Abstract
NK cells and diverse populations of unconventional T cells, such as MAIT cells, γδ T cells, invariant NKT cells, and DNTɑβ cells are important early effector lymphocytes. While some of these cells, such as NK cell and MAIT cells, have well-established roles in antiviral defense, the function of other populations remains more elusive. Here, we summarize and discuss current knowledge on NK cell and unconventional T cell responses to SARS-CoV-2 infection. Also covered is the role of these cells in the pathogenesis of severe COVID-19. Understanding the early, both systemic and local (lung), effector lymphocyte response in this novel disease will likely aid ongoing efforts to combat the pandemic.
Current Opinion in Virology 2021, 49:176–182
This review comes from a themed issue on Viral immunology
Edited by Antonio Bertoletti and Matteo Iannacone
For complete overview about the section, refer Viral immunology
Available online 19th June 2021
https://doi.org/10.1016/j.coviro.2021.06.005
1879-6257/© 2021 The Author(s). Published by Elsevier B.V. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Introduction
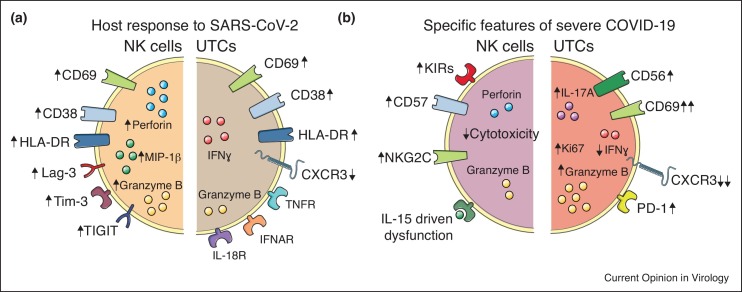
Since severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in late 2019 and spread throughout the world [1,2], we have in a short time gained in-depth understanding of both the virus, the host immune response against it, and factors contributing to coronavirus disease 2019 (COVID-19) [3]. In this Review, current knowledge of antiviral responses of natural killer (NK) cells and unconventional T cells in SARS-CoV-2 infection will be summarized followed by a discussion on the role of these cells in the aberrant immune response characteristic of severe COVID-19 (Figure 1 ). NK cells are innate lymphocytes abundant in peripheral blood but also found in many peripheral tissues including liver, lung, and uterus [4]. They respond without prior sensitization and are important in tumor surveillance, pregnancy, immune homeostasis, and as early (cytotoxic) effector cells during viral infections [5]. Indeed, the most compelling evidence for the importance of NK cells in antiviral immunity comes from patients with selective NK cell deficiencies who often suffer from life-threatening viral infections [6]. Unconventional T cells constitute an ever-expanding family of distinct T cell subpopulations that, instead of showing reactivity against conventional peptide — major histocompatibility complex (MHC) proteins, recognize a diverse set of ligands presented on MHC or MHC class I like molecules [7] (Box 1 ). Although perhaps less studied in the context of viral infections as compared to NK cells, unconventional T cells such as mucosal-associated invariant T cells (MAIT cells), CD1d-reactive natural killer T (NKT) cells, CD4 and CD8 double-negative αβ T cells (DNTαβ cells), and γδ T cells have also been reported to respond in SARS-CoV-2 infection [8••,9••,10••].
Figure 1.
Phenotypic and function alterations in NK cells and UTCs in SARS-CoV-2 infection.
(a) The host response of NK cells and UTCs to SARS-CoV-2 infection is illustrated with modulation of surface receptors (arrows indicate change in expression) as well as intracellular effector molecules. (b) Specific features of NK cells and UTCs that have been associated with severe COVID-19.
Box 1. Unconventional T cell populations.
The unconventional T cell populations covered in the current review are briefly introduced below. Except for these, other populations exist (reviewed in Ref. [7]) but have until now not been studied in COVID-19.
MAIT cells: Defined by co-expression of TCR-Vα7.2 and CD161 (or using a 5-OP-RU tetramer), predominantly CD4/CD8 double negative or CD8+, and recognizes riboflavin metabolites presented on MR1. Enriched at mucosal barriers and in the liver and have primarily been studied in the context of bacterial infections.
CD1d-restricted NKT cells: Displaying an invariant TCR composed of Vα24 typically paired with Vβ11 recognizing, among other things, endogenous glycolipids presented in CD1d. These cells have the capacity to produce a broad range of Th1, Th2, and Th17 cytokines as well as exhibiting cellular cytotoxicity.
