Abstract
Purpose
This study aimed to evaluate the associations between mitochondrial dynamics related genes -MFN1, MFN2 and DRP1 polymorphisms and risk of lung cancer.
Methods
Six polymorphisms of MFN1, MFN2 and DRP1 were genotyped in 600 cases and 600 controls using a MassARRAY platform.
Results
The MFN1 rs13098637-C and DRP1 rs879255689-A alleles were associated with an increased risk of lung cancer (prs13098637=0.004, prs879255689=0.005), while MFN2 rs4240897-A and rs2236058-G were related to a decreased risk of disease (p<0.001). The rs13098637-TC/CC and rs879255689-GA/AA were determined as risk genotypes for lung cancer (prs13098637=0.014, prs879255689=0.013), whereas the rs4240897-GA/AA and rs2236058-GG were identified as protective genotypes against lung cancer risk (p<0.001). Genetic model analysis showed that rs13098637 was correlated with an elevated risk of lung cancer in dominant and log-additive models (pdominant=0.007, plog-additive=0.004). Moreover, rs879255689 was associated with an increased risk of disease in all three models (pdominant=0.014, precessive=0.028, plog-additive=0.005). In contrast, rs4240897 and rs2236058 were related to reduced risk of disease in all three models (rs4240897: pall<0.001; rs2236058: pdominant=0.008, precessive<0.001, plog-additive<0.001). In addition, these associations were related to the smoking status and pathological type of lung cancer patients.
Conclusion
These results shed new light on the association between mitochondrial dynamics related genes and risk of lung cancer.
Keywords: lung cancer, gene polymorphisms, mitochondrial dynamics, MFN1, MFN2
Introduction
Lung cancer is the malignant tumor with the highest morbidity and mortality in the world.1 At present, it is believed that the interaction of genetic susceptibility, environmental factors, hormone levels and viral infections is the main pathogenic factor for lung cancer.2–4 Lung tissue damage caused by related pathogenic factors can lead to corresponding changes in genes, epigenetics and the entire transcriptome.5 And these changes will affect and gradually lead to the activation of abnormal molecular pathways and cell functions, which will cause precancerous lesions and further develop to lung cancer.6 In recent years, the targeted therapy of lung cancer has made great breakthroughs, but the 5-year survival rate of patients has only increased from 7% to 15%.7,8 The mechanism of the occurrence and development of lung cancer is complex and has not been fully elucidated so far. Therefore, there is an urgent need to explore the core molecules that regulate the occurrence and development of lung cancer, and design a more effective strategy for early prevention and targeted treatment of the disease.
Mitochondria are highly dynamic organelles that are regulated by many members of the GTPases superfamily.9 Generally, mitochondria are connected to each other to form a network structure in cells, and continue to divide and fuse to maintain their normal structure; however, the mitochondrial homeostasis is disturbed in the case of disease.10 The genes involved in mitochondrial dynamics mainly include dynamin-related protein-1 (DRP1), mitochondrial fusion protein 1, 2 (MFN1, MFN2) and so on.11 At present, there have been a large number of reports on the role of mitochondrial division and fusion disorders in tumors. Rehman et al found that the expression level of DRP1 in lung cancer tissues was significantly higher than that in adjacent tissues, while the expression level of MFN1 was significantly reduced in lung cancer tissues, and inhibition of DRP1 activity could significantly inhibit the growth rate of tumors in nude mice.12 Inoue-Yamauchi and Oda reported that inhibition of DRP1 can promote the release of cytochrome C and the apoptosis of colorectal cancer cells.13 In addition, Zhao et al also found that the use of DRP1 inhibitors can significantly inhibit the invasion and metastasis of breast cancer cells.14 These results suggested that mitochondrial division and fusion disorder played an important role in the occurrence and development of tumors. However, to date, little information is found about the single nucleotide polymorphism (SNP) of mitochondrial dynamics related genes in cancer, especially in lung cancer.
Six tag SNPs in mitochondrial dynamics related genes MFN1, MFN2 and DRP1 were selected as candidate SNPs. Rs13098637 and rs3976523 in MFN1 have been investigated in patients with myopia, and the C allele of rs13098637 has been identified as risk allele for low to moderate myopia.15 Rs4240897 in MFN2 was associated with decreased risk of tuberculosis in a genome-wide association study.16 Moreover, MFN2-rs2236058-GG genotype was correlated with reduced risk of thoracic aortic dissection.17 In addition, rs879255685 and rs879255689 in DRP1 were missense variants and associated with developmental delay, refractory epilepsy and altered function of peroxisomes and mitochondria.18–20 In the present study, we genotyped these SNPs in lung cancer patients and healthy controls and evaluated the associations between the SNPs and risk of lung cancer.
Materials and Methods
Participants
The subjects of this study included 600 lung cancer patients and 600 controls. All participants were of Chinese Han ethnicity and were recruited at Tangdu hospital. The patients were diagnosed with lung cancer by histopathological examination of biopsy specimens. The control group included randomly selected healthy individuals with no history of cancer. All participants provided written informed consent. This study was approved by the ethics committee of the hospital (No. 201003–52) and carried out in accordance with the World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects.
