Abstract
Background
Coronavirus disease 2019 (COVID-19) has been shown to cause serious health problems among them is the Acute Respiratory Distress syndrome (ARDS). Vitamin D receptor (VDR) signaling possibly partakes in the pathophysiology of this devastating complication.
Methods
In the current project, we have appraised expression levels of VDR, CYP27B1 and a number of associated lncRNAs in the circulation of COVID-19 patients versus healthy subjects using real-time PCR method.
Results
Expression of SNHG6 was considerably lower in COVID-19 patients compared with control subjects (Ratio of mean expression (RME) = 0.22, P value = 7.04E-05) and in both female and male COVID-19 patients compared with sex-matched unaffected individuals (RME = 0.32, P value = 0.04 and RME = 0.16, P value = 0.000679683, respectively). However, its expression was similar among ICU-hospitalized and non-ICU patients. Similarly, expression of SNHG16 was lower in in COVID-19 patients compared with controls (RME = 0.20, P value = 5.94E-05) and in both female and male patients compared with sex-matched controls (RME = 0.32, P value = 0.04 and RME = 0.14, P value = 0.000496435, respectively) with no significant difference among ICU-hospitalized and non-ICU hospitalized patients. Expression of VDR was lower in COVID-19 patients compared with controls (RME = 0.42, P value = 0.04) and in male patients compared with male controls (RME = 0.27, P value = 0.02). Yet, expression of VDR was statistically similar between female subgroups and between ICU-hospitalized and non-ICU hospitalized patients. Expression levels CYP27B, Linc00511 and Linc00346 were similar among COVID-19 patients and healthy subjects or between their subgroups. Significant correlations have been detected between expression levels of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 lncRNAs both among COVID-19 patients and among healthy controls with the most significant ones being SNHG6 and SNHG16 (r = 0.74, P value = 3.26e-17 and r = 0.81, P = 1.54e-22, respectively).
Conclusion
Combination of transcript levels of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 could differentiate patients from controls with AUC = 0.76, sensitivity = 0.62 and specificity = 0.81. The current data potentiate SNHG6, SNHG16 and VDR as possible contributors in COVID-19 infection but not in the severity of ARDS.
Keywords: COVID-19, Vitamin D receptor, VDR, lncRNA, SNHG6, SNHG16, Linc00511, Linc00346, CYP27B
Background
Coronavirus disease 2019 (COVID-19) has been shown to cause serious health problems. The most devastating complication of this disorder is the Acute Respiratory Distress syndrome (ARDS) [1] which is thought to be caused by a mixture of mechanisms among them are cytokine storm [2], abnormal activity of the renin-angiotensin apparatus [3], neutrophil stimulation [4] and enhanced coagulation [5]. This COVID-19 complication has been shown to be provoked by vitamin D deficiency and attenuated by induction of the vitamin D receptor (VDR) [6]. This observation is supported by the presence of VDR on immune cells, the regulatory role of the active vitamin D hormone on expression of the majority of cytokines and the role of this hormone on activation of immune defense responses while attenuation of the acquired immune responses [7–9]. Antimicrobial function of vitamin D is exerted through a cascade of events. Macrophages respond to microbial infection through pattern recognition receptors. Induction of these responses leads to activation of transcription of 1α-hydroxylase (CYP27B1) and VDR [10]. The impact of vitamin D on regulation of immune responses is modulated by availability of 25-hydroxyvitamin D, activation of CYP27B1 by the attacking pathogenic organisms and induction of 1,25-dihydroxyvitamin D in cellular compartments of the immune system [10, 11]. Moreover, expression of VDR has been shown to be affected by several mechanisms among them are long non-coding RNAs (lncRNAs) [12]. We have previously assessed expression of VDR-associated lncRNAs namely SNHG16, SNHG6, LINC00346 and LINC00511 in a number of disorders including epilepsy [13], lung cancer [14] and breast cancer [15]. In the current project, we have appraised expression levels of VDR, CYP27B1 and mentioned lncRNAs in the circulation of COVID-19 patients versus healthy subjects to unravel the role of these transcripts in the pathogenic processes during the course of COVID-19.
Methods
Enrolled individuals
This study is a pilot study to measure expression levels of VDR and related lncRNAs in COVID-19 cases and healthy controls. The current investigation was conducted on patients hospitalized to Nikan Hospital, Tehran, during March 2020 until April 2020. Patients have clinical manifestations of COVID-19 and the disorder was confirmed by a positive nasopharyngeal swab specimen. Patients were diagnosed to have moderate, severe and very severe disease courses and were assigned hospital treatment [16]. Control specimens were got from healthy persons without no clinical signs or recent exposure to patients with COVID-19. The study protocol was approved by ethical committee of Shahid Beheshti University of Medical Sciences. Informed consent was obtained from all patients. Paraclinical data was obtained from all hospitalized patients. Hematological tests were performed in Beckman Coulter MAXM AL Hematology Flow Cytometry System. C-reactive protein (CRP) was measured by latex-enhanced nephelometry.
