Abstract
Near the end of 2019, a new betacoronavirus started to efficiently transmit between humans, resulting in the current COVID-19 pandemic. Unprecedented worldwide efforts were made to identify and repurpose antiviral therapeutics from collections of approved drugs and known bioactive compounds. Typical pitfalls of this approach (promiscuous/cytotoxic compounds leading to false positives), combined with bypassing antiviral drug development parameters due to urgency have resulted in often disappointing outcomes. A flood of publications, press-releases, and media posts, created confusion in the general public and sometime mobilized precious resources for clinical trials with minimal prospect of success. Breakthroughs have been made, not in the laboratory but in the clinic, resulting from the empiric identification of mitigators of clinical signs such as the discovery of improved disease management through immunomodulators. This opinion piece will aim to capture some of the lessons that we believe the COVID-19 pandemic has taught about drug repurposing screens.
Current Opinion in Virology 2021, 49:183–193
This review comes from a themed issue on Engineering for viral resistance
Edited by Richard Plemper
For complete overview about the section, refer Engineering for viral resistance
Available online 19th June 2021
https://doi.org/10.1016/j.coviro.2021.06.004
1879-6257/Published by Elsevier B.V.
Introduction
In December 2019, a wave of viral pneumonia caused by a novel betacoronavirus was recorded in Wuhan, China, that expanded rapidly to pandemic level [1]. The ensuing coronavirus disease of 2019 (COVID-19) has since spread to most parts of the world and has been responsible for over 100 million cases and two million deaths worldwide as of January 2021. To fill the anticipated gap of 1–2 years between the emergence of a new viral disease and build-up of vaccine-induced herd immunity, major efforts have been made to identify readily applicable therapeutics for the treatment of COVID-19 through the repurpose screening of approved drugs and collections of known bioactive molecules. Compared with a timeframe of several years to identify and develop new drug candidates de novo, this approach promises to reduce screening to a very manageable panel of only a few thousand chemical entities for most of which clinical or preclinical data and safety profiles are available from other indications. In the context of the COVID-19 pandemic, substantial insight into the molecular biology and pathogenesis of the etiologic agent, SARS-CoV-2, was rapidly gained. Combined with previous knowledge of related betacoronaviruses such as the original SARS-CoV and Middle-Eastern Respiratory Syndrome (MERS) coronavirus, therapeutic strategies have pursued a dual goal: interfering with virus replication and improving disease management through symptom therapy. We will discuss the results of these efforts and some lessons learned for the design of drug repurposing campaigns against newly emerging viral challenges.
Screening of compound libraries
In order to quickly identify chemical compounds that may interfere with SARS-CoV-2 replication and are readily applicable or can be rapidly advanced into the clinic, the most pragmatic approach is to test small molecule libraries of approved drugs or known bioactives for which preclinical and/or clinical performance profiles are already available. Looking at small-molecule drugs only, this constitutes an encouragingly narrow collection; for instance, the U.S. Food and Drug Administration (FDA) has approved less than 2000 distinct molecular entities to date [2,3]. These can be assayed even in manual medium-throughput settings in only a few days and are also amenable to testing in more advanced, and more informative systems such as primary human tissue models [4••].
Core collections are available pre-plated from different commercial sources such as, the Prestwick library of 1520 compounds [5,6] or the Library of 1280 Pharmacologically Active Compounds (LOPAC1280) [7]. Other sets available to academia free-of-charge include the NCATS Pharmaceutical Collection (∼3000 compounds approved from several national agencies) [8] and collections that include investigational new drugs and subsets of bioactive preclinical compounds, such as the ReFRAME library (slightly larger at ∼12 000 compounds) [9] and the Pandemic Response-Boxes and COVID-19-Boxes (400 and 160 compounds, respectively). A product of recent antiviral discovery efforts, the latter collections are distributed on a case-by-case basis to avoid redundant screens and make screening and counterscreening results available online (i.e. ReFRAMEdb, PubChem, COVID19 open data portal). Although these initiatives fostered the rapid generation and distribution of performance information, one needs to keep in mind that collections of known bioactives largely comprise the failed developmental candidates of earlier drug development campaigns against unrelated indications. There is no clear rationale why these synthetically often heavily experienced and ultimately abandoned chemotypes should be expected to fare better against a novel indication such as SARS-CoV-2 than against the original developmental target.
A significant portion of known bioactive molecules and even of some approved drugs is made up of promiscuous, frequent-hitter chemotypes [10,11] that predictably emerged in many screens but are undevelopable. The motivation to rapidly identify potential inhibitors led to several of these compounds being reported as hits against COVID-19, straining wasting time and resources [11]. As it quickly turned out, the true mechanism of action of a large number of ‘antiviral candidates’ were, quite obvious, cytotoxic or, less obvious, cytostatic effects (Table 1 ).
Table 1.
