eg, fibrinogen, soluble receptor for advanced glycation [sRAGE])12,13 linked to particular COPD endotypes (eg, fibrinogen to COPD with frequent exacerbations, sRAGE to emphysema-predominant COPD); (5) metabolomic biomarkers such as sphingolipids,14 which have been linked to COPD exacerbations; and (6) microbiome signatures to identify bacterial diversity and communities (eg, Firmicutes phylum)15 present specifically in the airway and distal lung of individuals with COPD or sputum (eg, Veillonella species, Staphylococcus species) associated with 1-year survival and mortality, respectively, after hospitalization for COPD exacerbation.16
In this review we discuss the protein biomarkers that are associated with COPD clinical phenotypes such as those determined by spirometry (airflow obstruction assessed by FEV1, FEV1/FVC ratio), imaging (% emphysema, lung densitometry as estimated by 15th percentile density, or bronchial wall thickness of the inner perimeter of 10-mm airway on CT scan),17, 18, 19, 20, 21 or frequency of COPD exacerbations.
Advantages and Disadvantages of Protein Biomarkers Compared With Other Types of Molecular Biomarkers
The first biomarker, alpha-1 antitrypsin was discovered through serum protein electrophoresis in 1963.22 More recently, there has been a focus on genetics with the discovery of > 125 single nucleotide polymorphisms in SERPINA1 that have been associated with alpha-1 deficiency.23 Genetic research approaches are the first to come to mind for COPD personalized medicine and have yielded many novel COPD candidate genes beyond SERPINA124; however, one’s genotype is static throughout life and does not change with smoking or therapeutic interventions.2,12 Epigenetic and transcript approaches are closely related to the genome and have more potential as COPD biomarkers because they can change with time; however, most currently approved therapies in COPD and other diseases do not target the epigenome or transcriptome. Rather, they target proteins or enzymes and their products (eg, metabolites).12,25 Metabolites such as sphingolipids and leukotrienes could be excellent COPD biomarkers; nevertheless, there are currently few large-scale, replicated metabolomics studies in COPD.26,27 Technology has favored the search for protein biomarkers in COPD because of the latest development of large-scale protein arrays that can assay thousands of proteins simultaneously, cheaply, and quickly, and using low sample volume.28,29
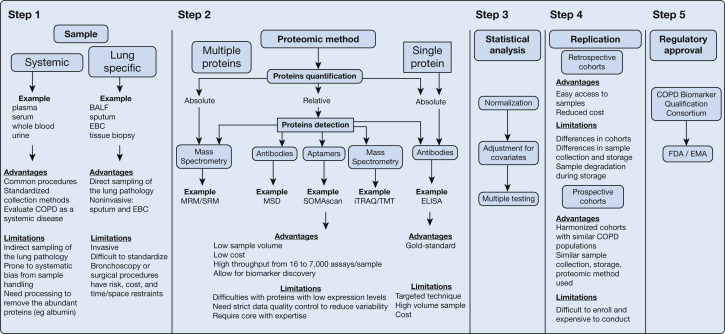
Major considerations for COPD biomarkers include the following: sample type, assays method, statistical analysis strategy, replication across similar populations in different studies, and whether a biomarker is likely to achieve regulatory approval (Fig 1). Some popular sample matrices for COPD biomarkers include plasma30 or urine,31 but also BAL fluid,32,33 sputum,34 exhaled breath condensate,35 and lung tissue.36,37 For many discovery studies, there is the potential to assay multiple proteins with different detection methods (antibodies, aptamers, or mass spectrometry) and quantification (relative or absolute). These technologic advances come with a major limitation that as the number of biomarkers (P) grows, it may significantly exceed the number of samples (N), which can lead to false discovery in small studies (where P >> N). Hence, we rely on statistical analysis to ensure data quality control and normalization to adjust for covariates and to apply multiple testing methods. Replication in retrospective or prospective cohorts helps biomarker validation and the regulatory approval steps. Regulatory approval typically focuses on consistency of existing data and adding assay parameters such as upper and lower limits of detection and coefficients of variation.
Figure 1.
Approach to judging a proteomic study. Brief description of the five steps to be considered when judging a well-designed proteomic study. Step 1: evaluation of samples used in the proteomic study with the advantages and limitations of systemic vs lung-specific samples. Step 2: evaluation of the proteomic method used in the study, including the platform (single vs multiplex), detection method (antibodies, aptamers, or mass spectrometry), and quantification (relative or absolute) with their advantages and limitations vs ELISA, the current criterion standard method. Step 3: evaluation of the statistical methods used with specific considerations for data quality control, normalization, and adjustment for covariates. Step 4: evaluation of the cohorts used for protein biomarker validation with advantages and limitations of retrospective vs prospective cohorts. Step 5: evaluation by the regulatory agencies involved in biomarker approval. BALF = BAL fluid; EBC = exhaled breath condensate; ELISA = enzyme-linked immunosorbent assay; EMA = European Medicines Agency; FDA = Food and Drug Administration; iTRAQ/TMT = isobaric tag for relative and absolute quantitation/tandem mass tags; MRM/SRM = multiple reaction monitoring/selective reaction monitoring; MSD = Meso Scale Discovery; SOMAscan = slow off-rate modified aptamers.