Expression assays
First, 4 ml of peripheral blood were collected from all hospitalized patients and healthy persons. Subsequently, total RNA was isolated from these specimens using the GeneAll Kit (Seoul, South Korea). Then, total RNA was transformed to cDNA using the OneStep RT-PCR Series Kit (BioFact™, Seoul, South Korea). Transcript quantities of VDR-associated genes were measured in specimens gathered from COVID-19 patients and healthy subjects using the RealQ Plus 2x Master Mix (Amplicon, Denmark). Primers Characteristics have been reported in our previous studies [13, 15].
Data analysis
R programming language was used for data analysis. Transcript quantities of VDR-associated genes were quantified from Ct values, considering B2M as the reference gene. The obtained values were log2 transformed and used for next steps. Expression levels of genes were compared between COVID-19 patients and healthy subjects and ICU-hospitalized and non-ICU hospitalized patients. The significance of difference in mean values of gene expression between two subgroups was computed using the t-test. Correlations between expression levels of genes were appraised via calculation of Spearman correlation coefficients. ROC curves were depicted using Bayesian Generalized Linear Model, Generalized Linear Model (GLM), and Linear Discriminant Analysis with 10-fold cross validation. Application of the GLM resulted in the most appropriate estimates. Youden’s J parameter was measured to find the optimum threshold. P value < 0.05 was considered as significant.
Results
Totally, 91 COVID-19 patients (38 females, 53 males) and 91 healthy subjects (39 females, 52 males) were included in the study. The mean age (±standard deviation) of the COVID-19 patients was 57.18 (±16.89) years. Thirty seven patients (40.6%) were hospitalized in the ICU. Table 1 displays the paraclinical variables obtained from medical records of the COVID-19 patients.
Table 1.
Paraclinical data of the COVID-19 patients
| Variable | Mean | Standard deviation |
|---|---|---|
| White Blood Cells (109/L) | 8.119 | 8.482 |
| Red Blood Cells (1012/L) | 4.681 | 0.768 |
| Hemoglobin (g/dL) | 12.704 | 2.182 |
| Hematocrit (%) | 39.267 | 6.595 |
| MCV (fl) | 83.985 | 5.702 |
| MCH (pg) | 27.154 | 2.330 |
| MCHC (g/dL) | 32.348 | 1.3720 |
| Platelet count (109/L) | 210.354 | 95.216 |
| Lymphocyte (%) | 21.043 | 11.323 |
| Neutrophil (%) | 69.098 | 13.087 |
| ESR (mm/hr) | 44.131 | 32.701 |
| CRP (mg/dL) | 73.256 | 69.540 |
Expression assays
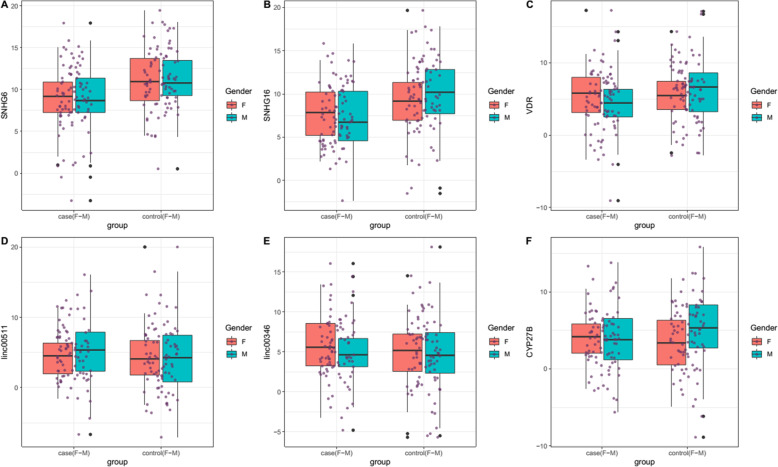
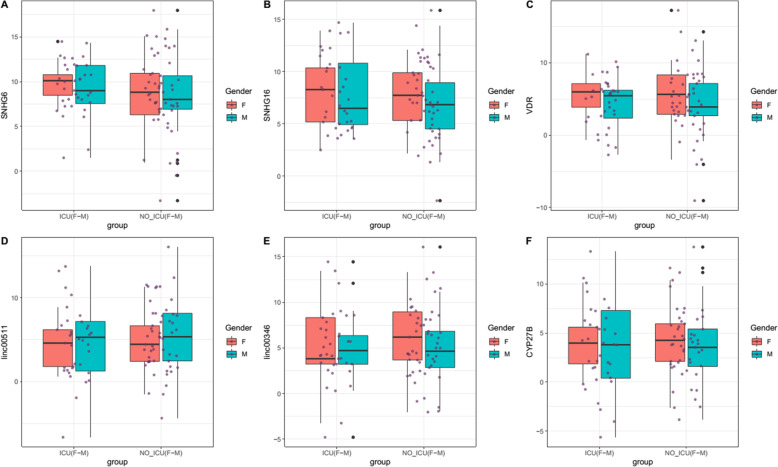
Figures 1 and 2 depict relative expressions of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 lncRNAs in total COVID-19 patients compared with healthy persons, and in ICU-hospitalized patients compared with non-ICU patients, respectively.
