Abstract
Purpose:
This study investigated the segmentation metrics of different segmentation networks trained on 730 manually annotated lateral lumbar spine X-rays to test the generalization ability and robustness which are the basis of clinical decision support algorithms.
Methods:
Instance segmentation networks were compared to semantic segmentation networks based on different metrics. The study cohort comprised diseased spines and postoperative images with metallic implants.
Results:
However, the pixel accuracies and intersection over union are similarly high for the best performing instance and semantic segmentation models; the observed vertebral recognition rates of the instance segmentation models statistically significantly outperform the semantic models' recognition rates.
Conclusion:
The results of the instance segmentation models on lumbar spine X-ray perform superior to semantic segmentation models in the recognition rates even by images of severe diseased spines by allowing the segmentation of overlapping vertebrae, in contrary to the semantic models where such differentiation cannot be performed due to the fused binary mask of the overlapping instances. These models can be incorporated into further clinical decision support pipelines.
Keywords: Convolutional neural networks, deep neural networks, instance segmentation, lumbar vertebrae, machine learning, postoperative image analysis, semantic segmentation, X-ray
INTRODUCTION
Various supervised and nonsupervised image segmentation algorithms are utilized in biomedical image segmentation;[1,2] typically, these are semantic segmentation or instance segmentation models.[3] Semantic segmentation algorithms are a subset of supervised learning algorithms that control the training of convolutional neural network classifiers from a set of labeled training image data. These types of algorithms produce only one label for every pixel in an input image. Instance segmentation models can serve multiple per-pixel labels, denoting overlapping instances from the same or differing class. This can be beneficial by X-rays, where due to the projection, vertebral instances can overlap each other, so the same pixel can belong to two different vertebral instances. X-rays are still one of the most commonly requested diagnostic imaging modalities, accounting for 1, 7 procedures on each inhabitant in Germany per year.[4] It is of utmost importance in the process of image analysis to acquire the position and the form of the vertebral bodies, since these often determine the pathology on the image: if relation or contour of the outer borders is deranged, it may represent fracture or tumor of the vertebral body. In order to create an automated system that performs morphometric analysis of the X-rays, the exact segmentation of the vertebrae is mandatory. There are degenerative conditions that modify the overall appearance of the vertebral body, making it difficult to delineate the contour of the vertebra such as fractures but also osteophytes that are present in almost every moderately or severely degenerated spine. In order to find a suitable model for this purpose, the performance of state-of-the-art semantic and instance segmentation algorithms was compared.
Material
For the training, validation, and testing, a set of 830 lateral lumbar spine radiographs were selected and retrieved from the picture archiving and communication system (PACS). All DICOM (Digital Imaging and Communications in Medicine) objects contained 12-bit depth image data and were saved as noncompressed grayscale PNG (Portable Network Graphics) images with histogram equalization. The image sizes ranged between 511 × 1177 and 1100 × 2870 pixel. The DICOM images contained preoperative and postoperative studies, before and after mono- or multisegmental interbody fusion and posterior instrumentation. Therefore, on the images, at least one but usually multiple severely diseased intervertebral spaces – i.e., many overlapping areas of vertebral instances on the lateral radiograph – were present.
The annotations were performed using the standalone, browser-based, open-source VIA (VGG annotation tool[5]) by trained radiologists and spine surgeons having experience of more than 5 years. Each vertebra was labeled with a polyline polygon up to 15 distinct points. Cages, screws, and instrumentation were also labeled, but these classes were not utilized for the present study. Figure 1 shows the graphical user interface of the annotation software. The authors chose to create two distinct classes, one for lumbar and one for sacral vertebrae instead of five distinct classes for each lumbar vertebra. The justification for this design decision is based on the similarity between the lumbar vertebrae and the differently shaped sacral vertebra. The resulting annotations were exported in JSON format and were further processed in Jupyter Notebook with pandas and OpenCV.
Figure 1.