Evidence for COPD Protein Biomarkers
Studies published in the last 10 years, heavily relying on plasma samples, have identified many candidate protein biomarkers. From our PubMed search performed in May 2020, we selected studies listing the terms “COPD” and “proteomic” in the key word section. These studies used a plethora of different proteomic platforms and biostatistical methods. Therefore, we decided to focus on plasma protein biomarkers that have been tested in studies with large numbers of patients (∼100 patients) and that have been replicated, defined as identifying proteins with a similar proteomic platform, in at least two independent cohorts. It is notable that many plasma protein biomarkers (Table 1) behave differently as a cross-sectional vs longitudinal biomarker. Additionally, many biomarkers are associated with multiple clinically relevant outcomes in COPD: FEV1, FEV1/FVC ratio, FEV1 decline, diffusing capacity for carbon monoxide (Dlco), emphysema distribution, and CT measurements of emphysema (lung densitometry as estimated by the 15th percentile density) or exacerbation frequency. Inflammatory biomarkers (C-reactive protein, fibrinogen, IL-6, IL-8, and monocyte chemoattractant protein-1), growth factors (vascular endothelial growth factor, epidermal growth factor receptor, apolipoprotein A1, hepatocyte growth factor, and bactericidal/permeability increasing fold-containing family B member 1), proteases/antiproteases (matrix metallopeptidase 8, matrix metallopeptidase 9, myeloperoxidase, tissue inhibitor of metalloproteinases 1, and alpha-1 antitrypsin), and lung-derived proteins (surfactant protein D and club cell secretory protein) correlate positively with clinically relevant outcomes. Antiinflammatory molecules (eg, sRAGE) tend to be inversely correlated with emphysema and airflow limitation. The most promising plasma biomarkers have been validated, defined as identifying proteins using different proteomic platforms in multiple cohorts. In addition, these biomarkers were associated not only with cross-sectional clinically relevant outcomes, but also with longitudinal outcomes (FEV1 decline, emphysema progression, and future exacerbations). The few biomarkers (apolipoprotein A1 and tissue inhibitor of metalloproteinases 1) found in the sputum have yet to be validated. To date, no biomarker has been validated in sputum and plasma to predict a COPD clinical outcome or disease progression. Some biomarkers have been associated with COPD diagnosis, severity, and progression; nevertheless, there is not one validated biomarker that is associated with all clinical outcomes and disease progression. Traditionally, the biomarkers associated with clinical outcomes pertinent to lung function in cross-sectional studies have been considered to reflect COPD severity (eg, sRAGE, emphysema severity); however, emerging data from longitudinal cohorts suggest that the same biomarker might reflect COPD activity considering that it was associated with emphysema progression (Table 2).
Table 1.
Lung and Plasma Proteomics Identify Candidate Cross-Sectional and Longitudinal Biomarkers
| Phenotype | Clinical Parameter | Biomarker |
|---|---|---|
| Cross-sectional | ||
| Lung function | Case/control |
|
| FEV1 | ||
| FEV1/FVC | ||
| Dlco | ||
| GOLD stages | ||
| Imaging | Emphysema | |
| LAA-950 | ||
| PD15 | ||
| Pi10 | ||
| Clinical phenotypes | Frequent exacerbations | |
| Longitudinal | ||
| Lung function | FEV1 decline | |
| Imaging | Emphysema progression | |
| Clinical phenotypes | Future exacerbations | |
AAT = alpha-1 antitrypsin; AHSG = alpha 2-HS glycoprotein; ALB = albumin; APOA1 = apolipoprotein A1; ATIII = antithrombin III; ATP = adenosine triphosphate; BPIFB1 = bactericidal/permeability-increasing-fold-containing family B member 1; C5a = complement C5a protein; C6orf58 = chromosome 6 open reading frame 58; CC16 = club cell secretory protein; CCL2 = C-C motif chemokine ligand 2; CCL11 = C-C motif chemokine ligand 11; CCL18 = C-C motif chemokine ligand 18; CCL20 = C-C motif chemokine ligand 20; CNDP1 = carnosine dipeptidase 1; CRP = C-reactive protein; CXCL9 = C-X-C motif chemokine ligand 9; CXCL10 = C-X-C motif chemokine ligand 10; Dlco = diffusing capacity for carbon monoxide; EGFR = epidermal growth factor receptor; FAS = Fas cell surface death receptor; GOLD = Global Initiative for Chronic Obstructive Lung Disease; GRP78 = 78-kDa glucose-regulated protein; HGF = hepatocyte growth factor; HRG = histidine-rich glycoprotein; ICAM-1 = intercellular adhesion molecule 1; IFNγ = interferon gamma; KRT19 = keratin 19; LAA-950 = low attenuation areas less than a threshold of -950 Hounsfield units; MCP-1 = monocyte chemoattractant protein-1; MDC = macrophage-derived chemokine; MIP3A = macrophage inflammatory protein 3 alpha; MMP8 = matrix metallopeptidase 8; MMP9 = matrix metalloproteinase 9; MMP10 = matrix metallopeptidase 10; MPO = myeloperoxidase; MSTP9 = putative macrophage-stimulating protein; PAI-II = plasminogen activator inhibitor-2; PD15 = 15th percentile lung density; Pi10 = inner perimeter of 10-mm airway; RAGE = receptor for advanced glycation endproducts; SAA = serum amyloid A; sCD163 = cluster of differentiation 163 protein; SERPINA7 = serpin family A member 7; SOD1 = superoxide dismutase; SP-D = surfactant protein D; sRAGE = soluble receptor for advanced glycation endproducts; TF = transferrin; TIMP1 = tissue inhibitor of metalloproteinases 1; TNFα = tumor necrosis factor alpha; TS1 = thymidylate synthase; VEGF = vascular endothelial growth factor.
Lung proteomics.
Plasma proteomics.
Validated using at least two methods in two cohorts.
Table 2.