Fig. 1.
Relative expressions of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 lncRNAs in total COVID-19 patients compared with controls
Fig. 2.
Relative expressions of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 lncRNAs in ICU-hospitalized patients compared with non-ICU patients
Expression of SNHG6 was remarkably lower in COVID-19 patients compared with controls (Ratio of mean expression (RME) = 0.22, P value = 7.04E-05) and in both female and male affected individuals compared with sex-matched controls (RME = 0.32, P value = 0.04 and RME = 0.16, P value = 0.000679683, respectively). However, its expression was statistically similar between ICU-hospitalized and non-ICU hospitalized patients. Similarly, expression of SNHG16 was lower in in COVID-19 patients compared with controls (RME = 0.20, P value = 5.94E-05) and in both female and male affected individuals compared with sex-matched healthy subjects (RME = 0.32, P value = 0.04 and RME = 0.14, P value = 0.000496435, respectively) with no significant difference among ICU-hospitalized and non-ICU hospitalized patients. Expression of VDR was lower in COVID-19 patients compared with controls (RME = 0.42, P value = 0.04) and in male patients compared with normal males (RME = 0.27, P value = 0.02). However, expression of VDR was statistically similar between female patients and normal females and between ICU-hospitalized and non-ICU hospitalized patients. Expression levels CYP27B, Linc00511 and Linc00346 were not different between COVID-19 patients and healthy subjects or between their subgroups (Table 2).
Table 2.
Detailed statistics of expression of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 lncRNAs in COVID-19 patients and controls (RME: Ratio of mean expression)
| SNHG6 | SNHG16 | VDR | Linc00511 | Linc00346 | CYP27B | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of Samples | SE | RME | P Value | 95% CI | SE | RME | P Value | 95% CI | SE | RME | P Value | 95% CI | SE | RME | P Value | 95% CI | SE | RME | P Value | 95% CI | SE | RME | P Value | 95% CI | |||||||
