ABSTRACT
Douglas Channel and the adjacent Hecate Strait (British Columbia, Canada) are part of a proposed route to ship diluted bitumen (dilbit). This study presents how two types of dilbit naturally degrade in this environment by using an in situ microcosm design based on dilbit-coated beads. We show that dilbit-associated n-alkanes were microbially biodegraded with estimated half-lives of 57–69 days. n-Alkanes appeared to be primarily degraded using the aerobic alkB, ladA and CYP153 pathways. The loss of dilbit polycyclic aromatic hydrocarbons (PAHs) was slower than of n-alkanes, with half-lives of 89–439 days. A biodegradation of PAHs could not be conclusively determined, although a significant enrichment of the phnAc gene (a marker for aerobic PAH biodegradation) was observed. PAH degradation appeared to be slower in Hecate Strait than in Douglas Channel. Microcosm-associated microbial communities were shaped by the presence of dilbit, deployment location and incubation time but not by dilbit type. Metagenome-assembled genomes of putative dilbit-degraders were obtained and could be divided into populations of early, late and continuous degraders. The majority of the identified MAGs could be assigned to the orders Flavobacteriales, Methylococcales, Pseudomonadales and Rhodobacterales. A high proportion of the MAGs represent currently unknown lineages or lineages with currently no cultured representative.
Keywords: dilbit, oil biodegradation, Douglas Channel, Hecate Strait, in situ microcosms
This study investigated the biodegradation of diluted bitumen and the associated microbial communities by using a novel in situ microcosm design deployed at the coast of British Columbia (Canada).
INTRODUCTION
Diluted bitumen (dilbit) is a crude oil product that consists of raw bitumen mixed with a diluent, which is added to reduce overall viscosity and thereby to enable dilbit transport through pipelines. Canadian bitumen is extracted from oil sands located in the western provinces of Alberta and Saskatchewan, Canada. The resulting dilbit is transported via pipelines to refineries in the USA or to export terminals on the Pacific coast of British Columbia (Canada). The town of Kitimat at the head of the Douglas Channel (Fig. 1A) was proposed as a new export terminal for shipping Canadian dilbit to global markets. From Kitimat dilbit tankers would follow the Douglas Channel fjord system into the adjacent Hecate Strait before entering the Pacific Ocean. This shipping route would create the risk of accidental dilbit spills into these two water bodies.
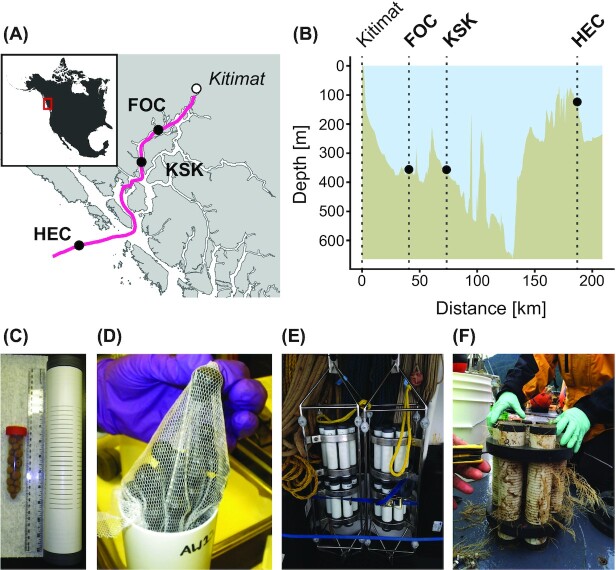
Figure 1.
Overview of experimental setting and setup. (A) Deployment locations of moorings containing the in situ microcosms (black dots). The town Kitimat is indicated by a white dot as a point of reference. The track shown in pink represents the main shipping route through Douglas Channel. (B) Bathymetric profile of the track indicated in pink in panel A. Deployment depths of the in situ microcosms are indicated with black dots. (C) Clay beads and corresponding PVC column for bead containment. The ruler shown for scale has a length of 30 cm. (D) Beads coated in AWB and contained in a Nylon mesh sack prior to loading the microcosm columns. (E) Bead-filled columns installed in the mooring setup. (F) Bead-filled columns after retrieval and after ca. 12 months of incubation. The shown microcosm columns were colonized by soft corals presumably belonging the genus Callogorgia.
In situ bioremediation by microbial hydrocarbon degraders contributes to natural oil weathering and represents a means to mitigate the effects of marine oil spills (e.g. Swannell, Lee and McDonagh 1996). Previous studies focusing on marine dilbit weathering have used flume tank mesocosms (King et al. 2014) or laboratory microcosms (Schreiber et al. 2019; Tremblay et al. 2019). These studies provided valuable first insights into dilbit weathering kinetics and the associated microbial communities. However, complementary in situ (field) experiments have so far not been carried out in the ocean.
The goal of this study was to obtain such complementary field data on in situ dilbit weathering and on the associated microbial communities. The main limitation of previous (ex situ) studies investigating marine dilbit degradation (King et al. 2014; Schreiber et al. 2019) are the relatively short time scale the experimental systems could be operated (in the order of days up to a month) before boundary effects create conditions that cause the systems to significantly deviate from natural, in situ conditions. Such boundary effects include: (i) the physical boundaries of the systems that represent an artificial surface for microbial colonization and hence create an disproportionately large seed bank of surface-associated bacteria, and (ii) a limited exchange of seawater in the systems that leads to a limited founder population, reduced recruitment to support natural species succession and finally an exhaustion of nutrients that would not be experienced under in situ conditions. In bottle microcosms nutrient limitation can be somewhat mitigated by artificial nutrient addition. This however comes at the cost of selecting against microorganisms adapted to in situ nutrient concentrations, which would potentially dominate microbial communities in the environment. The microcosm setup in this study was designed to specifically address the biases introduced by unrealistically nutrient concentrations and limited founder and recruitment populations. To this end, dilbit-coated clay beads were placed in columns with horizontal slits and this setup was then deployed at marine mooring stations. The slits in the containment columns of our setup allowed a continuous exchange of nutrients and microorganisms between the surrounding seawater and the dilbit-coated beads, and thereby very realistically simulated conditions that dilbit particles would experience in a marine pelagic environment.
The clay beads were coated with two common types of Canadian dilbit: Access Western Blend (AWB) and Cold Lake Blend (CLB). The bead-filled columns were deployed along the proposed dilbit shipping route at two stations in Douglas Channel (stations FOC and KSK) and one station in Hecate Strait (station HEC; Fig. 1A and B; Table 1). Fresh dilbit will float on seawater. However, a significant increase in density due to rapid weathering, the formation of oil–particle aggregates or strong mixing can all create conditions where dilbit can sink below the sea surface (Johannessen et al. 2020). In order to simulate such a scenario of subsurface dilbit, the bead-filled columns were positioned below the water surface layer close to the water bottom. Beads were retrieved after ca. 3 and 12 months of incubation. Dilbit weathering was determined by mass spectrometry for the analysis of aliphatic and aromatic hydrocarbons. Corresponding bead-associated microbial communities were characterized by amplicon and shotgun metagenomic sequencing, and compared to control communities of uncoated beads.
Table 1.
Details of mooring deployment stations. Abbreviations: n.d., no data.
| Mooring station | Latitude [°] | Longitude [°] | Depth [m] | Incubation times [days] | Temperature range [°C] | Salinity range* | NO3–1 / NO2–1 range [μmol/L] | PO4–3 range [μmol/L] | Dissolved oxygen [mL/L] | References |
|---|---|---|---|---|---|---|---|---|---|---|
| FOC | 53.736 | −129.030 | 358 m | 0, 97, 389 | 7–8 | 32.4–33.2 | 27–31 | 2.25 | 2.4–3.6 | Wright et al. (2016) |
| KSK | 53.480 | −129.209 | 361 m | 0, 95, 387 | 7–8 | 32.5–33.3 | n.d. | n.d. | 2.4–3.8 | Wright et al. (2016) |
| HEC | 52.821 | −129.846 | 125 m | 0, 80, 344 | 2–14 | 32–34 | 12–25 | 1.25–1.75 | 1.5–6 | Wright et al. (2015, 2016, 2017) |
Salinity is reported on the Practical Salinity Scale, PSS-78.
MATERIALS AND METHODS
General setup of experiment
This study used in situ microcosms to investigate how location (two sites in Douglas Channel and one site at Hecate Strait), dilbit type (CLB or AWB) and incubation time (3 or 12 months) affect microbial dilbit degradation and the associated microbial communities.
In situ microcosms consisted of PVC columns (6 cm OD, 32 cm long; PlastechPlus Inc., Laval, QC, Canada) filled with dilbit-coated or uncoated clay beads (porous, 8–16 mm diameter, 0.74±0.13 g/bead; Liapor GmbH, Hallerndorf-Pautzfeld, Germany). The columns featured threaded caps at each end and 1 mm wide slits over ca. 15 cm of the mid area (Fig. 1C). For this study, each bead-filled column represented an individual sample. The columns were attached to a mooring frame using body bands and collars (Fig. 1E). The mooring frames and body bands consisted of 316 stainless steel and were built by Strait Metal Ltd (Sidney, BC, Canada). The collars consisted of ultra high molecular weight polyethylene (UHMW) and were built by Eppic Waterjet Inc (Saanichton, BC, Canada).
The columns were incorporated into the mooring configuration of the World Class Tanker Safety Program (2014–2016) described previously (Wright et al. 2016). The moorings containing the microcosm columns were deployed in duplicate (two moorings at each site) from the CCGS John P. Tully in July 2014 at stations FOC (53.736°N and 129.030°W) and KSK (53.480°N and 129.209°W), both located in Douglas Channel, and in July 2015 at station HEC (52.821°N and 129.846°W), located in Hecate Strait near the entrance of Douglas Channel (Fig. 1A). The stations FOC, KSK and HEC of this study are identical to the stations FOC1, KSK1 and HEC1, respectively, described in more detail previously (Wright et al. 2015). The microcosm columns were deployed ca. 8.8 m above the sea floor (mid-point of the microcosm mooring frame), i.e. at 358 m depth for station FOC, 361 m for station KSK and 125 m for station HEC (Fig. 1B). An overview of station-related metadata is provided in Table 1.