Genotyping
Six SNPs in mitochondrial dynamics related genes MFN1, MFN2 and DRP1 were selected based on previous association studies. The minor allele frequencies (MAFs) of these SNPs are greater than 5% in East Asian populations according to the 1000 Genomes database. DNA was extracted using a QIAamp DNA Blood Midi Kit (QIAGEN, Germany). Primers were designed using Sequenom MassARRAY Assay Design 3.0 software. SNP genotyping was performed on Mass ARRAY iPLEX platform (Sequenom, San Diego, CA, USA).
Statistical Analysis
Statistical analysis was performed with SPSS package version 20.0 (SPSS, Chicago, IL, USA). MAFs of each SNP were checked for divergence from Hardy–Weinberg equilibrium (HWE). HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) was used to predict the potential functions of the SNPs. Allele and genotype frequencies in the cases and controls were evaluated using Chi-square tests. The association between SNPs and lung cancer risk was evaluated using SNPstats (https://www.snpstats.net/start.htm) and expressed by odds ratios (ORs) and 95% confidence intervals (CIs). Statistical significance was established when p < 0.05.
Results
The characteristics of the participants are presented in Table 1. The case group includes 384 males and 216 females, 381 smokers and 219 nonsmokers, with a mean age of 57.03 years; and the control group consists of 381 males and 219 females, 378 smokers and 222 nonsmokers, with a mean age of 56.45 years. No significant difference was observed in the distribution of sex, age, or smoking status between the two groups (p > 0.05). In addition, The lung cancer cases consist of 278 adenocarcinoma patients, 187 squamous cell carcinoma patients, 110 small cell lung cancer patients and 25 other types of lung cancer cases.
Table 1.
The Basic Information of the Participants
| Characteristics | Case (n=600) | Control (n=600) | χ2/t | p |
|---|---|---|---|---|
| Gender (%) | 0.032 | 0.858 | ||
| Male | 384 (64.0) | 381 (63.5) | ||
| Female | 216 (36.0) | 219 (36.5) | ||
| Age | 0.674 | 0.344 | ||
| Mean ±SD | 57.03±10.54 | 56.45±10.64 | ||
| Smoking (%) | 0.032 | 0.858 | ||
| Yes | 381 (63.5) | 378 (63.0) | ||
| No | 219 (36.5) | 222 (37.0) | ||
| Pathological types | ||||
| AC | 278 (46.3) | |||
| SCC | 187 (31.2) | |||
| SCLC | 110 (18.3) | |||
| Others | 25 (4.2) |
Abbreviations: AC, adenocarcinoma; SCC, squamous cell carcinoma; SCLC, small cell lung cancer.
The basic information for the candidate SNPs is listed in Table 2. The predicted function according to the HaploReg database showed that the four SNPs in MFN1 and MFN2 were involved in the regulation of the promoter or enhancer histone, changed motifs, and eQTL hits. Moreover, two SNPs in DRP1 were missense variants and led to changed amino acids.
Table 2.
Basic Information and Predicted Functions of Candidate SNPs
| SNP | Gene | Chromosome | Position | Allele | Role | Predicted Functions |
|---|---|---|---|---|---|---|
| rs13098637 | MFN1 | 3 | 179375026 | T>C | Intron | Promoter histone mark, motifs changed, eQTL hits |
| rs3976523 | MFN1 | 3 | 179381391 | A>C | Intron | Motifs changed, eQTL hits |
| rs4240897 | MFN2 | 1 | 11982698 | G>A | Intron | Enhancer histone mark, motifs changed, eQTL hits |
| rs2236058 | MFN2 | 1 | 12002304 | C>G | Intron | Enhancer histone mark, motifs changed, eQTL hits |
| rs879255685 | DRP1 | 12 | 32731019 | G>A | Missense Variant | Gly362Asp |
| rs879255689 | DRP1 | 12 | 32722602 | G>A | Missense Variant | Gly379Lys |
Abbreviations: SNP, single nucleotide polymorphism; eQTL, expression quantitative trait locus.
The MAFs of SNPs in cases and controls were described in Table 3. All of the SNPs were consistent with HWE (p > 0.05). Compared the MAFs of SNPs between cases and controls, we found that the minor allele C of MFN1-rs13098637 was associated with a 1.368-fold increased risk of lung cancer (95% CI: 1.106–1.691, p=0.004). In addition, the minor allele A of DRP1-rs879255689 was correlated with an 1.348-fold elevated risk of disease (95% CI: 1.094–1.662, p=0.005). In contrast, the minor alleles of MFN2 rs4240897-A and rs2236058-G were related to a decreased risk of disease (rs4240897: OR=0.681, 95% CI: 0.572–0.812, p<0.001; rs2236058: OR=0.718, 95% CI: 0.612–0.843, p<0.001).
Table 3.
The MAF and HWE of Candidate SNPs Between Lung Cancer Cases and Healthy Controls
| SNP | Gene | MAF-Cases | MAF-Controls | HWE p | OR (95% CI) | p |
|---|---|---|---|---|---|---|
| rs13098637 | MFN1 | 0.20 | 0.15 | 0.87 | 1.368(1.106–1.691) | 0.004* |
| rs3976523 | MFN1 | 0.36 | 0.33 | 0.78 | 1.138(0.962–1.347) | 0.132 |
| rs4240897 | MFN2 | 0.26 | 0.34 | 0.93 | 0.681(0.572–0.812) | <0.001* |
| rs2236058 | MFN2 | 0.45 | 0.53 | 0.41 | 0.718(0.612–0.843) | <0.001* |
| rs879255685 | DRP1 | 0.20 | 0.18 | 0.10 | 1.107(0.904–1.357) | 0.325 |
| rs879255689 | DRP1 | 0.20 | 0.16 | 0.44 | 1.348(1.094–1.662) | 0.005* |
Note: *p < 0.05 indicates statistical significance.