Examples of approved drugs with claimed anti-SARS-CoV-2 activity that are confounded by a very low (≤10) selectivity index
| Hit candidate | EC50 | CC50 | Selectivity index | Drug class |
|---|---|---|---|---|
| 2-deoxy-d-glucose | 9.1 mM [12] | 9.1 mM [12] | 1 | Hexokinase inhibitor |
| Amodiaquine dihydrochloride dihydrate, | 4.9 μM [6] | 34.4 μM [6] | 7 | Antimalarial |
| Amodiaquine hydrochloride | 5.6 μM [6] | >38.6 μM [6] | >6.8 | Antimalarial |
| Auranofin | 1.4 μM [82] | 5.7 μM [82] | 4.1 | Antirheumatic agent |
| Baloxavir acid | >100 μM [83] | 85.9 μM [83] | <1 | IAV Endonuclease inhibitor |
| Carmofur | 24.3 μM [84] | 133.4 μM [84] | 5.5 | Antineoplastic |
| Chloroquine | 4.7 μM [62•] 46.8 μM [6] (phosphate) | >50 μM [62•,6] >100 μM [4••] 45 μM [32] | >10 [62•] >1 [6] | Antimalarial |
| 3.9 μM [4••] 30 μM [32] | >25.6 [4••] | |||
| 1.5 [32] | ||||
| Chlorpromazine hydrochloride | 4 μM [6] | 11.9 μM [6] 48.9 μM [83] | 2.9 | Antipsychotic |
| 21.3 μM [83] | ||||
| Cycloheximide | 0.17 μM [12] 0.5 μM [NCATS OpenData Portal — SARS-CoV-2 cytopathic effect] | 1.7 μM [ReFRAMEdb] 0.8 μM [NCATS OpenData Portal — SARS-CoV-2 cytopathic effect] | 10 [ReFRAMEdb] 1.6 [NCATS OpenData Portal — SARS-CoV-2 cytopathic effect] | Protein synthesis inhibitor |
| Emetine | 0.47 μM [12] | 0.8 μM [ReFRAMEdb] | 1.7 | Protein synthesis inhibitor |
| Enisamium | 1 mM [85] | 10 mM [86] | 10 | Putative RNA polymerases inhibitor |
| Hydroxychloroquine | 11.2 μM [6] | >50 μM [6] | >4.5 | Antimalarial |
| Imatinib mesylate | 5.3 μM [6] 4.9 μM [4••] | >30.7 μM [6] >37.3 μM [4••] | >5.8 [6] 7.6 [4••] | Tyrosine kinase inhibitor |
| Ivermectin | 2.2 μM [60] 1.7 μM [62•] | 2.2 μM [62•] | 1−1.3 | Anthelmintics |
| Lopinavir | 26.6 μM [83] | 49.75 μM [83] | 1.9 [83] | HIV aspartic protease inhibitor |
| Mefloquine | 8.1 μM [6] | 18.5 μM [6] | 2.3 | Antimalarial |
| Nitazoxanide | 1.0 μM [62•] | 3.3 μM [62•] | 3.3 | Antiprotozoal |
| NMS873 | 25 nM [12] | 25 nM [12] | 1 | ATPase inhibitor |
| Pladienolide | 7 nM [12] | 7 nM [12] | 1 | Spliceosome inhibitor |
| Quinacrine dihydrochloride | 2.8 μM [4••] | 22 μM [4••] | 7.8 | IMPDH inhibitor |
| Ribavirin | 70 μM [12] | 70 μM [12] | 1 | IMPDH inhibitor |
| Ritonavir | 48.9 μM [83] | >100 μM [83] | >2 [83] | HIV aspartic protease inhibitor |
In addition to these conventional direct repurposing tests, some very innovative new screens were developed [4••,5,7,12] and applied to the bioactives collections for proof-of-concept. Unfortunately, this exercise in some cases only showcased the ‘least bad hits’ as bona fide therapeutic candidates, for instance even claiming a specific antiviral effect of cycloheximide [12]. To date, a broad panel of compounds has been selected based on repurposing screens or previously reported anti-coronavirus potential for clinical testing against COVID-19. Based on their general mechanism of action, this set can be subdivided into direct-acting and host-directed antivirals (Table 2 ), and we will consider their strengths and challenges individually.
Table 2.
Examples of repurposed or redirected drugs that have progressed to clinical trials
| Candidate drug | Sponsor | Phase | Outcome | Trial ID | Reference |
|---|---|---|---|---|---|
| AT-527 | Hoffmann-La Roche | Phase 2 | Completion date: Feb/May, 2021 | NCT04709835, NCT04396106 | |
| Atea Pharmaceuticals, Inc. | |||||
| Baricitinib | Eli Lilly and Company | Phase 3 | Completion date: June 2021 | NCT04421027 | |
| Baricitinib plus remdesivir | National Institute of Allergy and Infectious Diseases (NIAID) | Phase 3 | Reduce recovery time | NCT04401579 | [79] |
| Chloroquine/hydroxychloroquine | Multiple | Phase 3 | Not effective | ‘Solidarity’ clinical trial, NCT04501952 | [17••] |
| https://www.covid19treatmentguidelines.nih.gov/tables/table-2b/ | |||||
| CD24Fc | OncoImmune, Inc. | Phase 3 | Completion date: October 2020 | NCT04317040 | |
| Dexamethasone | University of Oxford | Phase 2/3 | Low 28-day mortality | NCT04381936 (Recovery Trial) | [87] |
| Emtricitabine and tenofovir | Universidad Nacional de Colombia | Phase 2/3 | Completion date: May 2021 | NCT04359095 | |
| Favipiravir | University of Pecs | Phase 3 | Completion date: June 2021 | NCT04600999 | |
| Interferon β-1a | Synairgen Research Ltd. | Phase 2 | Better recovery | NCT04385095 | [81] |
| Ivermectin | Clinica Universidad de Navarra, Universidad de Navarra | Phase 2 | Reduce viral loads in mild COVID-19 | NCT04390022 | [88] |
| https://www.covid19treatmentguidelines.nih.gov/antiviral-therapy/ivermectin/table--ivermectin-clinical-data/ | |||||
| Leflunomide | City of Hope Medical Center | Phase 1/2 | Completion date: September 2022 | NCT04532372 | |
| Lopinavir/ritonavir | University of Oxford | Phase 2/3 | Not effective | NCT04381936 (Recovery Trial) | [89] |
| Molnupiravir (MK-4482) | Merck & Co. | Phase 2/3 | Completion date: December, 2021 | NCT04575584, NCT04575597 | |
| PTC299 | PTC Therapeutics | Phase 2/3 | Completion date: July 2021 | NCT04439071 | |
| Remdesivir | Gilead Sciences | Phase 3 | Not effective, Completion date: April 2021 | ‘Solidarity’ clinical trial, NCT04501952 | [17••] |
| Ruxolitinib | Novartis Pharmaceuticals | Phase 3 | Did not meet endpoint | NCT04362137 | https://www.novartis.com/news/media-releases/novartis-provides-update-ruxcovid-study-ruxolitinib-hospitalized-patients-covid-19 |
| Sarilumab | Sanofi | Phase 3 | Did not meet endpoint | NCT04327388 | https://www.sanofi.com/en/media-room/press-releases/2020/2020-09-01-07-00-00 |
| Siltuximab | EusaPharma (UK) Limited | Phase 3 | Completion date: June, 2022 | NCT04616586 | |
| Sofosbuvir (plus Ledipasvir) | Almaza Military Fever Hospital | Phase 3 | Completion date: July, 2020 | NCT04530422 | |
| Tocilizumab | University of Oxford | Phase 2/3 | Improved survival | NCT04381936 (Recovery Trial) | [76] |
Direct-acting antivirals
Of all stages of the SARS-CoV-2 life cycle, processes associated with viral entry and replication in particular appear to be readily druggable (Figure 1 ). Repurposed drug candidates that interfere with the entry machinery predominantly belong to the host-directed antiviral group. However, SARS-CoV-2 replication critically relies on viral protease and RNA-dependent RNA polymerase (RdRP) activities that were considered attractive targets for repurposing of drugs that, for instance, successfully block the equivalent functions in the human immunodeficiency virus (HIV) and hepatitis C virus (HCV) replication cycles.