γδ T cells: First T cells to appear in the thymus during its ontogeny, human γδ T cells mainly express the Vδ2 (coupled with Vγ9) and, to a lesser extent, the Vδ1 TCR chains. Their TCR can recognize exogenous and endogenous molecules, including bacterial toxoids, viral proteins, microbial lipids (presented by CD1d), and phosphoantigens (through the expression of butyrophilins).
DNTɑβ cells: Mainly studied in the context of autoimmunity, they are also involved in anti-tumor immunity. In mice, DNTɑβ originate from the thymus and possess a polyclonal TCR repertoire, although distinct from other UTC subsets and conventional T cells. Their antigen specificity and TCR restriction is still unknown.
Alt-text: Box 1
NK cell and unconventional T cell responses against SARS-CoV-2
Since only a fraction of individuals infected with SARS-CoV-2 develop severe COVID-19 and aberrant immune responses have been associated with COVID-19 pathogenesis it is of importance to delineate beneficial and/or appropriate antiviral responses from other responses rather contributing to disease. Thus, in the upcoming paragraphs, beneficial responses from NK cells and unconventional T cells will first be discussed.
The early NK cell response to SARS-CoV-2 infection
NK cells are classically divided into cytokine producing CD56bright NK cells and cytotoxic CD56dim NK cells [4] and CD56dim NK cells can be further stratified in less or more differentiated subsets based on surface expression of receptors such as NKG2A, CD57, CD62L, and KIRs [11,12]. In acute SARS-CoV-2 infection, both the CD56bright and CD56dim NK cell subset drop in cell numbers in circulation [13••,14••,15] and this occurs even in mild infection [16]. This drop in numbers likely reflects active homing of NK cells from circulation to the lung since increased presence of NK cells in bronchioalveolar lavage (BAL) has been reported [17,18]. This homing is likely mediated by CXCR3, CXCR6, CCR5 on NK cells [19] and respective chemokines are increased in BAL of COVID-19 patients [17,20]. In this regard, it is interesting to note that CXCR6 is part of the major genetic risk loci for development of severe COVID-19 [21]. A role for CXCR3 and CCR5 for NK cell homing to lung is in line with what has been reported in acute influenza A virus (IAV) infection in mice [22]. However, similar chemokine receptor — chemokine combinations have also been suggested to direct NK cell homing to skin during acute dengue virus infection [23] suggesting them to not be specific for lung homing in acute SARS-CoV-2 infection.
Beyond reduced numbers of NK cells in peripheral blood and subsequent homing to lung tissue, NK cells also display a highly activated phenotype in acute SARS-CoV-2 infection, are actively proliferating, and have retained functional capacity [13••,14••,24]. NK cell activation was reported to be in particular pronounced among CD56bright NK cells and within CD56dim NK cell subsets displaying a less differentiated phenotype [13••]. It also occurred independently of NK cell education status [13••], a functional maturation process NK cells undergo regulated by inhibitory receptors recognizing cognate HLA molecules [25]. Such a response-profile is similar to what has previously been reported in several other acute viral infections, including those by dengue virus, tick-born encephalitis, West Nile virus, and attenuated yellow fever virus [23,26, 27, 28]. Altogether, this would suggest the NK cell response to be cytokine-driven in acute SARS-CoV-2 infection since CD56bright NK cells and less differentiated CD56dim NK cells are superior in responding to cytokines as compared to more differentiated CD56dim NK cells [11,12]. However, the exact signals driving NK cell activation in acute SARS-CoV-2 infection remains to be elucidated.
Beyond peripheral blood NK cells, lung also contains tissue-resident NK cells [4] and these have been shown to respond to IAV [29]. However, despite several studies utilizing single-cell RNA sequencing (scRNA-seq) on BAL of COVID-19 patients, through which it is possible to assess the local NK cell response [13••,17,18], a detailed mapping of the lung-resident NK cell compartment still remains to be performed. Additionally, if, and through which receptor ligand interactions, NK cells possibly recognize SARS-CoV-2 infected target cells should be studied in the future.
MAIT cells in SARS-CoV-2 infection
A wide range of viruses, including influenza A (IAV), hepatitis C (HCV), dengue and human immunodeficiency virus (HIV) have been reported to induce MAIT cell activation in humans (reviewed in detail in Refs. [30] and [31]). As viruses do not synthesize riboflavin (see Box 1), the functional regulation of MAIT cells in viral infections is believed to occur through TCR-independent mechanisms, particularly through interleukin (IL)-12, IL-18 and type I interferons (IFNɑ) [31].