During the deployment, seawater samples were collected in triplicate at the deployment depths using Sea-Bird carousel water samplers (Sea-Bird Scientific, Bellevue, WA) equipped with Niskin bottles. The seawater samples (2 L) were filtered at 10 psi, in triplicate onto Millipore 0.22 μm polyethersulfone membranes (Fisher Scientific, Ottawa, ON, Canada), and represented the microbial communities at T0. The filters were stored at −80°C for subsequent genomic analyses.
In October 2014, one set of in situ microcosms deployed at each of the stations FOC and KSK was recovered after ca. 3 months (97 days, FOC; 95 days, KSK) of incubation. The remaining replicate moorings at these stations were retrieved in July 2015 after ca. 12 months (389 days, FOC; 387 days, KSK) of incubation. Similarly, the first replicate mooring at station HEC was retrieved in October 2015 after 80 days (ca. 3 months) and the second in July 2016 after 344 days (ca. 12 months) of incubation.
Onboard the CCGS John P. Tully, bead samples were quickly transferred into sterile borosilicate bottles. Samples were stored at −80°C until processed further. Back in the laboratory, samples were thawed on ice and separated for genomics and chemistry analyses. For chemistry analyses, batches of ca. 60 g of beads (representing an estimated 50 beads) were transferred into 250 mL wide-mouth amber glass bottles with Teflon lined lids and frozen at −20°C. The remaining beads were transferred in 10 g batches into 50 mL tubes and stored at −80°C for subsequent genomic analyses.
Coating of clay beads and loading of columns
Clay beads were thoroughly rinsed twice with tap water for 1 min, and once with distilled water in a stainless steel sieve (mesh size 4.75 mm). The beads were transferred to a tray lined with paper towels and allowed to air dry. Once dried, the beads were re-washed with distilled water to remove traces of fine clay powder. The beads were air-dried as before and subsequently autoclaved in a glass bottle for 20 min.
Dilbit blends were provided by the Canadian Department of Fisheries and Oceans through the World Class Tanker Safety System (WCTSS) program. The beads (ca. 65 g, representing an estimated 90 beads and ca. 163 mL of volume) were soaked in either Cold Lake Blend (CLB) or Access Western Blend (AWB) dilbit for 3 h (mixing every 30 min). The coated beads were transferred onto trays lined with paper towels and located under a fume hood, and weathered at room temperature for 4 days. During this step, the beads were rolled gently on paper towels to accelerate surface drying and removal of excess dilbit.
Dilbit-coated beads and uncoated control beads were placed into Nylon mesh sacks (mesh size 2 mm) prior to loading into the PVC columns (Fig. 1D). The columns were prepared by first adding sterile uncoated spacer beads at the bottom, followed by the coated or control beads, and finally another layer of sterile uncoated spacer beads. This way, the mesh cloth sack was located in the middle area of the column where the slits for seawater exchange were located. The sacks further facilitated the removal of the beads following the in situ incubation.
Hydrocarbon quantification
Beads for hydrocarbon analyses were allowed to thaw prior to the addition of 100 mL of ‘distilled in glass’-grade dichloromethane (DCM) from Caledon (Georgetown, ON, Canada). The bottles were placed on a Wheaton R2P roller apparatus (Millville, NJ) set at 8 rpm for 1 h, after which the solvent was transferred to a 200 mL evaporation tube and concentrated on a TurboVap II (Biotage, Charlotte, NC) to a final volume of 10 mL. This process was repeated with 5 more rinses of 100 mL DCM. An aliquot of the concentrated solvent was then prepared for GC-MS analysis as described by Ortmann et al. (2020). Briefly, this included sample cleanup using a silica gel, the addition of internal standards and a solvent transfer to iso-octane.
A small subsample of the dilbit-coated beads was tested for the presence and quantity of dilbit hydrocarbons contained inside the beads as follows: after the beads had been processed to remove the outer dilbit coating as described above, they were placed in a Teflon bag. The bag was sealed and placed on a slate countertop located under a fume hood. A 3 lb sledgehammer was used to crush the beads into a fine power with individual pieces no larger than 1 mm in diameter. As the resulting powder was incompatible with the roller-based extraction method used for the oil fraction on the outside of the beads, the oil remaining in the crushed beads was extracted using a SOXHLET apparatus (Fisher Scientific). In detail, the crushed beads were transferred into a solvent rinsed cellulose thimble, which was amended with 5 g sodium sulfate and 2 g of copper, and spiked with 1 mL of surrogate recovery standard. The thimble was placed in the SOXHLET extractor, which was attached to a condenser and a 500 mL round-bottom flask containing 2 g of copper and 300 mL of DCM. The solvent was heated to a gentle boil (with the assistance of Teflon boiling chips) and extracted for 18 h. Following this extraction, the solvent was transferred to evaporation tubes, concentrated using the Turbo-Vap II and prepared for GC-MS analysis as described below. Previous method development showed that the roller-based and the SOXHLET-based extraction methods are both equally effective at extracting dilbit-associated hydrocarbons (data not shown).
Samples were prepared for GC-MS by liquid-liquid extraction using a modified version of U.S. Environmental Protection Agency method 3510C (Schreiber et al. 2019; Ortmann et al. 2020). Purified and concentrated extracts were analyzed using high-resolution gas chromatography (GC; 6890 GC; Agilent, Wilmington, DE) coupled to an Agilent 5975B mass selective detector operated in the selective ion-monitoring mode using the following GC (SLB-5ms column, 30 mby 0.25 mm inner diameter, 0.25-m film thickness; Supelco, Mississauga, ON, Canada) conditions: cool on-column injection with oven track mode (tracks 3°C higher than the oven temperature program), 85°C hold for 2 min, ramp at 4°C/min to 280°C and hold for 20 min. Quantification criteria for PAHs were as described previously (Tremblay et al. 2017).
Final hydrocarbon quantities were normalized to the weight of extracted beads. In order to account for the weight gain of beads during the incubation period, the calculated mass fractions were divided by a correction factor representing the ratio of bead weight prior to deployment and after recovery, i.e. mbeads(T = 0) / mbeads(T = x). Based on the hydrocarbon load per gram of beads, the coefficient of variation between replicate samples was 1–25%, which was considered acceptable.
Half-lives of n-alkanes and naphthalenes were conservatively estimated by linear regression based on loads at the start of the experiment and after ca. 3 months of incubation. It was not possible to estimate the half-lives of the PAH fractions by using a consistent approach across stations and dilbit types. Instead, half-lives of the PAH fractions were estimated with individual approaches that best accounted for the underlying PAH data (for details see Table S3, Supporting Information).
Characterization of microbial communities
Bead-associated microbial communities were characterized after ca. 3 and 12 months of incubation. Microbial communities present in corresponding seawater samples (i.e. from the same sampling site and a similar depth as the incubated beads) at the start of the microcosm deployment were considered the starting (T0) communities. In total our experimental design thus resulted in 81 samples intended for microbial community analysis (a detailed sample breakdown is presented in Figure S1, Supporting Information). Total nucleic acids of microbial communities were extracted using a hexadecyl-trimethyl-ammonium-bromide (CTAB) protocol as described previously (Tremblay et al. 2017). Extracted DNA samples were treated with RNase If (New England Biolabs, Whitby, ON, Canada) and purified using magnetic beads. DNA quantification was performed using the Quant-iT PicoGreen assay (Fisher Scientific Ltd, Edmonton, AB, Canada).
Basic characterization of microbial communities was carried out by 16S rRNA gene sequencing after PCR amplification using the primer set 515F (5 = -GTGCCAGCMGCCGCGGTAA-3 = ) and 806R (5 = -GGACTAC HVGGGTWTCTAAT-3 = ); the primer set represents the original primer set of the ‘Earth Microbiome’ project and targets both Archaea and Bacteria (Caporaso et al. 2011). PCR amplification was performed using the HotStarTaq Master Start polymerase kit (Qiagen, Toronto, ON, Canada). Reactions were performed in a 25-μL volume containing 510 ng of template DNA, 0.6 μM of each primer and 0.5 mg/mL of bovine serum albumin. Amplification was performed using an initial denaturation for 5 min at 95°C followed by 25 cycles of 30 s at 95°C, 30 s at 55°C, 45 s at 72°C and a final elongation for 7 min at 72°C. Success of the PCR amplification was evaluated by gel electrophoresis. PCR amplicons were purified using magnetic beads. PCR products were indexed using the KAPA HiFi HotStart Ready Mix (Roche, Laval, QC, Canada). The indexing PCR reaction was performed using an initial denaturation for 3 min at 95°C followed by 8 cycles of 30 s at 95°C, 30 s at 55°C, 30 s at 72°C and a final elongation for 5 min at 72°C. PCR amplicons were purified using magnetic beads and subsequently quantified using the QuantiT PicoGreen assay (Fisher Scientific Ltd). Equal amounts of indexed PCR products were pooled and sequenced using the Illumina MiSeq platform and the 500-cycle MiSeq reagent kit v2 (Illumina; San Diego, CA).
Sequencing data were processed using the AmpliconTagger pipeline (Tremblay and Yergeau 2019). OTUs classified to originate from chloroplasts or mitochondria and subsequently samples featuring fewer than 5000 remaining reads were removed from the raw OTU table. This filtering step removed six samples from the analysis: five samples of uncoated control beads and one T0 seawater sample from station FOC. Removal of rare OTUs as recommended previously (Bokulich et al. 2012) was performed prior to detailed analysis of community compositions: only OTUs with at least 0.05% relative abundance in at least three samples were retained. Microbial community compositions were compared based on principles of compositional data analysis (CoDA; Aitchison 1982). Dissimilarities between communities were expressed using the proportionality metric φ (Lovell et al. 2015) as calculated using the R package ‘propr’ (Quinn et al. 2017). Differences between communities were explored by carrying out a nonmetric multidimensional scaling (nMDS) ordination (as implemented in the R package ‘vegan’; Oksanen et al. 2010) based on the φ dissimilarity matrix. Marginal effects of factors on community dissimilarities were tested using permutational multivariate analysis of variance (PERMANOVA) as implemented in the ‘adonis2’ function of the R package ‘vegan’ (Oksanen et al. 2010).