Protein Biomarker Associated With Cross-sectional and Longitudinal COPD Outcomes
| Biomarker | Lung Function |
Imaging |
Exacerbations |
||||
|---|---|---|---|---|---|---|---|
| FEV1 or FEV1/FVC | Decline FEV1 | Dlco | Emphysema | Emphysema Progression |
Frequent and Previous | Future | |
| sRAGE | ↑b | ↑b | ↓a | ↓b | ↓b | ↑b | ↑a |
| IL-6 | ↑b | ↑a | NS | ↑a | ↑a | NS | NS |
| IL-8 | ↑b | NS | ↑a | NS | ↑a | ↑b | NS |
| Fibrinogen | ↓b | ↑b | NS | ↑b | ↓b | ↑b | ↑a |
| CRP | ↓b | ↑a | NS | ↑b | ↑a | ↑b | ↑a |
| SPD | ↓b | ↑a | NS | ↓b | ↑a | ↑b | NS |
| CC16 | ↑b | ↓b | NS | ↑a | ↓a | NS | ↑a |
| APOA1 | ↑a | NS | ↑a | NS | NS | NS | NS |
↑ = positive biomarker – clinical outcome association; ↓ = negative biomarker – clinical outcome association; APOA1 = apolipoprotein A1; CC16 = club cell secretory protein; CRP = C-reactive protein; Dlco = diffusing capacity for carbon monoxide; NS = not significant biomarker – clinical outcome association; SPD = surfactant protein D; sRAGE = soluble receptor for advanced glycation.
No replication.
Replication in two cohorts.
Judging Protein Biomarker Evidence
The sine qua non criterion for establishing biomarker validity is replication of associations across multiple independent populations. Similar to genetics, early biomarker studies were plagued by a lack of replication. Fortunately, major funding agencies have made coordinated efforts to build and maintain large COPD cohorts: Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE),53 SubPopulations and InteRmediate Outcome Measures In COPD Study (SPIROMICS), COPD Genetic Epidemiology (COPDGene),54 Treatment Of Emphysema With A Selective Retinoid Agonist (TESRA),55 and population cohorts with good lung phenotyping including the Framingham Heart Study (FHS), Multi-Ethnic Study of Atherosclerosis (MESA-Lung study), Cooperative Health Research in the Augsburg Region, (KORA), or those tailored toward specific COPD phenotypes (emphysema secondary to alpha-1 antitrypsin [AAT] deficiency), such as QUANTitative lung CT UnMasking emphysema progression in AATD (QUANTUM) and Genomic Research in Alpha-1 Antitrypsin Deficiency and Sarcoidosis (GRADS).56, 57, 58 The latest efforts aim to harmonize testing platforms and data analysis to validate promising biomarkers across multiple cohorts (Table 2).46
Other useful but not essential features that make a protein biomarker valuable include validation of the assay across multiple platforms using well-established standards. For clinical use, this may also include validating the range of the assay across concentrations that are likely to be encountered in real human populations. Stability of the assay measurements under different conditions (eg, time, method of collection, processing times) can affect assay stability and utility in real-world settings.59,60 Taking a long time to aliquot and rapid freezing of a sample may result in degradation of selected proteins. For example, RBC lysis from contact with ice may result in increased release of intracellular proteins, or at low centrifugation speeds, platelets and their contents may not separate from plasma.61 Less commonly, appreciated factors that can introduce bias are fasting time and time of day of sampling.62 There are also biological factors that can affect proteins such as exercise, age, sex, and sleep.63, 64, 65 All these features are important for biomarker discovery and can reduce the biomarker clinical utility by making it hard to perform consistent assays, but robust assays with the most clinical utility should not be greatly affected by these factors.
Other helpful but not essential features which improve the suitability of a biomarker include evidence for a mechanistic role (eg, AAT),27,57 association with disease progression or activity (eg, fibrinogen for exacerbations),66 and a known minimal clinically important difference (eg, changes of 4 units in the St. George’s Respiratory Questionnaire to detect a clinically significant change in the clinical outcome).67 Biomarkers of COPD progression with supporting mechanistic evidence are rare for COPD because progression typically occurs over decades; however, examples of biomarkers that are associated with disease progression include serum AAT level.68
An unmet need in the COPD biomarker field is discovery and validation of biomarkers associated with response to treatment. In monogenetic diseases such as AAT deficiency-serum levels of desmosine and isodesmosine, two markers of elastin degradation associated with clinical (FEV1) COPD outcomes, and morphologic (emphysema CT measurements) COPD outcomes, have been modified by weekly administration of augmentation therapy, intervention recognized to modify COPD progression in individuals with AAT deficiency.69 In smoking-related COPD, this task is not readily explored. First, there are few disease-modifying therapies in COPD to study their effect on biomarkers identified to correlate with COPD severity and activity. Second, individuals enrolled in COPD cohorts, both in the control or diseased arms, may be on non-COPD therapies (eg, statins, thiazolidinediones, angiotensin converting enzyme inhibitors, angiotensin II type 1 receptor antagonists) that have been shown to increase serum levels of COPD relevant biomarkers (eg, sRAGE).70 Stringent inclusion criteria and biostatistical methods can estimate the additive effect of these therapies to the biomarker variability.
Final considerations for a biomarker should include the target population. For the COPD population, these may include older age and current or former smokers, but also features such as sex and race, both of which are strongly associated with differential expression of blood biomarkers.63 Clear delineation of the population used to assess biomarker performance is essential when interpreting a biomarker test, but is often not considered (eg, D-dimer performs better in the diagnosis of pulmonary embolism in a high pretest probability population71 rather than all patients evaluated for chest pain).