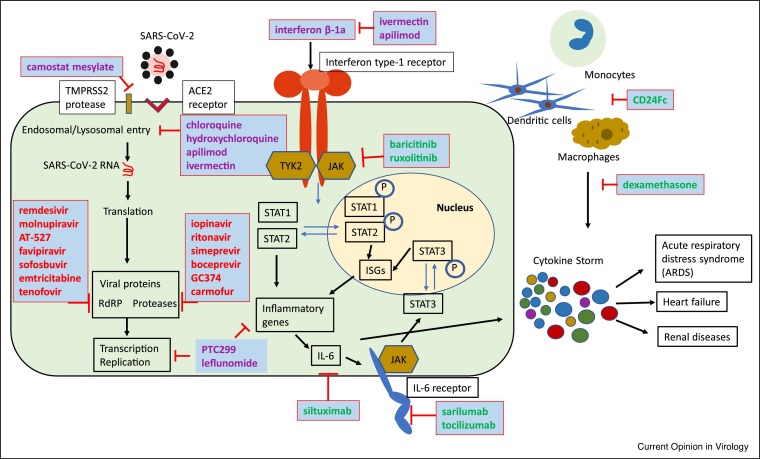
Figure 1.
Schematic of different stages of the SARS-CoV-2 cellular replication cycle that have been subjected to drug targeting attempts. Examples of approved drugs and advanced experimental therapeutics that have been tested are shown. Red: direct-acting antivirals; purple: host-targeted antivirals; green: immune-modulators for improved disease management.
The main SARS-CoV-2 protease Mpro in particular has been the target of repeated repurposing attempts including testing of HCV serine protease inhibitors (i.e. simeprevir and boceprevir [13,14]) and HIV aspartic protease inhibitors (i.e. lopinavir/ritonavir [14, 15, 16,17••,18]). SARS-CoV-2 Mpro is a cysteine protease, however, and these inhibitors designed against different protease classes returned at best marginal anti-coronavirus activity in cell culture and failed in clinical trials. An exception could be the serine protease inhibitor GC376, which has demonstrated in vitro activity against SARS-CoV-2 [14,19] and showed efficacy in vivo against a feline coronavirus [20,21]. The compound appears promising, but has yet to enter clinical development.
The viral RdRP complex has greater promise to serve as a valid target for existing drugs or advanced clinical candidates, based on its relatively high degree of conservation [22] and a tendency of nucleoside analog polymerase inhibitors for a broadened antiviral indication spectrum. It is therefore not surprising that repurposing attempts favored competitive substrate-analogs over allosteric polymerase inhibitors. Coronaviruses can be a challenging target for nucleoside analog inhibitors, however, due to a 3′-to-5′ exoribonuclease proofreading activity that is unusual among RNA viruses but can mediate resistance, for instance, to the nucleoside analog drug ribavirin [23, 24, 25]. Nevertheless, the only direct-acting inhibitor that has received emergency approval in the United States in the first year of the pandemic for clinical use against severe COVID-19 has been the broad-spectrum nucleoside analog remdesivir [26].
The drug is a phosphoamidate prodrug of the monophosphate form of GS-441524, a 1′-CN modified adenosine nucleoside. Originally developed against HCV and subsequently unsuccessfully repurposed against Ebola virus, anti-SARS-CoV-2 activity of remdesivir in cell culture was discovered at the onset of the pandemic and the drug has become standard-of-care since [27, 28, 29, 30]. Despite its success, the therapeutic impact of remdesivir is compromised by a requirement for intravenous administration and high tissue exposure in vivo in the liver, which was favorable for its original HCV indication but is less desirable for use against SARS-CoV-2 [26]. The lack of oral bioavailability restricts the recipient pool to hospitalized patients presenting with complicated disease, although greater therapeutic impact could without doubt be achieved by initiating treatment immediately after a positive test for SARS-CoV-2, ideally even before the development of clinical signs.
Repurposing attempts of orally available nucleoside analog drugs such as sofosbuvir (HCV) [31,32], emtricitabine and tenofovir (HIV) [32], or conditionally approved favipiravir (influenza virus) [31, 32, 33, 34, 35] failed in vitro and/or in the clinic, highlighting that nucleoside analogs have heightened potential for a broadened indication spectrum, but cross-antiviral activity cannot be taken for granted. However, novel orally available nucleoside analogs that were in earlier stages of formal development showed cross-activity against betacoronaviruses and have entered clinical trials since the onset of the pandemic. AT-527, a 2′-fluoro-2′-methyl guanosine analog prodrug, is currently being evaluated to treat HCV and SARS-CoV-2 infections. It has returned promising results in differentiated human airway primary cells and shown favorable pharmacokinetic profile after oral delivery to human and non-human primates [36]. Likewise, an orally available prodrug of the cytidine analog ß-d-N 4-hydroxycytidine, EIDD-2801 (molnupiravir) that was in development against influenza viruses [37••], was found to be orally efficacious against SARS-CoV-2 in the ferret infection model, reducing both virus burden and direct contact transmission rates [38••]. Phase I clinical trials demonstrated that molnupiravir is well tolerated and reaches antiviral plasma levels in humans that exceed efficacious concentrations in ferrets [37••,39]. Efficacy of the drug, which is currently in advanced Phase II/III clinical trials, was furthermore confirmed in mouse and hamster models of SARS-CoV-2 [40, 41, 42].