In SARS-CoV-2 infection, several groups have reported a substantial quantitative reduction of the circulating MAIT cell pool, in keeping with previous observations in other viral infections [30]. On the other hand, residual circulating MAIT cells showed a significant increase in expression of several activation markers (e.g. CD69 and granzyme B) [8••,9••,10••,32, 33, 34, 35]. Functionally, circulating MAIT cells are skewed towards a type 3 inflammatory state, and display impaired IFN-ɣ production potential and increased IL-17A release [9••,10••], similar to the Th17 skewing described for CD4+ T cells in severe COVID-19 [36]. Intriguingly, a recent study proposed that gender might have an impact on MAIT activation and lung infiltration [37], but further work is needed to further investigate such findings. MAIT cell depletion in peripheral blood is likely secondary to their migration to the inflamed lung, as supported by the increased MAIT cell frequencies in bronchoalveolar fluid (BALF), pleural fluid (PF), and endotracheal aspirates (ETA) of COVID-19 patients [9••,10••,32]. Chemokines such as CCL20, CXCL10/11 and CXCL16, have been proposed to drive MAIT cell recruitment, and scRNAseq data on putative ligand-receptor interactions indicate myeloid cells (i.e. neutrophils and macrophages) as a primary source of chemotactic signals in the lung [32]. In light of the occurrence of secondary bacterial infections in severe COVID-19 [38], a TCR-dependent MAIT cell activation mechanism is possible, through the MR1-dependent recognition of riboflavin derivatives of microbial origin [8••,39]. Moreover, work from Flament et al. showed an MR1-dependent MAIT cell activation after co-incubation with SARS-CoV-2 infected macrophages [8••], although the antigenic determinants of this activation remain unclear. On the other hand, TCR expression levels, usually decreased in case of TCR engagement, have been found unaltered in MAIT cells and other unconventional T cells from COVID-19 patients, suggesting a preferential cytokine-driven activation [10••].
Finally, in relation to vaccines against SARS-CoV-2, a seminal study investigating the mechanisms of immune response upon vaccination using adenoviral-based platforms (such as ChAdOx1) dissected a cytokine network where pDC and monocyte-derived factors (i.e. IFNɑ and TNF/IL-18) promote MAIT cell activation and subsequent release of anti-viral cytokines such as granzyme B and IFN-ɣ [40]. Importantly, genetic evidence from Mr1 −/− mice support the view of MAIT cells as crucial determinants of adenoviral-based vaccination efficacy, and future studies should determine whether this importance is conserved also in case of mRNA-based vaccination.
Responses from other unconventional T cell populations
The peripheral lymphodepletion coupled with an activated and proliferative state of residual circulating cells induced by acute SARS-CoV-2 infection applies also to other UTC subsets, in addition to MAIT cells. In ɣδ T cells, a specific reduction of the Vδ2 subset associated with COVID-19 severity has been confirmed in many studies, while the abundance of the Vδ1 subset was mainly unaffected by SARS-CoV-2 infection [10••,32,33,41, 42, 43]. Both Vδ1 and Vδ2 subsets are known to mediate wide anti-viral effects [44], but interestingly a selective expansion of the Vδ2 fraction was reported after SARS-CoV-1 infection during the 2003 outbreak [45]. In addition, Odlak et al. observed that naïve ɣδ T cells were selectively maintained in circulation, suggesting a specific recruitment of the mature/effector fraction to the lung [43]. However, ɣδ T cell frequency in ETA, BAL and PF was not different from that observed in blood [10••,32]. In line with the general immune recovery following virus clearance, ɣδ T cell abundance is normalized in convalescent patients [46], although follow-up studies would be important to assess eventual long-term effects of acute SARS-COV-2-infection.
Work specifically focused on iNKT cells is still largely lacking, and available evidence so far described conflicting results, likely due to different subset identification strategies. As an example, Jouan et al. observed a ≈10-fold reduction of iNKT cell frequency in severe COVID-19 patients, while we and others did not detect significant quantitative alterations in either mild or severe COVID-19 [9••,42]. Surprisingly, Stephenson et al. reported that the enrichment of iNKT cell abundance was positively associated with COVID-19 severity [34]. Thus, further work is needed to explain such discrepancies and investigate the functional alterations within the iNKT pool in SARS-CoV-2 infection.
While DNTɑβ have been mainly overlooked in the context of COVID-19, we reported a striking proliferative capacity in this subset, coupled with substantial induction of conventional activation markers such as CD38, but not CD69 [9••]. As for other UTC subsets, DNTɑβ downregulate CXCR3 in severe COVID-19 patients, suggesting a potential lung recruitment mediated by CXCL9/10/11 [9••]. Finally, a CyTOF-based longitudinal immuno-monitoring highlighted the tight co-regulation of DNTɑβ and neutrophil frequencies during COVID-19 recovery [46], presenting unexpected analogies with our previous observations in mice [47].