Why Do Protein Biomarkers Fail to Validate Across All Cohorts?
Many of the discovery biomarkers presented in Table 1 may not be generalizable to all COPD populations because of inherent differences between the cohorts. Inflammatory (C-reactive protein, fibrinogen, IL-6, and IL-8) and distal lung injury (sRAGE, surfactant protein D, club cell secretory protein, and apolipoprotein A1) biomarkers associated with clinical outcomes across multiple cohorts are shown in Table 2.13,46,47 The difficulties in replicating some biomarkers may be related to differences in the individuals recruited in these cohorts.
Even with a balanced number of men and women recruited in the cohorts, we are able to pin out protein biomarkers that might be sex-specific (eg, IL-16, VEGF).63
Because many cohorts are enriched for white, European-descendent individuals, fewer publications have reported on racially diversified COPD cohorts. COPDGene included approximately 33.5% non-Hispanic African Americans, but the plasma biomarkers identified within this subgroup have yet to replicated and validated in other cohorts with admixed backgrounds, such as SPIROMICS, MESA-Lung study, or Jackson Heart Study (JHS).48 Plasma endothelin-1 surfaced as a possible biomarker for heart failure and mortality in African Americans enrolled in the racially diverse Jackson Heart Study (JHS) cohort.72
SPIROMICS and COPDGene included smokers and COPD of all severity, with fewer healthy nonsmoker individuals; ECLIPSE included primarily white, European ancestry individuals with more severe COPD and rather fewer active smokers or patients with mild stage 1 COPD. Neither COPDGene nor ECLIPSE are population-based studies, but the Framingham Heart Study (FHS) and MESA-Lung study have been designed as population-based studies; their caveat is that they are not enriched for the population at risk for COPD and the effect of smoking or COPD development on any biomarker in these cohorts is skewed by the variability of the biomarker in the nondiseases subjects because of age, sex, race, and sample biases. The positive predictive value of a biomarker drops in population whose disease prevalence is low (eg, general population) vs a population enriched for COPD (eg, patients in the pulmonary clinic). Ideally, matching individuals enrolled for age, sex, race, and baseline lung function may overcome this conundrum, but this is difficult to achieve in longitudinal COPD studies where inclusion and follow-up of healthy, nonsmoker individuals are hindered by individuals’ cooperation, age-related storage material degradation, and costs usually underrecognized by the funding agencies.
Moving a Candidate Biomarker From Research to Clinical Practice
The COPD biomarkers described in Table 2 have made it far on the roadmap for novel biomarkers regulatory approval. Importantly, the COPD Biomarker Qualification Consortium, after reviewing the strong preclinical and clinical data, and the validation studies in multiple cohorts deemed plasma fibrinogen appropriate for FDA evaluation and approval as a biomarker to identify and predict the risk of future COPD exacerbations. Plasma fibrinogen benefits from a reproducible and widely available detection method; the availability of the testing method outside big research centers where most of the validation studies have been conducted is one big technical challenge. Subsequently, the regulatory agencies FDA and European Medicines Agency agreed that when plasma fibrinogen passes the last two relevant hurdles, interventional and prospectively designed studies, it will be ready for clinical use. The FDA/European Medicines Agency approved plasma fibrinogen as a biomarker of high risk for COPD exacerbation and all-cause mortality in COPD. Plasma fibrinogen dossier initiation, consultation, review, and approval stages took approximately 4 years.66 Next on the approvable biomarkers list are sRAGE46 and blood eosinophil count.
The Future of COPD Biomarkers: Where Do We Go From Here?
Phenotypes and Progression
Although there are now several encouraging biomarkers (eg, AAT, fibrinogen, sRAGE), there still remains significant gaps in our knowledge. For instance, there are few large studies that have investigated and replicated biomarkers of chronic bronchitis or disease progression, comorbidities, and death from COPD. These biomarkers will be crucial for identifying subjects who are at high risk and may benefit from an intervention. Furthermore, there are also no large studies which identify biomarkers of treatment response. These biomarkers will be essential in evaluating disease-modifying interventions because traditional metrics (FEV1, mortality, or emphysema progression) typically change over decades rather than months.
Single Biomarkers vs Biomarker Scores
Current evidence suggests that single biomarkers typically explain < 10% of the risk of COPD phenotypes.27,46 Similar to genetic risk scores, in which multiple genetic markers are weighted for a single total score, evidence suggests that multiple biomarker-derived scores significantly explain > 10% of risk. Examples of a multiple biomarker panel might be club cell secretory protein, fibrinogen, sRAGE, C-reactive protein, and surfactant protein D for FEV1.46 As with genetic risk scores, the disadvantage of protein risk scores is that they are likely to be sex-, phenotype-, and population-specific.