| Cases/ controls | |||||||||||||||||||||||||||||||
| Total | 91/91 | 0.54 | 0.22 | 0.00 | −3.29 | −1.14 | 0.56 | 0.20 | 0.00 | −3.40 | − 1.20 | 0.61 | 0.42 | 0.04 | −2.44 | −0.05 | 0.64 | 1.55 | 0.33 | −0.64 | 1.90 | 0.62 | 1.34 | 0.50 | −0.80 | 1.64 | 0.61 | 0.71 | 0.43 | −1.69 | 0.72 |
| F | 38/39 | 0.78 | 0.32 | 0.04 | −3.21 | −0.09 | 0.79 | 0.32 | 0.04 | −3.22 | −0.05 | 0.89 | 0.78 | 0.68 | −2.14 | 1.41 | 0.86 | 1.01 | 0.99 | −1.71 | 1.74 | 0.98 | 1.98 | 0.32 | −0.96 | 2.93 | 0.79 | 1.39 | 0.54 | −1.09 | 2.04 |
| M | 53/52 | 0.75 | 0.16 | 0.00 | −4.11 | −1.14 | 0.78 | 0.14 | 0.00 | −4.33 | −1.25 | 0.82 | 0.27 | 0.02 | −3.51 | −0.25 | 0.92 | 2.11 | 0.25 | −0.75 | 2.90 | 0.81 | 1.01 | 0.99 | −1.59 | 1.61 | 0.88 | 0.43 | 0.17 | −2.96 | 0.53 |
| ICU/ Non-ICU | |||||||||||||||||||||||||||||||
| Total | 37/54 | 0.75 | 1.74 | 0.29 | −0.69 | 2.30 | 0.75 | 1.49 | 0.44 | −0.91 | 2.06 | 0.84 | 0.90 | 0.85 | −1.82 | 1.51 | 0.87 | 0.60 | 0.40 | −2.47 | 0.99 | 0.84 | 0.73 | 0.59 | −2.12 | 1.21 | 0.85 | 0.79 | 0.68 | −2.03 | 1.34 |
| F | 13/25 | 0.97 | 2.18 | 0.26 | −0.85 | 3.10 | 1.09 | 1.11 | 0.90 | −2.12 | 2.41 | 1.29 | 0.90 | 0.91 | −2.78 | 2.48 | 1.01 | 0.62 | 0.51 | −2.74 | 1.38 | 1.46 | 0.59 | 0.61 | −3.77 | 2.25 | 1.03 | 0.92 | 0.91 | −2.22 | 1.99 |
| M | 24/29 | 1.10 | 1.64 | 0.52 | −1.49 | 2.92 | 1.04 | 1.93 | 0.37 | −1.14 | 3.04 | 1.12 | 1.05 | 0.95 | −2.18 | 2.33 | 1.27 | 0.52 | 0.46 | −3.50 | 1.61 | 1.03 | 0.90 | 0.88 | −2.23 | 1.92 | 1.23 | 0.71 | 0.69 | −2.98 | 1.98 |
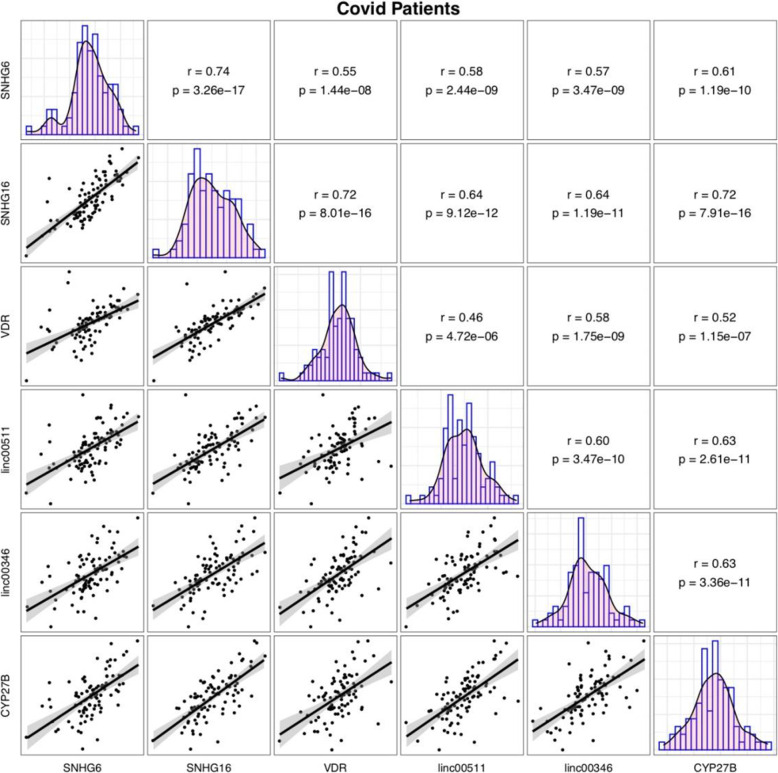
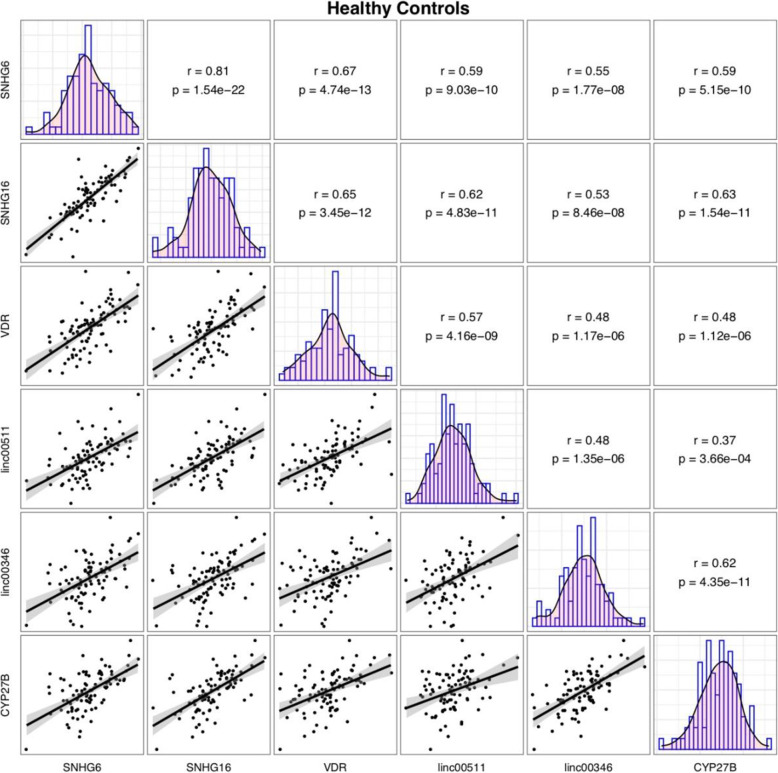
Significant correlations have been displayed between expression levels of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 lncRNAs both among COVID-19 patients and among healthy controls with the most significant ones being SNHG6 and SNHG16 (r = 0.74, P value = 3.26e-17 and r = 0.81, P = 1.54e-22, respectively) (Figs. 3 and 4).