Taken together, while compelling evidence have so far highlighted the deep alterations occurring within several members of the UTC family, many aspects of the cytokine-driven and, potentially, TCR-dependent UTC response to SARS-COV-2 infection and recovery remain elusive and require further investigation.
COVID-19 pathogenesis, NK cells, and unconventional T cells
Whilst most individuals infected with SARS-CoV-2 develop mild symptoms, a subgroup instead enters a severe (and sometimes fatal) disease course where inappropriate immune responses are thought to be major contributors. Below, current knowledge on the possible contribution of NK cells and UTCs to such outcomes are discussed.
NK cell dysfunction in severe COVID-19
NK cell hyperactivation, likely driven by IL-6, IL-6R, and IL-18, is a feature of severe COVID-19 as compared to mild or moderate disease [13••,48]. However, with prolonged hyperactivation comes dysfunction by which prolonged IL-15 stimulation has been suggested to contribute [49••,50]. In line with this, through scRNA-seq, genes involved in cytotoxic activity are suppressed in NK cells in severe COVID-19 [51]. Indeed, chronic stimulation with IL-15 has been shown to drive NK cell dysfunction, partly via epigenetic reprogramming [52]. Another feature of severe COVID-19, first reported by Maucourant et al. and later confirmed in two other independent studies, is the expansion of adaptive-like NK cells [13••,24,53]. Adaptive-like NK cell expansions were originally described in response to cytomegalovirus (CMV) infection [54], are characterized by high expression of NKG2C, CD57, and inhibitory self-KIR receptors [55] and undergo epigenetic reprogramming during their differentiation [56]. Except for in CMV infection, such expansions have previously also been reported in hantavirus [57], chikungunya virus [58], and HIV-1 infections [59], but always on a CMV-background similar to now in COVID-19 [13••]. However, whether adaptive-like NK cell expansions contribute to COVID-19 pathogenesis, instead target possible reactivation of CMV [60], or are part of an appropriate antiviral host-response needs to be determined in future studies. In this regard it is interesting to note that HLA-E, the ligand to the activating receptor NKG2C found on adaptive-like NK cells, is increased on lung parenchymal cells and immune cells from COVID-19 patients [13••] and that SARS-CoV-2, by itself, might induce this upregulation [61]. Finally, compared to in moderate COVID-19, NK cell numbers, as well as numbers of T cells, are reduced at the site of infection at the expense of granulocytic and myeloid-derived suppressor cells [17,18,62]. The interplay between NK cells and such cells in relation to COVID-19 pathogenesis should be assessed in future work.
MAIT cell activation associate with COVID-19 outcome
MAIT are by far the UTC subset showing the most prominent phenotypic alterations during acute SARS-CoV-2 infection, in terms of extent of their depletion and of induction of classical early activation markers, such as CD69 [9••,10••]. Up to 100% of MAIT cells can express CD69 in severe COVID-19 [9••,10••], and cytokines crucial for direct MAIT activation, such as IL-18, correlate to some extent with CD69 levels [8••,10••]. This correlation is likely to have functional relevance in vivo, given the high IL-18 receptor expression on MAIT cells [30] and the fact that IL-18 is part of the molecular signature of the inflammatory ‘misfiring’ observed in severe COVID-19 [63]. Along these lines, CD69 levels on MAIT cells (but not in other UTC subsets) were even further elevated in non-surviving COVID-19 patients, and positively correlated with several clinical parameters such as granulocyte numbers, PaO2/FiO2 ratio and CRP [8••,10••]. Indeed, models exclusively based on MAIT cell activation markers (i.e. expression of CD69, granzyme B, and IFN-γ) efficiently predicted the final outcome of SARS-CoV-2 infection [8••]. Hence, an exacerbated MAIT cell activation could contribute to severe COVID-19 symptomatology, especially considering that MAIT cells (together with ɣδ T cells) are amongst the immune subsets that mostly shape the overall cytokine milieu in blood and tissues in COVID-19 patients [32]. On the other hand, an adequate MAIT cell response to danger signals such as IFNα and IL-18 is essential for the optimal triggering of the adaptive immune response, as shown by vaccination studies [40]. Collectively, the delicate balance of a finely regulated activation of UTC, and particularly of MAIT cells, emerges as a crucial determinant of both efficient viral clearance and occurrence of SARS-CoV-2-derived immunopathological events.