Integration of Protein Biomarkers With Other Omics
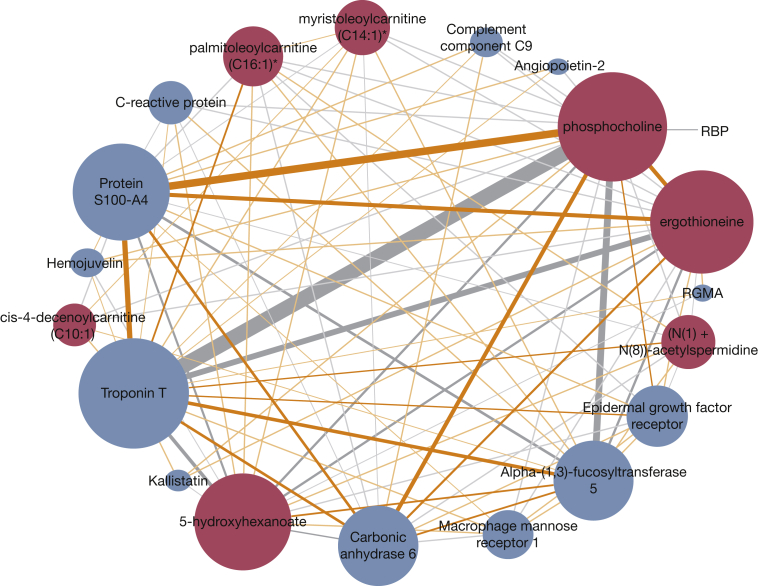
In biological systems, proteins do not act independently of other proteins or other classes of molecules (eg, nucleic acids, metabolites). For instance, we know that expression or measurement of most proteins are at least partially associated with genotype.73,74 Therefore, it is reasonable to assume that the best predictive models will include knowledge of multiple classes of molecules. Additional multiple proteins can be represented as networks and these networks can be integrated with other omics networks to better understand COPD. For instance, by integrating proteomics networks with metabolomics networks, we have been able to deconstruct several pathways that are associated with distinct COPD clinical phenotypes: oxidative phosphorylation with emphysema progression, antigen processing and presentation with exacerbation frequency, and glycerophospholipid metabolism with FEV1 and FEV1/FVC ratio (Fig 2).75 A network-based interactome facilitates a better understanding of the interplay between interconnected causative factors in a manner compounding the effects of any one factor. The interactome can lead to the identification of key hub molecules that might not be even evaluated by discovery platforms.
Figure 2.
Protein-metabolite network associated with FEV1. The network was identified after applying sparse multiple canonical correlation network (SmCCNet) to adjust proteomic and metabolomic data from the blood of 1,008 participants in the COPD Genetic Epidemiology study to study omics data association with FEV1. Edges thickness represents the level of association between metabolite-protein pairs relative to percent FEV1. The size of the network hubs and the nodes correspond to the highest connectivity (ie, number of edges connected to the node). The 13 proteins (blue) and seven metabolites (red) included in the network were individually associated with percent FEV1, but within the network the pair phosphocholine-troponin T has the highest pairwise correlation with percent FEV1, suggestive of a strong negative (gray) link between systemic inflammation (phosphocholine)-heart muscle strain (troponin T)-percent FEV1. Gray edges indicate negative correlation between the nodes and orange edges indicate positive correlation between the nodes. RBP = retinol-binding protein; RGMA = repulsive guidance molecule A. (Reprinted with permission from Mastej et al.75)
We are optimistic that harmonization of standard-of-practice procedures, storage methods, and data integration will pave the way for promising protein biomarkers and networks to become this decade’s contribution to personalized medicine in COPD.
Acknowledgments
Author contributions: K. A. S., K. A. P., and R. P. B. contributed substantially to the data collection and interpretation and the writing of the manuscript. K. A. S. and R. P. B. are responsible for the content of this manuscript. The manuscript has not, either in part or whole, presented or been reviewed before accepted for publication in CHEST.
Financial/nonfinancial disclosures: None declared.
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Other contributions: We thank Dr Maneesh Bhargava, MD, PhD (University of Minnesota) for his insightful comments.
Footnotes
FUNDING/SUPPORT: This study was funded by National Institutes of Health/National Heart, Lung, and Blood Institute 1R01HL137995 and 1R01HL152735-01 to Dr Bowler.
References
- 1.Han M.K., Martinez C.H., Au D.H. Meeting the challenge of COPD care delivery in the USA: a multiprovider perspective. Lancet Respir Med. 2016;4(6):473–526. doi: 10.1016/S2213-2600(16)00094-1. [DOI] [PubMed] [Google Scholar]
- 2.Regan E.A., Hersh C.P., Castaldi P.J. Omics and the search for blood biomarkers in chronic obstructive pulmonary disease. Insights from COPDGene. Am J Respir Cell Mol Biol. 2019;61(2):143–149. doi: 10.1165/rcmb.2018-0245PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wedzicha J.A., Brill S.E., Allinson J.P., Donaldson G.C. Mechanisms and impact of the frequent exacerbator phenotype in chronic obstructive pulmonary disease. BMC Med. 2013;11:181. doi: 10.1186/1741-7015-11-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wedzicha J.A., Rabe K.F., Martinez F.J. Efficacy of roflumilast in the COPD frequent exacerbator phenotype. Chest. 2013;143(5):1302–1311. doi: 10.1378/chest.12-1489. [DOI] [PubMed] [Google Scholar]
- 5.FDA-NIH Biomarker Working Group . Silver Spring; MD: 2016. BEST (Biomarkers, EndpointS, and other Tools) Resource. [PubMed] [Google Scholar]
- 6.Hobbs B.D., Hersh C.P. Integrative genomics of chronic obstructive pulmonary disease. Biochem Biophys Res Commun. 2014;452(2):276–286. doi: 10.1016/j.bbrc.2014.07.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakornsakolpat P., Prokopenko D., Lamontagne M. Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat Genet. 2019;51(3):494–505. doi: 10.1038/s41588-018-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosse Y., Lamontagne M., Gaudreault N. Early-onset emphysema in a large French-Canadian family: a genetic investigation. Lancet Respir Med. 2019;7(5):427–436. doi: 10.1016/S2213-2600(19)30056-6. [DOI] [PubMed] [Google Scholar]