Abbreviations: SNP, single nucleotide polymorphism; MAF, minor allele frequency; HWE, Hardy–Weinberg equilibrium.
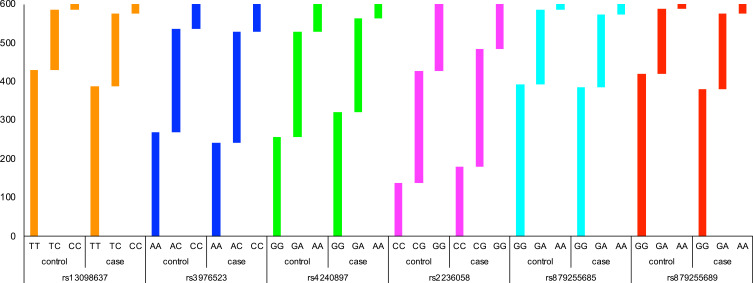
The genotype frequencies of SNPs in cases and controls are shown in Table 4 and Figure 1. Compared with the wild genotype TT, the TC and CC genotypes of MFN1-rs13098637 were associated with 1.34-fold and 2.04-fold increased risk of lung cancer (p=0.014). Similarly, the GA and AA genotypes of DRP1-rs879255689 exhibited 1.28-fold and 2.30-fold elevated risk of disease (p=0.013). However, the GA and AA genotypes of MFN2-rs4240897 were determined to be protective genotypes with 0.71-fold and 0.42-fold reduced risk of lung cancer (p<0.001). In addition, the GG genotype of MFN2-rs2236058 was also found to be protective genotype against lung cancer risk (OR=0.52, 95% CI: 0.37–0.71, p<0.001).
Table 4.
Genotype Frequency Distributions Between Lung Cancer Cases and Healthy Controls
| SNP | Genotype | Control | Case | OR (95% CI) | p |
|---|---|---|---|---|---|
| rs13098637 | T/T | 430 (71.7%) | 387 (64.5%) | 1 | 0.014* |
| T/C | 157 (26.2%) | 189 (31.5%) | 1.34 (1.04–1.73) | ||
| C/C | 13 (2.2%) | 24 (4%) | 2.04 (1.02–4.06) | ||
| rs3976523 | A/A | 269 (44.8%) | 243 (40.5%) | 1 | 0.300 |
| A/C | 268 (44.7%) | 285 (47.5%) | 1.18 (0.92–1.50) | ||
| C/C | 63 (10.5%) | 72 (12%) | 1.26 (0.86–1.85) | ||
| rs4240897 | G/G | 257 (42.8%) | 321 (53.5%) | 1 | <0.001* |
| G/A | 273 (45.5%) | 242 (40.3%) | 0.71 (0.56–0.90) | ||
| A/A | 70 (11.7%) | 37 (6.2%) | 0.42 (0.27–0.64) | ||
| rs2236058 | C/C | 138 (23%) | 179 (29.8%) | 1 | <0.001* |
| C/G | 289 (48.2%) | 306 (51%) | 0.82 (0.62–1.08) | ||
| G/G | 173 (28.8%) | 115 (19.2%) | 0.52 (0.37–0.71) | ||
| rs879255685 | G/G | 393 (65.5%) | 386 (64.3%) | 1 | 0.140 |
| G/A | 193 (32.2%) | 188 (31.3%) | 0.99 (0.77–1.26) | ||
| A/A | 14 (2.3%) | 26 (4.3%) | 1.91 (0.98–3.72) | ||
| rs879255689 | G/G | 421 (70.2%) | 381 (63.5%) | 1 | 0.013* |
| G/A | 167 (27.8%) | 194 (32.3%) | 1.28 (1.01–1.65) | ||
| A/A | 12 (2%) | 25 (4.2%) | 2.30 (1.14–4.65) |
Note: *p < 0.05 indicates statistical significance.
Abbreviations: SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Figure 1.
The genotype distributions of candidate SNPs between lung cancer cases and healthy controls.
The associations between SNPs and risk of disease were further evaluated under genetic models (Table 5). We found that the MFN1-rs13098637 was associated with an increased risk of lung cancer under dominant and log-additive models (pdominant=0.007, plog-additive=0.004). Moreover, DRP1-rs879255689 was correlated with an elevated risk of disease in all three models (pdominant=0.014, precessive=0.028, plog-additive=0.005). In contrast, MFN2 rs4240897 and rs2236058 were both related to reduced risk of disease in all three models (rs4240897: pall<0.001; rs2236058: pdominant=0.008, precessive<0.001, plog-additive<0.001).
Table 5.