Differential abundance analyses of OTUs
Differential abundance analysis of OTUs was performed using the ‘analysis of composition of microbiomes’ (ANCOM; Mandal et al. 2015) framework. The ANCOM analysis was executed using code version 2.0 (https://github.com/sidhujyatha/ANCOM) within the R environment. ANCOM analyses were performed using a significance level of 0.05 and a conservative 0.9 threshold for the W statistic. Differential abundances were determined between dilbit-coated and uncoated beads, and were calculated based on subsets only representing data from a single station and incubation time (e.g. separate subsets for FOC-3 months, FOC-12 months and HEC- 3 months, etc.). OTUs differentially more abundant on dilbit-coated beads were taxonomically classified using the SINA classifier (Pruesse, Peplies and Glöckner 2012) based on the SILVA Ref database release 138 (Quast et al. 2013) and using default parameters except for search-kmer-len = 8 and lca-quorum = 0.5.
Metagenomic analysis
A total of 81 metagenomic sequencing libraries (one library for each sample; see Figure S1, Supporting Information) were prepared with 10 ng of DNA of each sample using the Illumina Nextera XT library preparation protocol (Illumina). A pool of all libraries was sequenced using an Illumina HiSeq4000 in rapid mode 2 × 100 bp configuration at the Centre d'expertise et de services (Genome Quebec, Canada), which resulted in 129 Giga-bases (ca. 1.6 Gbp per sample) of sequencing data.
Sequencing adapters were removed from each read, and bases at the end of reads having a quality score less than 30 were trimmed (Trimmomatic v0.32; Bolger, Lohse and Usadel 2014) and scanned for sequencing adapter contamination using DUK (http://duk.sourceforge.net/). The resulting quality-controlled reads were co-assembled using Megahit v.1.1.2 (Li et al. 2015) with iterative k-mer sizes of 21, 31, 41, 51, 61, 71, 81 and 91 bases on a 3-Terabyte RAM compute node (Compute Canada; see Supplementary Information—Dataset S1 for assembly statistics). Gene coding sequences were predicted using Prodigal v2.6.2 (Hyatt et al. 2010) in ‘metagenome’ mode. QC-passed reads were mapped (BWA v0.7.15; http://bio-bwa.sourceforge.net) against assembled contigs to determine how much of the total metagenomic data was incorporated into the assembly and to obtain contig abundance profiles. On average 63% of the reads could be mapped onto the assembled contigs (see Supplementary Information—Dataset S2). Alignment files in BAM format were sorted by read coordinates using SAMtools v1.2 (Li et al. 2009) and only properly aligned read pairs were kept for downstream steps. Each BAM file (containing properly aligned paired-reads only) was analyzed for sequencing coverage of called genes and contigs, respectively, using BEDtools (v2.17.0; Quinlan and Hall 2010) and a custom BED file representing gene coordinates on each contig. Read counts assigned to the contig region of a gene location were considered read counts of the gene. To reduce the number of unambiguously mapped reads, only paired reads both overlapping their contig or gene were considered for read counts of genes. Coverage profiles of each sample were merged to generate a gene-abundance matrix (rows = genes, columns = samples). Read count summaries and mapping statistics are provided (Supplementary Information—Dataset S2).
The predicted genes were screened for key genes associated with microbial hydrocarbon degradation. Targeted genes for aerobic alkane degradation were the alkane 1-monooxygenase gene (alkB; Smits et al. 1999), the cytochrome P450 alkane hydroxylase gene (CYP153; van Beilen et al. 2006) and the long-chain alkane monooxygenase gene (ladA; Boonmak, Takahashi and Morikawa 2014), which are considered key genes for different alkane degradation pathways. The 1-methylalkyl succinate synthase/alkylsuccinate synthase gene (masD/assA; Gittel et al. 2015) was targeted as a key gene for anaerobic alkane degradation. Targeted key genes for PAH degradation were the gene coding for the iron sulfur protein large (α) subunit of the PAH initial dioxygenase (phnAc; Habe and Omori 2003) for aerobic PAH degradation, and the 2‐naphthoyl‐CoA reductase gene (ncr; Mouttaki, Johannes and Meckenstock 2012) for anaerobic PAH degradation. Putative genes associated with hydrocarbon degradation were identified based on BLAST score ratios (BSR) (Rasko, Myers and Ravel 2005) to bona fide reference genes listed previously: alkB (Nie et al. 2014), CYP153 (van Beilen et al. 2006; Nie et al. 2014), ladA (Boonmak, Takahashi and Morikawa 2014), masD/assA (Tan et al. 2014; Gittel et al. 2015), phnAc (Lozada et al. 2008; Ding et al. 2010), ncr (Morris et al. 2014). A list of all used reference genes is provided in the supplementary Dataset S3. BSR cut-off values (i.e. minimal values) were determined empirically by comparing BSR values (i) between bona fide reference genes, and (ii) between bona fide reference genes and closely related but functionally different genes. The determined BSR cut-off values were: alkB, 0.2; CYP153, 0.3; ladA, 0.2; masD/assA, 0.35; phnAc, 0.4; ncr, 0.35.
Detected hydrocarbon genes that were present in the generated MAGs (see below) were assigned the same taxonomic classification as the MAG. The MAG-based classification was extended to all genes not contained in a MAG but with a sequence identity of ≥0.86 (i.e. genus-level cut-off) with a MAG-associated gene. All remaining hydrocarbon genes were taxonomically classified based on a comparison to microbial genomes contained in the GenomeDB database release 86 (Dong and Strous 2019) and using the BLASTP mode of DIAMOND (Buchfink, Xie and Huson 2015). A consensus classification for each gene was calculated as follows (R code available at https://github.com/lschreib/dilbit_beads2014): Of all DIAMOND BLASTP hits only hits with a bitscore within 95% of the bitscore of the best hit were retained (i.e. top-% filter). Consensus classification for a given taxonomic rank was only assigned if the classifications of all retained hits agreed (i.e. strict consensus classification) otherwise the corresponding rank was classified as ‘Unknown’.
Differential abundance analysis of hydrocarbon-degradation genes between dilbit-coated and uncoated genes was carried out using the ALDEx2 pipeline (Fernandes et al. 2014) in R. In short, the per-gene technical variation for each sample was inferred by generating 128 Monte-Carlo instances drawn from the sample's Dirichlet distribution. The generated Monte-Carlo instances were transformed using the centered log-ratio (clr) transformation. The resulting clr values were compared to identify general community patterns. Genes more relatively abundant on dilbit-coated beads compared to uncoated beads (from hereon referred to as dilbit-associated genes) were determined using a one-tailed Student's t-test. All calculated P-values were adjusted using the Benjamini–Hochberg procedure. Differential gene abundances with an adjusted P-value of ≤0.05 were considered statistically significant. Intersections of sets of differentially abundant genes between stations were determined using R.
Metagenome assembled genomes (MAGs) were generated from the assembled data using Metabat2 (version 2.12.1; Kang et al. 2019). The MAGs were classified using GTDB-Tk (version 1.1.0; Chaumeil et al. 2019) based on release 89 of the Genome Taxonomy Database (GTDB; Parks et al. 2020) and as implemented in KBase (U.S. Department of Energy, USA; Arkin et al. 2018). Relative abundances of MAGs expressed in RPKM were calculated by normalizing the total read count of MAG-associated contigs by genome size (in kilobasepair) and by library size (in millions). MAGs most likely involved in dilbit-degradation were identified based on (i) an increased relative abundance in metagenomes of dilbit-coated beads, and (ii) by screening for the presence of dilbit-associated hydrocarbon degradation genes as described above.
Data visualization
Geographic maps were created in R with the packages ‘raster’ (Hijmans 2019), ‘maps’ (Becker et al. 2018) and ‘ggplot2’ (Wickham 2016), and based on data from Natural Earth (https://www.naturalearthdata.com) and GADM (https://gadm.org). All other plots for this manuscript were created in R using the ‘ggplot2’ package (Wickham 2016), and subsequently annotated and arranged using Adobe Illustrator (Adobe).
Data availability
All raw sequence reads generated for this study have been submitted to NCBI's SRA and are available under the BioProject accession number PRJNA629517. The generated 16S rRNA amplicon reads are available under SAMN15949[457–537] and the shotgun metagenomics reads are available under SAMN15950[781–873]. The metagenome-assembled genomes of putative dilbit-degraders are available under SAMN19594[111-146]. The metagenomic assembly is available on the zenodo.org file repository (https://doi.org/10.5281/zenodo.4009541). All bioinformatics scripts used, as well as OTU and gene count tables, are available upon request.
RESULTS
Experimental setup
Bottom water salinities at the two Douglas Channel stations (i.e. FOC and KSK) and the Hecate Strait station (HEC) were similar and >32 (Practical Salinity Scale, PSS-78) throughout the incubations (Wright et al. 2016). The water temperature at stations FOC and KSK was 6–8°C during the incubation period (Wright et al. 2016). Based on records from 2013 to 2016, the water temperature at station HEC was 2–14°C during the incubation period (Wright et al. 2015, 2016, 2017).
Upon retrieval of the moorings, after 3 or 12 months, it was noted that macrobiota, including soft corals, had colonized the microcosm columns (Fig. 1F). It was further noted that the mass of dilbit-coated beads had increased by 68–94% during the incubation (data not shown), with no apparent trend with incubation time. The nature of the weight gain was not investigated further. It might have represented a combination of biofouling and seawater uptake.
Fate of bead-associated dilbit
The coating procedure of the clay beads created a stable coating of heavily weathered (>25%) dilbit, which was considerably depleted of alkanes and 2- to 6-ring polycyclic aromatic hydrocarbons (PAHs; Figures S2 and S3, Supporting Information).
After 3 months of incubation, the quantified hydrocarbons had on average decreased to 38–75% of the original amount (Figure S4, Supporting Information). After the full 12 months of incubation, only 28–37% of the quantified hydrocarbons were still present on the clay beads (Figure S4, Supporting Information). For stations FOC and KSK, the greatest hydrocarbon loss occurred within the first 3 months of incubation. In contrast, at station HEC the greatest loss occurred between 3 and 12 months of incubation. Hydrocarbon losses were not significantly different between the two tested dilbit types (Figure S4, Supporting Information).
A closer inspection of losses of the individual hydrocarbon compounds revealed different loss rates between n-alkanes, naphthalenes and PAHs with three and more rings (Fig. 2). The half-lives were 57–69 days for the alkane fraction and 52–87 days for the naphthalene fraction (Fig. 3A; Tables S1 and S2, Supporting Information). Losses of PAH fractions were on average slower than those of n-alkanes and naphthalenes (Fig. 3C and Table S3, Supporting Information). Additionally, the PAH fraction at station HEC showed no significant decrease after ca. 3 months of incubation (Fig. 3C). The estimated half-lives of the PAH fractions were 89–439 days (Table S3, Supporting Information).