- 9.Barnes P.J. Reduced histone deacetylase in COPD: clinical implications. Chest. 2006;129(1):151–155. doi: 10.1378/chest.129.1.151. [DOI] [PubMed] [Google Scholar]
- 10.Lam H.C., Cloonan S.M., Bhashyam A.R. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. J Clin Invest. 2013;123(12):5212–5230. doi: 10.1172/JCI69636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vucic E.A., Chari R., Thu K.L. DNA methylation is globally disrupted and associated with expression changes in chronic obstructive pulmonary disease small airways. Am J Respir Cell Mol Biol. 2014;50(5):912–922. doi: 10.1165/rcmb.2013-0304OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowler R.P., Wendt C.H., Fessler M.B. New strategies and challenges in lung proteomics and metabolomics. An Official American Thoracic Society Workshop Report. Ann Am Thorac Soc. 2017;14(12):1721–1743. doi: 10.1513/AnnalsATS.201710-770WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng D.T., Kim D.K., Cockayne D.A. Systemic soluble receptor for advanced glycation endproducts is a biomarker of emphysema and associated with AGER genetic variants in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;188(8):948–957. doi: 10.1164/rccm.201302-0247OC. [DOI] [PubMed] [Google Scholar]
- 14.Cruickshank-Quinn C.I., Jacobson S., Hughes G. Metabolomics and transcriptomics pathway approach reveals outcome-specific perturbations in COPD. Sci Rep. 2018;8(1):17132. doi: 10.1038/s41598-018-35372-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez F.J., Erb-Downward J.R., Huffnagle G.B. Significance of the microbiome in chronic obstructive pulmonary disease. Ann Am Thorac Soc. 2013;10(suppl):S170–S179. doi: 10.1513/AnnalsATS.201306-204AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leitao Filho F.S., Alotaibi N.M., Ngan D. Sputum microbiome is associated with 1-year mortality after chronic obstructive pulmonary disease hospitalizations. Am J Respir Crit Care Med. 2019;199(10):1205–1213. doi: 10.1164/rccm.201806-1135OC. [DOI] [PubMed] [Google Scholar]
- 17.Celli B.R., Cote C.G., Marin J.M. The body-mass index, airflow obstruction, dyspnea, and exercise capacity index in chronic obstructive pulmonary disease. N Engl J Med. 2004;350(10):1005–1012. doi: 10.1056/NEJMoa021322. [DOI] [PubMed] [Google Scholar]
- 18.Meguro M., Barley E.A., Spencer S., Jones P.W. Development and validation of an improved, COPD-specific version of the St. George Respiratory Questionnaire. Chest. 2007;132(2):456–463. doi: 10.1378/chest.06-0702. [DOI] [PubMed] [Google Scholar]
- 19.Troosters T., Vilaro J., Rabinovich R. Physiological responses to the 6-min walk test in patients with chronic obstructive pulmonary disease. Eur Respir J. 2002;20(3):564–569. doi: 10.1183/09031936.02.02092001. [DOI] [PubMed] [Google Scholar]
- 20.Schroeder J.D., McKenzie A.S., Zach J.A. Relationships between airflow obstruction and quantitative CT measurements of emphysema, air trapping, and airways in subjects with and without chronic obstructive pulmonary disease. AJR Am J Roentgenol. 2013;201(3):W460–W470. doi: 10.2214/AJR.12.10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Celli B.R., MacNee W., Force A.E.T. Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur Respir J. 2004;23(6):932–946. doi: 10.1183/09031936.04.00014304. [DOI] [PubMed] [Google Scholar]
- 22.Laurell C.B., Eriksson S. Hypo-alpha-1-antitrypsinemia [in German] Verh Dtsch Ges Inn Med. 1964;70:537–539. [PubMed] [Google Scholar]
- 23.DeMeo D.L., Silverman E.K. Alpha1-antitrypsin deficiency. 2: genetic aspects of alpha(1)-antitrypsin deficiency: phenotypes and genetic modifiers of emphysema risk. Thorax. 2004;59(3):259–264. doi: 10.1136/thx.2003.006502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shrine N., Guyatt A.L., Erzurumluoglu A.M. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat Genet. 2019;51(3):481–493. doi: 10.1038/s41588-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agusti A. The path to personalised medicine in COPD. Thorax. 2014;69(9):857–864. doi: 10.1136/thoraxjnl-2014-205507. [DOI] [PubMed] [Google Scholar]
- 26.Kan M., Shumyatcher M., Himes B.E. Using omics approaches to understand pulmonary diseases. Respir Res. 2017;18(1):149. doi: 10.1186/s12931-017-0631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stockley R.A., Halpin D.M.G., Celli B.R., Singh D. Chronic obstructive pulmonary disease biomarkers and their interpretation. Am J Respir Crit Care Med. 2019;199(10):1195–1204. doi: 10.1164/rccm.201810-1860SO. [DOI] [PubMed] [Google Scholar]
- 28.Raffield L.M., Dang H., Pratte K.A. Comparison of proteomic assessment methods in multiple cohort studies. Proteomics. 2020;20(12) doi: 10.1002/pmic.201900278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Candia J., Cheung F., Kotliarov Y. Assessment of variability in the SOMAscan assay. Sci Rep. 2017;7(1):14248. doi: 10.1038/s41598-017-14755-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zarei S., Mirtar A., Morrow J.D., Castaldi P.J., Belloni P., Hersh C.P. Subtyping chronic obstructive pulmonary disease using peripheral blood proteomics. Chronic Obstr Pulm Dis. 2017;4(2):97–108. doi: 10.15326/jcopdf.4.2.2016.