Association Between SNPs and Risk of Lung Cancer in Genetic Models
| SNP | Model | Genotype | Control | Case | OR (95% CI) | p |
|---|---|---|---|---|---|---|
| rs13098637 | Dominant | T/T | 430 (71.7%) | 387 (64.5%) | 1 | 0.007* |
| T/C-C/C | 170 (28.3%) | 213 (35.5%) | 1.40 (1.09–1.78) | |||
| Recessive | T/T-T/C | 587 (97.8%) | 576 (96%) | 1 | 0.068 | |
| C/C | 13 (2.2%) | 24 (4%) | 1.87 (0.94–3.70) | |||
| Log-additive | — | — | — | 1.37 (1.11–1.70) | 0.004* | |
| rs3976523 | Dominant | A/A | 269 (44.8%) | 243 (40.5%) | 1 | 0.130 |
| A/C-C/C | 331 (55.2%) | 357 (59.5%) | 1.19 (0.95–1.50) | |||
| Recessive | A/A-A/C | 537 (89.5%) | 528 (88%) | 1 | 0.410 | |
| C/C | 63 (10.5%) | 72 (12%) | 1.16 (0.81–1.67) | |||
| Log-additive | — | — | — | 1.14 (0.96–1.35) | 0.130 | |
| rs4240897 | Dominant | G/G | 257 (42.8%) | 321 (53.5%) | 1 | <0.001* |
| G/A-A/A | 343 (57.2%) | 279 (46.5%) | 0.65 (0.52–0.82) | |||
| Recessive | G/G-G/A | 530 (88.3%) | 563 (93.8%) | 1 | <0.001* | |
| A/A | 70 (11.7%) | 37 (6.2%) | 0.49 (0.32–0.75) | |||
| Log-additive | — | — | — | 0.67 (0.56–0.80) | <0.001* | |
| rs2236058 | Dominant | C/C | 138 (23%) | 179 (29.8%) | 1 | 0.008* |
| C/G-G/G | 462 (77%) | 421 (70.2%) | 0.71 (0.54–0.91) | |||
| Recessive | C/C-C/G | 427 (71.2%) | 485 (80.8%) | 1 | <0.001* | |
| G/G | 173 (28.8%) | 115 (19.2%) | 0.59 (0.45–0.77) | |||
| Log-additive | — | — | — | 0.72 (0.61–0.85) | <0.001* | |
| rs879255685 | Dominant | G/G | 393 (65.5%) | 386 (64.3%) | 1 | 0.680 |
| G/A-A/A | 207 (34.5%) | 214 (35.7%) | 1.05 (0.83–1.33) | |||
| Recessive | G/G-G/A | 586 (97.7%) | 574 (95.7%) | 1 | 0.048* | |
| A/A | 14 (2.3%) | 26 (4.3%) | 1.92 (0.99–3.72) | |||
| Log-additive | — | — | — | 1.11 (0.90–1.36) | 0.320 | |
| rs879255689 | Dominant | G/G | 421 (70.2%) | 381 (63.5%) | 1 | 0.014* |
| G/A-A/A | 179 (29.8%) | 219 (36.5%) | 1.35 (1.06–1.72) | |||
| Recessive | G/G-G/A | 588 (98%) | 575 (95.8%) | 1 | 0.028* | |
| A/A | 12 (2%) | 25 (4.2%) | 2.14 (1.06–4.30) | |||
| Log-additive | — | — | — | 1.36 (1.10–1.68) | 0.005* |
Note: *p < 0.05 indicates statistical significance.
Abbreviations: SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Stratified analysis was carried out in the aspect of smoking status (Table 6) and pathological type of lung cancer (Table 7). We found that MFN1-rs13098637 was associated with risk of lung cancer in both smokers and nonsmokers; while DRP1-rs879255689 was only related to risk of disease in nonsmokers (p<0.05). Moreover, MFN2-rs4240897 was correlated with declining risk of disease in both smokers and nonsmokers, while MFN2-rs2236058 was only significant in smokers (p<0.05). In addition, MFN1-rs13098637 was associated with increased risk of adenocarcinoma and small cell lung cancer, and DRP1-rs879255689 was related to elevated risk of squamous cell carcinoma and small cell lung cancer (p<0.05). In contrast, MFN2-rs4240897 was protective variant for all three types of lung cancer (p<0.05). However, MFN2-rs2236058 was not significant in any types of disease, which may due to the limited sample size.
Table 6.