Screenshot of the VIA annotation tool with manual annotations of a lumbar vertebra including implants
METHODS
The aim of the study was to compare the performance of different segmentation models. The pynetdicom[6] library running on the portable local WinPython[7] with the anaconda environment was used for training with different libraries (Keras, TensorFlow, or PyTorch). For the qualitative statistical analysis, four pixel-based metrics based on the merged binary masks of the model predictions and two observation-based metrics were collected. For the statistical analysis, the open-source SciPy and Statsmodels[8] packages were used. ANOVA was performed on continuous variables where applicable. The assumption of normal distribution was tested with the Shapiro–Wilk test of normality. For nonnormal distributed variables and binary data nonparametric tests: Kruskal–Wallis test and post hoc either Tukey's multiple-comparison-method or Holm–Bonferroni method were performed.
Data retrieval and preparation
As shown in the workflow, Figure 2, the first step was data retrieval from the PACS. Eight hundred and thirty images were retrieved, anonymized, and annotated manually from the archive. Each model required different input formats (JSON, image masks with two distinct classes, or raw image material), and the resolution constraints also had to be fixed, which are features and special requirements of each model.
Figure 2.
Workflow of the data retrieval and processing
Model training
The authors used a GeForce RTX 2080 Ti graphic card for the k-fold training. The entire dataset was split into 730 images for training and validation, and the remaining 100 images remained always the same for testing. The k-fold validation was performed 5 times with splitting the 730 images: 80% for the training set and the remaining 20% for the validation set. We have selected 5 different segmentation models for the training of the dataset, each of them trained and tested with the same seed for each k-fold validation training. The models are categorized into semantic and instance segmentation models. U-Net,[9,10] PSPNet,[11,12] and DeepLabv3[13] are semantic segmentation models, whereas Mask R-CNN[14,15,16] and YOLACT[17] are instance segmentation models. For further information about the models see Supplementary Material 1 (54.1KB, pdf) .
Results and inference
Once all the models had completed training (training time varied from 10 to 20 h for each fold), the predictions were made for the test set. The tests for each fold for each model (so 25 altogether) were performed one after another, and different metrics for comparison studies were obtained for each test image which are described further down in this article.
Metrics used
In order to evaluate the segmentation models, we have used different performance metrics inspired by Long et al.[18] from the repository of Martin Keršner.[19] Since these pixel-based metrics do not mirror the clinical usability of the models, a simple recognition rate was utilized to determine which model is superior in recognizing the distinct, nonoverlapping vertebrae on an image and another recognition metric was used that excluded from these images the ones where the model falsely segmented the vertebrae as nonoverlapping areas on the predictions, however, the vertebrae were overlapping on the projection (ground truth) [Table 1].
Table 1.
Metrics
| Metric\model | Mask R-CNN | U-Net | DeepLabv3 | YOLACT | PSPNet |
|---|---|---|---|---|---|
| Mean IoU average (%) (SD) | 85.87 (4.45) | 82.20 (4.75) | 83.23 (4.40) | 87.77 (3.59) | 80.97 (4.69) |
| Pixel accuracy average (%) (SD) | 96.27 (1.39) | 95.39 (3.17) | 95.48 (2.94) | 96.94 (1.12) | 95.13 (3.10) |
| Mean accuracy average (%) (SD) | 92.47 (4.00) | 91.00 (4.32) | 91.56 (4.02) | 94.15 (2.77) | 91.34 (4.00) |
| Frequency weighted IoU (%) average (SD) | 93.74 (1.86) | 94.49 (2.11) | 94.53 (1.92) | 94.33 (2.01) | 93.97 (2.14) |
| Recognition rate (%) | 99.0 | 91.0 | 70.0 | 98.0. | 43.0 |
| Composite recognition rate (%) | 89.0 | 71.0 | 55.0 | 82.0 | 36.0 |
SD-Standard deviation, IoU-Intersection over union
Pixel accuracy
Pixel accuracy can be calculated as the percentage of pixels in the generated labeled mask that are classified correctly when compared with the original labeled mask.