Fig. 3.
Correlation between expressions of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 in COVID-19 patients
Fig. 4.
Correlation between expressions of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 in controls
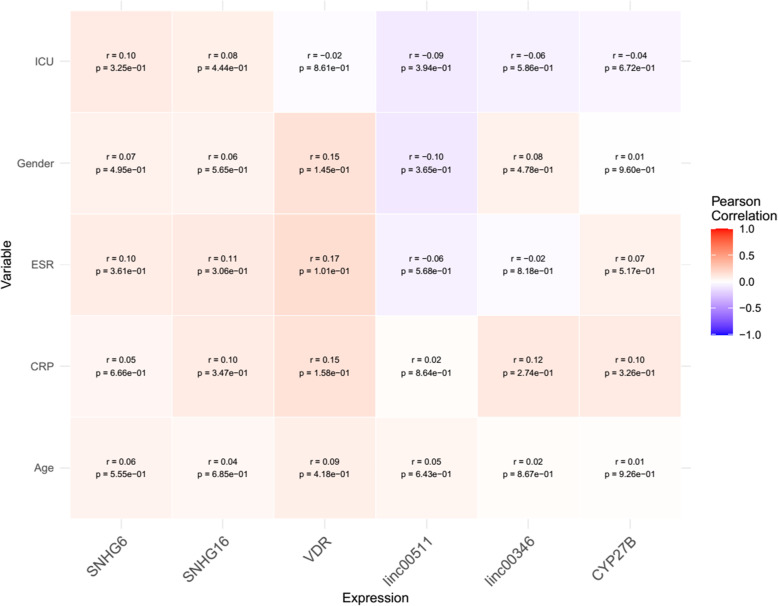
Next, we appraised correlation between expression levels of mentioned genes and patients’ gender and age as well as some paraclinical parameters demonstrating no robust correlations (Fig. 5).
Fig. 5.
Association between expression levels of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 and clinical/demographic data
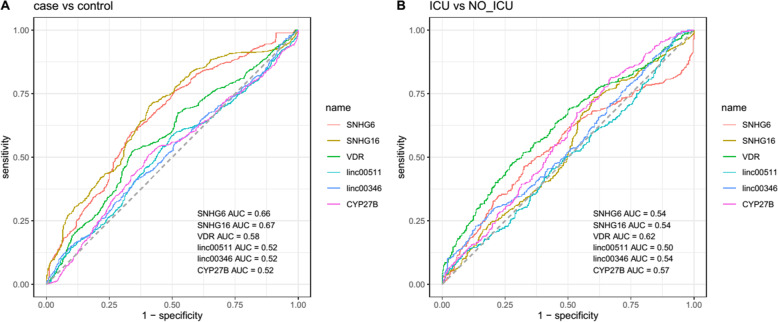
Finally, the diagnostic power of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 has been appraised in distinguishing COVID-19 patients from healthy controls and in distinguishing ICU-hospitalized patients from the other group of patients (Fig. 6).
Fig. 6.
ROC curves demonstrating the diagnostic power of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 in distinguishing COVID-19 patients from healthy controls (A) and in distinguishing ICU-hospitalized patients from the other group of patients (B)
While none of the individual genes could appropriately distinguish COVID-19 patients from healthy controls or ICU-hospitalized patients from non-ICU-hospitalized one, combination of transcript levels of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 could differentiate patients from controls with AUC = 0.76, sensitivity = 0.62 and specificity = 0.81. However, this type of analysis revealed no suitable diagnostic power for distinguishing ICU-hospitalized patients from non-ICU-hospitalized one (Table 3).
Table 3.