Figure 2.
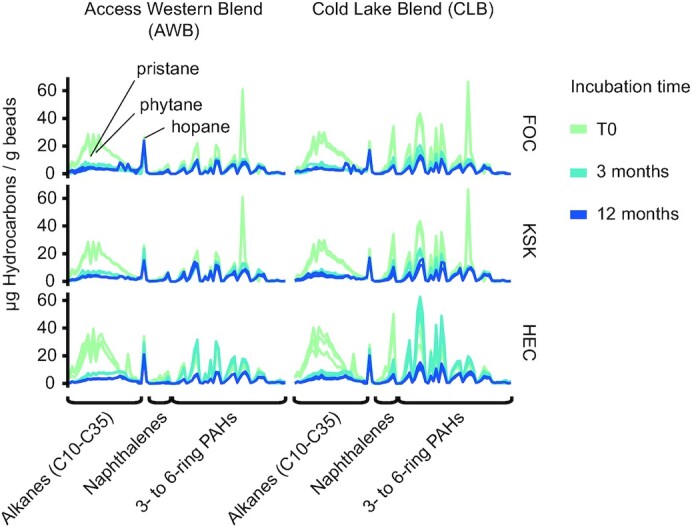
Changes of dilbit composition over time. Quantity of resolved hydrocarbon compounds normalized to the weight of extracted beads. Each line represents a single sample. The T0 samples of stations FOC and KSK are identical, and the corresponding data are hence duplicated between the panels of both stations. Incubation times of beads are color-coded. Abbreviations: PAHs—polycyclic aromatic hydrocarbons.
Figure 3.
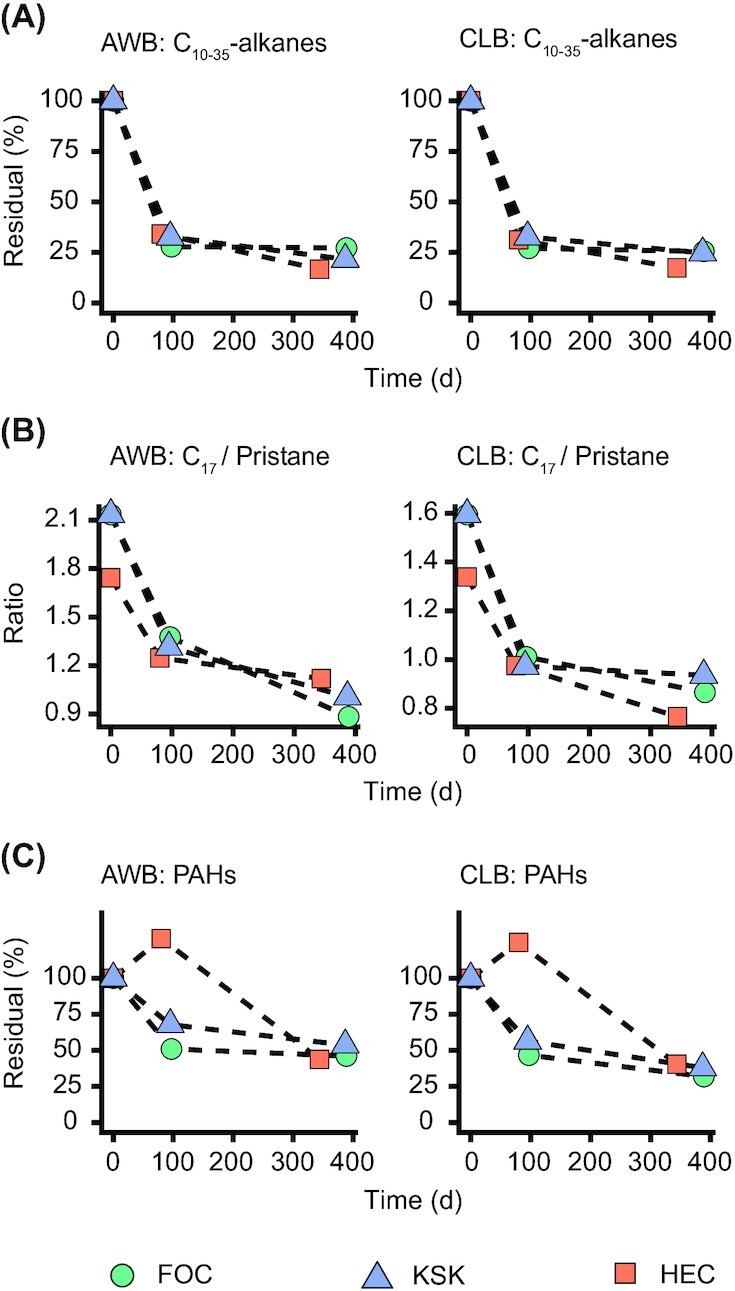
Loss of n-alkanes and PAHs over time. (A) Mean (n = 3) residual percentages of n-alkanes (sum of C10–C35 n-alkanes) over time. (B) Ratio of heptadecane (C17) to pristane over time. Decreasing ratios over time are an indicator for biodegradation of heptadecane. (C) Mean (n = 3) residual percentages of PAHs (sum of quantified methylated and non-methylated PAHs with 3–5 rings) over time. Deployment stations are shape- and color-coded. Dashed lines connecting data points were added to visualize trends. Stations FOC and KSK share the same T0 samples, the data points for these stations are hence identical at T0. Abbreviations: PAHs—polycyclic aromatic hydrocarbons; AWB—Access Western Blend dilbit; CLB—Cold Lake Blend dilbit.
Possible loss mechanisms of dilbit from the beads include bulk dilbit desorption, dissolution of dilbit components into the surrounding seawater and microbial biodegradation of dilbit components. The ratio between n-heptadecane (C17) and pristane decreased significantly over time irrespective of the sampling station or dilbit type (Fig. 3B). This indicates that significant biodegradation of n-alkanes took place during the incubations (Blumer and Sass 1972). Although the amount of PAHs also decreased significantly during the incubations, it could not conclusively be determined how much of this decrease was caused by biodegradation.
A spot check of coated beads incubated at station KSK revealed that a fraction of the dilbit coating had migrated into the inside of the porous beads during the incubation. After 12 months of incubation this inside fraction accounted for 11–30% of the quantified hydrocarbon fraction (Figure S5B, Supporting Information). The C17/pristane ratio of the inner and outer hydrocarbon fractions were not significantly different (Student's t test; P value threshold ≤ 0.05) after 12 months of incubation (Figure S5C, Supporting Information). This indicates that the inner and outer hydrocarbon fractions were likely subjected to similar weathering processes.
Microbial communities associated with dilbit-coated beads
Using 16S rRNA gene amplicon sequencing, we characterized the composition of microbial communities of dilbit-coated and uncoated beads as well as the compositions of communities present in seawater at the start of the incubations. An initial comparison with microbial communities from dilbit-amended bottle microcosms from the same sampling sites (Schreiber et al. 2019) showed that the bead-associated microbial communities were significantly different from bottle-associated communities (Figure S6, Supporting Information). Consequently, we did not further compare results from the present study with those from corresponding bottle microcosms of Douglas Channel and Hecate Strait.
The communities that had developed on dilbit-coated beads were significantly different from those of the initial seawater environment and those of uncoated beads (Fig. 4, Table 2 and Figure S7, Supporting Information). Our analyses further identified incubation time and deployment location as additional significant drivers of community composition (Table 2). Focusing only on communities of dilbit-coated beads indicated that dilbit type (i.e. AWB or CLB) on the other hand did not significantly influence the community composition (Fig. 4 and Table S4, Supporting Information).
Figure 4.
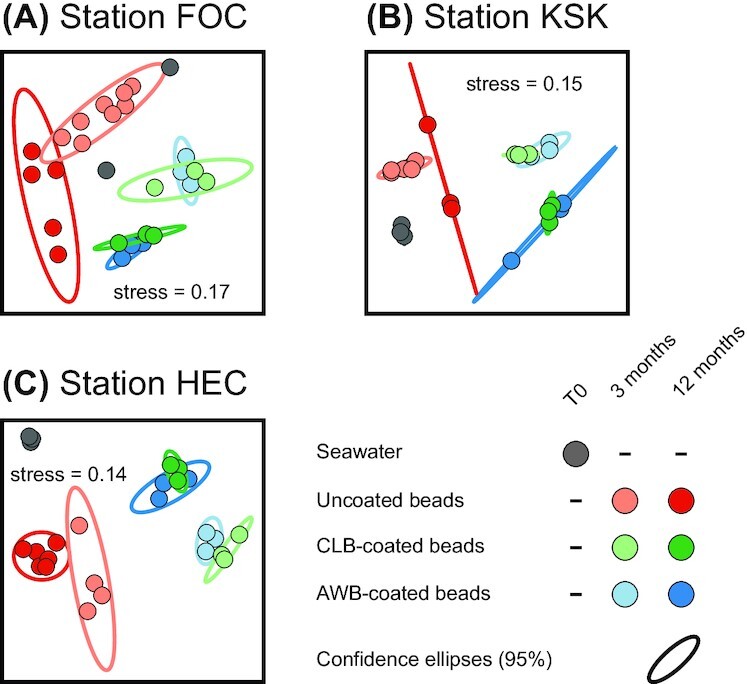
Microbial communities associated with beads. Nonmetric multidimensional scaling (nMDS) ordination of microbial communities from deployment locations FOC (A), KSK (B) and HEC (C). Bead-coatings and incubation times are color-coded. Community dissimilarities are based on the proportionality metric φ. Ellipses indicate 95% confidence range. Each data point represents a microbial community as characterized by 16S rRNA gene amplicon sequencing.
Table 2.