Conclusions
As apparent from studies reviewed above, it is clear that both NK cells and UTCs robustly contribute to the early antiviral immune response against SARS-CoV-2. They further have the capacity to relocate to the site of infection through distinct chemokine — chemokine receptor pathways where they can likely target infected cells but also interact with other recruited immune cells. In parallel, both NK cells and MAIT cells might also contribute to pathogenesis in severe COVID-19. However, although we in record time have learnt immensely about these cells in COVID-19, as outlined above throughout the text, many important outstanding questions remain to be answered.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
•• of outstanding interest
Acknowledgements
The authors thank present and past members of their group for their contributions towards the understanding of antiviral NK cell and unconventional T cell responses. Their work has been supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 948692), the Swedish Research Council, the Swedish Cancer Society, the Swedish Foundation for Strategic Research, Knut and Alice Wallenberg Foundation, the Center for Innovative Medicine at Karolinska Institutet, Region Stockholm, and Karolinska Institutet. The authors apologize to colleagues whose work has not been cited owing to space constraints.
References
- 1.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S.C., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho T., Krammer F., Iwasaki A. The first 12 months of COVID-19: a timeline of immunological insights. Nat Rev Immunol. 2021;21:245–256. doi: 10.1038/s41577-021-00522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Björkström N.K., Ljunggren H.-G., Michaëlsson J. Emerging insights into natural killer cells in human peripheral tissues. Nat Rev Immunol. 2016;16:310–320. doi: 10.1038/nri.2016.34. [DOI] [PubMed] [Google Scholar]
- 5.Vivier E., Tomasello E., Baratin M., Walzer T., Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 6.Mace E.M., Orange J.S. Emerging insights into human health and NK cell biology from the study of NK cell deficiencies. Immunol Rev. 2019;287:202–225. doi: 10.1111/imr.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godfrey D.I., Uldrich A.P., McCluskey J., Rossjohn J., Moody D.B. The burgeoning family of unconventional T cells. Nat Immunol. 2015;16:1114–1123. doi: 10.1038/ni.3298. [DOI] [PubMed] [Google Scholar]
- 8••.Flament H., Rouland M., Beaudoin L., Toubal A., Bertrand L., Lebourgeois S., Rousseau C., Soulard P., Gouda Z., Cagninacci L., et al. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat Immunol. 2021;22:322–335. doi: 10.1038/s41590-021-00870-z. [DOI] [PubMed] [Google Scholar]; In this study large cohorts of patients with less and more severe COVID-19 were studied and a link between MAIT cell activation and cytotoxicity and outcome of infection was reported.
- 9••.Parrot T., Gorin J.-B., Ponzetta A., Maleki K.T., Kammann T., Emgård J., Perez-Potti A., Sekine T., Rivera-Ballesteros O., Karolinska COVID-19 Study Group, et al. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abe1670. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of the first more comprehensive reports on the MAIT cell response in SARS-CoV-2 reporting on MAIT cell redistribution from circulation to lungs and identifying an immunotype of CD69hi CXCR3low MAIT cells associating with severe disease.
- 10••.Jouan Y., Guillon A., Gonzalez L., Perez Y., Boisseau C., Ehrmann S., Ferreira M., Daix T., Jeannet R., Francois B., et al. Phenotypical and functional alteration of unconventional T cells in severe COVID-19 patients. J Exp Med. 2020;217 doi: 10.1084/jem.20200872. [DOI] [PMC free article] [PubMed] [Google Scholar]; First paper to thoroughly map the response of several unconventional T cell populations in severe COVID-19 including MAIT cells, iNKT cells, and ɣδ T cells.
- 11.Björkström N.K., Riese P., Heuts F., Andersson S., Fauriat C., Ivarsson M.A., Björklund A.T., Flodström-Tullberg M., Michaëlsson J., Rottenberg M.E., et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. 2010;116:3853–3864. doi: 10.1182/blood-2010-04-281675. [DOI] [PubMed] [Google Scholar]
- 12.Juelke K., Killig M., Luetke-Eversloh M., Parente E., Gruen J., Morandi B., Ferlazzo G., Thiel A., Schmitt-Knosalla I., Romagnani C. CD62L expression identifies a unique subset of polyfunctional CD56dim NK cells. Blood. 2010;116:1299–1307. doi: 10.1182/blood-2009-11-253286. [DOI] [PubMed] [Google Scholar]
- 13••.Maucourant C., Filipovic I., Ponzetta A., Aleman S., Cornillet M., Hertwig L., Strunz B., Lentini A., Reinius B., Brownlie D., et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]; First paper to dissect the NK cell response to SARS-CoV-2 infection in detail including reporting a general activation of NK cells in the infection as well as expansion of adaptive-like NK cell in patients with severe COVID-19.