Intersection over union
The most common metric for the evaluation for segmentation models is the intersection over union (IoU) (also known as the Jaccard Index), calculated as the overlapping area between the predicted labeled mask and the original labeled mask [Figure 3]. For a multi-class segmentation such as ours, the mean-IoU is calculated by averaging the IoU of each class.
Figure 3.

Visual explanation of the intersection over union
Mean accuracy
Mean accuracy is defined as the mean of number of pixels of class i predicted to class i over the total number of pixels of class i for all classes i.
Frequency weighted intersection over union
In frequency weighted IoU, the IoU for each class is computed first and then average over all the classes is computed.
Recognition rate
A radiologist with 10 years of practice selected all the images where the model recognized all lumbar and one sacral vertebra as a distinct, unique area on the image. All the images were labeled binary (1 if every vertebra present and 0 if at least one is not recognized). Note that the recognition rate does not score the quality of the annotations. This means it does not reflect how well each vertebra is delineated or whether the distinct vertebrae are detected as nonoverlapping areas. To address this problem, a further criterion was added:
Composite recognition rate
Additionally to the above criteria, all the images were labeled as 0 if overlapping vertebral instances falsely as distinct instances appeared on the binary mask.
RESULTS
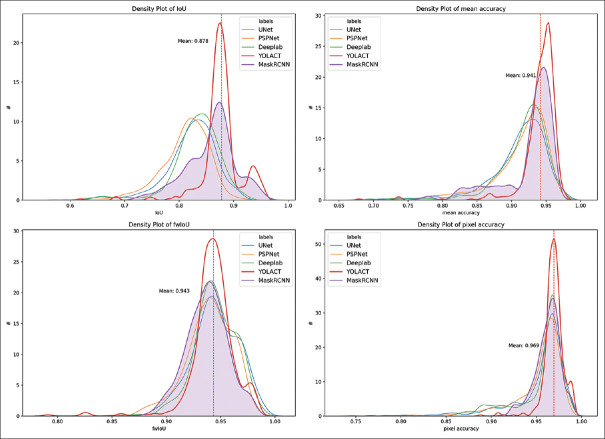
The density plot of the four pixel-based metrics is shown in Figure 4. Moreover, the density plots for each model are visible in electronic Supplementary Material 2 (1.2MB, pdf) . The test for normal distribution failed in at least one of the model's variables by every metric, so the nonparametric Kruskal–Wallis test was performed for testing whether samples originate from the same distribution. All of the tests for all the metrics showed statistically significant differences within the models [Table 2]. The results of the post hoc analysis are shown in electronic Supplementary Material. Post hoc analysis of the IoU showed statistically significant difference (P < 0.01) between all models with relatively wide range of the results (80.97%–87.77%). Statistically significant result was observed even between the best performing DeepLabv3 and YOLACT in this metric (P < 0.001). By frequency weighted IoU, however, no statistically significant difference was shown between these two models (P = 0.294). Mean accuracy and pixel accuracy showed no statistically difference between these two models either. The results for the pairwise comparison are in electronic Supplementary Material 2 (1.2MB, pdf) .
Figure 4.
Density plots of the metrics
Table 2.
Metric analytical statistics
| Metric | Kruskal-Wallis Test |
|---|---|
| Mean IoU average | Statistics=787.299, P<0.001 Different distributions (reject H0) |
| Pixel accuracy average | Statistics=170.028, P<0.001 Different distributions (reject H0) |
| Mean accuracy average | Statistics=383.056, P<0.001 Different distributions (reject H0) |
| Frequency weighted IoU | Statistics=62.343, P<0.001 Different distributions (reject H0) |
IoU-Intersection over union
In Figure 5, we show two cases: (a) where the semantic models deliver better delineation of the vertebrae, and failure of Mask R-CNN due to an “extra vertebrae” as an example for the sources of failure. In case (b), all models but YOLACT deliver an under average pixel accuracy, whereas the reasons are two-fold: first, there are two overlapping vertebrae that had been recognized correctly in the instance segmentation models, and as one large fused blob in the semantic models, and second, an extra, thoracic vertebra (11th thoracic vertebra) that had been at least partially delineated on all models but YOLACT. Only vertebrae caudally from Th12 are present on the ground truth masks. In consequence, if there are more vertebrae in the predicted mask than in the ground truth mask, it lowers the accuracy.