Influence of factors on bead-associated microbial communities. Statistically significant factors are marked with an asterisk (*).
| Treatment factor | Explained variation (r2) | P value |
|---|---|---|
| Dilbit coating (Yes/No)* | 0.54 | 0.0002 |
| Incubation time (3 or 12 months)* | 0.12 | 0.0002 |
| Station (FOC, KSK or HEC)* | 0.04 | 0.0045 |
| Residual | 0.29 | – |
Across deployment stations, incubation times and dilbit types, 238 out of a total of 20 851 Operational Taxonomic Units (OTUs) were identified as significantly more abundant on dilbit-coated beads compared to uncoated beads (Fig. 5A and B). A proportion of these dilbit-associated OTUs were classified to belong to genera that have previously been demonstrated to be capable of hydrocarbon degradation, e.g. the genera Cycloclasticus and Pseudomonas (Fig. 5C). A comparison of dilbit-associated OTUs across stations resulted in only four OTUs (two OTUs at 3 months and two OTUs at 12 months of incubation) that were consistently identified as differentially abundant (Fig. 5A and B). This low number of shared dilbit-associated OTUs between stations confirms a location-specific community response to the addition of dilbit. A closer inspection of the relative abundances of the dilbit-associated OTUs showed that the dilbit-associated OTUs represented a significant proportion (40–90%) of the microbial communities of dilbit-coated beads (Fig. 5C).
Figure 5.
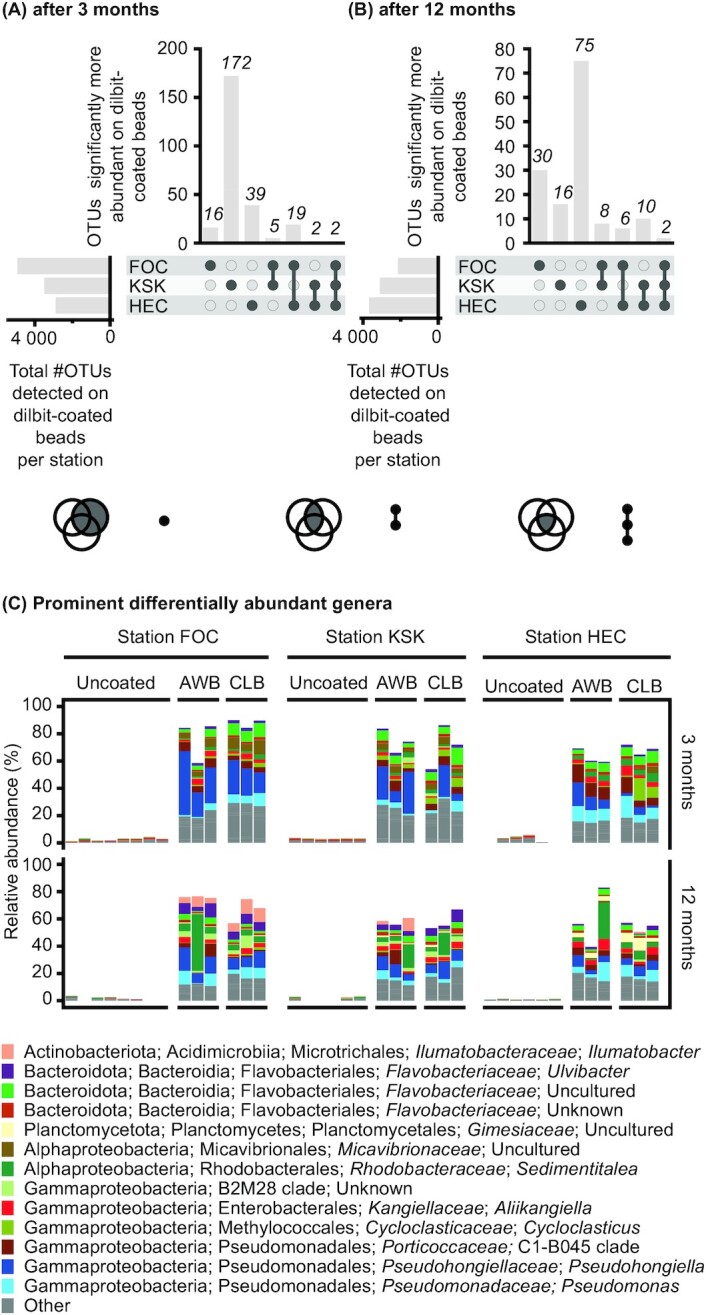
OTUs differentially more abundant on dilbit-coated beads. Number of OTUs differentially more abundant on dilbit-coated beads in comparison to uncoated beads after 3 months (A) and 12 months (B) of incubation as determined by ANCOM testing. (C) Relative abundances and taxonomic affiliations of OTUs differentially more abundant on dilbit-coated beads. The 13 most prominent genera (highest mean relative abundance across communities from dilbit-coated beads) are color-coded. The OTU data are based on 16S rRNA gene amplicon sequencing.
Putative microbial dilbit degraders
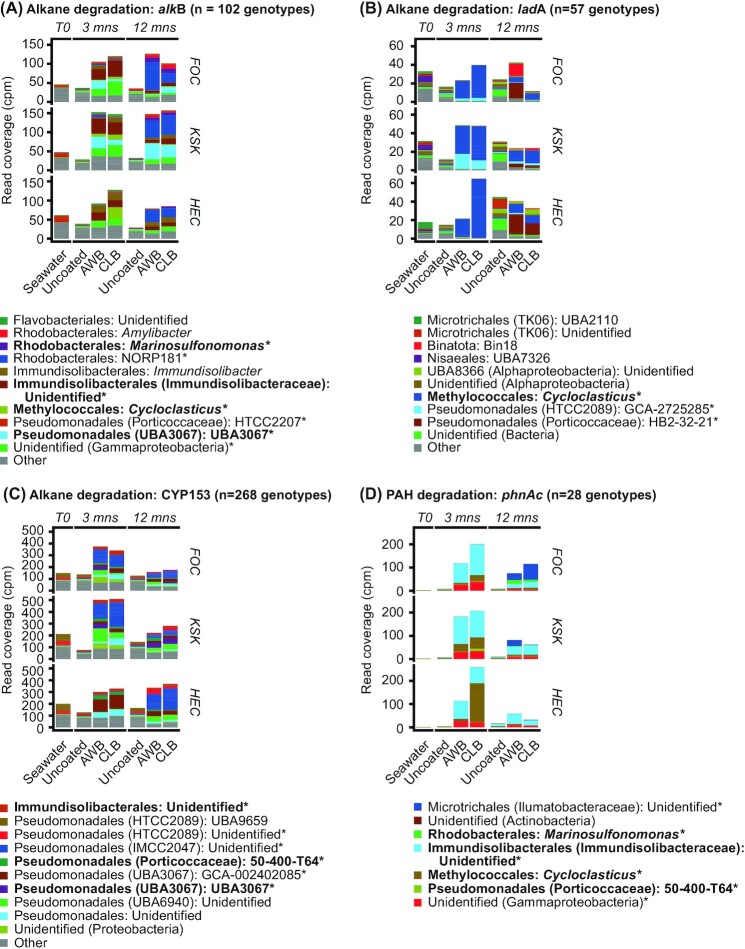
The characterization of the microbial communities based on 16S rRNA gene sequencing was supplemented with a shotgun metagenomic analysis to elucidate the role of microorganisms in the degradation of dilbit hydrocarbons. We detected a diversity of genotypes (defined within the framework of this study as gene orthologs of different taxonomic origin) associated with the aerobic microbial degradation of alkanes in our metagenomes. In detail, 102 genotypes of the alkB gene, 57 genotypes of the ladA gene and 268 genotypes of the CYP153 gene were detected (Fig. 6). In contrast, no genes associated with the anaerobic masD/assA pathway for alkane degradation were detected. In general, the relative abundances of alkB and CYP153 genes were significantly higher in dilbit-associated metagenomes than in metagenomes of seawater (T = 0 community) and uncoated beads (Fig. 6A and C). No such clear trend was discernable for the ladA gene (Fig. 6B).
Figure 6.
Relative abundances and taxonomic classifications of putative hydrocarbon degraders carrying genes associated with alkane (alkB, CYP153 and ladA) and PAH (phnAc) degradation. Relative abundances of alkB (A), ladA (B), CYP153 (C) and phnAc(D) genes are expressed in counts per million reads [cpm]). Taxonomic identities of the most prominent (highest mean relative cpm values across all communities) genotypes are color-coded. Source genera of putative hydrocarbon degraders that harbor more than one kind of degradation gene are shown in boldface. Genotypes without an assigned genus were either not unambiguously classified (‘Unknown’) or originate from uncultured lineages (e.g. ‘UBA3067’). The shown abundances are based on metagenomic shotgun data and in general represent the mean values of n = 3 (seawater, AWB, CLB) or n = 6 (uncoated beads) biological replicates. Genotypes marked with an asterisk (*) represent genotypes associated with dilbit-associated MAGs. Abbreviations: mns, months.
In addition to key genes for aerobic alkane degradation, a total of 28 genotypes of the key gene for aerobic PAH degradation, the phnAc gene, were detected. Similar to what was observed for the alkane degradation genes, no key genes of the corresponding anaerobic pathway (i.e. the degradation of PAHs under anaerobic conditions) were detected in the obtained metagenomes. Similarly to what was observed for the alkB and CYP153 genes, the relative abundance of the phnAc gene was significantly higher in metagenomes of dilbit-coated beads than in those from uncoated beads (Fig. 6D).
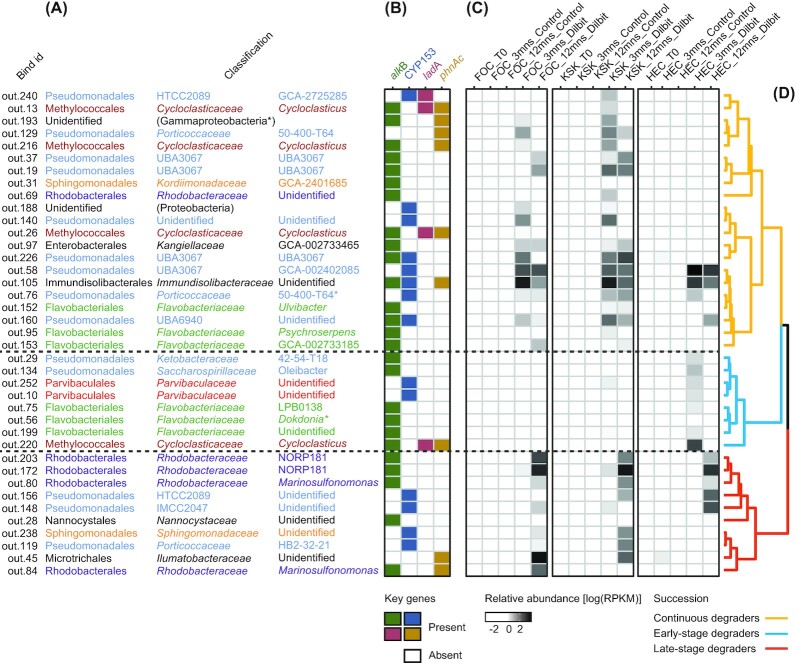
A subset of the detected putative hydrocarbon degradation genes could be linked to 75 (out of a total of 252) metagenome-assembled genomes (MAGs). An inspection of the abundance profiles of these 75 MAGs (Figure S8, Supporting Information) led to the identification of 39 MAGs (Fig. 7) that showed a significant enrichment on dilbit-coated beads compared to uncoated beads. A separate analysis at the single gene level confirmed a significant (one-tailed t-test, P ≤ 0.05) enrichment of hydrocarbon degradation genes of these MAGs in metagenomes of dilbit-coated beads (Figures S9 and S10, Supporting Information), and thereby supports the conclusion that microorganisms associated with these MAGs were enriched as a result of the dilbit-coating. Unfortunately, none of the dilbit-associated MAGs contained genes coding for ribosomal RNA, so that it was not possible to link the MAGs to the results of our OTU-based analysis.
