Abstract
T cell activation is sensitive to the mechanical properties of an activating substrate. However, there are also contrasting results on how substrate stiffness affects T cell activation, including differences between T cells of mouse and human origin. Towards reconciling these differences, this report examines the response of primary human T cells to polyacrylamide gels with stiffness between 5–110 kPa presenting activating antibodies to CD3 and CD28. T cell proliferation and IL-2 secretion exhibited a biphasic functional response to substrate stiffness, which can be shifted by changing density of activating antibodies and abrogated by inhibition of cellular contractility. T cell morphology was modulated by stiffness at early time points. RNA-seq indicates that T cells show differing monotonic trends in upregulated genes and pathways towards both ends of the stiffness spectrum. These studies provide a framework of T cell mechanosensing and suggest an effect of ligand density that may reconcile different, contrasting patterns of stiffness sensing seen in previous studies.
Keywords: T cell activation, mechanosensing, biphasic response, stiffness sensing
Introduction
T cells have emerged as a powerful tool in the treatment of cancer, allowing for high specificity, extended efficacy, and reduced off-target effects. Adoptive T cell therapy has shown particular promise in the treatment of blood cancers [1, 2]. However, a current barrier to wide use of this approach is the reliable production of T cells that comprise the therapeutic agent. In particular, this production involves the ex vivo expansion of a small, starting population of cells derived from the individual undergoing treatment [3, 4]. Expansion can be initiated by activating T cells using a synthetic system, most commonly polystyrene beads that present antibodies to the CD3 and CD28 receptors on the T cell surface; this commercial Dynabeads system provides signaling normally associated with Antigen Presenting Cells (APCs) in a manufactured format [5]. While successful, this system faces difficulty in activating T cells isolated from patients undergoing cancer treatment, in which T cells exhibit an exhausted phenotype [6].
One approach towards improving T cell expansion arose from the discovery that T cells can sense the mechanical properties of an activating substrate. O’Connor et al. demonstrated that mixed CD4+/CD8+ populations of primary human T cells activated on flat polydimethylsiloxane (PDMS) elastomer surfaces presenting anti-CD3 and anti-CD28 antibodies exhibit greater expansion on soft (Young’s modulus E ~100 kPa) compared to stiff (E ~2 MPa) surfaces. The use of soft PDMS to enhance T cell expansion was confirmed in other formats such as beads and scaffolds, demonstrating new capabilities for cell production, including rescue of expansion for exhausted T cells [7, 8]. However, key aspects of this mechanosensing behavior are not well understood. In particular, an opposite trend of activation was seen for mouse CD4+ T cells responding to polyacrylamide (PA) gels ranging from 10–200 kPa in Young’s Modulus, with higher activation observed on higher stiffness [9]. Moreover, the ranges of elastic moduli of the PDMS materials used in previous reports are much higher than that associated with physiological tissues (few to hundreds of kPa). A later report by Saitakis et al. confirmed that T cells respond to substrate stiffness in this nominally superphysiological range, but also showed increased short-term function (migration and cytokine secretion) of human T cells with increasing stiffness [10]. Together, these studies suggest that T cell mechanosensing may be biphasic, exhibiting a peak between a range of substrate elastic moduli; the existence of and range for this peak may be dependent on the specific type of substrate and activating ligands that are presented to T cells. This concept is supported by a later study by Wahl et al. that demonstrated biphasic spreading as a function of substrate stiffness [11]. However, that study was limited to early cell responses and focused predominantly on the Jurkat cell line, which exhibits multiple signaling defects in comparison to normal human T cells [12]. These studies also did not link early T cell responses and subsequent functionality of activated T cells.
Towards an improved understanding of T cell mechanosensing, this report examines the response of human CD4+/CD8+ T cells activated on PA gels, controlling both substrate stiffness and density of activating ligands. Our results demonstrate a biphasic response of T cell expansion with respect to these parameters. Notably, short-term T cell responses showed limited correlation with expansion. We explore the transcriptome of mechanosensitive T cell activation to determine how stiffness induces different activation programs that regulates observed functional differences.
Materials and methods
Method Details
Polyacrylamide gel fabrication and protein preparation
Polyacrylamide gels were prepared following established methods [9, 13]. Gel stiffness was controlled by varying amounts of acryl monomers with bis-acrylamide crosslinker. Gel precursor solution was mixed from 40% acryl (Sigma) and 2% bis-acrylamide (Fisher) in deionized water, and gels were polymerized by adding 10% ammonium persulfate (APS) and Tetramethylethylenediamine (TEMED) at 1:100 and 1:1000 ratio to precursor solution, and mixed with streptavidin-acrylamide for biotinylated antibody attachment. Final concentrations of acrylamide, bis-acrylamide, and streptavidin-acrylamide in Table S1. Gels polymerized between a functionalized and a cleaned coverslip. Gels were made and stored overnight in PBS in 4 C one day before antibody coating procedures.
