Abstract
Wenzhou mammarenavirus (WENV) is a zoonotic pathogen newly discovered in east and southeast Asia. WENV has been found in wild rodent animals around the world while its standing is barely understood in Guangzhou city, where is known as a region of outbreak hotspot for zoonotic emerging infectious diseases. To investigate the prevalence and genomic characteristics of mammarenavirus in Guangzhou City, lung tissue samples from wild rodent species were collected from five districts of Guangzhou City in the year 2015 and 2016. The viral RNA was extracted and then subjected to mammarenavirus-specific PCR. The result revealed approximately 1.0% (3/306) nucleic acid positivity for lung tissue samples obtained from three rodent species: Mus musculus, Rattus flavipectus, and Rattus norvegicus. Viral metagenomic sequencing of three samples was then carried out and two full segment L and three full segment S sequences were obtained. Phylogenetics analysis indicated the sequences of the new mammarenavirus strain have 76.2% - 94.4% similarity to known WENV encoded genes, with the highest similarity to the WENV 9–24 strain. Population structure analysis grouped all known WENV into seven lineages, and this WENV Guangzhou strain was grouped with WENV 9–24 as well. Though the seroprevalence result was not available, our data provides the first nucleic acid evidence of circulating WENV in Guangzhou city, and it suggested WENV had a broader host tropism than previously known.
Keywords: Wenzhou mammarenavirus, Genomic characteristics, Wild rodent, Phylogenetic analysis, Population structure, Guangzhou city
1. Introduction
Wild rodent is known as one of the primary natural reservoirs for zoonotic pathogens. In the past decades, the number of incidences of emerging infectious diseases (EIDs) transmitted by rodents are arising around the globe, causing tremendous burdens on global public health and economic stability of societies, and unfortunately its upwards trending is likely to continue [1,2]. Unplanned urbanization and expanded human activities in the wild field have promoted the human-rodent interactions and consequentially amplified the opportunities for zoonotic transmission. At least 66 identified zoonotic pathogenic agents, including Mammarenavirus, Orthohantavirus, and Orthohepevirus were frequently identified in wild rodent species [[3], [4], [5]]. The genus Mammarenavirus is a group of predominantly rodent-borne viruses commonly infected people in Africa and Latin America [6,7], consisting of several infamous lethal zoonotic viruses. For instance, the Lassa virus from the Mammarenavirus genus causes proximal 500,000 incidences of infection and 5000 deaths each year [8,9]. With fears of being weaponized and used in bioterrorism, several Mammarenaviruses were even listed as category A bioterrorism agents [10]. Since new viral species discovered in the genus Mammarenavirus may present similar pathogenicity and lethality, additional concerns about improving surveillance and diagnosis of them are accordingly desired.
Five domestic viral species of Mammarenaviruses (Alxa Mammarenavirus, Chevrier Mammarenavirus, lymphocytic choriomeningitis virus, Ryukyu mammarenavirus, and Wenzhou mammarenavirus (WENV)) have been documented in China so far [5,[11], [12], [13], [14]]. Among them, WENV is the newest member firstly discovered in Wenzhou, a city in eastern China in 2015 [13]. WENV and its variants like Cardamones virus, Xingyi virus, and Haikou virus, have only been detected in East and Southeast Asia [13,[15], [16], [17], [18]]. Similar to other Mammarenavirus, the WENV genome also contains two segments (segment L and segment S) encoding 4 proteins: Nucleoprotein (NP), Glycoprotein (GP), Zinc finger matrix protein (Z), and RNA-dependent RNA polymerase (RdRp) [18]. With regard to host range, WENV has been previously identified in brown rats (Rattus norvegicus), Pacific rats (Rattus exulans), yellow-breasted rats (Rattus flavipectus), black rats (Rattus rattus), lesser rice-field rats (Rattus losea), white-bellied rats (Niviventer niviventer), and the Asian house shrew (Suncus murinus) [13,15,19,20]. Also, Blasdell et al. have reported Cardamones virus can lead to dengue-like or influenza-like symptoms in human [15]. Moreover, recent studies investigated the prevalence of WENV antibodies in 4.6% (29/636) of healthy adults in Beijing and Shandong province, and 2.8% (23/828) of patients with unknown fever in Yunnan, indicating WENV may cause human infection or diseases [21,22].
Guangzhou city is the provincial capital of province of Guangdong and one of the most populous cities with more than 15 million residents. Due to its subtropical climate and the extensive international and domestic human traffic, Guangzhou city historically had a high incidence of a variety of EIDs outbreaks, including the rodent-borne scrub typhus and hemorrhagic fever with renal syndrome [[23], [24], [25]]. Given the vulnerability for exposure to zoonotic infectious diseases of the city, local public health departments have implemented routine measurements to examine the rodent animals with pathogen detection and surveillance for monitoring and control of potential rodent-borne EIDs [26,27]. While little attention has been paid to Mammarenavirus carried by rodents in the city, although the emerge of Mammarenavirus can be detrimental to the public health and often deadly to people with direct exposure.
In our study, a total of 306 lung tissue samples from five wild rodent species in Guangzhou were collected. Molecular detection and Illumina sequencing were used to determine the nucleic acid presence and genomic characteristics of WENV. We envision the findings provide a meaningful baseline of the prevalence of WENV for wild rodents in Guangzhou and will shed light on new thinking for zoonotic disease surveillance and prevention.