An inherent problem of broad-spectrum nucleoside analogs is their recognition and incorporation by host polymerases, which can result in undesirable off-target effects. Because of a lack of proofreading activity of host cell mitochondrial RNA polymerases, mitochondrial toxicity is frequently observed [43] and must be carefully monitored during development. Despite these potential liabilities, broad-spectrum nucleoside analogs have emerged in the pandemic as the antiviral drug class with perhaps the highest potential to provide a first line defense against a newly emerged pandemic pathogen.
Host-directed antivirals
The idea of repurposing host-directed drugs for antiviral therapy has experienced a renaissance in the past two decades, based on the promise that host-directed antivirals may combine a broad antiviral indication spectrum with a heightened barrier against the emergence of viral resistance. Screening campaigns for host-directed antivirals typically deliver an abundance of seemingly attractive antiviral ‘hits’, many of which come from the cancer therapeutics and metabolic disease drugs subgroups [11]. Two decades of repurposing attempts of these drug classes against changing viral targets (i.e. SARS-CoV, pandemic 2009 influenza virus, Ebola virus, Zika virus, SARS-CoV-2) have unfortunately highlighted some fundamental limitations to their use for the treatment of acute RNA virus infections. Major liabilities include severe cytotoxic and/or cytostatic effects that are often the basis for in vitro ‘antiviral activity’ but can be counterproductive when trying to control a viral infection in vivo, side effects unacceptable for an antiviral indication, and pharmacokinetic properties and/or tissue exposure levels that do not meet those required for antiviral activity.
An additional potential caveat of this approach that was less appreciated pre-pandemic but amplified by COVID-19 arises from flaws in screening protocol design and assay interpretation, which can lead to the selection of host targets with little physiological relevance in the context of a natural infection. A prominent example of this problem are the anti-malaria and anti-rheumatoid arthritis therapeutics chloroquine and hydroxychloroquine. These drugs are polypharmacological, but one mechanism of action is interference with host cell lysosomal activity and autophagy. As one may anticipate, chloroquine showed a clear, albeit moderate, anti-SARS-CoV-2 in vitro, using as a host system the VeroE6 cell line that is derived from African green monkey kidney tissue [27,44]. Based on this minimal cell culture-derived evidence, the drug has since been tested against COVID-19 in a large number of clinical trials, which have consistently demonstrated that it lacks efficacy [45, 46, 47, 48]. Despite this overwhelming clinical information, significant public attention was focused for months on a claimed benefit supposedly revealed in poorly designed clinical tests that were not conducted in compliance with accepted proper scientific standards [49,50].
In hindsight, the molecular basis for the clinical failure of chloroquine is inherent in its interference with the cellular endocytotic pathway, since it prevents the cathepsin-mediated maturation of SARS-CoV-2 spike (S) protein. However, SARS-CoV-2 entry depends on cathepsin activity only in cells such as VeroE6, which lack the host serine protease TMPRSS2 [51••,52•], but is of limited relevance in vivo. Accordingly, chloroquine lacks anti-SARS-CoV-2 activity in engineered VeroE6 cells expressing TMPRSS2 [51••,52•] and is equally inactive in any naturally TMPRSS2-positive cell line [51••,52•], differentiated primary human bronchial epithelial cells [53•], hamsters [34], ferrets [54] and non-human primates [53•,55]. The claimed anti-SARS-CoV-2 activity of chloroquine is emblematic for overinterpretation of a single assay that — in an attempt to accelerate the drug discovery process — was not subjected to a rigorous counterscreen of biological relevance in informative systems such as, disease-relevant primary cells. As a consequence, precious resources were directed to unproductive clinical trials in a time of need, trial participants were needlessly subjected to a conceptually flawed therapeutic approach, and people could even be harmed by the side effects of an ineffective drug.
As an interesting counterpoint to the chloroquine failure, camostat mesylate blocks TMPRSS2 and was therefore missed as an anti-SARS-CoV-2 candidate in screens conducted on VeroE6 cells. However, the compound showed antiviral activity when tested against SARS-CoV-2 in TMPRSS2 expressing cells including human airway organoids [51••,56], and has justifiably been advanced to clinical testing [57,58].
Whereas chloroquine appears to have run its course in the second year of the pandemic, a controversy surrounding another anti-parasitic drug, ivermectin, is still playing out. Ivermectin has exquisite, nanomolar potency against its original indication [59••]. Standing at the center of debate is a very basic dose-response virus inhibition assay that was again conducted on VeroE6 cells and returned modest anti-SARS-CoV-2 activity in the low micromolar range [60]. Similar antiviral effects of ivermectin have been previously reported for a range of other viruses [61]. However, two basic parameters of antiviral drug development are overlooked in these studies: i) the allegedly ‘active’ antiviral concentration of ivermectin is roughly equivalent to the known cytotoxic concentration in many cell lines, including Vero cells [61,62•], which confounds interpretation of a specific antiviral effect; and ii) human pharmacokinetic data for ivermectin reveal that this antiviral (and thus, cytotoxic) concentration is not reached in human plasma, which also explains the excellent safety profile of the drug when used as approved [59••,63,64].
Without doubt, the availability of clinical and advanced preclinical data typically comprising pharmacokinetics, pharmacodynamics, tissue distribution, and tolerability in different species including humans is a fundamental advantage of drug repurposing over de novo development. However, this advantage becomes tangible only if the available knowledge is applied to a critical evaluation of whether a seemingly exciting antiviral effect observed in cell culture has any therapeutic relevance. Ideally, this assessment should be carried out before human trials for the new indication are considered.