Association Between SNPs and Risk of Lung Cancer in Smokers and Nonsmokers
| SNP | Model | Genotype | Smokers | Nonsmokers | ||
|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | |||
| rs13098637 | Dominant | T/T | 1 | 0.028* | 1 | 0.130 |
| T/C-C/C | 1.41 (1.04–1.93) | 1.36 (0.91–2.02) | ||||
| Recessive | T/T-T/C | 1 | 0.980 | 1 | 0.004* | |
| C/C | 1.01 (0.42–2.46) | 5.06 (1.42–18.03) | ||||
| Log-additive | — | 1.31 (1.00–1.72) | 0.052 | 1.47 (1.04–2.07) | 0.026* | |
| rs3976523 | Dominant | A/A | 1 | 0.320 | 1 | 0.230 |
| A/C-C/C | 1.16 (0.87–1.54) | 1.26 (0.87–1.84) | ||||
| Recessive | A/A-A/C | 1 | 0.520 | 1 | 0.550 | |
| C/C | 1.17 (0.73–1.87) | 1.19 (0.67–2.10) | ||||
| Log-additive | — | 1.12 (0.90–1.40) | 0.290 | 1.18 (0.89–1.56) | 0.240 | |
| rs4240897 | Dominant | G/G | 1 | 0.014* | 1 | 0.004* |
| G/A-A/A | 0.70 (0.52–0.93) | 0.57 (0.39–0.84) | ||||
| Recessive | G/G-G/A | 1 | 0.039* | 1 | 0.002* | |
| A/A | 0.60 (0.37–0.98) | 0.30 (0.13–0.68) | ||||
| Log-additive | — | 0.73 (0.59–0.91) | 0.005* | 0.57 (0.42–0.78) | <0.001* | |
| rs2236058 | Dominant | C/C | 1 | 0.420 | 1 | 0.180 |
| C/G-G/G | 0.88 (0.64–1.20) | 1.40 (0.99–3.11) | ||||
| Recessive | C/C-C/G | 1 | 0.012* | 1 | 0.230 | |
| G/G | 0.65 (0.46–0.91) | 1.35 (0.46–2.79) | ||||
| Log-additive | – | 0.82 (0.67–0.99) | 0.049* | 1.81 (0.36–2.41) | 0.140 | |
| rs879255685 | Dominant | G/G | 1 | 0.930 | 1 | 0.550 |
| G/A-A/A | 1.01 (0.75–1.36) | 1.13 (0.76–1.68) | ||||
| Recessive | G/G-G/A | 1 | 0.680 | 1 | 0.052 | |
| A/A | 1.19 (0.51–2.80) | 3.78 (1.22–11.71) | ||||
| Log-additive | — | 1.03 (0.79–1.34) | 0.840 | 1.26 (0.90–1.76) | 0.170 | |
| rs879255689 | Dominant | G/G | 1 | 0.110 | 1 | 0.042* |
| G/A-A/A | 1.28 (0.95–1.72) | 1.52 (1.01–2.30) | ||||
| Recessive | G/G-G/A | 1 | 0.100 | 1 | 0.140 | |
| A/A | 2.20 (0.83–5.85) | 2.08 (0.77–5.65) | ||||
| Log-additive | — | 1.30 (0.99–1.70) | 0.056 | 1.46 (1.03–2.06) | 0.030* | |
Note: *p < 0.05 indicates statistical significance.
Abbreviations: SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Table 7.
Association Between SNPs and Risk of Adenocarcinoma, Squamous Cell Carcinoma and Small Cell Lung Cancer
| SNP | Model | Genotype | Adenocarcinoma | Squamous Cell Carcinoma | Small Cell Lung Cancer | |||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | |||
| rs13098637 | Dominant | T/T | 1 | 0.019* | 1 | 0.180 | 1 | 0.009* |
| T/C-C/C | 1.44 (1.06–1.95) | 1.28 (0.90–1.83) | 1.78 (1.16–2.71) | |||||
| Recessive | T/T-T/C | 1 | 0.028* | 1 | 0.760 | 1 | 0.420 | |
| C/C | 2.41 (1.11–5.25) | 1.18 (0.41–3.38) | 1.63 (0.52–5.13) | |||||
| Log-additive | — | 1.44 (1.11–1.87) | 0.006* | 1.23 (0.90–1.69) | 0.2 | 1.62 (1.13–2.33) | 0.011* | |
| rs3976523 | Dominant | A/A | 1 | 0.071 | 1 | 0.066 | 1 | 0.047 |
| A/C-C/C | 1.31 (0.98–1.75) | 1.38 (0.98–1.94) | 0.66 (0.44–1.00) | |||||
| Recessive | A/A-A/C | 1 | 0.670 | 1 | 0.820 | 1 | 0.520 | |
| C/C | 1.10 (0.70–1.73) | 1.06 (0.62–1.82) | 1.23 (0.66–2.29) | |||||
| Log-additive | — | 1.19 (0.96–1.47) | 0.120 | 1.21 (0.94–1.56) | 0.140 | 0.83 (0.60–1.13) | 0.230 | |
| rs4240897 | Dominant | G/G | 1 | 0.011* | 1 | 0.029* | 1 | 0.015* |
| G/A-A/A | 0.69 (0.52–0.92) | 0.69 (0.49–0.96) | 0.60 (0.40–0.91) | |||||
| Recessive | G/G-G/A | 1 | 0.009* | 1 | 0.006* | 1 | 0.053 | |
| A/A | 0.50 (0.28–0.86) | 0.41 (0.20–0.82) | 0.45 (0.19–1.05) | |||||
| Log-additive | — | 0.70 (0.56–0.88) | 0.002* | 0.68 (0.52–0.89) | 0.004* | 0.63 (0.45–0.88) | 0.006* | |
| rs2236058 | Dominant | C/C | 1 | 0.150 | 1 | 0.082 | 1 | 0.086 |
| C/G-G/G | 1.84 (0.89–2.62) | 1.41 (0.95–2.10) | 1.96 (0.76–3.31) | |||||
| Recessive | C/C-C/G | 1 | 0.260 | 1 | 0.220 | 1 | 0.130 | |
| G/G | 1.46 (0.65–2.02) | 1.27 (0.87–1.84) | 1.43 (0.91–2.26) | |||||
| Log-additive | — | 1.44 (0.77–1.77) | 0.054 | 1.24 (0.98–1.56) | 0.072 | 1.45 (0.78–1.94) | 0.052 | |
| rs879255685 | Dominant | G/G | 1 | 0.610 | 1 | 0.860 | 1 | 0.720 |
| G/A-A/A | 1.08 (0.80–1.46) | 1.03 (0.73–1.46) | 1.08 (0.71–1.65) | |||||
| Recessive | G/G-G/A | 1 | 0.470 | 1 | 0.172 | 1 | 0.250 | |
| A/A | 1.38 (0.59–3.25) | 3.13 (0.94–7.01) | 1.91 (0.67–5.44) | |||||
| Log-additive | — | 1.10 (0.84–1.43) | 0.500 | 1.18 (0.88–1.59) | 0.280 | 1.14 (0.79–1.65) | 0.490 | |
| rs879255689 | Dominant | G/G | 1 | 0.230 | 1 | 0.023* | 1 | 0.045* |
| G/A-A/A | 1.21 (0.89–1.65) | 1.50 (1.06–2.12) | 1.55 (1.02–2.36) | |||||
| Recessive | G/G-G/A | 1 | 0.230 | 1 | 0.120 | 1 | 0.018* | |
| A/A | 1.71 (0.72–4.03) | 2.21 (0.84–5.80) | 3.46 (1.32–9.08) | |||||
| Log-additive | — | 1.22 (0.93–1.60) | 0.150 | 1.48 (1.09–2.01) | 0.013* | 1.60 (1.12–2.29) | 0.011* | |
Note: *p < 0.05 indicates statistical significance.