2. Materials and methods
2.1. Study sites and sample collection
Ethical approval for this study was obtained from the Ethics Committee of the Guangzhou Center for Disease Control and Prevention (Approval number: GZCDC-ECAR-2015A0005). The rodents were captured and coordinated using live traps provided by Centers for Disease Control and Prevention (CDC) at Baiyun, Conghua, Huadu, Haizhu, and Zengcheng Districts in Guangzhou between April and November 2016. Rodent was assessed for species identification by experts in CDC and information on animal species, gender, mass, and sampling settings were recorded. To investigate the presence and genomic characteristics of WENV, the animals were euthanized, and lung tissue were collected and stored at −80 °C for further analysis.
2.2. PCR screening
The lower lobe of lung tissues (150–200 mg) from Bandicota indica (B. indica), Rattus flavipectus (R. flavipectus), Rattus norvegicus (R. norvegicus), Rattus rattus (R. rattus), and the total lung tissue from Mus musculus (M. musculus) were resuspended and lysed by addition of 1.0 ml Hank's balanced salt solution (HBSS) buffer followed by homogenization using a Tissuelyser LT (Qiagen, Germany) (two steel balls, 5 mm, 5 min, 50 Hz). Total viral genome DNA and RNA were extracted using the TGuide S32 Magnetic Viral DNA/RNA kit (Tiangen Biotech, Beijing, China). The viral RNA was reversed into cDNA by using FastKing one-step RT-PCR kit (Tiangen Biotech, Beijing, China), and the cDNA was used as a template DNA for PCR verification using primers LVL-3359D/G 5′-AGAATYAGTGAAAGGGARAGYAATTC-3′, LVL-3754A/D 5′-CACATCATTGGTCCCCATTTACTRTGATC-3′ [28]. The PCR products were ligated into the pGEM-T vector (Promega, USA) and transformed into DH5α competent cells (Tiangen Biotech, Beijing, China), and the positive clones were sequenced (Sangon Biotech, Shanghai, China).
2.3. Metagenome sequencing
The homogenates were also filtered through a 0.45-μm polyvinylidene difluoride filter (Millipore, Germany) to remove eukaryotic and bacterium-sized particles. The filtered samples were then centrifuged at 8000 ×g for 30 min at 4 °C, and digested in a cocktail of DNase and RNase enzymes to remove naked DNA and RNA, including Turbo DNase (ThermoFisher, USA), Universal nuclease (ThermoFisher, USA), and RNase One (Sigma, USA) at 37 °C for 1.5 h. The viral genomic nucleic acids were isolated using a QIAamp MinElute Virus Spin kit (QIAGEN, USA). The viral first-strand cDNA was synthesized using primer K-8 N (5′-GACCATCTAGCGACCTCCA CNNNNNNNN-3′) and the Superscript III system (ThermoFisher, USA). To synthesis the first-strand cDNA into dsDNA, the cDNA was incubated at 37 °C for 1 h in the presence of Klenow fragment (TAKARA, China) and amplified by primer K (5′-GACCATCTAGCGACCTCCAC-3′) and Phusion High-Fidelity PCR Master Mix (ThermoFisher, USA). The PCR products were purified with a MinElute Gel Extraction kit (QIAGEN, USA) to obtain a DNA fragment mixture with a target fragment size of >150 bp. About 5 μg samples were prepared and sent to BGI Co., Ltd. (Wuhan, China) for NGS sequencing. Amplified viral nucleic acid libraries were analyzed using an Illumina HiSeq X-ten sequencer (Illumina, USA) for a single read of 150 bp in length. All FASTQ files were assessed using FastQC to assess overall quality [29]. The clean reads were mapped to WENV segments S (GenBank NC_026018.1) and L (GenBank NC_026019.1) using the medium sensitivity/Fast mode and iterate up to five times using Geneious prime 2020.0.3 software (https://www.geneious.com/) [30]. The mapped sequences were used to generate consensus sequences to obtain the primary genome sequences, and then the sequences were manually checked individually.
2.4. Phylogenetic analyses
The sequences were aligned with representative sequences of other Mammarenavirus (Table S1). Multiple sequence alignment was performed using MAFFT version 7 (https://mafft.cbrc.jp/alignment/server/) [31]. The alignment was manually checked and end-trimmed to match to the newly obtained RdRp, Z, GPC, and NP gene sequences. The final multiple sequence alignment was used for maximum likelihood (ML) phylogenetic analysis with GTR + G + I (RdRp gene, segment L), TN93 + G (GPC gene), TN93 + G (NP gene), K2 + G + I (Z gene) and T92 + G + I (segment S) as the best-fit model of nucleotide substitution and 1000 bootstrap resampling by using Mega X [32].
2.5. Population structure
We analyzed the WENV population structure using the program STRUCTURE (version 2.3.4), which applied a Bayesian statistical model to cluster genotypes into populations without prior information about their genetic relatedness, in which the whole population is divided into K subpopulations characterized by a set of allele frequencies at each locus [33]. To run STRUCTURE, map distances were set equal to the PI site physical distances. The optimal number of populations was determined by running the model for K values from 1 to 12. For each K, ten runs were performed with MCMC run lengths of 50,000 and 20,000 burn-in. Evanno's method [34] the trend of (LnPr(X|K), and STRUCTURE documentation [35] were used to select the optimal K with STRUCTURE HARVESTER [36]. The results of independent runs were merged by permutating clusters using CLUMPP [40] to generate a Q-value matrix. To evaluate the contribution of the ancestral component to each PI site, a run with the optimal K was performed with the SITEBYSITE option selected.