Functionalized coverslips were treated with base piranha, 5% (3-Aminopropyl)triethoxysilane (APTES), and 5% glutaraldehyde. Cleaned coverslips were washed in detergent solution. The solution was sandwiched between a functionalized coverslip and cleaned coverslip, polymerized at room temperature for 20 minutes, and the cleaned coverslip removed using tweezers.
Young’s modulus (E) was measured by using a custom-built indentation apparatus. Slabs of polyacrylamide gels with thickness of ~10 mm were deformed using a flat cylindrical head. A calibrated mass was applied, producing a deflection. Hertzian contact between the head and gel was assumed, which allows for the estimation of the material’s Young’s modulus from the head diameter (D), deflection (h), weight (m), gravitational constant (g), and Poisson ratio (ν) (0.457 for polyacrylamide gels). Young’s modulus was calculated with the following equation:
For antibody attachment, biotinylated Goat-anti-Mouse (Biolegend) was pooled onto the substrates, unless otherwise stated, at 1 μg/mL in PBS, for 2 hours at room temperature. After washing, 5% BSA (Sigma) was pooled onto the substrate for blocking for 2 hours at room temperature. After washing, a cocktail of 10 μg/mL of anti-CD3 (clone OKT3) and 40 μg/mL of anti-CD28 (clone 9.3, Bio X cell, Fisher) in PBS was pooled onto the substrate overnight for 16 hours at 4 C, before rinsing with PBS and media.
To asses and normalize surface concentration of antibody on the substrates, OKT3 was modified with Alexa Fluor 488 NHS Ester (ThermoFisher) and used in the coating process. Confocal microscopy (Leica) was used to determine uniformity of coating and length of the coating layer on top of the gels.
T cell isolation and culture
Human T cells were isolated from Leukapheresis packs derived from healthy adult donors (New York Blood Center) using RosetteSep Human T cell Enrichment kit (Stemcell Technologies) and Ficoll gradient centrifugation. Complete culture media consisted of RPMI 1640 (Thermo) supplemented with 10 mM HEPES (Gibco), 10 mM L-glutamine (Gibco), 10% (v/v) fetal bovine serum (FBS; Gibco), 0.34% (v/v) β-mercaptoethanol (Sigma-Aldrich), and 10 mM penicillin-streptomycin (Gibco). Cells were frozen in 40% FBS + 10% DMSO in complete media and thawed, resting for 16 hours before use in experiments. Cells were kept under standard culture conditions (37° C, 5% CO2 / 95% air).
Long term culture expansion
Briefly, cells were thawed and rested overnight for 16 hours. Cells were diluted to 1×106 cells/mL of complete media and 1 mL cell solution were seeded on to the coated polyacrylamide gels in 24 well plates at density of 5660 cells/mm2. For positive controls, 1×106 cells were mixed with 3×106 Dynabeads Human T activator CD3/CD28 (Thermo) and seeded onto sterile 24 well plate.
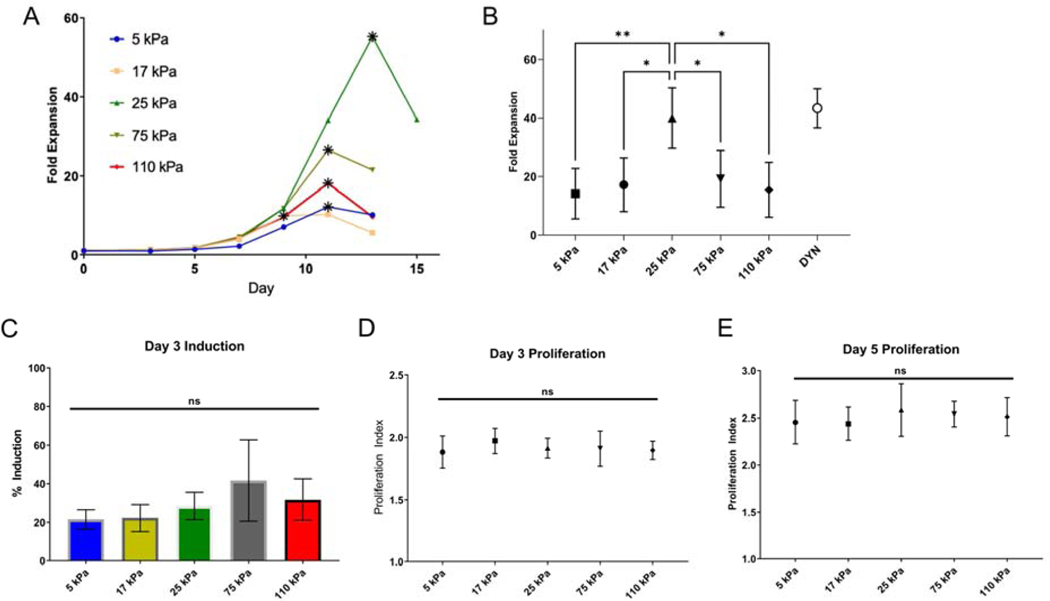
At 72 hours after seeding, cells were removed from the substrates with gentle pipetting and removed from Dynabeads with magnetic holder provided by the vendor. Cell solution was counted with hemocytometer with Trypan blue dilution for dead cell exclusion, and cell size measured using a Scepter cell counter (Millipore). Cell solution were diluted with complete media to maintain a density of 8×105 cells/mL, and reseeded onto new uncoated tissue culture wells. Enumeration, sizing, and dilution was repeated every 48 hours thereafter for up to 15 days. Specifically, each experiment was continued until a decrease in cell number was observed at one of the 48 hour intervals. The maximum fold expansion of a starting population of cells, which typically occurred between 7 and 15 days post seeding and represented the last measurement before a decrease in cell number was observed, was used as single measure for comparison between conditions. This measurement is illustrated by data points in Figure 2A denoted by an asterisk (*).