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oelsner E.C., Balte P.P., Grams M.E. Albuminuria, lung function decline, and risk of incident chronic obstructive pulmonary disease. The NHLBI Pooled Cohorts Study. Am J Respir Crit Care Med. 2019;199(3):321–332. doi: 10.1164/rccm.201803-0402OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ropcke S., Holz O., Lauer G. Repeatability of and relationship between potential COPD biomarkers in bronchoalveolar lavage, bronchial biopsies, serum, and induced sputum. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0046207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leiten E.O., Eagan T.M.L., Martinsen E.M.H. Complications and discomfort after research bronchoscopy in the MicroCOPD study. BMJ Open Respir Res. 2020;7(1) doi: 10.1136/bmjresp-2019-000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baraniuk J.N., Casado B., Pannell L.K. Protein networks in induced sputum from smokers and COPD patients. Int J Chron Obstruct Pulmon Dis. 2015;10(18):1957–1975. doi: 10.2147/COPD.S75978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franciosi L., Govorukhina N., Fusetti F. Proteomic analysis of human epithelial lining fluid by microfluidics-based nanoLC-MS/MS: a feasibility study. Electrophoresis. 2013;34(18):2683–2694. doi: 10.1002/elps.201300020. [DOI] [PubMed] [Google Scholar]
- 36.Lee E.J., In K.H., Kim J.H. Proteomic analysis in lung tissue of smokers and COPD patients. Chest. 2009;135(2):344–352. doi: 10.1378/chest.08-1583. [DOI] [PubMed] [Google Scholar]
- 37.Steiling K., Kadar A.Y., Bergerat A. Comparison of proteomic and transcriptomic profiles in the bronchial airway epithelium of current and never smokers. PLoS One. 2009;4(4):e5043. doi: 10.1371/journal.pone.0005043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Titz B., Sewer A., Schneider T. Alterations in the sputum proteome and transcriptome in smokers and early-stage COPD subjects. J Proteomics. 2015;128:306–320. doi: 10.1016/j.jprot.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Nicholas B.L., Skipp P., Barton S. Identification of lipocalin and apolipoprotein A1 as biomarkers of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;181(10):1049–1060. doi: 10.1164/rccm.200906-0857OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verrills N.M., Irwin J.A., He X.Y. Identification of novel diagnostic biomarkers for asthma and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183(12):1633–1643. doi: 10.1164/rccm.201010-1623OC. [DOI] [PubMed] [Google Scholar]
- 41.Pinto-Plata V., Toso J., Lee K. Profiling serum biomarkers in patients with COPD: associations with clinical parameters. Thorax. 2007;62(7):595–601. doi: 10.1136/thx.2006.064428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walter R.E., Wilk J.B., Larson M.G. Systemic inflammation and COPD: the Framingham Heart Study. Chest. 2008;133(1):19–25. doi: 10.1378/chest.07-0058. [DOI] [PubMed] [Google Scholar]
- 43.Dickens J.A., Miller B.E., Edwards L.D. COPD association and repeatability of blood biomarkers in the ECLIPSE cohort. Respir Res. 2011;12:146. doi: 10.1186/1465-9921-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohler M., Sandberg A., Kjellqvist S. Gender differences in the bronchoalveolar lavage cell proteome of patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2013;131(3):743–751. doi: 10.1016/j.jaci.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 45.Merali S., Barrero C.A., Bowler R.P. Analysis of the plasma proteome in COPD: novel low abundance proteins reflect the severity of lung remodeling. COPD. 2014;11(2):177–189. doi: 10.3109/15412555.2013.831063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zemans R.L., Jacobson S., Keene J. Multiple biomarkers predict disease severity, progression and mortality in COPD. Respir Res. 2017;18(1):117. doi: 10.1186/s12931-017-0597-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bradford E., Jacobson S., Varasteh J. The value of blood cytokines and chemokines in assessing COPD. Respir Res. 2017;18(1):180. doi: 10.1186/s12931-017-0662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ngo D., Peterson B., Keyes M., Gao Y. The proteomic profile associated with lung function and COPD in the Jackson Heart Study. Am J Respir Crit Care Med. 2020;201:A6136. [Google Scholar]
- 49.Carolan B.J., Hughes G., Morrow J. The association of plasma biomarkers with computed tomography-assessed emphysema phenotypes. Respir Res. 2014;15:127. doi: 10.1186/s12931-014-0127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coxson H.O., Dirksen A., Edwards L.D. The presence and progression of emphysema in COPD as determined by CT scanning and biomarker expression: a prospective analysis from the ECLIPSE study. Lancet Respir Med. 2013;1(2):129–136. doi: 10.1016/S2213-2600(13)70006-7. [DOI] [PubMed] [Google Scholar]
- 51.Bozinovski S., Hutchinson A., Thompson M. Serum amyloid a is a biomarker of acute exacerbations of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177(3):269–278. doi: 10.1164/rccm.200705-678OC. [DOI] [PubMed] [Google Scholar]
- 52.Keene J.D., Jacobson S., Kechris K. Biomarkers predictive of exacerbations in the SPIROMICS and COPDGene cohorts. Am J Respir Crit Care Med. 2017;195(4):473–481. doi: 10.1164/rccm.201607-1330OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vestbo J., Anderson W., Coxson H.O. Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) Eur Respir J. 2008;31(4):869–873. doi: 10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- 54.Regan E.A., Hokanson J.E., Murphy J.R. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7(1):32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones P.W., Rames A.D. Tesra (treatment Of Emphysema With A Selective Retinoid Agonist) Study Results. Am J Respir Crit Care Med. 2011;183:2011. A641. [Google Scholar]