Detailed parameters of ROC curve assessment for appraisal of the diagnostic power of genes in distinguishing COVID-19 patients from healthy controls and in distinguishing ICU-hospitalized patients from the non-ICU patients (Se: sensitivity, Sp: specifciicty)
| SNHG6 | SNHG16 | VDR | Linc00511 | Linc00346 | CYP27B | All | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of Samples | AUC | Se | Sp | AUC | Se | Sp | AUC | Se | Sp | AUC | Se | Sp | AUC | Se | Sp | AUC | Se | Sp | AUC | Se | Sp | |
| Total patients/ controls | ||||||||||||||||||||||
| Total | 91/91 | 0.66 | 0.60 | 0.66 | 0.67 | 0.70 | 0.59 | 0.58 | 0.52 | 0.66 | 0.52 | 0.58 | 0.50 | 0.52 | 0.41 | 0.65 | 0.52 | 0.51 | 0.59 | 0.76 | 0.62 | 0.81 |
| ICU/Non_ICU | ||||||||||||||||||||||
| Total | 37/54 | 0.54 | 0.46 | 0.67 | 0.54 | 0.74 | 0.40 | 0.62 | 0.48 | 0.72 | 0.50 | 0.97 | 0.08 | 0.54 | 0.23 | 0.87 | 0.57 | 0.81 | 0.33 | 0.53 | 0.25 | 0.84 |
Discussion
Vitamin D deficiency and defects in activation of VDR have been found to intensify respiratory syndrome in COVID-19 through inducing a wounding response in stellate cells of the respiratory system [17]. Moreover, vitamin D levels have been found to be associated with the number of COVID-19 cases/million as well as number of deaths from COVID-19/ million [18]. Based on the presence of extensive evidence regarding the role of VDR signaling in the pathogenic course of COVID-19 and its devastating complication i.e. ARDS, we have appraised expression levels of VDR, CYP27B1 and some VDR-associated lncRNAs in the peripheral blood of COVID-19 patients versus healthy subjects. We reported lower expressions of SNHG6 and SNHG16 in COVID-19 patients of both sexes compared with sex-matched controls. In addition, expression of VDR was lower in COVID-19 patients compared with controls and in male patients compared with male controls. Conversely, expression of VDR was statistically similar between female patients and normal females. Expression of none of genes was different between ICU-hospitalized and non-ICU hospitalized patients. SNHG6 has been shown to activate TGF-β/Smad signaling pathway [19]. The interaction between coronaviruses and this pathway is complicated. For instance, SARS-associated coronavirus (SARS-CoV) nucleocapsid protein has been shown to enhance TGF-β-associated expression of PAI-1 but decreasing Smad3/Smad4-asociated apoptosis of lung epithelial cells [20]. On the other hand, SARS-CoV-2 infection has been shown to increase TGF-β production [21]. Moreover, in severe COVID-19, SARS-CoV-2 has been found to induce TGF-β-dominated chronic immune responses that do not target SARS-CoV-2 antigens [22]. Several viruses alter TGF-β signaling to inhibit cell apoptosis and enhance proliferation of fibroblasts and differentiation of myofibroblasts [23]. Over-production of TGF- β has been reported in acute-phase SARS and COVID-19 in relation with the development of lung fibrosis. Moreover, this phenomenon has been detected in autopsy samples and a considerable percentage of SARS, MERS and COVID-19 survivors [17]. Therefore, up-regulation of TGF-β in COVID-19 patients might contribute in the lung fibrosis [21]. The observed down-regulation of SNHG6 in the peripheral blood of COVID-19 patients might be due to a regulatory feedback loop between this lncRNA and TGF-β.
SNHG16 has been shown to activate TGF-β1/SMAD5 pathway via miR-16–5p/SMAD5-regulatory cascade, therefore activating CD73 expression in γδ1 T cells [24]. Down-regulation of SNHG16 in COVID-19 patients might result in decreased proportion of γδ1 T cells, thus activation of pro-inflammatory cascades and pulmonary fibrosis, since some subsets of γδ T cells regulate immunosuppressive functions and induce immune tolerance in certain contexts [25].
It is not clear whether the observed decreased level of VDR in COVID-19 is the cause or effects of COVID-19. However, previous studies have indicated that certain polymorphisms in the VDR gene might have adverse impact on the outcome of patients with COVID-19 [26, 27]. Therefore, we suggest simultaneous assessment of functional polymorphisms within this gene and its expression in larger populations of COVID-19 patients to distinguish the possible correlations. Moreover, VDR has been found to transcriptionally silence TGF-β signaling via genomic competition with Smad3 recruitment on pro-fibrotic and pro-inflammatory genes [28]. Therefore, the observed down-regulation of VDR in COVID-19 cases might result in up-regulation of TGF-β signaling and related pathologic events.
Expression levels CYP27B, Linc00511 and Linc00346 were not different between COVID-19 patients and healthy subjects or between their subgroups, implying their independence from COVID-19 infection or disease course.