3. Results
3.1. Study populations
A total of 306 wild rodent animals were lively trapped for sample collection, including 16 B. indica, 14 M. musculus, 62 R. flavipectus, 195 R. norvegicus, and 19 R. rattus, with average weight of 250.0, 28.8, 172.4, 241.6 and 288.7 g respectively. The rodent animals were collected from five districts (Baiyun, Conghua, Huadu, Haizhu, and Zengcheng districts) in Guangzhou City. Distribution and characteristics of the sampling population are presented in Table 1.
Table 1.
Characteristics of the study population.
| Varrible | Sample size (N) | % Male | Average mass (g) | Positive (n) | Positive (%) |
|---|---|---|---|---|---|
| Species | |||||
| B. indica | 16 | 25.0 | 250.0 | 0 | 0 |
| M. musculus | 14 | 35.7 | 28.8 | 1 | 7.1 |
| R. flavipectus | 62 | 35.5 | 172.4 | 1 | 1.6 |
| R. norvegicus | 195 | 43.1 | 241.6 | 1 | 0.5 |
| R. rattus | 19 | 36.8 | 288.7 | 0 | 0 |
| District | |||||
| Baiyun | 15 | 73.3 | 184.7 | 0 | 0 |
| Conghua | 79 | 35.4 | 216.5 | 3 | 3.8 |
| Haizhu | 101 | 32.7 | 287.8 | 0 | 0 |
| Huadu | 97 | 39.2 | 186.6 | 0 | 0 |
| Zengcheng | 14 | 28.6 | 129.0 | 0 | 0 |
| Location | |||||
| Residential area | 167 | 34.7 | 221.9 | 0 | 0 |
| Farmers' market | 115 | 40.0 | 230.9 | 3 | 2.6 |
| Wild field | 24 | 41. 7 | 271.1 | 0 | 0 |
| Classification | |||||
| Urban | 163 | 38.7 | 254.8 | 1 | 0.6 |
| Rural | 143 | 41.3 | 193.1 | 2 | 1.4 |
| Total | 306 | 39.9 | 221.2 | 3 | 1.0 |
Abbreviation: BY, Baiyun; CH, Conghua; HD, Huadu; HZ, Haizhu; ZC, Zengcheng.
3.2. PCR screening
The mammarenavirus-specific PCR primers were used for screening a total of 306 lung samples for the presence of mammarenavirus RNA [28]. The sequence fragment was successfully amplified and strong bands was shown in the gel for three samples (Fig. 1). As indicated, around 1.0% (3 out of 306) animal samples showed mammarenavirus nucleic acid positivity, including sample CH27 from M. musculus (7.1%), sample CH50 from R. flavipectus (1.6%), and sample CH31 from R. norvegicus (0.5%) (Table 2). Interestingly, these positive rodents were all from the Conghua district. CH27 and CH31 were collected from rural areas, while CH50 was from urban area (Table S2).
Fig. 1.

PCR detection of mammarenavirus. M: DNA Marker; 1. Positive plasmid control (containing partial RDRP gene); 2, Sample no. ch27; 3, Sample no. ch31; 4, sample no. ch50; 5, sample no. ch01 (negative sample); 6, Blank control.
Table 2.
PCR detection of WENV in wild rodents.
| Species | Common name | N | % (n/N) | Place type of positive rodents |
|---|---|---|---|---|
| B. indica | Greater bandicoot rat | 16 | 0 (0/16) | – |
| M. musculus | House mouse | 14 | 7.1 (1/14) | Farmer's market |
| R. flavipectus | Yellow-breasted rat | 62 | 1.6 (1/62) | Farmer's market |
| R. norvegicus | Brown rat | 195 | 0.5 (1/195) | Farmer's market |
| R. rattus | Roof rat | 19 | 0 (0/19) | – |
| Total | 306 | 1.0 (3/306) | – |
3.3. Species identification
After sequencing, three 397 bp long sequence fragments were obtained from the PCR product of the above three samples, which shared around 98% similarity. The sequences and its counterpart of WENV isolate Rn-242 (GenBank: NC_026019.1), which was isolated from Zhejiang Province, had a sequence similarity of 90.7 (360/397) - 91.7% (364/397). Gene sequences of several closely related mammarenavirus species were downloaded from the GenBank database. Phylogenetic analysis was performed to align these three sequences with other reference sequences, as shown in Fig. 2. The result showed that Guangzhou strains CH27, CH31, and CH50 were grouped into the WENV branch based on the phylogenetic tree.
Fig. 2.
Species identification based on phylogenetic analysis of PCR amplified products (partial RDRP gene). OW, Old-world arenaviruses; NW, New-world arenaviruses.