In addition to cytotoxic compounds, drugs and bioactives targeting host metabolic pathways have also been rediscovered in screening campaigns against SARS-CoV-2. Many of these have an impressive literature history of in vitro inhibitory activity against numerous viral targets. A prime example for this group and the challenges associated with their antiviral use is the large and structurally diverse family of cellular dihydroorotate dehydrogenase (DHODH) inhibitors that has been discovered over the years. These compounds act in cytostatic and antiviral manners through interference with the de novo pyrimidine biosynthesis pathway [65]. Representatives include the drug leflunomide, approved to treat rheumatoid arthritis, and the experimental inhibitor PTC299 [66, 67, 68, 69]. DHODH blockers are known for exquisite broad-spectrum antiviral activity in cell culture [68,70,71]. In vivo, they can reduce expression of inflammatory cytokines, but universally lack antiviral activity because of the pyrimidine salvage pathway that is available to cells in a living organism but insignificant in cell culture [65]. Despite this well understood ‘deal-breaker’ that prevents in vivo efficacy, DHODH inhibitors were advanced to clinical testing against SARS-CoV-2 [66, 67, 68, 69]. Again, these trials bound resources and burdened participants despite better knowledge and without much scientifically grounded prospect of success.
Disease management
Whereas repurposing attempts of host-directed drugs with the goal of blocking virus replication have largely not succeeded in the COVID-19 pandemic, improving management of severe disease through an empirical evaluation of the effect of immune-modulators in the clinic has been successful. Unlike antiviral therapeutics that are most effective when treatment is initiated at the earliest stage of infection, the highly dynamic nature of acute viral disease mandates careful timing of immune-modulating treatment to limit risks of a compromised initial host antiviral response and secondary nosocomial infections as a result of impaired immune function. Early characterization of COVID-19 in hospitalized patients revealed a biphasic pattern of disease progression, starting with a period of relatively moderate clinical signs that could advance at a second stage to acute respiratory distress syndrome (ARDS), systemic inflammatory responses, and a ‘cytokine storm’ [72,73]. These hallmarks of severe COVID-19 triggered repurposing attempts of different classes of immunoactive drugs. To date, the low-cost corticosteroid dexamethasone is still one of the most effective treatments of severe COVID-19 symptoms identified. Treatment reduced patient mortality in the phase 3 ReCOVERY study [74••], confirming a role of late-stage hyperinflammation in case fatalities.
Although the COVID-19-associated ‘cytokine storm’ has not yet been fully characterized, interleukin-6 (IL-6) is known as a major driver of hyperinflammatory responses. Potent inhibitors of IL-6 and its receptor such as siltuximab and tocilizumab showed mild benefits in phase 3 clinical trials [75, 76, 77]. Targeting downstream Janus kinase (JAK) signaling pathways with baricitinib in combination with remdesivir furthermore showed clear benefit compared to remdesivir with placebo in reducing severe illness [78,79], granting this combination treatment emergency use approval by the FDA in November 2020.
The fusion protein CD24Fc provides protection against simian immunodeficiency virus induced pneumonia in rhesus monkeys by fortifying the immune checkpoint CD24–Siglec 10 interaction to reduce inflammation [80]. Repurposing this approach against COVID-19 appeared also effective, since preliminary evidence reported a 50% reduction in mortality and respiratory failures in a still active phase 3 study. By comparison, antiviral interferon β-1a had little to none effect on hospitalized patients in the large Solidarity trial [17••]. However, patients improved more rapidly when an aerosolized form of interferon β-1a was administered at an earlier disease stage [81]. Current standard-of-care and ongoing trials have thus consistently confirmed the importance of immunomodulators as crucial therapeutic tools to mitigate severe COVID-19.
Conclusions
Many repurposing screens carried out in response to the COVID-19 pandemic have had very little impact because some basic principles of drug development were overlooked or may have been considered irrelevant in the face of a global pandemic. Looking forward, we believe that avoiding known caveats in two critical areas of hit discovery specifically will help to rapidly separating distractions from viable therapeutic candidates:
-
i)Assay design:
-
•Permissive immortalized cell lines such as VeroE6 cells offer a readily available system for initial screens. However, as steps of the viral replication cycle might be fundamentally altered in these cell lines, the risk of false positives (i.e. chloroquine) and false negatives (i.e. camostat mesylate) is high. Meaningful counterscreens must be integrated into any discovery campaign that validate the physiological relevance of results (i.e. disease-relevant primary cells or organoid models [4••]).
-
•
-
ii)Contextualization of data:
-
•Basic cytotoxicity/cytostatic assays must be part of any antiviral screen, and very low SI values are a warning that the observed effect is likely off-target. Rigorous assay evaluation is critical, since especially cytostatic effects can easily be masked by improper assay design. For repurposed drugs, historic cytotoxicity data are usually plentiful in the literature and online databases (Table 3 ) provide a path to instant virtual counterscreens.
-
•Minute antiviral effects must not be overinterpreted, since virus growth is logarithmical in nature. This problem is often exaggerated by a narrow focus solely on EC50 (rather than more robust EC90 or EC99) values of inhibitor candidates that do not necessarily reflect true antiviral impact.
-
•Known pharmaceutical parameters of repurposed drugs such as pharmacokinetics, pharmacodynamics, tissue distribution, and tolerability must be included in the evaluation of whether an antiviral effect observed in cell culture has clinical potential (i.e. ivermectin). Approved drug status should not be falsely interpreted as a guarantee of safety in humans at any concentration.
-
•
Table 3.
Collection of some online resources for COVID-19 antiviral research
Considering these basic parameters during the COVID-9 crisis could have avoided unnecessary clinical trials that burdened trial participants unnecessarily and bound resources. This experience also raises the fundamental question of how likely drug repurposing screens are to deliver on their core promise to identify a rapid pharmacological first-line defense against a newly emerged viral pathogen.