Significant correlations have been displayed between expression levels of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 lncRNAs both among COVID-19 patients and among normal controls with the most significant ones being SNHG6 and SNHG16. These two lncRNAs have also been shown to correlate with each other in other disease contexts [13], implying their functional interactions in different situations. Expression of these genes were not correlated with paraclinical data indicating the independence of these transcripts from these parameters particularly inflammation markers.
Consistent with the similar levels of expressions of genes between ICU-hospitalized patients and other patients, these genes could not distinguish these two subgroups of patients. However, combination of transcript levels of VDR, CYP27B and SNHG6, SNHG16, Linc00511 and Linc00346 could differentiate patients from controls with moderate power.
Taken together, the current data potentiate SNHG6, SNHG16 and VDR as possible contributors in the COVID-19 infection but not in the severity of ARDS. Future studies should attempt to uncover the fundamental mechanism of this possible contribution and unravel the molecular mediators in this pathway. However, we do not have the data of 25(OH) D levels of patients and controls. We state this point as a limitation of our study.
Acknowledgements
Not applicable.
Abbreviations
- COVID-19
Coronavirus disease 2019
- ARDS
Acute Respiratory Distress syndrome
- VDR
Vitamin D receptor
- lncRNAs
long non-coding RNAs
- CRP
C-reactive protein
- SARS-CoV
SARS-associated coronavirus
Authors’ contributions
SGF and MT wrote the manuscript and contributed in study design. LMR, BMH and FN collected the data and confirmed the patient’s diagnosis. AS analyzed the data. All authors approved the manuscript.
Funding
Not applicable.
Availability of data and materials
The analysed data sets generated during the study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participant
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent forms were obtained from all study participants. Informed consent forms were obtained from all study participants. The study protocol was approved by the ethical committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.RETECH.REC.1399.592). All methods were performed in accordance with the relevant guidelines and regulations.
Consent of publication
Not applicable.
Competing interests
The authors declare they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Arezou Sayad, Email: ar.sayad@yahoo.com.
Soudeh Ghafouri-Fard, Email: s.ghafourifard@sbmu.ac.ir.
References
- 1.Ghafouri-Fard S, Noroozi R, Vafaee R, Branicki W, Poṡpiech E, Pyrc K, et al. Effects of host genetic variations on response to, susceptibility and severity of respiratory infections. Biomed Pharmacother. 2020;128:110296. doi: 10.1016/j.biopha.2020.110296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim J, Choi SM, Lee J, Park YS, Lee CH, Yim JJ, Yoo CG, Kim YW, Han SK, Lee SM. Effect of renin-angiotensin system blockage in patients with acute respiratory distress syndrome: a retrospective case control study. Korean J Crit Care Med. 2017;32(2):154–163. doi: 10.4266/kjccm.2016.00976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gouda MM, Shaikh SB, Bhandary YP. Inflammatory and fibrinolytic system in acute respiratory distress syndrome. Lung. 2018;196(5):609–616. doi: 10.1007/s00408-018-0150-6. [DOI] [PubMed] [Google Scholar]
- 6.Quesada-Gomez JM, Entrenas-Castillo M, Bouillon R. Vitamin D receptor stimulation to reduce acute respiratory distress syndrome (ARDS) in patients with coronavirus SARS-CoV-2 infections: Revised Ms SBMB 2020_166. J Steroid Biochem Mol Biol. 2020;202:105719. doi: 10.1016/j.jsbmb.2020.105719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouillon R, Marcocci C, Carmeliet G, Bikle D, White JH, Dawson-Hughes B, Lips P, Munns CF, Lazaretti-Castro M, Giustina A, Bilezikian J. Skeletal and Extraskeletal actions of vitamin D: current evidence and outstanding questions. Endocr Rev. 2019;40(4):1109–1151. doi: 10.1210/er.2018-00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martens PJ, Gysemans C, Verstuyf A, Mathieu C. Vitamin D’s effect on immune function. Nutrients. 2020;12(5):1248. [DOI] [PMC free article] [PubMed]