Figure 5.
Example of the inference results for the models for the same images. In Row (a) failure of Mask R-CNN to correctly identify a lumbar vertebra. In Row (b) all semantic models fail to delineate both vertebral instances and showing only a fused mask for the lumbar 4th and 5th vertebrae
Since the internal metrics of each model are based on slightly different methods, for the current study a common metric was chosen based on a merged binary mask of the prediction. This method is imperfect and in many cases results in accuracy loss for instance segmentations. This shortcoming gets particularly obvious in cases where the predicted binary masks of the vertebral instance are overlapping. To overcome this limitation, we visually inspected the vertebral instances and excluded the falsely identified contours: (a) wrong labeling as overlapping (=recognition rate) and (b) false marking as nonoverlapping (=composite recognition rate). There was no difference in the recognition rate between the folds within the same model, so these were not statistically tested. The recognition rates between the models were compared with Kruskal–Wallis test to test for differences between the groups. Due to the statistically significant (H = 142.96, P < 0.001) result of the test, post hoc testing was used to determine which groups are significantly different from others [Table 3]. Holm–Bonferroni test was performed that showed statistically significant difference between the best performing semantic model in the recognition (U-Net) and the best performing instance segmentation model (Mask R-CNN), whereas no statistical difference could be shown between U-Net and YOLACT and between Mask R-CNN and YOLACT in terms of recognition rate. Since the Kruskal–Wallis test on the composite recognition rate also showed statistically significant difference between the models (H = 82.067, P < 0.001), here also a post hoc Holm–Bonferroni test was performed [Table 4]. This showed no statistically significant difference between the Mask R-CNN and YOLACT but strong statistical significance between all other semantic and instance segmentation models, thus also mathematically proving the observed superior performance of the instance segmentation models. The contours of the vertebrae annotations are irregular by the YOLACT model (a) and smoother by the U Net model (b) [Figure 6], also smooth contours were observed by the outputs of the Mask R CNN segmentations to (not illustrated).
Table 3.
Recognition rate
| Model 1 | Model 2 | Stat | pval | pval_corr | Reject H0 |
|---|---|---|---|---|---|
| DeepLab | Mask R-CNN | −6.36 | 0.00 | 0.00 | True |
| DeepLab | PSPNet | 5.30 | 0.00 | 0.00 | True |
| DeepLab | U-Net | −4.85 | 0.00 | 0.00 | True |
| DeepLab | YOLACT | −5.92 | 0.00 | 0.00 | True |
| Mask R-CNN | PSPNet | 11.23 | 0.00 | 0.00 | True |
| Mask R-CNN | U-Net | 2.93 | 0.00 | 0.01 | True |
| Mask R-CNN | YOLACT | 0.58 | 0.57 | 0.57 | False |
| PSPNet | U-Net | −9.56 | 0.00 | 0.00 | True |
| PSPNet | YOLACT | −10.58 | 0.00 | 0.00 | True |
| U-Net | YOLACT | −2.15 | 0.03 | 0.07 | False |
Table 4.