3.4. Sequence comparison of WENV Guangzhou strains with other Chinese strains
The three samples (CH27, CH31, and CH50) with PCR positivity were then subjected to metagenomics sequencing. Two segment L and three segment S sequences were assembled (GenBank accessions: MZ272057 - MZ272061, Table S2). To characterize the new strains and to determine the relationship of these strains with other Chinese strains, we next compared the nucleotide and amino acid sequences of RdRp, Z, NP, and GPC open reading frames (ORFs) from the three Guangzhou strains to those of previously characterized members of the mammarenavirus genus identified in China (Table 3). The sequence analysis revealed a close relationship of the Guangzhou strains, with 95.2% - 98.7% in GPC ORF, 96.0% - 99.1% in NP ORF, 98.5% - 98.8% in Z ORF, and 99.1% in RdRp ORF. The Guangzhou strains had the highest similarity to WENV strain 9–24 (GenBank accession: MF414207.1 and MF414208.1), with RdRp, Z, NP, and GPC ORFs sequence identities with 91.6% - 92.0%, 94.1% -94.4%, 93.0% - 93.7%, and 91.8% - 91.9%, respectively (Table 3). According to the species demarcation criteria approved by the International Taxonomy Committee on Viruses (ICTV), the Guangzhou strains CH27, CH31, and CH50 identified in our study belongs to the species of Wenzhou mammarenavirus.
Table 3.
Sequence comparison of WENV Guangzhou strains with other Chinese mammarenavirus strains.
| Strains/place | nt | WENV CH27 |
WENV CH31 |
WENV CH50 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Z ORF | NP ORF | GPC | RdRp | Z ORF | NP ORF | GPC | RdRp | Z ORF | NP ORF | GPC | ||
| WENV/Hainan | nt | 92.0 | 89.4–89.7 | 87.9–88.8 | 90.3 | 91.2 | 89.3–89.4 | 87.7–88.6 | 90.0 | 91.4 | 88.8–89.2 | 87.9–88.8 |
| WENV/Guizhou | nt | 84.7 | 85.0 | 85.6–85.7 | 82.2 | 85.0 | 84.6 | 85.1–85.2 | 82.0 | 84.7 | 85.1 | 85.1 |
| WENV/Strain 9–24a | nt | 94.4 | 93.3 | 91.8 | 92.0 | 94.1 | 93.0 | 91.8 | 91.6 | 94.4 | 93.7 | 91.9 |
| WENV/Shandong | nt | 90.6 | 89.5 | 88.5 | 88.8 | 90.3 | 89.3 | 88.3 | 88.5 | 90.0 | 89.8 | 88.0 |
| WENV /Xinjiang | nt | 88.2 | 86.3 | 86.0 | 86.4 | 87.9 | 85.9 | 86.0 | 86.2 | 87.6 | 86.8 | 85.3 |
| WENV/Yunnan | nt | 87.0–89.4 | 77.0–87.5 | 85.8–87.1 | 86.1–86.4 | 87.3–90.3 | 77.0–87.0 | 85.7–87.1 | 86.0–86.2 | 86.4–90.0 | 76.3–87.5 | 85.4–86.3 |
| WENV/Zhejianga | nt | 86.1–92.9 | 84.1–89.5 | 86.4–89.5 | 89.8–90.5 | 85.3–92.6 | 83.8–89.4 | 86.2–89.4 | 89.5–90.2 | 85.5–92.3 | 83.9–89.5 | 86.6–89.6 |
| WENV/Cambodia | nt | 90.0–90.3 | 88.1–89.4 | 83.0–86.6 | 88.8–88.9 | 90.3–90.6 | 88.7–89.7 | 83.0–86.6 | 88.5–88.6 | 89.4–89.7 | 89.0 | 86.6 |
| WENV/Malaysia | nt | 89.4 | 88.6 | 86.7 | 88.3 | 89.7 | 88.4 | 86.7 | 88.0 | 88.8 | 88.8 | 86.6 |
| ALXV/Inner Mogonlia | nt | 62.2 | 59.1 | 57.7 | 56.4 | 62.2 | 58.7 | 57.8 | 56.2 | 61.9 | 59.4 | 58.0 |
| LCMV/Jilin | nt | 60.5–61.1 | 62.9–63.1 | 55.3–57.0 | 56.1 | 60.2–60.8 | 62.5–62.7 | 55.6–57.3 | 56.0 | 59.9–60.5 | 62.9–63.4 | 56.0–57.4 |
| LIJV/Yunan | nt | 65.8 | 74.8 | 70.9 | 66.2 | 65.8 | 74.5 | 70.9 | 66.3 | 65.8 | 74.5 | 70.9 |
Note: WENV indicates Wenzhou virus; ALXV indicates Alxa virus; LCMV indicates Lymphocytic choriomeningitis virus; LIJV indicates Lijiang virus, belonging to Chevrier mammarenavirus.
Strain from Yunnan or Guizhou province.