- 14••.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; An extensive study based on single cell RNA sequencing mapping the response of circulating immune cells in severe COVID-19.
- 15.Jiang Y., Wei X., Guan J., Qin S., Wang Z., Lu H., Qian J., Wu L., Chen Y., Chen Y., et al. COVID-19 pneumonia: CD8+ T and NK cells are decreased in number but compensatory increased in cytotoxic potential. Clin Immunol. 2020;218 doi: 10.1016/j.clim.2020.108516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neeland M.R., Bannister S., Clifford V., Dohle K., Mulholland K., Sutton P., Curtis N., Steer A.C., Burgner D.P., Crawford N.W., et al. Innate cell profiles during the acute and convalescent phase of SARS-CoV-2 infection in children. Nat Commun. 2021;12:1084–1085. doi: 10.1038/s41467-021-21414-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 18.Huang W., Li M., Luo G., Wu X., Su B., Zhao L., Zhang S., Chen X., Jia M., Zhu J., et al. The inflammatory factors associated with disease severity to predict COVID-19 progression. J Immunol. 2021;206:1597–1608. doi: 10.4049/jimmunol.2001327. [DOI] [PubMed] [Google Scholar]
- 19.Brownlie D., Rødahl I., Varnaite R., Asgeirsson H., Glans H., Falck-Jones S., Vangeti S., Buggert M., Ljunggren H.-G., Michaëlsson J., et al. Distinct lung-homing receptor expression and activation profiles on NK cell and T cell subsets in COVID-19 and influenza. bioRxiv. 2021 doi: 10.1101/2021.01.13.426553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T., et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 21.Severe Covid-19 GWAS Group, Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernández J., Prati D., Baselli G., et al. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlin L.E., Hemann E.A., Zacharias Z.R., Heusel J.W., Legge K.L. Natural killer cell recruitment to the lung during influenza A virus infection is dependent on CXCR3, CCR5, and virus exposure dose. Front Immunol. 2018;9:781. doi: 10.3389/fimmu.2018.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmer C.L., Cornillet M., Solà-Riera C., Cheung K.-W., Ivarsson M.A., Lim M.Q., Marquardt N., Leo Y.S., Lye D.C., Klingström J., et al. NK cells are activated and primed for skin-homing during acute dengue virus infection in humans. Nat Commun. 2019;10 doi: 10.1038/s41467-019-11878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varchetta S., Mele D., Oliviero B., Mantovani S., Ludovisi S., Cerino A., Bruno R., Castelli A., Mosconi M., Vecchia M., et al. Unique immunological profile in patients with COVID-19. Cell Mol Immunol. 2020;395:497. doi: 10.1038/s41423-020-00557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anfossi N., André P., Guia S., Falk C.S., Roetynck S., Stewart C.A., Breso V., Frassati C., Reviron D., Middleton D., et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 26.Yao Y., Strauss-Albee D.M., Zhou J.Q., Malawista A., Garcia M.N., Murray K.O., Blish C.A., Montgomery R.R. The natural killer cell response to West Nile Virus in young and old individuals with or without a prior history of infection. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blom K., Braun M., Pakalniene J., Lunemann S., Enqvist M., Dailidyte L., Schaffer M., Lindquist L., Mickiene A., Michaëlsson J., et al. NK cell responses to human tick-borne encephalitis virus infection. J Immunol. 2016;197:2762–2771. doi: 10.4049/jimmunol.1600950. [DOI] [PubMed] [Google Scholar]
- 28.Marquardt N., Ivarsson M.A., Blom K., Gonzalez V.D., Braun M., Falconer K., Gustafsson R., Fogdell-Hahn A., Sandberg J.K., Michaëlsson J. The human NK cell response to yellow fever virus 17D is primarily governed by NK cell differentiation independently of NK cell education. J Immunol. 2015;195:3262–3272. doi: 10.4049/jimmunol.1401811. [DOI] [PubMed] [Google Scholar]
- 29.Scharenberg M., Vangeti S., Kekäläinen E., Bergman P., Al-Ameri M., Johansson N., Sondén K., Falck-Jones S., Färnert A., Ljunggren H.-G., et al. Influenza A virus infection induces hyperresponsiveness in human lung tissue-resident and peripheral blood NK cells. Front Immunol. 2019;10:1116. doi: 10.3389/fimmu.2019.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ussher J.E., Willberg C.B., Klenerman P. MAIT cells and viruses. Immunol Cell Biol. 2018;96:630–641. doi: 10.1111/imcb.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Godfrey D.I., Koay H.F., McCluskey J., Gherardin N.A. The biology and functional importance of MAIT cells. Nat Immunol. 2019;20:1110–1128. doi: 10.1038/s41590-019-0444-8. [DOI] [PubMed] [Google Scholar]