Composite recognition rate
| Model 1 | Model 2 | Stat | pval | pval_corr | Reject H0 |
|---|---|---|---|---|---|
| DeepLab | Mask R-CNN | −7.14 | 0.00 | 0.00 | True |
| DeepLab | PSPNet | 4.09 | 0.00 | 0.00 | True |
| DeepLab | U-Net | −4.34 | 0.00 | 0.00 | True |
| DeepLab | YOLACT | −5.77 | 0.00 | 0.00 | True |
| Mask R-CNN | PSPNet | 10.57 | 0.00 | 0.00 | True |
| Mask R-CNN | U-Net | 4.66 | 0.00 | 0.00 | True |
| Mask R-CNN | YOLACT | 1.97 | 0.052 | 0.052 | False |
| PSPNet | U-Net | −7.30 | 0.00 | 0.00 | True |
| PSPNet | YOLACT | −8.83 | 0.00 | 0.00 | True |
| U-Net | YOLACT | −3.19 | 0.00 | 0.00 | True |
Figure 6.

Example for differences between the instance (a) and semantic (b) models' segmentation results
An interesting feature found during testing different images was that the YOLACT model was able to partially identify beforehand unseen and as such untrained objects, such as vertebral body replacement, however, wrongly delineated [Figure 7]. Another surprising finding was the ability of the YOLACT and partially of the Mask R-CNN model to delineate fractured vertebrae, even though these were also not part of the training set [Figure 8]. This behavior shows the universal nature of neural networks.
Figure 7.

YOLACT identifying unseen and untrained object (vertebral body replacement implant) as existing class (Cage)
Figure 8.

Mask R-CNN model delineates fractured vertebrae, even though these were not part of the training set
DISCUSSION
The recent development of open-source segmentation networks has enabled researchers to experiment with the automation and analysis of biomedical images.[20] Thus, creating semi- or fully automated image processing workflows for segmentation became fast and feasible. The human-level spinal vertebral segmentation is the first and essential step for many computational spine analysis tasks, for example, different spatial relations of vertebral bodies (Cobb angle or lordosis angle) and shape characterization. There are only a few published articles that utilize the power of convolutional neural networks in the segmentation of spine X-rays; recently, Cho et al.[21] utilized a U-Net-based model with very good performance on segmenting the lumbar vertebrae on their own set although there were some very important limitations in their study. First of all, they did not consider to include any kind of implants such as cages, which limits the utility of the system for practical day-to-day purposes in outcome evaluation of surgical treatment. Further limitation is that no severe deformities of the vertebral bodies and intervertebral spaces were included. This implies that their algorithm is not applicable for the majority of the patients. In our study, we overcome these limitations by training on a sample containing over 70% of images depicting implants (multiple cage systems, screws, rods, and also cement augmentations). In addition to that, we included the full spectrum of degeneratively diseased intervertebral spaces, which increases the segmentation difficulties due to the numerous overlapping areas of the same class. Despite this, our metrics show excellent pixel accuracies and IoU for all models.
Chan-Pang et al.[22] used a three-step system that included spine region of interest (RoI) detection, vertebral RoI detection, and vertebral segmentation on anteroposterior whole-body images. The image processing steps helped to find the RoIs for each vertebra and a U-Net performed the segmentation of the vertebral instances within the RoI. The image segmentation was thus performed on a cropped image region that resulted in better pixel accuracies. In our study, the instance segmentation models accepted the full-resolution images and we performed no RoI cropping, thus reducing the detection steps. Ruhan et al.[23] used Faster R-CNN object detection as the first step toward automatically identifying landmarks from spine X-ray images. In their work, intervertebral spaces were detected without the contour detection of the vertebrae themselves. Although our study focuses on the detection of the contour of the vertebrae, the delineated endplates draw the exact borders of the intervertebral spaces. The extraction and analysis of these intervertebral spaces can be simply derived from the predicted masks by classical morphological operations such as dilation, erosion, or Boolean operations. The fact that the best-ranking instance segmentation model performed better than all other semantic segmentation models is fortunate since the results of an instance segmentation model can be used without any extensive postprocessing step, given no vertebrae is missed. Removing overlapping class predictions for the same class is with nonmaximal suppression is possible.