Figure 2. T cell expansion shows biphasic response to substrate stiffness.
(A) Long term proliferation of T cells activated on coated hydrogels. The data in this panel are from a representative experiment, comparing expansion of cells from a single donor as a function of substrate stiffness. For each substrate, the data point corresponding to maximum fold expansion is indicated with an asterisk (*). (B) Maximum fold expansion of T cells activated on hydrogels. Data are mean ± s.d., n= 3 – 8 across at least 3 independent experiments for each condition, *p<0.05, **p<0.01, One-way ANOVA, Tukey multiple comparison test. (C) T cell induction, measured as the percentage of cells that exhibit at least one cell division by 3 days of seeding. Percentage of starting cell population induced to divide 3 days post seeding. Proliferation, defined as average number of divisions per dividing cell, of activated T cells, 3 days (D) and 5 days (E) post seeding, as measured by CFSE dilution. Data are mean ± s.d., n=5 across 3 independent experiments, One-way ANOVA (α = 0.05).
Cell assays
CFSE dye dilution assay
Commercial CellTrace CFSE Cell Proliferation Kit (ThermoFisher) was used to quantify percent induction and activation. Briefly, prior to activation, cells were stained with 100 μM CFSE for 5 minutes at room temperature, and washed twice with complete media. Cells were collected and analyzed 3 and 5 days after stimulation.
Interleukin-2 secretion assay
IL-2 secretion was measured on a cell-by-cell basis using a surface capture approach (IL-2 secretion detection kit APC, Miltenyi Biotec). Briefly, cells were incubated with surface bound IL-2 catch reagent prior to seeding onto PA gel substrates prepared as previously described. 0.5 mL of cell solution at 5×105 cells/mL were seeded onto substrates at density of ~1415 cells/mm2 for 4 hours. Cells were then incubated with APC conjugated detection antibody against IL-2 and fluorescent antibody against CD8a (BioLegend), fixed, and analyzed with flow cytometry (BD FACSCanto) with minimum of 10000 events. Median mean fluorescence was normalized by signal from positive control (Dynabeads) in each experiment.
For inhibitor experiments, blebbistatin dissolved in 90% DMSO was added to cell solution immediately prior to seeding onto substrates. DMSO control groups had equal concentration of DMSO in solution.
Flow Cytometry
All flow cytometry was performed on a FACSCanto II (BD Biosciences) with minimum of 10000 gated events. Analysis of percent induction and proliferation were performed on FlowJo 7.6 (FlowJo) and all other analysis were performed on FCS Express V6 (De Novo).
Cell morphology
T cells were seeded on coated substrates as previously described. At indicated time points, samples were removed from the culture dish, washed, and fixed in 4% PFA for 15 minutes at room temperature. Samples were washed, then permeabilized with 0.1% Triton X for 10 minutes at room temperature. Samples were blocked with 5% BSA in PBS for one hour, and then incubated with Alexa Fluor 568 Phalloidin (ThermoFisher) (1:40 of manufacturer’s recommended stock solution to 5% BSA in PBS) for one hour at room temperature. Samples were imaged within 48 hours.
Image analysis was performed in ImageJ, using background subtraction, thresholding, and the Analyze Particles function to determine the cell area, circularity, and roundness. 15–50 cells were analyzed for each sample at each timepoint.
Statistics
Expansion, IL-2, and cell morphology experimental data were analyzed with standard one- and two-way ANOVA with Tukey multiple comparison on GraphPad Prism 7 (GraphPad Software). A p value <0.05 was considered statistically significant. All error bars are standard deviation unless otherwise stated.
RNA-sequencing and data analysis
Following 4 hour activation on PA gels of varying stiffness, cells were collected and lysed with Qiazol cell lysis buffer (Qiagen) by vortexing for 1 minute. The supernatant was collected and frozen at −80 C. Total RNA extraction was performed by the Shared Resource at Molecular Pathology Core at Columbia Irving Cancer Center. RNA quality is assessed via RNA Integrity Number (RIN) measured by an Agilent Bioanalyzer. Only samples with RIN scores >8 are used for RNA-sequencing.
RNA-sequencing was performed by the JP Sulzberger Columbia Genome Center. Poly-A pulldown was used to enrich mRNA from total RNA samples, and library construction was prepared using Illumina TruSeq chemistry. 20 million reads (2×100bp) were sequenced for each sample on the Illumina Novaseq 6000 (Illumina).