- 32.Ren X., Wen W., Fan X., Hou W., Su B., Cai P., Li J., Liu Y., Tang F., Zhang F., et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184:1895–1913.e19. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carissimo G., Xu W., Kwok I., Abdad M.Y., Chan Y.-H., Fong S.-W., Puan K.J., Lee C.Y.-P., Yeo N.K.-W., Amrun S.N., et al. Whole blood immunophenotyping uncovers immature neutrophil-to-VD2 T-cell ratio as an early marker for severe COVID-19. Nat Commun. 2020;11:5243–5412. doi: 10.1038/s41467-020-19080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephenson E., Reynolds G., Botting R.A., Calero-Nieto F.J., Morgan M.D., Tuong Z.K., Bach K., Sungnak W., Worlock K.B., Yoshida M., et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat Med. 2021;27:904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deschler S., Kager J., Erber J., Fricke L., Koyumdzhieva P., Georgieva A., Lahmer T., Wiessner J.R., Voit F., Schneider J., et al. Mucosal-Associated Invariant T (MAIT) cells are highly activated and functionally impaired in COVID-19 patients. Viruses. 2021;13:241. doi: 10.3390/v13020241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L., Gozzi L., Iannone A., Tartaro Lo D., Mattioli M., et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020;11 doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu C., Littleton S., Giroux N.S., Mathew R., Ding S., Kalnitsky J., Yang Y., Petzold E., Chung H.A., Rivera G.O., et al. Mucosal Associated Invariant T (MAIT) cell responses differ by sex in COVID-19. Med. 2021;2:755–772.e5. doi: 10.1016/j.medj.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koay H.F., Fulford T.S., Godfrey D.I. An unconventional view of COVID-19 T cell immunity. J Exp Med. 2020;217 doi: 10.1084/jem.20201727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Provine N.M., Amini A., Garner L.C., Spencer A.J., Dold C., Hutchings C., Silva Reyes L., FitzPatrick M.E.B., Chinnakannan S., Oguti B., et al. MAIT cell activation augments adenovirus vector vaccine immunogenicity. Science. 2021;371:521–526. doi: 10.1126/science.aax8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rijkers G., Vervenne T., van der Pol P. More bricks in the wall against SARS-CoV-2 infection: involvement of γ9δ2 T cells. Cell Mol Immunol. 2020;17:771–772. doi: 10.1038/s41423-020-0473-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laing A.G., Lorenc A., del Molino del Barrio I., Das A., Fish M., Monin L., oz-Ruiz M.M.X., McKenzie D.R., Hayday T.S., Francos-Quijorna I., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 43.Odak I., Barros-Martins J., Bošnjak B., Stahl K., David S., Wiesner O., Busch M., Hoeper M.M., Pink I., Welte T., et al. Reappearance of effector T cells is associated with recovery from COVID-19. EBioMedicine. 2020;57 doi: 10.1016/j.ebiom.2020.102885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yazdanifar M., Mashkour N., Bertaina A. Making a case for using γδ T cells against SARS-CoV-2. Crit Rev Microbiol. 2020;46:689–702. doi: 10.1080/1040841X.2020.1822279. [DOI] [PubMed] [Google Scholar]
- 45.Poccia F., Agrati C., Castilletti C., Bordi L., Gioia C., Horejsh D., Ippolito G., Chan P.K.S., Hui D.S.C., Sung J.J.Y., et al. Anti-severe acute respiratory syndrome coronavirus immune responses: the role played by V gamma 9V delta 2 T cells. J Infect Dis. 2006;193:1244–1249. doi: 10.1086/502975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez L., Pekkarinen P.T., Lakshmikanth T., Tan Z., Consiglio C.R., Pou C., Chen Y., Mugabo C.H., Nguyen N.A., Nowlan K., et al. Systems-level immunomonitoring from acute to recovery phase of severe COVID-19. Cell Rep Med. 2020;1 doi: 10.1016/j.xcrm.2020.100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ponzetta A., Carriero R., Carnevale S., Barbagallo M., Molgora M., Perucchini C., Magrini E., Gianni F., Kunderfranco P., Polentarutti N., et al. Neutrophils driving unconventional T cells mediate resistance against murine sarcomas and selected human tumors. Cell. 2019;178:346–360.e24. doi: 10.1016/j.cell.2019.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koutsakos M., Rowntree L.C., Hensen L., Chua B.Y., van de Sandt C.E., Habel J.R., Zhang W., Jia X., Kedzierski L., Ashhurst T.M., et al. Integrated immune dynamics define correlates of COVID-19 severity and antibody responses. Cell Rep Med. 2021;2 doi: 10.1016/j.xcrm.2021.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Liu C., Martins A.J., Lau W.W., Rachmaninoff N., Chen J., Imberti L., Mostaghimi D., Fink D.L., Burbelo P.D., Dobbs K., et al. Time-resolved systems immunology reveals a late juncture linked to fatal COVID-19. Cell. 2021;184:1836–1857.e22. doi: 10.1016/j.cell.2021.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; A systems immunology paper longitudinally assessing the immune response to SARS-CoV-2 infection identifying IL-15 driven NK cell dysfunction in fatal COVID-19 cases.