Abbreviations: SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Discussion
Abnormal division and fusion of mitochondria not only lead to altered morphology and function but also closely related to the occurrence and development of diseases and tumors.21,22 In this study, we genotyped six SNPs in three mitochondrial dynamics related genes MFN1, MFN2 and DRP1, in a case-control cohort and found that two SNPs (MFN1-rs13098637 and DRP1-rs879255689) associated with an increased risk of lung cancer and two SNPs (MFN2 rs4240897 and rs2236058) associated with a reduced risk of the disease.
MFN1 gene is located at chromosome 3 q25-26, with a molecular weight of 84kDa and consists of 742 amino acid. MFN1 is located in the outer mitochondrial membrane, and its N-terminus and C-terminus are both exposed to the cytoplasm. There is a GTPase domain at its N-terminus, which participates in the oligomerization of mitochondrial fusion-related proteins and promotes the fusion of adjacent mitochondrial outer membranes.23 Moreover, MFN1 is also involved in many physiological functions such as maintaining the number of healthy mitochondria in the cell and reducing the content of intracellular ROS.24 Li et al found that MFN1 was lowly expressed in osteosarcoma tissue and related to the poor prognosis, and overexpression of MFN1 can lead to osteosarcoma cell cycle arrest, inhibit cell proliferation and promote apoptosis.25 Zhao et al reported that knockdown of MFN1 in breast cancer cell lines can lead to mitochondrial fragmentation and promote breast cancer cell metastasis, while overexpression of MFN1 can inhibit the formation of lamellipodia in breast cancer cells and reduce the recruitment of mitochondria to the lamina area.14 Huang et al demonstrated that the ratio of DRP1/MFN1 was significantly up-regulated in liver cancer tissues, with increased mitochondrial division and decreased fusion, thereby promoting mitochondrial autophagy and inhibiting mitochondrial-dependent apoptosis, and ultimately promoting the proliferation and growth of liver cancer cells.26 In this study, we identified that the minor allele C of MFN1-rs13098637 was associated with an increased risk of lung cancer, suggesting that rs13098637 polymorphism may have an influence on the development of lung cancer via disturbing the mitochondrial homeostasis in the human body.
MFN2 gene is located at chromosome 1 and it has 80% similar protein sequence with MFN1. MFN2 is also an outer mitochondrial membrane GTPase and involved in mitochondrial dynamics and function. In addition, MFN2 could affects the interaction between endoplasmic reticulum (ER) and mitochondria and ER stress response, which is distinct from MFN1.27 Abnormal expression of MFN2 has been associated with variable types of disease, including Alzheimer’s disease, Parkinson’s disease, obesity, diabetes/insulin resistance, and cardiomyopathy.27 In addition, previous studies have shown that MFN2 expression was downregulated in lung, liver, colorectal and breast cancers.12,13,28,29 The decreased MFN2 levels affected the mitochondrial fragmentation, and the mitochondrial fragmentation was proven to be a protective factor against Ca2+-dependent apoptosis.30 In our study, we identified that MFN2 rs4240897 and rs2236058 polymorphisms associated with decreased risk of lung cancer, which is consistent with previous association study on tuberculosis and thoracic aortic dissection,16,17 respectively. However, the underlying molecular mechanism is still need to be investigated in further study.