Figure 7.
Metagenome assembled genomes (MAGs) of putative dilbit-degraders. Only MAGs that contain key genes for hydrocarbon degradation and also showed a significant enrichment on dilbit-coated beads are shown. (A) Taxonomic classification of MAGs based on detected phylogenetic marker genes. Bins marked with an asterisk (*) did not contain marker genes and were instead classified based on the taxonomic classification of the detected key genes for hydrocarbon degradation. Taxonomic groups that are present more than once are color-coded. (B) Presence of key hydrocarbon genes in the MAGs. (C) Mean relative abundances of MAGs expressed as the log10 of the MAGs' RPKM (Reads Per Kilobase of genome size per Million mapped reads) values. (D) Cluster analysis based on the Pearson distance between the relative abundances of the MAGs, i.e. MAGs with similar abundance profiles will cluster together. Populations of ‘early degraders’, ‘late degraders and ‘continuous degraders" were identified based on the formed clusters and are color-coded.
Overall, the identified dilbit-associated MAGs represented a large proportion of the total community of putative dilbit degraders (Figure S12, Supporting Information). In detail, the identified MAGs represent on average 65% of the alkB community, 77% of the CYP153 community and 81% of the phnAc community. In contrast, the dilbit-associated MAGs represent only 25% of the ladA community.
The majority of the dilbit-associated MAGs represent members of the orders Flavobacteriales, Methylococcales, Pseudomonadales and Rhodobacterales (Fig. 7). In total, 10 of the dilbit-associated MAGs could be classified to the genus level and represent members of the genera Cycloclasticus, Dokdonia, Marinosulfonomonas, Oleibacter, Psychroserpens and Ulvibacter. The remaining dilbit-associated MAGs could either not be unambiguously classified to the genus level (14 MAGs) or were classified to represent microorganisms that currently have no cultured representative (15 MAGs).
Based on gene-presence profiles, 37 out of 39 dilbit-associated MAGs carry at least one gene indicative of alkane degradation. Of these the alkB and the CYP153 genes were the most widespread and were present in 26 (alkB) and 14 (CYP153) MAGs. In contrast, the ladA gene was only detected in three MAGs classified as the genus Cycloclasticus and a single MAG closely related to a currently uncultured species of the Pseudomonadales. Similar to the ladA gene, the phnAc gene also only showed a limited distribution among the dilbit-associated MAGs and was only detected in 9 MAGs. In more detail, phnAc genes were detected in MAGs classified as Cycloclasticus (4 MAGs), an unidentified species of Immundisolibacteraceae, a currently uncultured species of Porticoccaceae, an unidentified species of Illumatobacteraceae, a MAG classified as Marinosulfonomonas and finally an unidentified gammaproteobacterial species. The majority (7 out of 9) of MAGs containing phnAc genes also contain key genes for alkane degradation. Especially noteworthy in this context are three Cycloclasticus MAGs that contain the alkB, the ladA and the phnAc genes.
Based on a cluster analysis of abundance profiles, we could assign the dilbit- associated MAGs to three populations: MAGs primarily abundant after 3 months (i.e. ‘early degraders’), MAGs primarily abundant after 12 months (‘late degraders’) and MAGs abundant at both sampling points (‘continuous degraders’). After 3 months of incubation MAGs classified as ‘continuous degraders’ represented the most relatively abundant population (85%) of dilbit degraders (Figure S13, Supporting Information). In contrast ‘early degraders’ and ‘late degraders’ only represented 12% and 3%, respectively, of the putative dilbit-degraders at this time point. After 12 months of incubation the relative abundance of putative dilbit degraders classified as ‘late degraders’ had increased to 56%. At this time, ‘continuous degraders’ and ‘early degraders’ represented 42% and 2%, respectively, of the putative dilbit degraders. Except for an apparent overrepresentation of Rhodobacteraceae MAGs in the population of ‘late degraders’, we were unable to identify a pattern between taxonomic and successional groups, or between the presence of specific degradation genes and successional groups.
DISCUSSION
This study introduces a novel in situ microcosm design to study the biodegradation of crude oil under realistic in situ conditions, and most importantly in the absence of an actual oil spill. Using this design, we investigated the degradation of dilbit in Canadian Pacific coastal waters.
Like other types of crude oil, dilbit consists of saturates, aromatic hydrocarbons, resins and asphaltenes (King 2019). Resins and asphaltenes are considered to be highly resistant to biodegradation (Leahy and Colwell 1990) and are assumed to not induce toxicity in aquatic organisms (Khan 2008). In contrast, saturates, such as n-alkanes, and aromatics have been shown to be more readily biodegradable (Leahy and Colwell 1990), and are, in the case of polycyclic aromatic hydrocarbons (PAHs), also of ecotoxicological importance (Hylland 2006). The present study focused specifically on these two hydrocarbon fractions of dilbit. In unweathered dilbit, biodegradable n-alkanes and PAHs represent with ca. 1 wt% only a very small portion of all hydrocarbon compounds (Environment and Climate Change Canada 2017). Biodegradation alone would hence be insufficient to remove dilbit in the scenario of a spill, and would only be relevant as a mitigation of the ecotoxicological consequences of such a spill.
This study provides the first estimates of in situ degradation kinetics of dilbit-associated n-alkanes and PAHs in Canadian Pacific coastal waters. Importantly, in terms of total hydrocarbon losses there were no significant differences between the tested AWB and CLB dilbit types (Fig. 4). The degradation of dilbit hydrocarbons was observed at all three tested sites and after already 3-month of incubation. Dilbit-associated n-alkanes degraded with estimated half-lives of 57–69 days, with no significant differences between mooring stations or dilbit type. These estimates are conservative (representing maximum bounds); due to the limited time resolution of this study, kinetic values were calculated by linear regression. If degradation were actually non-linear with time, the half-lives would likely be shorter. Previous studies of dilbit-degradation kinetics simulating marine conditions in comparison reported half-lives of 4–35 days for alkane fractions (King et al. 2014; Schreiber et al. 2019). Polycyclic aromatic hydrocarbons were on average lost more slowly than n-alkanes with (conservative) half-lives of 89–439 days. Since the estimated half-lives represent maximum bounds, they are consistent with results from previous studies of half-lives of PAH fractions of dilbit under simulated marine conditions (26–58 days, King et al. 2014; > 28 days, Schreiber et al. 2019).
The dilbit PAH fraction showed a higher initial loss rate in Douglas Channel than in Hecate Strait. Due to entrainment of bottom water and river discharge, the Douglas Channel system features a seasonally-fluctuating gradient of decreasing nutrients along the transect from station FOC to station HEC (Johannessen, Wright and Spear 2016; Wright et al. 2017). As microbial degradation of crude oil depends on the availability of sufficient amounts of nutrients (Atlas and Bartha 1973), the higher PAH loss rates might have been the result of this nutrient gradient.
Alternatively, the observed pattern could be explained by differences in the degree of pre-adaptation for the degradation of PAHs. Baseline concentrations of PAHs are higher in Douglas Channel than in Hecate Strait, due to inputs of PAHs from an aluminum smelter located at Kitimat (Simpson et al. 1998; Yang et al. 2017). These elevated PAH levels could have primed microbial communities of Douglas Channel for PAH degradation. However, we did not observe an elevated abundance of genes responsible for microbial PAH degradation at the Douglas Channel stations at T0 or throughout the incubations (Fig. 6D), which would argue against this hypothesis.
The diagnostic C17/pristane ratios decreased with time in all of the in situ microcosm setups and thereby indicate active microbial degradation of n-alkanes. A significant enrichment of genes involved in n-alkane oxidation in metagenomes from dilbit-coated beads in contrast to uncoated beads further supports this conclusion (Fig. 6). Although the concentration of dilbit-associated PAHs also decreased over time in the microcosms, it is was not possible to determine what proportion of these PAHs was lost due to biodegradation. A comparison of loss kinetics between isomers of methylated PAHs (Wang et al. 1998) could be used as an indicator of PAH biodegradation. Unfortunately however, the hydrocarbon data generated in this study did not feature the necessary isomer-resolution. Although we were unable to define the extent of microbial PAH degradation, a significant enrichment of the phnAc gene, a key gene of the microbial pathway for aerobic PAH degradation (Habe and Omori 2003), in dilbit-associated metagenomes indicates the microbial PAH biodegradation did take place.
Metagenome-assembled genomes (MAGs) of putative dilbit-degraders were generated and could be divided into the successional groups of early, late and continuous degraders. Although the group of early degraders showed its highest relative abundance after 3 months, it still only represented a small proportion of the total degrader community at this time (Figure S13, Supporting Information). This could indicate that this group represents early colonizers of the dilbit beads, which were sampled during the declining stage of their lifecycle. The group of continuous degraders showed a high relative abundance after both 3 and 12 months of incubation. However, the contribution of this group to the overall degrader community appeared to have declined after 12 months to the benefit of a significantly increased contribution of the late degraders. All three successional groups are composed of MAGs from a diversity of taxonomic origins, and we were unable to discern a general link between taxonomic and successional groups. A noteworthy exception to this are Rhodobacteraceae MAGs representing the genus Marinosulfonomonas and the NORP181 clade, which represent four out of the ten MAGs of the group of late degraders. Interestingly, neither the NORP181 clade nor the genus Marinosulfonomonas have so far been shown to be capable of degrading hydrocarbons associated with crude oil.