Real Time Analysis (Illumina) was used for base calling and bcl2fastq (version 2.20) was used for converting Binary Base Call files to fastq format, with adaptor trimming. Pseudoalignment was performed using kallisto [14] and aligned to a kallisto index from Human: GRCh38 transcriptome, generating kallisto abundance files.
For differential gene analysis, R library tximport was used to convert kallisto transcript counts into aggregated gene counts. Differential analysis of gene count data and rlog transformation was performed using the DESeq2 package [15]. Volcano plots were generated using EnhancedVolcano package [16]. Functional pathway analysis using overrepresentation analysis (ORA) and gene set enrichment analysis (GSEA) was performed using clusterProfiler [17] and fgsea package [18]. Venn diagrams were compiled using BioVenn [19].
Results
T cell expansion is biphasic with respect to substrate stiffness
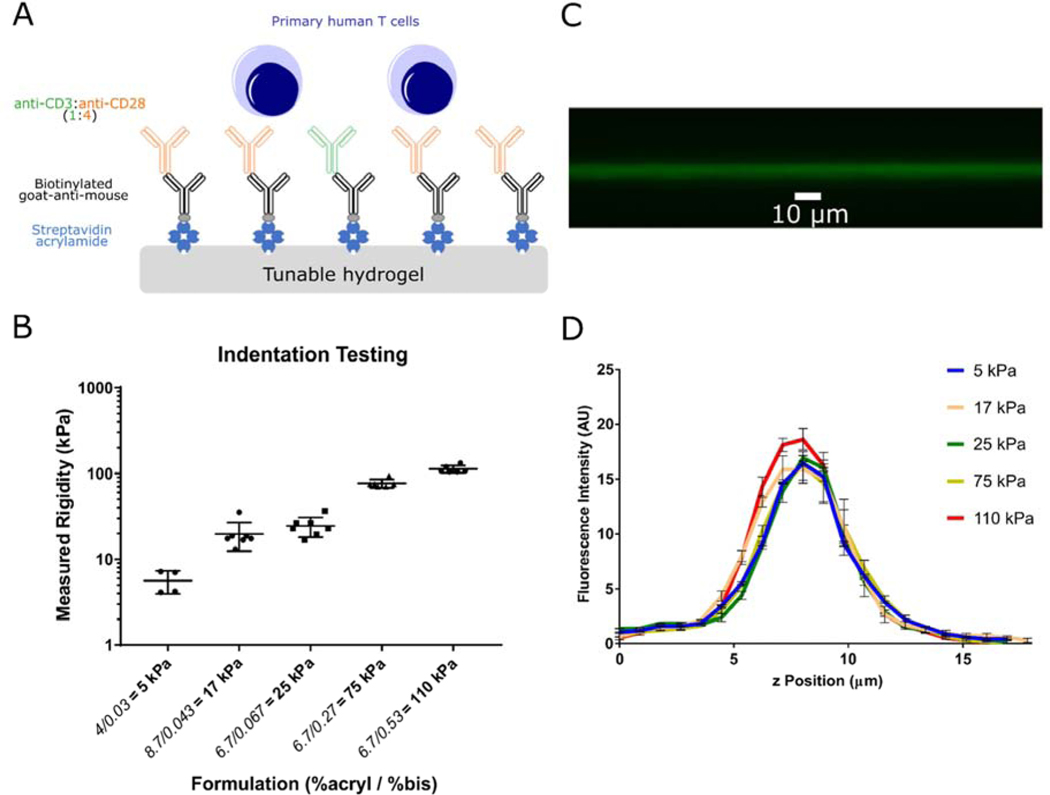
A series of PA hydrogels covering Young’s Moduli of 5 to 110 kPa was prepared using established techniques (Table S1) [9]. Acrylamide-conjugated streptavidin was copolymerized into these gels to provide binding sites for biotinylated antibodies, which were applied in subsequent incubation steps (Fig. 1A). The Young’s modulus (E) of each formulation was measured by indentation (Fig. 1B); the modulus associated with each composition was used to identify the gels throughout the remainder of this report. Captured activating antibodies to CD3 and CD28 (clones OKT3 and 9.3, respectively) were restricted to the gel surface (Fig. 1C). The concentration of acrylamide-streptavidin was adjusted (denoted as 1X coating) to provide a consistent level of coating across different gel compositions (Fig. 1D). CD4+/CD8+ primary human T cells showed a complex pattern of activation and expansion as a function of substrate stiffness. Across all activating substrates, T cells showed an initial period of increasing cell volume (Fig. S1 and rapid expansion that plateaued around 9–13 days as cells came to rest (Fig. 2A). However, the duration of this period of rapid growth and the peak number of cells generated from identical starting populations was sensitive to substrate stiffness; cells activated on 25 kPa gels showed the greatest fold increase in cell number (Fig. 2B), which within each experiment was associated with the longest period of expansion (Fig. 2A). These measures of cell expansion decreased as substrate stiffness either increased (towards 110 kPa) or decreased (towards 5 kPa) (Fig. 2B). As such, these results capture a biphasic response of cell expansion on substrates of a uniform chemistry (and covering a range of modulus attainable with this system), supporting the idea that previous studies were examining opposite sides of a single peak. Notably, differences in cell proliferation as a function of substrate stiffness were not evident early in the expansion process. Examination of induction (the percentage of cells that have undergone at least one division) and proliferative potential (the average number of divisions exhibited by cells that divided at least once) using a CFSE dye-dilution assay revealed no differences at earlier time points of 3 and 5 days post seeding (Fig. 2C–E).