3.5. Phylogenetic analysis
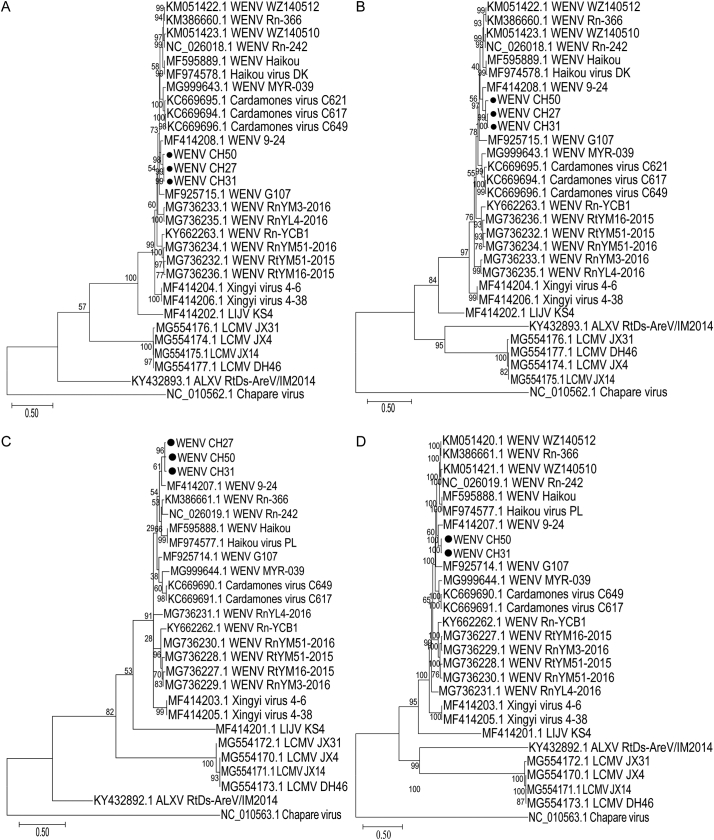
To analysis the inter-species evolution, phylogenetic analysis of four encoded WENV genes (NP, GPC, Z, and RdRp) identified in the rodents obtained in Guangzhou showed that these strains exhibited a close relationship to WENV isolate 9–24 (GenBank accessions: MF414207.1 and MF414208.1), forming a distinct lineage in accordance with their geographical distribution as well (Fig. 3A, B, C and D).
Fig. 3.
Phylogenetic trees based on four encoded gene. Phylogenies of the NP (A), GPC (B), Z (C) and RDRP (D) Genes were constructed using mega X and the reference chapare virus sequence (L: NC_010562.1; S: NC_010562.1) as the outgroup.
3.6. Population structure and lineages Classification
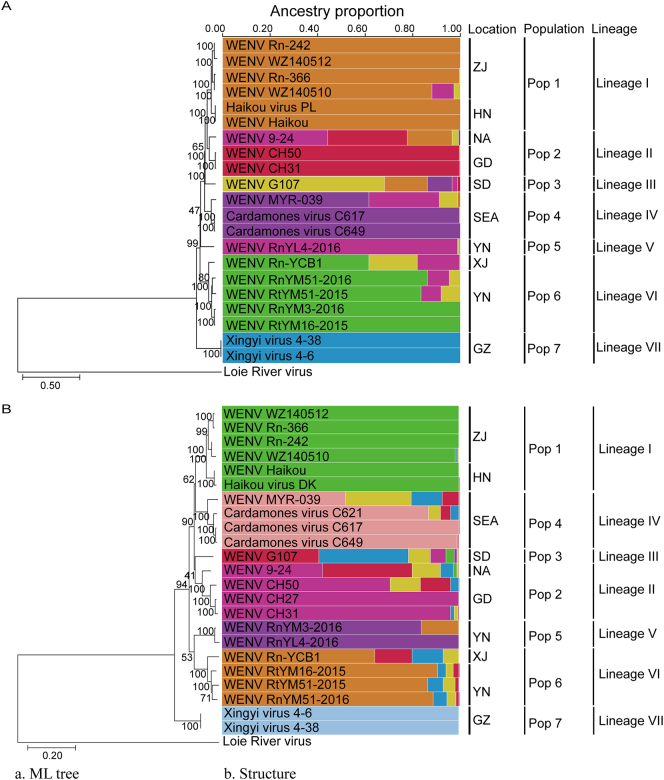
To estimate the optimal number of subpopulations in the WENV dataset, STRUCTURE was run for values of K from 1 to 12. The results showed that the new segment L yielded a major peak at K = 7 and the new segment S yielded a major peak at K = 9 (Fig. S1), indicating the optimal subpopulation number was 7 (segment L) and 9 (segment S).
Analysis of ancestry components was next performed for K = 7 (segment L) and K = 9 (segment S), and genomes were plotted according to their geographical origin (Fig. 4A, B). The results showed the clustering trend of ancestral population was highly site-specific (e.g., pop 1 occurs in ZJ and HN, pop2 occurs in NA and GD). The pop 2 for Guangzhou strains and WENV 9–24 included WENV genomes sampled in different provinces of Zhejiang, Hainan, Shandong, Yunnan isolates, indicating this population contributed in varying proportion to genomes from other areas.
Fig. 4.
Population structure of wenzhou virus (WENV). (A) Bar plot representing the proportion of ancestral population components from the structure linkage model for K = 7 based on segment L. each column represents a WENV genome, the virus name is marked in the column. Genomes are ordered by population. (B) Bar plot representing the proportion of ancestral population components from the structure linkage model for K = 9 based on segment S. Lineages classification based on phylogenetic trees. Phylogenies of the segment L (A) and S (B) were constructed using mega X and the reference loie river virus sequence. The detailed information of the virus used in this figure was in Supplementary Table 1. GD, Guangdong Province; GZ, Guizhou Province; HN, Hainan Province; SD, Shandong Province; XJ, Xinjiang Uygur autonomous region; YN, Yunnan Province; ZJ, Zhejiang Province; SEA, South-Eastern Asia, including countries Cambodia and Malaysia; NA, information not available, but the region is either Guizhou or Yunnan Province.
According to the major ancestry components inferred by STRUCTURE, the WENV sequences could be clustered into seven lineages (Fig. 4A, B): Lineage I included WENV Rn-242, WZ140510, Rn-366, WZ140512, Haikou virus PL, Haikou virus DK, and Haikou; Lineage II included WENV 9–24, CH27, CH31, and CH50; Lineage III included WENV G107. Lineage IV included WENV MYR-039 and Cardamones virus C617, C621, and C649; Lineage V included WENV RnYL4-2016; Lineage VI included WENV Rn-YCB1, RnYM51-2016, RtYM51-2015, and RtYM16-2015; Lineage VII included Xingyi virus 4–6 and 4–38. As indicated, Guangzhou strains and WENV 9–24 were clustered into the same lineage II.