We hypothesize that the successional groups reflect compositional changes of the dilbit coating as it transitions to a dominance of more and more recalcitrant hydrocarbons whose biodegradation requires more specialized microorganisms. In the case of alkanes, this would result in a transition from an early biodegradation of short-chain alkanes to a biodegradation of long-chain alkanes during the later stages. Results of our analysis of MAGs appear to contradict this conclusion, as none of the MAGs classified as late degraders contain the ladA key gene for long-chain alkane degradation. A closer inspection of the community of putative dilbit degraders carrying the ladA gene revealed that ladA gene was generally not well represented by the MAGs. Thus in contrast to the MAG-based analysis, at the single gene level the emergence of several ladA genotypes affiliated with the Binatota phylum, the Microtrichales, Dehalococcoidales or the alphaproteobacterial UBA8366 clade could be observed after 12 months of incubation (Figure S11, Supporting Information).
Interestingly, MAGs associated with the family Immundisolibacteraceae, the genus Marinosulfonomonas and the genus Cycloclasticus appeared to be involved in the biodegradation of both alkanes and PAHs. Immundisolibacter, the currently only described genus of the Immundisolibacteraceae, has so far primarily been described as a degrader of PAHs in soil environments (Corteselli, Aitken and Singleton 2017; Brzeszcz et al. 2020). A recent study (Somee et al. 2021) and the present study, however, suggest that representatives of this family could also be involved in the degradation of hydrocarbons in a marine setting. Immundisolibacter has further only been shown to be capable of PAH degradation (Corteselli, Aitken and Singleton 2017). The capability of alkane degradation by this genus has so far not been demonstrated and is inferred solely from genomic evidence. Representatives of the genus Cycloclasticus are known for their ability to degrade PAHs (Dyksterhouse et al. 1995) and are often encountered in association with marine oil spills (Maruyama et al. 2003; Yang et al. 2016). The genomic and proteomic potential for degradation of alkanes by representatives of the genus Cycloclasticus has been reported previously (Rubin-Blum et al. 2017) and at least one representative of the genus has been shown to be capable of alkane degradation (Gutierrez et al. 2018). The genus Marinosulfonomonas has so far only been described as a methylotroph (Holmes et al. 1997) and has, to our knowledge, not yet been shown to be capable of degrading alkanes or PAHs.
Our metagenomic analysis detected a high proportion of MAGs (29 out of 39) of putative dilbit degraders that either had no closely-related representative in public sequence databases or were affiliated with genomes from currently undescribed genera, i.e. single- cell-derived and metagenome- derived genomes. This observation highlights the difficulties of deducing in situ degradation rates and their associated microbial communities based on lab incubations. The lack of cultured representatives of the identified putative dilbit-degraders, further suggests that these organisms cannot easily be cultured in a lab context and that their metabolic and growth characteristics therefore differs from those of currently described hydrocarbon degraders. This conclusion is supported by the significant differences between dilbit-associated microbial communities observed in this study compared to those observed in corresponding bottle microcosms (Figure S6, Supporting Information; Schreiber et al. 2019).
Our microbial community analysis indicates that there were no significant differences between communities of beads coated with AWB dilbit and beads coated with CLB dilbit. As the overall hydrocarbon losses were also comparable between both dilbit types, this suggests that the results of this study could also be representative for other types of dilbit. Additionally, these results raise the question how much influence the exact type of spilled crude oil would truly have both on the biodegradation rates of oil hydrocarbons and on the associated degrader communities.
In summary, using a potential shipping route for dilbit as an example, this study presents novel data on the degradation of dilbit in coastal waters under realistic in situ conditions and on the microbial communities involved in this degradation. We show that the location along this shipping route can have a significant influence on the degradation of dilbit hydrocarbons and the corresponding microbial communities. In contrast, the specific type of dilbit appeared to not be a significant factor in this context. This study identified several microorganisms potentially involved in the in situ biodegradation of dilbit hydrocarbons and successfully generated metagenome-assembled genomes for a subset of these microorganisms. The identified microorganisms could be assigned to different successional populations and for a large part represent currently unknown lineages or lineages with currently no cultured representative.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Canadian Coast Guard and specifically the crews of the CCGS John P. Tully for their participation and assistance during fieldwork. We also thank Lucius Perreault, Ron Lindsay and Roger Savoie from the Institute of Ocean Sciences (Fisheries and Oceans Canada; Sidney, BC, Canada) for their support in the deployment and recovery of our moorings during the various missions at sea. We are grateful to Alison Thomas and Graeme Soper from the Bedford Institute of Oceanography (Dartmouth, NS, Canada) for their contribution to chemical analyses. We want to acknowledge the excellent technical support of Danielle Ouellette, Julie Champagne, Erica Ma and Katrina Dingle. We acknowledge Compute Canada (www.computecanada.ca) for access to the University of Waterloo High Performance Computing (HPC) infrastructure (GP4/Graham system). This work was supported through the World Class Tanker Safety Program of the Government of Canada.
Contributor Information
Lars Schreiber, Energy, Mining and Environment Research Center, National Research Council of Canada (NRC), 6100 Royalmount Ave, Montreal, QC H4P 2R2, Canada.
Nathalie Fortin, Energy, Mining and Environment Research Center, National Research Council of Canada (NRC), 6100 Royalmount Ave, Montreal, QC H4P 2R2, Canada.
Julien Tremblay, Energy, Mining and Environment Research Center, National Research Council of Canada (NRC), 6100 Royalmount Ave, Montreal, QC H4P 2R2, Canada.
Jessica Wasserscheid, Energy, Mining and Environment Research Center, National Research Council of Canada (NRC), 6100 Royalmount Ave, Montreal, QC H4P 2R2, Canada.
Sylvie Sanschagrin, Energy, Mining and Environment Research Center, National Research Council of Canada (NRC), 6100 Royalmount Ave, Montreal, QC H4P 2R2, Canada.
Jennifer Mason, Centre for Offshore Oil, Gas and Energy Research (COOGER), Bedford Institute of Oceanography, Fisheries and Oceans Canada (DFO), 1 Challenger Drive, P.O. Box 1006, Dartmouth, NS B2Y 4A2, Canada.
Cynthia A Wright, Institute of Ocean Sciences, Fisheries and Oceans Canada (DFO), 9860 West Saanich Road, P.O. Box 6000, Sidney, BC V8L 4B2, Canada.
David Spear, Institute of Ocean Sciences, Fisheries and Oceans Canada (DFO), 9860 West Saanich Road, P.O. Box 6000, Sidney, BC V8L 4B2, Canada.
Sophia C Johannessen, Institute of Ocean Sciences, Fisheries and Oceans Canada (DFO), 9860 West Saanich Road, P.O. Box 6000, Sidney, BC V8L 4B2, Canada.
Brian Robinson, Centre for Offshore Oil, Gas and Energy Research (COOGER), Bedford Institute of Oceanography, Fisheries and Oceans Canada (DFO), 1 Challenger Drive, P.O. Box 1006, Dartmouth, NS B2Y 4A2, Canada.
Thomas King, Centre for Offshore Oil, Gas and Energy Research (COOGER), Bedford Institute of Oceanography, Fisheries and Oceans Canada (DFO), 1 Challenger Drive, P.O. Box 1006, Dartmouth, NS B2Y 4A2, Canada.
Kenneth Lee, Ecosystem Science, Fisheries and Oceans Canada (DFO), 200 Kent St, Ottawa,ON K1A 0E6, Canada.
Charles W Greer, Energy, Mining and Environment Research Center, National Research Council of Canada (NRC), 6100 Royalmount Ave, Montreal, QC H4P 2R2, Canada; Department of Natural Resource Sciences, McGill University, Macdonald-Stewart Building, McGill, 21111 Lakeshore Road, Sainte-Anne-de-Bellevue, QC H9X 3V9, Canada.
AUTHOR CONTRIBUTIONS
CWG, NF, KL, TLK, BR and DS designed the study. DS designed the microcosm mooring frame. NF developed the experimental standard operating procedures. NF carried out bead coating, bead loading and bead preparation for downstream analyses. DS, SCJ, CAW and NF organized the logistics of the study, and deployed and retrieved the microcosms. JT and JW assisted with bioinformatics analyses. JM and BR generated and curated the hydrocarbon data. SS extracted DNA and performed amplicon and metagenomic sequencing. LS analyzed and integrated all data. LS wrote the manuscript with significant contribution by NF for the Methods section. All co-authors commented on and provided substantial edits to the manuscript.
Conflicts of interest
None declared.