Figure 1. Characterization of the hydrogel system for T cell activation.
(A) Schematic of antibody coated hydrogels as T cell activation platform. Polyacrylamide gels were polymerized with streptavidin-acrylamide, then coated with biotinylated Goat-anti-mouse and secondary activating antibodies to CD3 and CD28. Mixed CD4+/CD8+ polyclonal primary human T cells were stimulated on activating substrates. (B) Mechanical testing via indentation indicate hydrogels with varying ratios of acrylamide and bis-acrylamide have Young’s modulus between 5 to 110 kPa. (Data are mean ± s.d., n=4 gels for 5 kPa, n=7 gels for all other formulations) (C) Confocal microscopy imaging of fluorescent antibodies coated on hydrogels indicate attachment of antibodies to hydrogel surface. (D) Streptavidin-acrylamide concentrations were varied to obtain similar coating of antibodies on hydrogels, as verified by fluorescence intensities on the surface of gels. (n=3 gels for each formulation)
T cell activation is modulated by substrate stiffness and ligand density
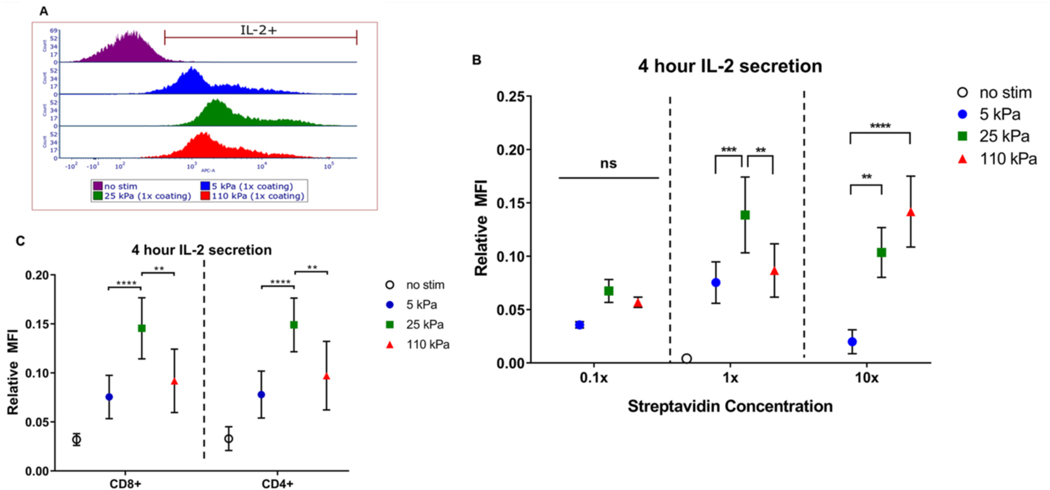
We next examined earlier effects of stiffness on T cell function, comparing secretion of Interluekin-2 (IL-2) over the first 4 hours following seeding onto hydrogels of different stiffnesses (Fig. 3A) [9, 20, 21]. IL-2 secretion is a hallmark of early T cell activation, and we observe the 25 kPa gel induces the highest secretion compared to the 5 kPa and 110 kPa (Fig. 3B), across both CD4+ and CD8+ populations (Fig. 3C). IL-2 secretion thus followed the long-term response seen for expansion as a function of substrate stiffness, and was used in subsequent experiments to investigate T cell mechanosensing.
Figure 3. Short term IL-2 secretion modulated by substrate stiffness and ligand density.
(A) IL-2 secretion of T cells characterized by flow cytometry. (B) IL-2 secretion of T cells activated for 4 hours on hydrogels with standardized coating (n=12 across 5 independent experiments), 0.1x coating (n=3 across 2 independent experiments), and 10x coating. Data are mean ± s.d., n=6 across 2 independent experiments. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, 2-way ANOVA, Tukey multiple comparison test. (C) IL-2 secretion of CD4+ and CD8+ T cell subsets. Data are mean ± s.d., n=12 across 5 independent experiments. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, 2-way ANOVA, Tukey multiple comparison test.
The density of activating antibodies on a cell or artificial surface has been shown to play a key role in affecting T cell activation [22, 23]. To test how ligand density affects T cell mechanosensing, the concentration of acrylamide-streptavidin incorporated was varied across gels with moduli of 5, 25, and 110 kPa. Activation of T cells using gels with ten-fold decrease in acrylamide-streptavidin concentration (denoted 0.1X coating) reduced overall IL-2 secretion (Fig. 3B). Conversely, activation using gels with ten-fold increase in acrylamide-streptavidin concentration (denoted 10X coating) shifted the peak in IL-2 secretion to higher stiffness (Fig. 3B, S2). Intriguingly, the results obtained at this higher ligand density are in accordance to previous results in which mice CD4+ T cells show higher IL-2 secretion with increasing substrate stiffness [9].