- 9.Bikle DD, Patzek S, Wang Y. Physiologic and pathophysiologic roles of extra renal CYP27b1: case report and review. Bone Rep. 2018;8:255–267. doi: 10.1016/j.bonr.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilezikian JP, Bikle D, Hewison M, Lazaretti-Castro M, Formenti AM, Gupta A, et al. Mechanisms in endocrinology: vitamin D and COVID-19. Eur J Endocrinol. 2020;183(5):R133–RR47. doi: 10.1530/EJE-20-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noroozi R, Branicki W, Pyrc K, Łabaj PP, Pospiech E, Taheri M, Ghafouri-Fard S. Altered cytokine levels and immune responses in patients with SARS-CoV-2 infection and related conditions. Cytokine. 2020;133:155143. doi: 10.1016/j.cyto.2020.155143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang YJ, Bikle DD. LncRNA: a new player in 1α, 25(OH)(2) vitamin D(3) /VDR protection against skin cancer formation. Exp Dermatol. 2014;23(3):147–150. doi: 10.1111/exd.12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazdeh M, Zamani M, Eftekharian MM, Komaki A, Arsang-Jang S, Taheri M, Ghafouri-Fard S. Expression analysis of vitamin D receptor-associated lncRNAs in epileptic patients. Metab Brain Dis. 2019;34(5):1457–1465. doi: 10.1007/s11011-019-00446-9. [DOI] [PubMed] [Google Scholar]
- 14.Gheliji T, Oskooei VK, Hafez AA, Taheri M, Ghafouri-Fard S. Evaluation of expression of vitamin D receptor related lncRNAs in lung cancer. Noncoding RNA Res. 2020;5(3):83–7. [DOI] [PMC free article] [PubMed]
- 15.Kholghi Oskooei V, Geranpayeh L, Omrani MD, Ghafouri-Fard S. Assessment of functional variants and expression of long noncoding RNAs in vitamin D receptor signaling in breast cancer. Cancer Manag Res. 2018;10:3451–3462. doi: 10.2147/CMAR.S174244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Son K-B, Lee T-J, Hwang S-S. Disease severity classification and COVID-19 outcomes, Republic of Korea. Bull World Health Organ. 2021;99(1):62–66. doi: 10.2471/BLT.20.257758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans RM, Lippman SM. Shining light on the COVID-19 pandemic: a vitamin D receptor checkpoint in defense of unregulated wound healing. Cell Metab. 2020;32(5):704–709. doi: 10.1016/j.cmet.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yadav D, Birdi A, Tomo S, Charan J, Bhardwaj P, Sharma P. Association of vitamin D status with COVID-19 infection and mortality in the Asia Pacific region: a cross-sectional study. Indian J Clin Biochem. 2021:1–6. [DOI] [PMC free article] [PubMed]
- 19.Zhu M, Wang X, Gu Y, Wang F, Li L, Qiu X. MEG3 overexpression inhibits the tumorigenesis of breast cancer by downregulating miR-21 through the PI3K/Akt pathway. Arch Biochem Biophys. 2019;661:22–30. doi: 10.1016/j.abb.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Zhao X, Nicholls JM, Chen Y-G. Severe acute respiratory syndrome-associated coronavirus nucleocapsid protein interacts with Smad3 and modulates transforming growth factor-β signaling. J Biol Chem. 2008;283(6):3272–3280. doi: 10.1074/jbc.M708033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, Guo D, Hu W, Yang J, Tang Z, Wu H, Lin Y, Zhang M, Zhang Q, Shi M, Liu Y, Zhou Y, Lan K, Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira-Gomes M, Kruglov A, Durek P, Heinrich F, Tizian C, Heinz GA, et al. SARS-CoV-2 in severe COVID-19 induces a TGF-β-dominated chronic immune response that does not target itself. Nat Commun. 2021;12(1):1–14. doi: 10.1038/s41467-021-22210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuyama T, Kubli SP, Yoshinaga SK, Pfeffer K, Mak TW. An aberrant STAT pathway is central to COVID-19. Cell Death Differ. 2020:1–17. [DOI] [PMC free article] [PubMed]
- 24.Ni C, Fang Q-Q, Chen W-Z, Jiang J-X, Jiang Z, Ye J, et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+ γδ1 Treg cells. Signal Transduct Target Ther. 2020;5(1):1–14. doi: 10.1038/s41392-020-0129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul S. Shilpi, Lal G. role of gamma-delta (γδ) T cells in autoimmunity. J Leukoc Biol. 2015;97(2):259–271. doi: 10.1189/jlb.3RU0914-443R. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell F. Vitamin-D and COVID-19: do deficient risk a poorer outcome? Lancet Diabetes Endocrinol. 2020;8(7):570. [DOI] [PMC free article] [PubMed]
- 27.Mohan M, Cherian JJ, Sharma A. Exploring links between vitamin D deficiency and COVID-19. PLoS Pathog. 2020;16(9):e1008874-e. doi: 10.1371/journal.ppat.1008874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding N, Ruth TY, Subramaniam N, Sherman MH, Wilson C, Rao R, et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell. 2013;153(3):601–613. doi: 10.1016/j.cell.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analysed data sets generated during the study are available from the corresponding author on reasonable request.