- 50.Sahoo D., Katkar G.D., Khandelwal S., Behroozikhah M., Claire A., Castillo V., Tindle C., Fuller M., Taheri S., Rogers T.F., et al. AI-guided discovery of the invariant host response to viral pandemics. bioRxiv. 2020 doi: 10.1101/2020.09.21.305698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao C., Bora S.A., Parimon T., Zaman T., Friedman O.A., Palatinus J.A., Surapaneni N.S., Matusov Y.P., Cerro Chiang G., Kassar A.G., et al. Cell-type-specific immune dysregulation in severely Ill COVID-19 patients. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2020.108590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merino A., Zhang B., Dougherty P., Luo X., Wang J., Blazar B.R., Miller J.S., Cichocki F. Chronic stimulation drives human NK cell dysfunction and epigenetic reprograming. J Clin Invest. 2019;129:3770–3785. doi: 10.1172/JCI125916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rendeiro A.F., Casano J., Vorkas C.K., Singh H., Morales A., DeSimone R.A., Ellsworth G.B., Soave R., Kapadia S.N., Saito K., et al. Profiling of immune dysfunction in COVID-19 patients allows early prediction of disease progression. Life Sci Alliance. 2021;4 doi: 10.26508/lsa.202000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gumá M., Angulo A., Vilches C., Gómez-Lozano N., Malats N., López-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 55.Béziat V., Liu L.L., Malmberg J.-A., Ivarsson M.A., Sohlberg E., Björklund A.T., Retière C., Sverremark-Ekström E., Traherne J., Ljungman P., et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. 2013;121:2678–2688. doi: 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlums H., Cichocki F., Tesi B., Theorell J., Béziat V., Holmes T.D., Han H., Chiang S.C.C., Foley B., Mattsson K., et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. 2015;42:443–456. doi: 10.1016/j.immuni.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Björkström N.K., Lindgren T., Stoltz M., Fauriat C., Braun M., Evander M., Michaëlsson J., Malmberg K.-J., Klingström J., Ahlm C., et al. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med. 2011;208:13–21. doi: 10.1084/jem.20100762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petitdemange C., Becquart P., Wauquier N., Béziat V., Debré P., Leroy E.M., Vieillard V. Unconventional repertoire profile is imprinted during acute chikungunya infection for natural killer cells polarization toward cytotoxicity. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gumá M., Cabrera C., Erkizia I., Bofill M., Clotet B., Ruiz L., López-Botet M. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J Infect Dis. 2006;194:38–41. doi: 10.1086/504719. [DOI] [PubMed] [Google Scholar]
- 60.Le Balc’h P., Pinceaux K., Pronier C., Seguin P., Tadié J.-M., Reizine F. Herpes simplex virus and cytomegalovirus reactivations among severe COVID-19 patients. Crit Care. 2020;24:530–533. doi: 10.1186/s13054-020-03252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bortolotti D., Gentili V., Rizzo S., Rotola A., Rizzo R. SARS-CoV-2 spike 1 protein controls natural killer cell activation via the HLA-E/NKG2A pathway. Cells. 2020;9 doi: 10.3390/cells9091975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kvedaraite E., Hertwig L., Sinha I., Ponzetta A., Hed Myrberg I., Lourda M., Dzidic M., Akber M., Klingström J., Folkesson E., et al. Major alterations in the mononuclear phagocyte landscape associated with COVID-19 severity. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2018587118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]