Substrate stiffness affects T cell morphology
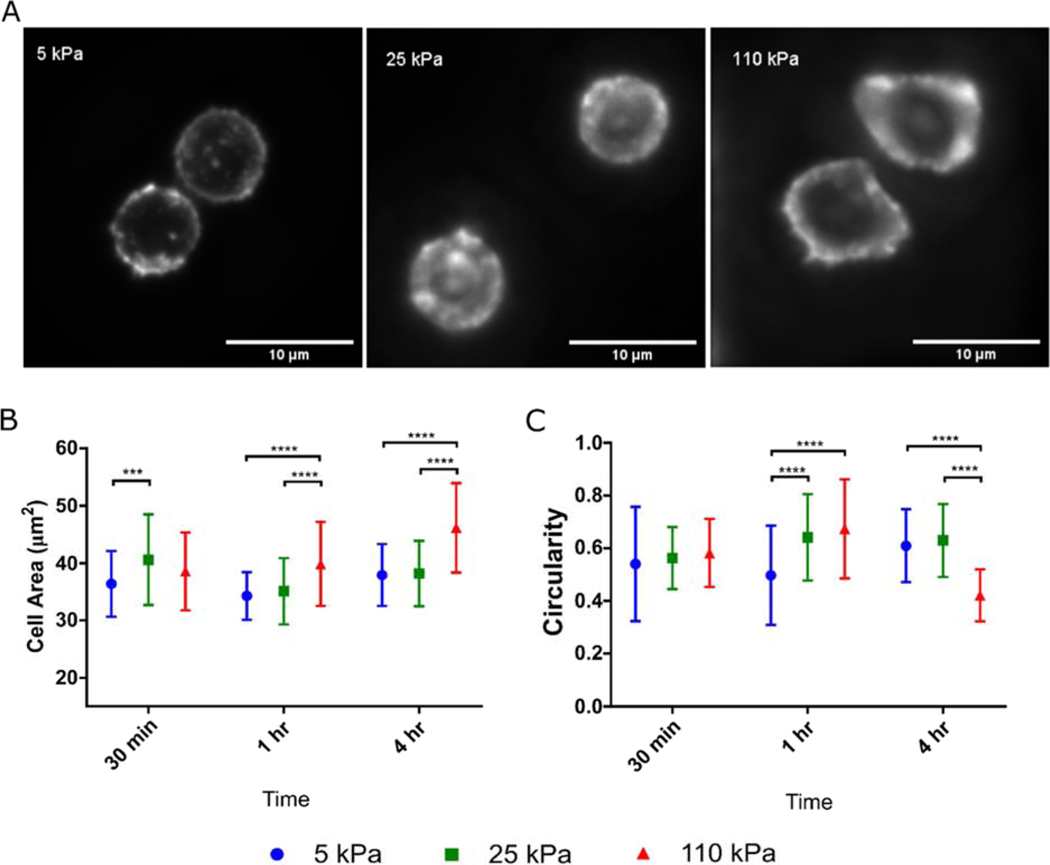
Looking towards even earlier cell responses, we compared T cell spreading on activating substrates of different stiffnesses (Fig. 4A). At 30 minutes post seeding, cells showed the highest spreading area on the 25 kPa gels, suggesting that initial engagement of a surface by T cells also exhibits a biphasic behavior (Fig. 4B), mirroring that of IL-2 secretion and later expansion. However, this response subsequently shifts. At the 1 and 4 hour time points, cell spreading was highest on the 110 kPa substrates. Cell circularity was also dependent on substrate stiffness and time (Fig. 4C). The 5 kPa induced cells exhibit the lowest circularity at 1 hour, while the 110 kPa induced cells exhibit the lowest circularity at 4 hours (Fig. 4C). Together, these results indicate that while key cellular functions including expansion, IL-2 secretion, and early cell spreading follow a biphasic sensitivity to substrate modulus, other cellular functions can concurrently show monotonic responses over the range of stiffness included in this study.
Figure 4. T cell spreading is a function of substrate stiffness.
(A) T cell morphology was assayed by staining for actin. This image illustrates cells on hydrogels 1 hr after seeding. (B) Spreading area and (C) circularity of T cells on hydrogels at 30 minutes, 1 hour, and 4 hour timepoints post seeding, characterized by membrane stain. Data pooled from at least 40 cells across 6 independent substrates for each condition. Data are mean ± s.d., *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, 2-way ANOVA, Tukey multiple comparison test.