The semantic segmentation masks contain fused binary masks of multiple vertebral instances. The pixels only belong to one class, they are not separated onto instances, that makes the labeling of each vertebral instance difficult and in every case needs morphological postprocessing, especially in severe degenerative cases or when cages are present [Figure 9]. The postprocessing code of a semantic segmentation model in such cases is nontrivial and was not addressed in this article. With the results of the instance segmentation, the vertebrae can be labeled automatedly, beginning on the lower part of the image if an instance of the sacral vertebra is present. The consecutive lumbar vertebral instances toward the top of the image represent the fifth, fourth, third, second, and the first lumbar vertebrae, respectively. Special cases may arise in instance segmentation when the segmentation model skips predicting an instance (due to low prediction score) or predicts multiple overlapping classes or instances. The first case can be addressed by lowering the threshold score for the prediction in the model. In case of multiple overlapping instances, the prediction with the higher score can be chosen with nonmaximum suppression.[24] We observed that the more severe degenerations are present, the better the prediction of the instance segmentation models is, due to the correct identification of the overlaps. In cases where less degenerative changes are present, the semantic segmentation models achieve better accuracies, but these observed differences were not addressed for statistical analytical testing.
Figure 9.

Example for overlapping vertebral instances
With further conventional morphological postprocessing, one will be able to derive clinically important information from the postoperative images; this information can serve as a decision support tool for clinicians. Every degeneration or pathological alteration that causes morphological change of the anatomical structures is computable by further analysis. This is promising in regard to changes in intervertebral space, osteophyte formation, dislocation, or failure of metallic implants, especially since it allows analysis of those features over time. In Figure 10, possible steps of further information extraction are shown: (a) contour segmentation, (b) osteophyte and corner detection with erosion, dilation, and subtraction, and (c7) endplate approximation. A further step will be the analysis of the interobserver agreement of the derived clinically important information (like Cobb angle) with human observers. This could pave the way for the development of a clinical decision support algorithm for the automated assessment of postoperative postural changes of the vertebrae.
Figure 10.

Possible further processing steps for the integration into clinical decision support and computer-aided diagnosis pipelines (a) contour segmentation, (b) osteophyte and corner detection, (c) endplate approximation
CONCLUSION
In this article, we have compared semantic segmentation and instance segmentation models for automated segmentation of vertebral instances in standard lateral X-rays of a real-life clinical cohort with severe degenerative diseases. To our knowledge, such an analysis has never been reported for real-life cohort with degenerative spine. Instance segmentation models on lumbar spine X-ray perform superior to semantic segmentation models regarding recognition rates by allowing the segmentation of overlapping vertebrae. This feature puts it well ahead of semantic models where such differentiation cannot be performed due to the fused mask of the overlapping instances. Instance segmentation therefore can be incorporated into further clinical decision support and computer-aided diagnosis pipelines.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
COMPLIANCE WITH ETHICAL STANDARDS
The present study is based on an institutional retrospective analysis. By decision of the ethics committee in charge for this type of study, formal consent is not required. All procedures performed in studies involving human participants were in accordance with the ethical standards of the national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.[25]
SUPPLEMENTARY MATERIALS
REFERENCES
- 1.Fourcade A, Khonsari RH. Deep learning in medical image analysis: A third eye for doctors. J Stomatol Oral Maxillofac Surg. 2019;120:279–88. doi: 10.1016/j.jormas.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys. 2019;29:102–27. doi: 10.1016/j.zemedi.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Shervin M, Boykov Y, Porikli F, Plaza A, Kehtarnavaz N, Terzopoulos D. Image segmentation using deep learning: A survey. ArXiv. 2020;2001:5566. doi: 10.1109/TPAMI.2021.3059968. [DOI] [PubMed] [Google Scholar]
- 4.Nekolla EA, Schegerer AA, Griebel J, Brix G. Frequency and doses of diagnostic and interventional Xray applications: Trends between 2007 and 2014. Radiologe. 2017;57:555–62. doi: 10.1007/s00117-017-0242-y. [DOI] [PubMed] [Google Scholar]
- 5.Dutta A, Zisserman A. Proceedings of the 27th ACM International Conference on Multimedia (MM ’19) New York, NY, USA: ACM; 2019. The VIA annotation software for images, audio and video; pp. 2276–9. [Google Scholar]
- 6. [Last accessed on 2019 Apr 01]. Available from: https://github.com/pydicom/pynetdicom .