Well conducted drug repurposing strategies have saved lives, as demonstrated by the examples of direct-acting remdesivir and the immune modulators. By contrast, repurposing searches for applicable host-directed antivirals and rushed efforts to test compendiums of known bioactives against SARS-CoV-2 were unsuccessful, as were similar earlier activities against, for instance, pandemic 2009 influenza virus, Ebola virus, MERS, and zika virus. This disappointing outcome is not surprising, given that a large proportion of the known bioactives is made up of failed developmental candidates of yesterday’s campaigns that were presumably abandoned for good reason. However, the COVID-19 pandemic has channeled the development of exciting new tools (such as the REFRAME initiative) to make comprehensive profiles of known drug properties rapidly available to investigators. This information can greatly accelerate the discovery process if there is willingness in the research community to accept that an exciting repurposing ‘hit’ may simply represent a cytotoxic compound unsuitable for antiviral therapy.
In our view, the greatest potential for successful repurposing against a newly emerged viral challenge has been the selective testing of approved or advanced-stage experimental drugs with known broadened-spectrum direct antiviral activity. We believe that it is no coincidence that a known broad-spectrum antiviral, remdesivir, is the only small molecule drug that has received emergency approval for anti-SARS-CoV-2 therapy to date and that two promising candidates currently in clinical trials, molnupiravir and AT-527, had likewise demonstrated broad spectrum direct acting antiviral activity before the COVID-19 pandemic struck.
COVID-19 is without doubt not the last global viral pandemic. We believe that efforts are best directed at expanding the broad-spectrum antiviral arsenal in interpandemic times, which requires that viable non-pandemic indications are identified that open up a path to clinical approval of new drugs before a crisis unfolds. Whereas nucleoside analog inhibitors have high promise of meeting the broad-spectrum requirement among the small-molecule drug classes, liabilities such as off-target effects, mitochondrial toxicity, and mutagenic and/or teratogenic potential must be understood and, if possible, mitigated. The magic drug that meets the needs of all patient groups and addresses all future viral challenges may remain perpetually elusive. However, in reach may be the development of a reasonably sized subset of antiviral drugs with diverse strengths and limitations, and overlapping but not identical broadened indication spectra that could drastically shorten first-line response times to a newly emerged viral challenge, fundamentally improving the global status of pandemic preparedness.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was supported, in part, by Public Health Service grants AI071002 (to RKP) and AI141222 (to RKP), from the N.I.H./NIAID. Opinions expressed are those of the authors alone. The funders had no role in manuscript design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Given his role as Section Editor, Richard Plemper had no involvement in the peer-review of this article and has no access to information regarding its peer-review. Full responsibility for the editorial process for this article was delegated to Jasmine Tomar.
References
- 1.Zhu N., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kinch M.S., et al. An overview of FDA-approved new molecular entities: 1827-2013. Drug Discov Today. 2014;19:1033–1039. doi: 10.1016/j.drudis.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Mullard A. 2020 FDA drug approvals. Nat Rev Drug Discov. 2021;20:85–90. doi: 10.1038/d41573-021-00002-0. [DOI] [PubMed] [Google Scholar]
- 4••.Han Y., et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study the authors screened a collection of drugs against SARS-CoV-2 entry using two organoids cultures relevant for SARS-CoV-2 infection in humans, instead of the usual VeroE6 cell line.
- 5.Touret F., et al. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci Rep. 2020;10 doi: 10.1038/s41598-020-70143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weston S., et al. Broad anti-coronavirus activity of food and drug administration-approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo. J Virol. 2020;94 doi: 10.1128/JVI.01218-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riva L., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang R., et al. The NCATS pharmaceutical collection: a 10-year update. Drug Discov Today. 2019;24:2341–2349. doi: 10.1016/j.drudis.2019.09.019. [DOI] [PubMed] [Google Scholar]
- 9.Janes J., et al. The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. Proc Natl Acad Sci U S A. 2018;115:10750–10755. doi: 10.1073/pnas.1810137115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baell J.B., Holloway G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 11.Plemper R.K., Cox R.M. Biology must develop herd immunity against bad-actor molecules. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bojkova D., et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo H.S., et al. Simeprevir potently suppresses SARS-CoV-2 replication and synergizes with remdesivir. ACS Cent Sci. 2021;7:792–802. doi: 10.1021/acscentsci.0c01186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu L., et al. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat Commun. 2020;11 doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheahan T.P., et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeon S., et al. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob Agents Chemother. 2020;64 doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.WHO Solidarity Trial Consortium Repurposed antiviral drugs for Covid-19 - interim WHO solidarity trial results. N Engl J Med. 2020;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]; This large scale clinical study provided long-awaited answers on a panel of repurposed drugs with controversial preliminary results, demonstrating the lack of efficacy of hydroxychloroquine and lopinavir/ritonavir.