DRP1 gene is located at chromosome 12 and encodes the most important mitochondrial fission related protein. DRP1 is also a cytosolic GTPase, which can be recruited to the outer mitochondrial membrane and exert its function.31 Mitochondrial fission is critical for tissue development and function, and organelle Ca2+ homeostasis and cell apoptotic signaling.30,32 Therefore, DRP1 has been widely involved in the development of variable types of diseases and cancers. Yu et al reported that the expression of DRP1 was significantly upregulated in lung cancer tissues and associated with poor prognosis of patients.33 Hu et al revealed that ROS-mediating CaMKII/DRP1 signaling played a crucial role in the regulation of mitochondrial fission and apoptosis in triple-negative breast cancer cells.34 Moreover, Deng et al found that the anti-tumor effect of baicalein in lung cancer depended on the DRP1-mediated mitochondrial fission to a large extent.35 In addition, Liang et al demonstrated that DRP1 was upregulated in pancreatic cancer and let to more mitochondrial fission and enhanced aerobic glycolysis, which resulting in cancer cell growth and metastasis.36 In the present study, we identified that DRP1-rs879255689 was related to elevated risk of lung cancer. Rs879255689 is a missense variant and led to Gly379Lys, therefore, we speculated that this variant may change the mitochondrial fission and dynamics of lung cancer patients and participant in the development of the disease.
To assess the interaction between SNPs and smoking and the associations in different types of lung cancer, we performed a stratification analysis. We found that MFN1-rs13098637 and MFN2-rs4240897 have associations with risk of lung cancer in both smokers and nonsmokers, suggesting that these polymorphisms have strong relationships with risk of disease. However, the MFN2-rs2236058 was only significant in smokers, suggesting that rs2236058 polymorphisms may have interaction with smoking. In addition, MFN1-rs13098637 and DRP1-rs879255689 was related to different types of lung cancer, which further demonstrated that different types of cancer can be caused by different mechanisms.
To sum up, we found that MFN1-rs13098637 and DRP1-rs879255689 polymorphisms were associated with an increased risk of lung cancer, while MFN2 rs4240897 and rs2236058 were protective variants against the risk of the disease. Our results shed new light on the association between mitochondrial dynamics related genes and risk of lung cancer.
Acknowledgments
We are grateful to the patients and control subjects for their participation in this study.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Malhotra J, Malvezzi M, Negri E, La Vecchia C, Boffetta P. Risk factors for lung cancer worldwide. Eur Respir J. 2016;48(3):889–902. doi: 10.1183/13993003.00359-2016 [DOI] [PubMed] [Google Scholar]
- 3.Argirion I, Weinstein SJ, Männistö S, Albanes D, Mondul AM. Serum insulin, glucose, indices of insulin resistance, and risk of lung cancer. Cancer Epidemiol Biomarkers Prev. 2017;26(10):1519–1524. doi: 10.1158/1055-9965.EPI-17-0293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belluomini L, Caldart A, Avancini A, et al. Infections and immunotherapy in lung cancer: a bad relationship? Int J Mol Sci. 2020;22(1):42. doi: 10.3390/ijms22010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida K, Gowers KHC, Lee-Six H, et al. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature. 2020;578(7794):266–272. doi: 10.1038/s41586-020-1961-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Łagiedo M, Sikora J, Kaczmarek M. Damage-associated molecular patterns in the course of lung cancer–a review. Scand J Immunol. 2015;82(2):95–101. doi: 10.1111/sji.12308 [DOI] [PubMed] [Google Scholar]
- 7.Yi L, Zhang W, Zhang H, et al. Systematic review and meta-analysis of the benefit of celecoxib in treating advanced non-small-cell lung cancer. Drug Des Devel Ther. 2018;12:2455–2466. doi: 10.2147/DDDT.S169627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L. Short- and long-term outcomes in elderly patients with locally advanced non-small-cell lung cancer treated using video-assisted thoracic surgery lobectomy. Ther Clin Risk Manag. 2018;14:2213–2220. doi: 10.2147/TCRM.S175846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Bliek AM, Shen Q, Kawajiri S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb Perspect Biol. 2013;5(6):a011072. doi: 10.1101/cshperspect.a011072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie LL, Shi F, Tan Z, Li Y, Bode AM, Cao Y. Mitochondrial network structure homeostasis and cell death. Cancer Sci. 2018;109(12):3686–3694. doi: 10.1111/cas.13830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8(11):870–879. doi: 10.1038/nrm2275 [DOI] [PubMed] [Google Scholar]
- 12.Rehman J, Zhang HJ, Toth PT, et al. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26(5):2175–2186. doi: 10.1096/fj.11-196543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue-Yamauchi A, Oda H. Depletion of mitochondrial fission factor DRP1 causes increased apoptosis in human colon cancer cells. Biochem Biophys Res Commun. 2012;421(1):81–85. doi: 10.1016/j.bbrc.2012.03.118 [DOI] [PubMed] [Google Scholar]