Effect of stiffness sensing on T cell transcription
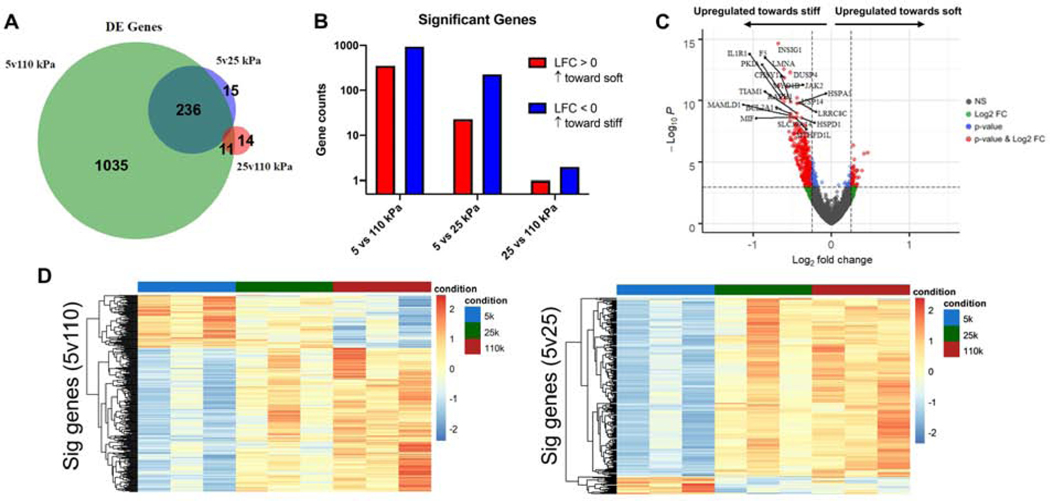
Towards deeper understanding of T cells response to substrate stiffness, we carried out transcriptomic analysis of T cells that were activated for 4 hours on hydrogel substrates of 5, 25, and 110 kPa stiffness, values chosen to capture the biphasic response of cell expansion. Comparison of cells activated on gels of 5 vs. 110 kPa revealed a large set of differentially expressed genes (DEGs) (Fig. 5A&B). A greater number of DEGs showed increased expression on the 110 kPa substrates (Log2Fold Change, LFC < 0) than those that showed decreased expression (Fig. 5B&C). Including the intermediate substrate modulus, a subset of these DEGs were also observed to change between cells activated on the 5 and 25 kPa samples (Fig. 5A&B). A much smaller and distinct subset of genes were differentially expressed between cells activated on the 25 and 110 kPa substrates (Fig. 5A&B). The larger overlap between DEGs identified for the 5 vs. 110 kPa and 5 vs 25 kPa comparisons suggests that gene transcription changes predominantly as Young’s modulus increases from 5 to 25 kPa, with different genes increasing or decreasing monotonically. This is reflected in a transcriptome heat map of DEGs identified for the 5 vs 110 kPa comparison (Fig. 5D), in which hierarchal clustering reveals one distinct group that increases with substrate stiffness and a second that decreases.
Figure 5. Stiffness sensing induces gradient of gene expression.
(A) Number of Differentially Expressed (DE) genes between for pairwise comparisons. (B) Number of genes with p-adjusted value < 0.1 based on DESeq2 algorithm (Log2Fold Change > 0 indicates upregulation towards lower stiffness, Log2Fold Change < 0 indicates upregulation towards higher stiffness). (C) Volcano plot for DE genes between 5 vs 110 kPa conditions. (D) Heatmap of significant genes between 5 vs 110 kPa and 5 vs 25 kPa conditions.
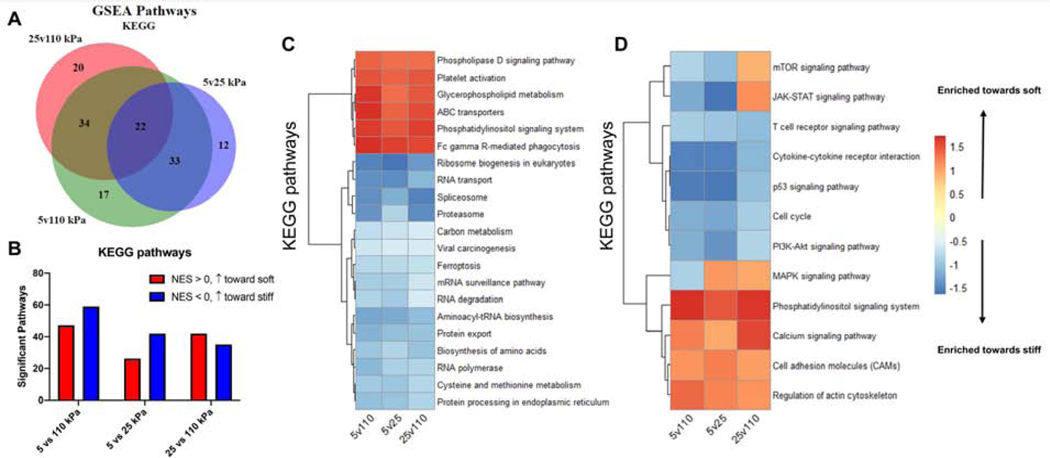
Recognizing that transcriptional changes in individual genes provide limited insight into cell state, gene set enrichment analysis (GSEA) was carried out using the KEGG (Kyoto Encyclopedia of Genes and Genomes) dataset [24]. Similar to transcriptional changes in individual genes, the highest number of enriched pathways was observed in the 5 vs. 110 kPa comparison (Fig. 6A&B), with a greater number of pathways increasing with higher stiffness (Fig. 6B). Within the top ranked pathways, we focused on those associated with T cell activation and mechanosensing and observe a majority of pathways are enriched monotonically (Fig. 6C). Activation on higher stiffness induced enrichment of processes of pathways closely associated to T cell growth such as TCR signaling, cytokine-receptor interaction, p53, cell cycle, and PI3K-Akt signaling. Activation on substrates of lower stiffness induced enrichment in processes involved in mechanosensitive functions, most notably regulation of actin cytoskeleton and cell adhesion molecules, as well as canonical hallmarks of short term activation such as calcium signaling.