- 18.Cao B., et al. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vuong W., et al. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat Commun. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y., et al. Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedersen N.C., et al. Efficacy of a 3C-like protease inhibitor in treating various forms of acquired feline infectious peritonitis. J Feline Med Surg. 2018;20:378–392. doi: 10.1177/1098612X17729626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.te Velthuis A.J. Common and unique features of viral RNA-dependent polymerases. Cell Mol Life Sci. 2014;71:4403–4420. doi: 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogando N.S., et al. The curious case of the nidovirus exoribonuclease: its role in RNA synthesis and replication fidelity. Front Microbiol. 2019;10:1813. doi: 10.3389/fmicb.2019.01813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minskaia E., et al. Discovery of an RNA virus 3'->5' exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith E.C., et al. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho A., et al. Synthesis and antiviral activity of a series of 1'-substituted 4-aza-7,9-dideazaadenosine C-nucleosides. Bioorg Med Chem Lett. 2012;22:2705–2707. doi: 10.1016/j.bmcl.2012.02.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pizzorno A., et al. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell Rep Med. 2020;1 doi: 10.1016/j.xcrm.2020.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruijssers A.J., et al. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beigel J.H., et al. Remdesivir for the treatment of Covid-19 - final report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zandi K., et al. Repurposing nucleoside analogs for human coronaviruses. Antimicrob Agents Chemother. 2020;65 doi: 10.1128/AAC.01652-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Shufeng. Evaluation of 19 antiviral drugs against SARS-CoV-2 infection. biorxiv. 2020 doi: 10.1101/2020.04.29.067983. [DOI] [Google Scholar]
- 33.Shannon A., et al. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat Commun. 2020;11 doi: 10.1038/s41467-020-18463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaptein S.J.F., et al. Favipiravir at high doses has potent antiviral activity in SARS-CoV-2-infected hamsters, whereas hydroxychloroquine lacks activity. Proc Natl Acad Sci U S A. 2020;117:26955–26965. doi: 10.1073/pnas.2014441117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lou Y., et al. Clinical outcomes and plasma concentrations of baloxavir marboxil and favipiravir in COVID-19 patients: an exploratory randomized, controlled trial. Eur J Pharm Sci. 2021;157 doi: 10.1016/j.ejps.2020.105631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Good S.S., et al. AT-527, a double prodrug of a guanosine nucleotide analog, is a potent inhibitor of SARS-CoV-2 in vitro and a promising oral antiviral for treatment of COVID-19. Antimicrob Agents Chemother. 2021;65 doi: 10.1128/AAC.02479-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37••.Toots M., et al. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors demonstrate that a nucleoside analog, molnupiravir, is well tolerated at plasma levels that exceed efficacious concentrations against influenza viruses.
- 38••.Cox R.M., Wolf J.D., Plemper R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat Microbiol. 2021;6:11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors demonstrate that a nucleoside analog, molnupiravir, is able to abrogate in less than 24 hours SARS-CoV-2 shedding from infected ferrets and blocks transmission.
- 39.Painter W.P., et al. Human safety, tolerability, and pharmacokinetics of molnupiravir, a novel broad-spectrum oral antiviral agent with activity against SARS-CoV-2. Antimicrob Agents Chemother. 2021;65 doi: 10.1128/AAC.02428-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wahl A., et al. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591:451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenke K., et al. Orally delivered MK-4482 inhibits SARS-CoV-2 replication in the Syrian hamster model. Nat Commun. 2021;12 doi: 10.1038/s41467-021-22580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdelnabi Rana, et al. Molnupiravir (EIDD-2801) inhibits SARS-CoV-2 replication and enhances the efficacy of favipiravir in a Syrian hamster infection model. biorxiv. 2020 doi: 10.1101/2020.12.10.419242. [DOI] [Google Scholar]
- 43.Feng J.Y., et al. Role of mitochondrial RNA polymerase in the toxicity of nucleotide inhibitors of hepatitis C virus. Antimicrob Agents Chemother. 2016;60:806–817. doi: 10.1128/AAC.01922-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J., et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borba M.G.S., et al. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: a randomized clinical trial. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Self W.H., et al. Effect of hydroxychloroquine on clinical status at 14 days in hospitalized patients with COVID-19: a randomized clinical trial. JAMA. 2020;324:2165–2176. doi: 10.1001/jama.2020.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boulware D.R., et al. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid-19. N Engl J Med. 2020;383:517–525. doi: 10.1056/NEJMoa2016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fiolet T., et al. Effect of hydroxychloroquine with or without azithromycin on the mortality of coronavirus disease 2019 (COVID-19) patients: a systematic review and meta-analysis. Clin Microbiol Infect. 2021;27:19–27. doi: 10.1016/j.cmi.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gautret P., et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Gautret P., Raoult D. Nullane salus extra ecclesiam. New Microbes New Infect. 2020;37 doi: 10.1016/j.nmni.2020.100714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Hoffmann M., et al. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585:588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]; In this study the authors demonstrate that chloroquine acts on a pathway for viral entry that is not active in lung cells by using cells that express the host protease TMPRSS2.
- 52•.Ou T., et al. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study deciphers the molecular basis for the inactivity of chloroquine in TMPRSS2-expressing cells.
- 53•.Maisonnasse P., et al. Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature. 2020;585:584–587. doi: 10.1038/s41586-020-2558-4. [DOI] [PubMed] [Google Scholar]; This study demonstrates clearly the lack of efficacy of chloroquine against SARS-CoV-2 in human primary cells and non-human primates, the two most relevant models to study SARS-CoV-2 entry and replication.
- 54.Park S.J., et al. Antiviral efficacies of FDA-approved drugs against SARS-CoV-2 infection in ferrets. mBio. 2020;11 doi: 10.1128/mBio.01114-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenke K., et al. Hydroxychloroquine prophylaxis and treatment is ineffective in macaque and hamster SARS-CoV-2 disease models. JCI Insight. 2020;5 doi: 10.1172/jci.insight.143174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mykytyn A.Z., et al. SARS-CoV-2 entry into human airway organoids is serine protease-mediated and facilitated by the multibasic cleavage site. eLife. 2021;10 doi: 10.7554/eLife.64508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi Jae-Phil. Foistar®(Camostat mesylate) associated with the significant decrease in CRP levels compared to Kaletra®(Lopinavir/Ritonavir) treatment in Korean mild COVID-19 pneumonic patients. medrxiv. 2020 doi: 10.1101/2020.12.10.20240689. [DOI] [Google Scholar]
- 58.Hofmann-Winkler H., et al. Camostat mesylate may reduce severity of coronavirus disease 2019 sepsis: a first observation. Crit Care Explor. 2020;2 doi: 10.1097/CCE.0000000000000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59••.Bray M., et al. Ivermectin and COVID-19: a report in antiviral research, widespread interest, an FDA warning, two letters to the editor and the authors’ responses. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104805. [DOI] [PMC free article] [PubMed] [Google Scholar]; The initial claims that Ivermectin could have an antiviral effect against SARS-CoV-2 led to many reactions, which are well represented in these letters to the editor, explaining clearly the contrast between the putative activity range of Ivermectin and the known achievable levels in humans.