- 14.Zhao J, Zhang J, Yu M, et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32(40):4814–4824. doi: 10.1038/onc.2012.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou YC, Lei JH, Wang Y, Xu S. Correlation between polymorphisms in the MFN1 gene and myopia in Chinese population. Int J Ophthalmol. 2015;8(6):1126–1130. doi: 10.3980/j.issn.2222-3959.2015.06.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi H, Zhang YB, Sun L, et al. Discovery of susceptibility loci associated with tuberculosis in Han Chinese. Hum Mol Genet. 2017;26(23):4752–4763. doi: 10.1093/hmg/ddx365 [DOI] [PubMed] [Google Scholar]
- 17.Han J, Liu J, Zhou Q, Nie S, Liu J, Wen S. Single nucleotide polymorphisms (SNPs) genotyping reveals that Mfn2 polymorphisms are associated with thoracic aortic dissection in Han Chinese population. Med Sci Monit. 2019;25:2419–2428. doi: 10.12659/MSM.915272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang CR, Manlandro CM, Arnoult D, et al. A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J Biol Chem. 2010;285(42):32494–32503. doi: 10.1074/jbc.M110.142430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanstone JR, Smith AM, McBride S, et al. DNM1L-related mitochondrial fission defect presenting as refractory epilepsy. Eur J Hum Genet. 2016;24(7):1084–1088. doi: 10.1038/ejhg.2015.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao YH, Robak LA, Xia F, et al. Missense variants in the middle domain of DNM1L in cases of infantile encephalopathy alter peroxisomes and mitochondria when assayed in Drosophila. Hum Mol Genet. 2016;25(9):1846–1856. doi: 10.1093/hmg/ddw059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan DC. Mitochondrial dynamics and its involvement in disease. Annu Rev Pathol. 2020;15:235–259. doi: 10.1146/annurev-pathmechdis-012419-032711 [DOI] [PubMed] [Google Scholar]
- 22.Chen H, Chan DC. Mitochondrial dynamics in regulating the unique phenotypes of cancer and stem cells. Cell Metab. 2017;26(1):39–48. doi: 10.1016/j.cmet.2017.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zorzano A, Liesa M, Sebastián D, Segalés J, Palacín M. Mitochondrial fusion proteins: dual regulators of morphology and metabolism. Semin Cell Dev Biol. 2010;21(6):566–574. doi: 10.1016/j.semcdb.2010.01.002 [DOI] [PubMed] [Google Scholar]
- 24.Ishihara N, Otera H, Oka T, Mihara K. Regulation and physiologic functions of GTPases in mitochondrial fusion and fission in mammals. Antioxid Redox Signal. 2013;19(4):389–399. doi: 10.1089/ars.2012.4830 [DOI] [PubMed] [Google Scholar]
- 25.Li X, Wang FS, Wu ZY, Lin JL, Lan WB, Lin JH. MicroRNA-19b targets Mfn1 to inhibit Mfn1-induced apoptosis in osteosarcoma cells. Neoplasma. 2014;61(3):265–273. doi: 10.4149/neo_2014_034 [DOI] [PubMed] [Google Scholar]
- 26.Huang Q, Cao H, Zhan L, et al. Mitochondrial fission forms a positive feedback loop with cytosolic calcium signaling pathway to promote autophagy in hepatocellular carcinoma cells. Cancer Lett. 2017;403:108–118. doi: 10.1016/j.canlet.2017.05.034 [DOI] [PubMed] [Google Scholar]
- 27.Filadi R, Pendin D, Pizzo P. Mitofusin 2: from functions to disease. Cell Death Dis. 2018;9(3):330. doi: 10.1038/s41419-017-0023-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W, Lu J, Zhu F, et al. Pro-apoptotic and anti-proliferative effects of mitofusin-2 via Bax signaling in hepatocellular carcinoma cells. Med Oncol. 2012;29(1):70–76. doi: 10.1007/s12032-010-9779-6 [DOI] [PubMed] [Google Scholar]
- 29.Cheng X, Zhou D, Wei J, Lin J. Cell-cycle arrest at G2/M and proliferation inhibition by adenovirus-expressed mitofusin-2 gene in human colorectal cancer cell lines. Neoplasma. 2013;60(6):620–626. doi: 10.4149/neo_2013_080 [DOI] [PubMed] [Google Scholar]
- 30.Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ, Rizzuto R. Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol Cell. 2004;16(1):59–68. doi: 10.1016/j.molcel.2004.09.026 [DOI] [PubMed] [Google Scholar]
- 31.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12(8):2245–2256. doi: 10.1091/mbc.12.8.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Favaro G, Romanello V, Varanita T, et al. DRP1-mediated mitochondrial shape controls calcium homeostasis and muscle mass. Nat Commun. 2019;10(1):2576. doi: 10.1038/s41467-019-10226-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu L, Xiao Z, Tu H, Tong B, Chen S. The expression and prognostic significance of Drp1 in lung cancer: a bioinformatics analysis and immunohistochemistry. Medicine (Baltimore). 2019;98(48):e18228. doi: 10.1097/MD.0000000000018228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J, Zhang Y, Jiang X, et al. ROS-mediated activation and mitochondrial translocation of CaMKII contributes to Drp1-dependent mitochondrial fission and apoptosis in triple-negative breast cancer cells by isorhamnetin and chloroquine. J Exp Clin Cancer Res. 2019;38(1):225. doi: 10.1186/s13046-019-1201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng X, Liu J, Liu L, Sun X, Huang J, Dong J. Drp1-mediated mitochondrial fission contributes to baicalein-induced apoptosis and autophagy in lung cancer via activation of AMPK signaling pathway. Int J Biol Sci. 2020;16(8):1403–1416. doi: 10.7150/ijbs.41768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang J, Yang Y, Bai L, Li F, Li E. DRP1 upregulation promotes pancreatic cancer growth and metastasis through increased aerobic glycolysis. J Gastroenterol Hepatol. 2020;35(5):885–895. doi: 10.1111/jgh.14912 [DOI] [PubMed] [Google Scholar]