- 56.Rennard S.I. The Promise of Observational Studies (ECLIPSE, SPIROMICS, and COPDGene) in achieving the goal of personalized treatment of chronic obstructive pulmonary disease. Semin Respir Crit Care Med. 2015;36(4):478–490. doi: 10.1055/s-0035-1555609. [DOI] [PubMed] [Google Scholar]
- 57.Beiko T., Janech M.G., Alekseyenko A.V. Serum proteins associated with emphysema progression in severe alpha-1 antitrypsin deficiency. Chronic Obstr Pulm Dis. 2017;4(3):204–216. doi: 10.15326/jcopdf.4.3.2016.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strange C., Senior R.M., Sciurba F. Rationale and design of the genomic research in alpha-1 antitrypsin deficiency and sarcoidosis study. Alpha-1 protocol. Ann Am Thorac Soc. 2015;12(10):1551–1560. doi: 10.1513/AnnalsATS.201503-143OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tuck M.K., Chan D.W., Chia D. Standard operating procedures for serum and plasma collection: early detection research network consensus statement standard operating procedure integration working group. J Proteome Res. 2009;8(1):113–117. doi: 10.1021/pr800545q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geyer P.E., Voytik E., Treit P.V. Plasma proteome profiling to detect and avoid sample-related biases in biomarker studies. EMBO Mol Med. 2019;11(11) doi: 10.15252/emmm.201910427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rikkert LG, Coumans FAW, Hau CM, Terstappen L, Nieuwland R. Platelet removal by single-step centrifugation [published online ahead of print June 17, 2020]. Platelets. https://doi.org/10.1080/09537104.2020.1779924. [DOI] [PubMed]
- 62.Depner C.M., Melanson E.L., McHill A.W., Wright K.P., Jr. Mistimed food intake and sleep alters 24-hour time-of-day patterns of the human plasma proteome. Proc Natl Acad Sci U S A. 2018;115(23):E5390–E5399. doi: 10.1073/pnas.1714813115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Torres J.P., Casanova C., Pinto-Plata V. Gender differences in plasma biomarker levels in a cohort of COPD patients: a pilot study. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0016021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jammes Y., Steinberg J.G., Ba A., Delliaux S., Bregeon F. Enhanced exercise-induced plasma cytokine response and oxidative stress in COPD patients depend on blood oxygenation. Clin Physiol Funct Imaging. 2008;28(3):182–188. doi: 10.1111/j.1475-097X.2008.00795.x. [DOI] [PubMed] [Google Scholar]
- 65.Agusti A., Hedner J., Marin J.M., Barbe F., Cazzola M., Rennard S. Night-time symptoms: a forgotten dimension of COPD. Eur Respir Rev. 2011;20(121):183–194. doi: 10.1183/09059180.00004311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller B.E., Tal-Singer R., Rennard S.I. Plasma fibrinogen qualification as a drug development tool in chronic obstructive pulmonary disease. Perspective of the Chronic Obstructive Pulmonary Disease Biomarker Qualification Consortium. Am J Respir Crit Care Med. 2016;193(6):607–613. doi: 10.1164/rccm.201509-1722PP. [DOI] [PubMed] [Google Scholar]
- 67.Jones P.W. St. George's Respiratory Questionnaire: MCID. COPD. 2005;2(1):75–79. doi: 10.1081/copd-200050513. [DOI] [PubMed] [Google Scholar]
- 68.Campbell E.J., Campbell M.A., Boukedes S.S., Owen C.A. Quantum proteolysis by neutrophils: implications for pulmonary emphysema in alpha 1-antitrypsin deficiency. J Clin Invest. 1999;104(3):337–344. doi: 10.1172/JCI6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma S., Lin Y.Y., Turino G.M. Measurements of desmosine and isodesmosine by mass spectrometry in COPD. Chest. 2007;131(5):1363–1371. doi: 10.1378/chest.06-2251. [DOI] [PubMed] [Google Scholar]
- 70.Sukkar M.B., Ullah M.A., Gan W.J. RAGE: a new frontier in chronic airways disease. Br J Pharmacol. 2012;167(6):1161–1176. doi: 10.1111/j.1476-5381.2012.01984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kearon C., de Wit K., Parpia S. Diagnosis of pulmonary embolism with d-Dimer adjusted to clinical probability. N Engl J Med. 2019;381(22):2125–2134. doi: 10.1056/NEJMoa1909159. [DOI] [PubMed] [Google Scholar]
- 72.Jankowich M.D., Wu W.C., Choudhary G. Association of elevated plasma endothelin-1 levels with pulmonary hypertension, mortality, and heart failure in african american individuals: the Jackson Heart Study. JAMA Cardiol. 2016;1(4):461–469. doi: 10.1001/jamacardio.2016.0962. [DOI] [PubMed] [Google Scholar]
- 73.Sun W., Kechris K., Jacobson S. Common genetic polymorphisms influence blood biomarker measurements in COPD. PLoS Genet. 2016;12(8) doi: 10.1371/journal.pgen.1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun B.B., Maranville J.C., Peters J.E. Genomic atlas of the human plasma proteome. Nature. 2018;558(7708):73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mastej E., Gillenwater L., Zhuang Y., Pratte K.A., Bowler R.P., Kechris K. Identifying protein-metabolite networks associated with COPD phenotypes. Metabolites. 2020;10(4):124. doi: 10.3390/metabo10040124. [DOI] [PMC free article] [PubMed] [Google Scholar]