- 60.Caly L., et al. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mastrangelo E., et al. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug. J Antimicrob Chemother. 2012;67:1884–1894. doi: 10.1093/jac/dks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62•.Sales-Medina Douglas Ferreira, Ferreira Ludmila Rodrigues Pinto, Romera Lavínia M.D., Gonçalves Karolina Ribeiro, Guido Rafael V.C., Courtemanche Gilles, Buckeridge Marcos S., Durigon Édison L., Moraes Carolina B., Freitas-Junior Lucio H. Discovery of clinically approved drugs capable of inhibiting SARS-CoV-2 in vitro infection using a phenotypic screening strategy and network-analysis to predict their potential to treat covid-19. biorxiv. 2020 doi: 10.1101/2020.07.09.196337. [DOI] [Google Scholar]; In this study, the authors test the antiviral potential of a panel of drugs with potential to be repurposed, including Ivermectin, with a systematic look at effects driven by a cytotoxic effect, observing as expected that Ivermectin does not show a specific antiviral effect against SARS-CoV-2.
- 63.Schmith V.D., Zhou J.J., Lohmer L.R.L. The approved dose of ivermectin alone is not the ideal dose for the treatment of COVID-19. Clin Pharmacol Ther. 2020;108:762–765. doi: 10.1002/cpt.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Momekov G., Momekova D. Ivermectin as a potential COVID-19 treatment from the pharmacokinetic point of view: antiviral levels are not likely attainable with known dosing regimens. Biotechnol Biotechnol Equip. 2020;34:469–474. [Google Scholar]
- 65.Luthra P., et al. Inhibiting pyrimidine biosynthesis impairs Ebola virus replication through depletion of nucleoside pools and activation of innate immune responses. Antiviral Res. 2018;158:288–302. doi: 10.1016/j.antiviral.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hahn F., et al. IMU-838, a developmental DHODH inhibitor in phase II for autoimmune disease, shows anti-SARS-CoV-2 and broad-spectrum antiviral efficacy in vitro. Viruses. 2020;12:12. doi: 10.3390/v12121394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu K., et al. A small-scale medication of Leflunomide as a treatment of COVID-19 in an open-label blank-controlled clinical trial. Virol Sin. 2020;35:725–733. doi: 10.1007/s12250-020-00258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luban J., et al. The DHODH inhibitor PTC299 arrests SARS-CoV-2 replication and suppresses induction of inflammatory cytokines. Virus Res. 2021;292 doi: 10.1016/j.virusres.2020.198246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiong R., et al. Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2. Protein Cell. 2020;11:723–739. doi: 10.1007/s13238-020-00768-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoffmann H.-H., et al. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc Natl Acad Sci U S A. 2011;108:5777–5782. doi: 10.1073/pnas.1101143108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krumm S.A., et al. Potent host-directed small-molecule inhibitors of myxovirus RNA-dependent RNA-polymerases. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen J., et al. Clinical progression of patients with COVID-19 in Shanghai, China. J Infect. 2020;80:e1–e6. doi: 10.1016/j.jinf.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sundararaj Stanleyraj J., et al. Treating COVID-19: are we missing out the window of opportunity? J Antimicrob Chemother. 2021;76:283–285. doi: 10.1093/jac/dkaa442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74••.The RECOVERY Collaborative Group, et al. Dexamethasone in hospitalized patients with Covid-19 - preliminary report. N Engl J Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]; This clinical study demonstrated the potential of immune modulators such as dexamethasone to manage COVID-19 and reduce mortality.
- 75.Salama C., et al. Tocilizumab in patients hospitalized with Covid-19 pneumonia. N Engl J Med. 2021;384:20–30. doi: 10.1056/NEJMoa2030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.The RECOVERY Collaborative Group Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2021;397:1637–1645. doi: 10.1016/S0140-6736(21)00676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guaraldi G., et al. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020;2:e474–e484. doi: 10.1016/S2665-9913(20)30173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoang T.N., et al. Baricitinib treatment resolves lower-airway macrophage inflammation and neutrophil recruitment in SARS-CoV-2-infected rhesus macaques. Cell. 2020;184:460–475. doi: 10.1016/j.cell.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kalil A.C., et al. Baricitinib plus remdesivir for hospitalized adults with Covid-19. N Engl J Med. 2020;384:795–807. doi: 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tian R.R., et al. CD24Fc protects against viral pneumonia in simian immunodeficiency virus-infected Chinese rhesus monkeys. Cell Mol Immunol. 2020;17:887–888. doi: 10.1038/s41423-020-0452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Monk P.D., et al. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir Med. 2021;9:196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rothan H.A., et al. The FDA-approved gold drug auranofin inhibits novel coronavirus (SARS-COV-2) replication and attenuates inflammation in human cells. Virology. 2020;547:7–11. doi: 10.1016/j.virol.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choy K.T., et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jin Z., et al. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 85.Holubovska O., et al. Enisamium is an inhibitor of the SARS-CoV-2 RNA polymerase and shows improvement of recovery in COVID-19 patients in an interim analysis of a clinical trial. medRxiv. 2021 doi: 10.1101/2021.01.05.21249237. [DOI] [Google Scholar]
- 86.Boltz D., et al. Activity of enisamium, an isonicotinic acid derivative, against influenza viruses in differentiated normal human bronchial epithelial cells. Antivir Chem Chemother. 2018;26 doi: 10.1177/2040206618811416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gordon D.E., et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370 doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chaccour C., et al. The effect of early treatment with ivermectin on viral load, symptoms and humoral response in patients with non-severe COVID-19: A pilot, double-blind, placebo-controlled, randomized clinical trial. EClinicalMedicine. 2021;32 doi: 10.1016/j.eclinm.2020.100720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.The RECOVERY Collaborative Group Lopinavir-ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2020;396:1345–1352. doi: 10.1016/S0140-6736(20)32013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]