REFERENCES
- Aitchison J. The statistical analysis of compositional data. J R Stat Soc Ser B (Methodological). 1982;44:139–77. [Google Scholar]
- Arkin AP, Cottingham RW, Henry CSet al. KBase: the United States Department of Energy Systems Biology Knowledgebase. Nat Biotechnol. 2018;36:566–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlas RM, Bartha R. Stimulated biodegradation of oil slicks using oleophilic fertilizers. Environ Sci Technol. 1973;7:538–41. [DOI] [PubMed] [Google Scholar]
- Becker RA, Wilks AR, Brownrigg Ret al. maps: Draw Geographical Maps. 2018; https://CRAN.R-project.org/package=maps. [Google Scholar]
- Blumer M, Sass J. Oil pollution: persistence and degradation of spilled fuel oil. Science. 1972;176:1120–2. [DOI] [PubMed] [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJet al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2012;10:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonmak C, Takahashi Y, Morikawa M. Cloning and expression of three ladA-type alkane monooxygenase genes from an extremely thermophilic alkane-degrading bacterium Geobacillus thermoleovorans B23. Extremophiles. 2014;18:515–23. [DOI] [PubMed] [Google Scholar]
- Brzeszcz J, Kapusta P, Steliga Tet al. Hydrocarbon removal by two differently developed microbial inoculants and comparing their actions with biostimulation treatment. Molecules. 2020;25:661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WAet al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci. 2011;108:4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil P-A, Mussig AJ, Hugenholtz Pet al. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019;36:1925–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corteselli EM, Aitken MD, Singleton DR. Description of Immundisolibacter cernigliae gen. nov., sp. nov., a high-molecular-weight polycyclic aromatic hydrocarbon-degrading bacterium within the class Gammaproteobacteria, and proposal of Immundisolibacterales ord. nov. and Immundisolibacteraceae fam. nov. Int J Syst Evol Microbiol. 2017;67:925–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G-C, Heuer H, Zühlke Set al. Soil type-dependent responses to phenanthrene as revealed by determining the diversity and abundance of polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenase genes by using a novel PCR detection system. Appl Environ Microbiol. 2010;76:4765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Strous M. An integrated pipeline for annotation and visualization of metagenomic contigs. Front Genetics. 2019;10:999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyksterhouse SE, Gray JP, Herwig RPet al. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int J Syst Evol Microbiol. 1995;45:116–23. [DOI] [PubMed] [Google Scholar]
- Environment and Climate Change Canada (ECCC) . Physiochemical Properties of Petroluem Products. URL: https://open.canada.ca/data/en/dataset/53c38f91-35c8-49a6-a437-b311703db8c5, 2017. [Google Scholar]
- Fernandes AD, Reid JN, Macklaim JMet al. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome. 2014;2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittel A, Donhauser J, Røy Het al. Ubiquitous presence and novel diversity of anaerobic alkane degraders in cold marine sediments. Front Microbiol. 2015;6:1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez T, Morris G, Ellis Det al. Hydrocarbon-degradation and MOS-formation capabilities of the dominant bacteria enriched in sea surface oil slicks during the Deepwater Horizon oil spill. Mar Pollut Bull. 2018;135:205–15. [DOI] [PubMed] [Google Scholar]
- Habe H, Omori T. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci Biotechnol Biochem. 2003;67:225–43. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ. raster: Geographic Data Analysis and Modeling, 2019; https://CRAN.R-project.org/package=raster. [Google Scholar]
- Holmes AJ, Kelly DP, Baker SCet al. Methylosulfonomonas methylovora gen. nov., sp. nov., and Marinosulfonomonas methylotropha gen. nov., sp. nov.: novel methylotrophs able to grow on methanesulfonic acid. Arch Microbiol. 1997;167:46–53. [DOI] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, LoCascio PFet al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylland K. Polycyclic aromatic hydrocarbon (PAH) ecotoxicology in marine ecosystems. J Toxicol Environ Health Part A. 2006;69:109–23. [DOI] [PubMed] [Google Scholar]
- Johannessen SC, Greer CW, Hannah CGet al. Fate of diluted bitumen spilled in the coastal waters of British Columbia, Canada. Mar Pollut Bull. 2020;150:110691. [DOI] [PubMed] [Google Scholar]
- Johannessen SJ, Wright CA, Spear D. Seasonality and physical control of water properties and sinking and suspended particles in Douglas Channel, British Columbia. Canadian Data Report of Hydrography and Ocean Sciences. p 26. Cat. No.: Fs97-18/308E, Department of Fisheries and Oceans; Canada. 2016. [Google Scholar]
- Kang DD, Li F, Kirton Eet al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7:e7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan CW. Identification of components in crude oil that are chronically toxic to the early life stages of fish. M.Sc. Thesis. Queen's University, Kingston, ON, Canada, 2008. URI: http://hdl.handle.net/1974/956. [Google Scholar]
- King TL, Robinson B, Boufadel Met al. Flume tank studies to elucidate the fate and behavior of diluted bitumen spilled at sea. Mar Pollut Bull. 2014;83:32–7. [DOI] [PubMed] [Google Scholar]
- King TL. Physical properties dynamics of oil sands products and their influence on spill response. Ph.D. Thesis. Saint Mary's University, Halifax, NS, Canada, 2019. URI:http://library2.smu.ca/handle/01/28226. [Google Scholar]
- Leahy JG, Colwell RR. Microbial degradation of hydrocarbons in the environment. Microbiol Mol Biol Rev. 1990;54:305–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C-M, Luo Ret al. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker Aet al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell D, Pawlowsky-Glahn V, Egozcue JJet al. Proportionality: a valid alternative to correlation for relative data. PLoS Comput Biol. 2015;11:e1004075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozada M, Riva Mercadal JP, Guerrero LDet al. Novel aromatic ring-hydroxylating dioxygenase genes from coastal marine sediments of Patagonia. BMC Microbiol. 2008;8:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Van Treuren W, White RAet al. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis. 2015;26:27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama A, Ishiwata H, Kitamura Ket al. Dynamics of microbial populations and strong selection for Cycloclasticus pugetii following the Nakhodka oil spill. Microb Ecol. 2003;46:442–53. [DOI] [PubMed] [Google Scholar]
- Morris BEL, Gissibl A, Kümmel Set al. A PCR-based assay for the detection of anaerobic naphthalene degradation. FEMS Microbiol Lett. 2014;354:55–9. [DOI] [PubMed] [Google Scholar]
- Mouttaki H, Johannes J, Meckenstock RU. Identification of naphthalene carboxylase as a prototype for the anaerobic activation of non-substituted aromatic hydrocarbons. Environ Microbiol. 2012;14:2770–4. [DOI] [PubMed] [Google Scholar]
- Nie Y, Chi C-Q, Fang Het al. Diverse alkane hydroxylase genes in microorganisms and environments. Sci Rep. 2014;4:4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt Ret al. vegan: community ecology package. 2010; https://CRAN.R-project.org/package=vegan. [Google Scholar]
- Ortmann AC, Cobanli SE, Wohlgeschaffen Get al. Measuring the fate of different diluted bitumen products in coastal surface waters. Mar Pollut Bull. 2020;153:111003. [DOI] [PubMed] [Google Scholar]
- Parks DH, Chuvochina M, Chaumeil P-Aet al. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat Biotechnol. 2020;38:1079–86. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz Pet al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn TP, Richardson MF, Lovell Det al. propr: an R-package for identifying proportionally abundant features using compositional data analysis. Sci Rep. 2017;7:16252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Myers GSA, Ravel J. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics. 2005;6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin-Blum M, Antony CP, Borowski Cet al. Short-chain alkanes fuel mussel and sponge Cycloclasticus symbionts from deep-sea gas and oil seeps. Nat Microbiol. 2017;2:17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber L, Fortin N, Tremblay Jet al. Potential for microbially mediated natural attenuation of diluted bitumen on the coast of British Columbia (Canada). Appl Environ Microbiol. 2019;85:e00086–00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CD, Harrington CF, Cullen WRet al. Polycyclic aromatic hydrocarbon contamination in marine sediments near Kitimat, British Columbia. Environ Sci Technol. 1998;32:3266–72. [Google Scholar]
- Smits THM, Röthlisberger M, Witholt Bet al. Molecular screening for alkane hydroxylase genes in Gram-negative and Gram-positive strains. Environ Microbiol. 1999;1:307–17. [DOI] [PubMed] [Google Scholar]
- Somee MR, Dastgheib SMM, Shavandi Met al. Distinct microbial communities along the chronic oil pollution continuum of the Persian Gulf converge with oil spill accidents. Sci Rep. 2021;11:11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swannell RP, Lee K, McDonagh M. Field evaluations of marine oil spill bioremediation. Microbiol Rev. 1996;60:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B, Charchuk R, Li Cet al. Draft genome sequence of uncultivated Firmicutes (Peptococcaceae SCADC) single cells sorted from methanogenic alkane-degrading cultures. Genome Announc. 2014;2:e00909–00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay J, Fortin N, Elias Met al. Metagenomic and metatranscriptomic responses of natural oil degrading bacteria in the presence of dispersants. Environ Microbiol. 2019;21:2307–19. [DOI] [PubMed] [Google Scholar]
- Tremblay J, Yergeau E, Fortin Net al. Chemical dispersants enhance the activity of oil- and gas condensate-degrading marine bacteria. ISME J. 2017;11:2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay J, Yergeau E. Systematic processing of ribosomal RNA gene amplicon sequencing data. GigaScience. 2019;8:giz146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beilen JB, Funhoff EG, van Loon Aet al. Cytochrome P450 alkane hydroxylases of the CYP153 family are common in alkane-degrading eubacteria lacking integral membrane alkane hydroxylases. Appl Environ Microbiol. 2006;72:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Fingas M, Blenkinsopp Set al. Comparison of oil composition changes due to biodegradation and physical weathering in different oils. J Chromatogr A. 1998;809:89–107. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer: New York, 2016. [Google Scholar]
- Wright CA, Vagle S, Hannah Cet al. Physical, chemical and biological data collected in Douglas Channel and approaches to Kitimat, October 2015-July 2016. Canadian Data Report of Hydrography and Ocean Sciences. Cat.No.: Fs97-16/202E. Department of Fisheries and Oceans. Canada. 2017. [Google Scholar]
- Wright CA, Vagle S, Hannah Cet al. Physical, chemical and biological oceanographic data collected in Douglas Channel and the approaches to Kitimat, June 2013-July 2014. Physical, chemical and biological oceanographic data collected in Douglas Channel and the approaches to Kitimat, June 2013-July 2014. Cat.No.: Fs97-16/196E. Department of Fisheries and Oceans. Canada. 2015. [Google Scholar]
- Wright CA, Vagle S, Hannah Cet al. Physical, chemical and biological oceanographic data collected in Douglas Channel and the approaches to Kitimat, October 2014-July 2015. Canadian Data Report of Hydrography and Ocean Sciences. Cat.No.: Fs97-16/200E. Department of Fisheries and Oceans. Canada. 2016. [Google Scholar]
- Yang T, Speare K, McKay Let al. Distinct bacterial communities in surficial seafloor sediments following the 2010 Deepwater Horizon blowout. Front Microbiol. 2016;7:1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Hollebone BP, Laforest Set al. Occurrence, source and ecological assessment of baseline hydrocarbons in the intertidal marine sediments along the shoreline of Douglas Channel to Hecate Strait in British Columbia. Mar Pollut Bull. 2017;122:450–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequence reads generated for this study have been submitted to NCBI's SRA and are available under the BioProject accession number PRJNA629517. The generated 16S rRNA amplicon reads are available under SAMN15949[457–537] and the shotgun metagenomics reads are available under SAMN15950[781–873]. The metagenome-assembled genomes of putative dilbit-degraders are available under SAMN19594[111-146]. The metagenomic assembly is available on the zenodo.org file repository (https://doi.org/10.5281/zenodo.4009541). All bioinformatics scripts used, as well as OTU and gene count tables, are available upon request.