- 7. [Last accessed on 2019 Apr 01]. Available from: https://github.com/winpython/winpython/issues/764 .
- 8.Skipper S, Perktold J. Statsmodels: Econometric and statistical modeling with python. Proceedings of the 9th Python in Science Conference. 2010;Vol. 57:61. [Google Scholar]
- 9.Olaf R, Fischer P, Brox T. International Conference on Medical Image Computing and Computer - Assisted Intervention. Cham: Springer; 2015. U-Net: Convolutional networks for biomedical image segmentation; pp. 234–41. [Google Scholar]
- 10. [Last accessed 2019 Jul 01]. Available from: https://github.com/qubvel/segmentation_models .
- 11.Hengshuang Z, Shi J, Qi X, Wang X, Jia J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Pyramid scene parsing network; pp. 2881–90. [Google Scholar]
- 12. [Last accessed on 2019 Jun 01]. Available from: https://github.com/divamgupta/image-segmentationkeras .
- 13.Chen LC, Papandreou G, Schroff F, Adam H. [Last accessed on 2019 Jul 01];Rethinking atrous convolution for semantic image segmentation. Available from https://arxiv.org/pdf/1706.05587 . [Google Scholar]
- 14.Kaiming H, Gkioxari G, Dollár P, Girshick R. Proceedings of the IEEE International Conference on Computer Vision. 2017. Mask R-CNN; pp. 2961–9. [Google Scholar]
- 15. [Last accessed on 2019 Oct 07]. Available from: https://github.com/matterport/Mask_RCNN .
- 16.Ren S, He K, Girshick R, Sun J. Faster R-CNN: Towards Real-time Object Detection with Region Proposal Networks. IEEE Trans Pattern Anal Mach Intell. 2016;39:1137–49. doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 17.Bolya D, Zhou C, Xiao F, Lee YJ. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2019. Yolact: Real-time instance segmentation; pp. 9157–66. [Google Scholar]
- 18.Long J, Jonathan, Shelhamer E, Darrell T. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015. Fully convolutional networks for semantic segmentation; pp. 3431–40. [Google Scholar]
- 19. [Last accessed on 2019Jun 01]. Available from: https://github.com/martinkersner/py_img_seg_eval .
- 20. [Last accessed on 2019 Aug 01]. Available from: https://towardsdatascience.com/review-u-netbiomedical-image-segmentation-d02bf06ca760 .
- 21.Cho BH, Kaji D, Cheung ZB, Ye IB, Tang R, Ahn A, et al. Automated measurement of lumbar lordosis on radiographs using machine learning and computer vision. Global Spine J. 2020;10:611–8. doi: 10.1177/2192568219868190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan-Pang K, Fu MJ, Lin CJ, Horng MH, Sun YN. Proceedings of the 2018 International Conference on Information Hiding and Image Processing. Association for Computing Machinery. New York: United States; 2018. Vertebrae Segmentation from X-ray Images Using Convolutional Neural Network; pp. 57–61. [Google Scholar]
- 23.Ruhan S, Owens W, Wiegand R, Studin M, Capoferri D, Barooha K, et al. 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) Piscataway, NJ, United States: IEEE; 2017. Intervertebral disc detection in X-ray images using faster R-CNN; pp. 564–7. [DOI] [PubMed] [Google Scholar]
- 24. [Last accessed on 2019 Aug 01]. Available from: https://towardsdatascience.com/non-maximumsuppression-nms-93ce178e177c .
- 25.World Medical Association. World medical association declaration of Helsinki: Ethical principles form medical research involving human subjects. JAMA. 2013;310:21914. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.