4. Discussion
WENV was first discovered in Zhejiang province of eastern China in 2015 [13]. Ever since then, WENV was detected or isolated in five other Chinese provinces, including Guizhou, Hainan, Shandong, Xinjiang, and Yunnan, indicating its wide geographical distribution in China [13,16,[18], [19], [20]]. However, there is limited genomic evidence of WENV in Guangdong province, especially in the capital city of Guangzhou. In present study, we investigated WENV infections in wild rodents in the capital city of Guangdong Province, Southern China. Our results demonstrated that the nucleic acid presence of WENV in three of the five rodent species and in one of the five districts of Guangzhou city. For the first time, M. musculus was identified as a new host for WENV in this study, which expands the host range of WENV. We found that WENV detected in approximately 1.0% of samples but all came from the Conghua district, suggesting the distribution of WENV may currently be limited to a small area and rodent population. Thus, our data rings the bell to local CDC for WENV routine surveillance in wild rodents, as well as other natural reservoirs.
Sequence analysis of partial RdRp gene of Guangzhou strains CH27, CH29, and CH50 showed that these viruses are closely related to WENVs, indicating a common ancestor. The phylogenetic tree had a different shape from the tree based on all four genes, e.g., NP, GPC, Z and RdRp, suggesting more complex evolution of WENV. By estimating the drift level from a hypothetical common population, the applied STRUCTURE model allows inference about the most likely original location of the sampled genomes. Overall, our data indicated that WENV lineages diverged from an ancestral population circulating in Zhejiang/Hainan, Shandong, southeast Asia, Yunnan (within two ancestral populations), and Guizhou, indicating a regional distribution of WENV. The ancestral component of Segment L in the three Guangzhou strains was shared with strain 9–24 but was not detected in other strains. The ancestral lineage may be less common in rodent populations in other locations. Ancestral components of Segment S were detected in multiple groups, suggesting greater transfer of Segment S than that of Segment L.
Zoonotic diseases that originate from wild animals are a significant concern of One Health, a global strategy that pursues a comprehensive, multidisciplinary, and multisectoral approach to attain optimal health for humans, animals, and the environment [37,38]. Understanding the distribution of WENV and other viruses in wildlife is essential for the accurate prediction of the impact of emerging zoonoses, especially in wildlife species with broad human-animal interfaces. The rodents were trapped in residential areas, such as farmers markets and other areas close to human settlements. However, samples tested WENV-positive were all collected from two farmers' market, where is typically an area with high pedestrian flow and vehicular traffic. Current serological survey and clinical data indicate that WENV has the properties of causing human disease [15,21,22,39], which means there is a risk of WENV spreading to humans. Studying the source and route of WENV transmission from animal to the human population in Guangzhou or other cities is of key important to public health base on One Health strategies, indicating to further zoonotic surveillance on WENV. To implement the One Health initiative, future work should explore the infection risk of occupational workers in places with frequent rodent activities, such as retail and wholesale markets, animal farms, and other related settings. Human medical departments should work with veterinary departments to implement WENV routine surveillance in wild rodent and occupational populations to expand current rodent-borne disease surveillance efforts.
The present study has several limitations: 1) we only performed viral detection with lung samples, potentially providing the limited information for the prevalence of WENV in rodents in Guangzhou, China. 2) The efficiency of PCR amplification varied for different amplified fragments due to the bias of PCR amplification, resulting in different depth of amplified fragments. 3) It is also notable that we were unable to perform a serological screening for WENV infection, which is critical for diagnose in many circumstances along with nucleic acid testing as positive nucleic acid test result doesn't equal to an infection, and more importantly, serological screening can help assess the prevalence of past WENV infection in the rodent population.
5. Conclusion
This is the first study to investigate the genomic characterization of WENV among wild rodents in a super metropolitan city with over 15 million residents. WENV were detected in three rodent species (M. musculus, R. flavipectus and R. norvegicus) which was reported for the first time in Guangzhou City. The WENV Guangzhou strain was different from the previously reported strains isolated in China and southeast Asia, and only clustered with strain WENV 9–24. For future experiment, virus isolation is desired to characterize the pathogenic features and understand potential zoonotic characteristic in human infections.
The following are the supplementary data related to this article.
Supplementary Figure 1.
Figure S1. The optimal K value of Segment L and Segment S
Supplementary material 1
Supplementary material 2
Funding information
This work was supported by the National Key Research and Development Project (2018YFE0208000), the Key-Area Research and Development Program of Guangdong Province (2018B020241002), the National Science and Technology Major Project (2018ZX10101002-001-001), and the Guangdong Provincial Science and Technology Project (2018B020207013).
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
We are indebted to the staffs of Baiyun, Conghua, Huadu, Haizhu and Zengcheng District Centers of Diseases Control and Prevention for capturing the wild rodents in this study.
Contributor Information
Jian-Yong Wu, Email: chienyung@foxmail.com.
Cheng Guo, Email: cg2984@cumc.columbia.edu.
Yao Xia, Email: xiayao0125@outlook.com.
Hui-Min Bao, Email: bhm1022@163.com.
Yan-Shan Zhu, Email: zhuyansh@mail2.sysu.edu.cn.
Zhong-Min Guo, Email: guozhm@mail.sysu.edu.cn.
Yue-Hong Wei, Email: gzcdc_weiyh@gz.gov.cn.
Jia-Hai Lu, Email: lujiahai@mail.sysu.edu.cn.
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Shi Z.-L. SARS-CoV-2 spillover events. Science. 2021;371(6525):120–122. doi: 10.1126/science.abf6097. [DOI] [PubMed] [Google Scholar]
- 3.Han B.A., Schmidt J.P., Bowden S.E., Drake J.M. Rodent reservoirs of future zoonotic diseases. P. Natl. Acad. Sci. USA. 2015;112(22):7039–7044. doi: 10.1073/pnas.1501598112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sridhar S., Yip C.C., Wu S., Chew N.F., Leung K.H., Chan J.F., Zhao P.S., Chan W.M., Poon R.W., Tsoi H.W., Cai J.P., Chan H.S., Leung A.W., Tse C.W., Zee J.S., Tsang O.T., Cheng V.C., Lau S.K., Woo P.C., Tsang D.N., Yuen K.Y. Transmission of rat hepatitis E virus infection to humans in Hong Kong: a clinical and epidemiological analysis. Hepatology. 2020;73(1):10–22. doi: 10.1002/hep.31138. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L., Li S., Huang S.J., Wang Z.D., Wei F., Feng X.M., Jiang D.X., Liu Q. Isolation and genomic characterization of lymphocytic choriomeningitis virus in ticks from northeastern China. Transbound. Emerg. Dis. 2018;65(6):1733–1739. doi: 10.1111/tbed.12946. [DOI] [PubMed] [Google Scholar]
- 6.Fatima S.H., Zaidi F., Adnan M., Ali A., Jamal Q., Khisroon M. Rat-bites of an epidemic proportion in Peshawar vale; a GIS based approach in risk assessment. Environ. Monit. Assess. 2018;190(4):233. doi: 10.1007/s10661-018-6605-7. [DOI] [PubMed] [Google Scholar]
- 7.Kausrud K.L., Viljugrein H., Frigessi A., Begon M., Davis S., Leirs H., Dubyanskiy V., Stenseth N.C. Climatically driven synchrony of gerbil populations allows large-scale plague outbreaks. Proc. Biol. Sci. 2007;274(1621):1963–1969. doi: 10.1098/rspb.2007.0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez J.-P., Souris M., Valdivia-Granda W. Global spread of Hemorrhagic fever viruses: predicting pandemics. In: Salvato M.S., editor. Hemorrhagic Fever Viruses: Methods and Protocols. Springer New York; New York, NY: 2018. pp. 3–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ilori E.A., Frank C., Dan-Nwafor C.C., Ipadeola O., Krings A., Ukponu W., Womi-Eteng O.E., Adeyemo A., Mutbam S.K., Musa E.O., Lasuba C.L.P., Alemu W., Okogbenin S., Ogbaini E., Unigwe U., Ogah E., Onoh R., Abejegah C., Ayodeji O., Ihekweazu C. Increase in Lassa fever cases in Nigeria, January-March 2018. Emerg. Infect. Dis. 2019;25(5):1026–1027. doi: 10.3201/eid2505.181247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention (U.S.) Bioterrorism Agents/Diseases. 2018. https://emergency.cdc.gov/agent/agentlist-category.asp (Accessed May 26 2021)
- 11.Wu Z., Du J., Lu L., Yang L., Dong J., Sun L., Zhu Y., Liu Q., Jin Q. Detection of hantaviruses and Arenaviruzses in three-toed jerboas from the Inner Mongolia autonomous region, China. Emerg. Microb. Infect. 2018;7(1):35. doi: 10.1038/s41426-018-0036-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Committee on Taxonomy of Viruses Genus: Mammarenavirus. 2020. https://talk.ictvonline.org/ictv-reports/ictv_online_report/negative-sense-rna-viruses/w/arenaviridae/1117/genus-mammarenavirus (Accessed January 14 2021)
- 13.Li K., Lin X.-D., Wang W., Shi M., Guo W.-P., Zhang X.-H., Xing J.-G., He J.-R., Wang K., Li M.-H., Cao J.-H., Jiang M.-L., Holmes E.C., Zhang Y.-Z. Isolation and characterization of a novel arenavirus harbored by rodents and shrews in Zhejiang province, China. Virology. 2015;476:37–42. doi: 10.1016/j.virol.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 14.Wu Z., Lu L., Du J., Yang L., Ren X., Liu B., Jiang J., Yang J., Dong J., Sun L., Zhu Y., Li Y., Zheng D., Zhang C., Su H., Zheng Y., Zhou H., Zhu G., Li H., Chmura A., Yang F., Daszak P., Wang J., Liu Q., Jin Q. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome. 2018;6(1):178. doi: 10.1186/s40168-018-0554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blasdell K., Duong V., Eloit M., Chretien F., Ly S., Hul V., Deubel V., Morand S., Buchy P. Evidence of human infection by a new mammarenavirus endemic to Southeastern Asia. eLife. 2016;5 doi: 10.7554/eLife.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Rao L.Y., Lv G., Zhang L., Fu R.J., Wang S.S., Wu Y., Cui X.J., Yin F.F. Complete genome sequence of a Mammarenavirus Harbored by rodents on Hainan Island, China. Microbiol. Res. Announc. 2018;6(10) doi: 10.1128/genomeA.00129-18. e00129-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Z., Han Y., Liu B., Li H., Zhu G., Latinne A., Dong J., Sun L., Su H., Liu L., Du J., Zhou S., Chen M., Kritiyakan A., Jittapalapong S., Chaisiri K., Buchy P., Duong V., Yang J., Jiang J., Xu X., Zhou H., Yang F., Irwin D.M., Morand S., Daszak P., Wang J., Jin Q. Decoding the RNA viromes in rodent lungs provides new insight into the origin and evolutionary patterns of rodent-borne pathogens in mainland Southeast Asia. Microbiome. 2021;9(1):18. doi: 10.1186/s40168-020-00965-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu M.M., Li L.L., Wang X.F., Duan Z.J. Complete genome sequence of a novel variant of Wenzhou Mammarenavirus. Microbiol. Res. Announc. 2017;5(47):e01303–e01317. doi: 10.1128/genomeA.01303-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J.X., Yang X.L., Liu H.Z., Wang L., Zhou J.H., Han X., Zhu Y., Yang W.H., Pan H., Zhang Y.Z., Shi Z.L. Prevalence of Wenzhu virus in small mammals in Yunnan Province, China. PLoS. Neglect. Trop. D. 2019;13(2) doi: 10.1371/journal.pntd.0007049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan Z.Z., Yu H.J., Xu L., Zhao Z.H., Zhang P.S., Qu Y.G., He B., Tu C.C. Virome profiling of rodents in Xinjiang Uygur autonomous region, China: Isolation and characterization of a new strain of Wenzhou virus. Virology. 2019;529:122–134. doi: 10.1016/j.virol.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Guo L., Liu S., Song J., Han L., Zhang H., Wu C., Wang C., Zhou H., Wang J. Seroprevalence of Wenzhou virus in China. Biosafety Health. 2020;2(3):152–156. doi: 10.1016/j.bsheal.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Jiang M., Yang Y., Dong N., Liu J., Zhang W. Human cases of Wenzhou virus infection confirmed by pseudo-virus neutralization test in Yunnan province, China. Eur. J. Immunol. 2019;49:1060. [Google Scholar]
- 23.Wei Y., Wang Y., Li X., Qin P., Lu Y., Xu J., Chen S., Li M., Yang Z. Meteorological factors and risk of hemorrhagic fever with renal syndrome in Guangzhou, southern China, 2006-2015. PLoS. Neglect. Trop. D. 2018;12(6) doi: 10.1371/journal.pntd.0006604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peckham R. Routledge; Plague and the City: 2018. Epidemics, Photography, and the Ruined City; pp. 101–125. [Google Scholar]
- 25.Sun Y., Wei Y.H., Yang Y., Ma Y., de Vlas S.J., Yao H.W., Huang Y., Ma M.J., Liu K., Li X.N., Li X.L., Zhang W.H., Fang L.Q., Yang Z.C., Cao W.C. Rapid increase of scrub typhus incidence in Guangzhou, southern China, 2006-2014. BMC Infect. Dis. 2017;17(1):13. doi: 10.1186/s12879-016-2153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li T., Yang Z., Dong Z., Wang M. Meteorological factors and risk of scrub typhus in Guangzhou, southern China, 2006–2012. BMC Infect. Dis. 2014;14(1):139. doi: 10.1186/1471-2334-14-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Z., Yang Z., Guo R., Gao Y., Ren W., Xiaoning L., Li C. An analysis on surveillance of plague host animails in Guangzhou city from 2006 to 2007. Chin. J. Contr. Endem. Diseas. 2008;23:372–373. (In Chinese) [Google Scholar]
- 28.Leski T.A., Stockelman M.G., Moses L.M., Park M., Stenger D.A., Ansumana R., Bausch D.G., Lin B. Sequence variability and geographic distribution of Lassa virus, Sierra Leone. Emerg. Infect. Dis. 2015;21(4):609–618. doi: 10.3201/eid2104.141469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews S. FastQC. 2020. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed March 25 2018)
- 30.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Misawa K., Katoh K., K. K-I, M. T MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falush D., Stephens M., Pritchard J. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evanno G., Regnaut S., Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 2005;14(8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 35.Janes J.K., Miller J.M., Dupuis J.R., Malenfant R.M., Gorrell J.C., Cullingham C.I., Andrew R.L. The K = 2 conundrum. Mol. Ecol. 2017;26(14):3594–3602. doi: 10.1111/mec.14187. [DOI] [PubMed] [Google Scholar]
- 36.Earl D.A., von Holdt B.M. Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour. 2012;4(2):359–361. [Google Scholar]
- 37.Ribeiro C.D., van de Burgwal L.H.M., Regeer B.J. Overcoming challenges for designing and implementing the one health approach: a systematic review of the literature. One Health. 2019;7:100085. doi: 10.1016/j.onehlt.2019.100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohamed A. Bovine tuberculosis at the human-livestock-wildlife interface and its control through one health approach in the Ethiopian Somali pastoralists: a review. One Health. 2020;9:100113. doi: 10.1016/j.onehlt.2019.100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu S., Zhang H., Wu C., Guo L., Wang J. Seroprevalence of Wenzhou virus in China. Eur. J. Immunol. 2019;49:1036. doi: 10.1016/j.bsheal.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2