Figure 6. Monotonic enrichment of activation pathways.
(A) Number of Differentially Expressed (DE) genes between for pairwise comparisons. (B) Number of significantly enriched pathways (FDR < 0.1) from KEGG gene sets with for each pairwise comparison (NES > 0 is enrichment towards lower stiffness, NES < 0 is enrichment towards higher stiffness). (C) Normalized enrichment score (NES) of KEGG pathways shared by pairwise comparisons (FDR < 0.1 in 5vs110 kPa comparison). (D) NES of T cell activation related pathways shared by pairwise comparisons. (Top 50 pathways, ranked by FDR)
Effect of cytoskeletal protein inhibitors on mechanosensitive T cell activation
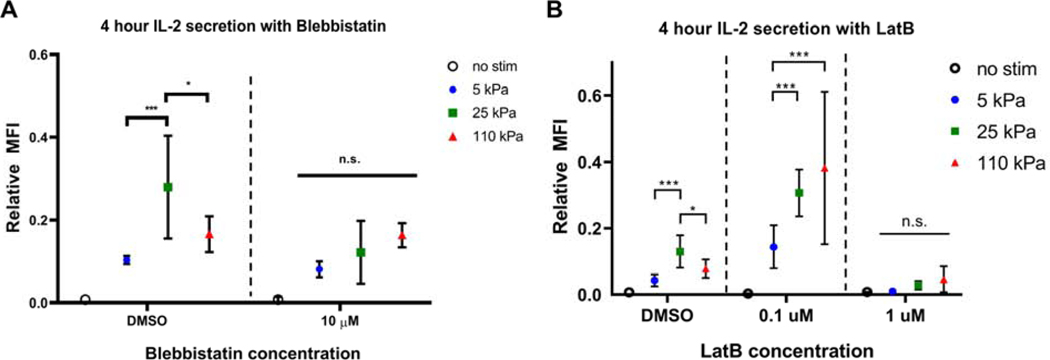
Following on the observation that gene pathways associated with actin cytoskeletal regulation respond strongly to substrate modulus, we used pharmacological inhibitors to examine the role of this structure in T cell mechanosensing. Specifically, 4-hour secretion of IL-2 was compared for T cells in the presence of the actomyosin contractility inhibitor blebbistatin, and the actin assembly inhibitor latrunculin-B. At a concentration of 10 μM, blebbistatin abrogated the mechanosensing response; IL-2 secretion was similar across all hydrogel substrates, but higher than the non-stimulated control (Fig. 7A).
Figure 7. Inhibition of proteins associated with cytoskeletal contractility affects T cell stiffness sensing.
(A) IL-2 secretion of T cells activated for 4 hours on hydrogels in presence of Blebbistatin compared to DMSO controls. Data are mean ± s.d., n=7 across 3 independent experiments. *p<0.05, **p<0.01, ***p<0.001, 2-way ANOVA, Tukey multiple comparison test. (B) IL-2 secretion of T cells activated for 4 hours on hydrogels in presence of Labtrunculin B compared to DMSO controls. Data are mean ± s.d., n=6 across 3 independent experiments. *p<0.05, **p<0.01, ***p<0.001, 2-way ANOVA, Tukey multiple comparison test.
At 0.1 μM of latrunculin B, we observed a shift in IL-2 secretion pattern from a biphasic response shown by the vehicle control to a monotonic increase in activation with increasing stiffness. At 1 μM, we see a complete loss of activation across all conditions (Fig. 7B). Together, these experiments validate the role of actin cytoskeletal function in modulating T cell mechanosensing.
Discussion
Mechanical forces play increasingly recognized roles in T cell activation and subsequent function [9, 10, 25–28]. Early studies in this area demonstrated that application of forces to T cells through substrate-immobilized antibodies targeting CD3 led to cell activation [29, 30]. Subsequent studies demonstrated that T cells can exert force on their extracellular environment through TCR/CD3 [27, 31–34]. Bringing these two activities together, it was demonstrated that T cells can carry out sensing of the mechanical stiffness of an activating substrate [9, 10, 22, 35]. These and a rapidly growing number of complementary studies are shedding new light into how mechanical forces modulate communication between T cells and their targets in vivo through the cell-cell contact area termed the immune synapse (IS). Notably, studies in this area examine a wider set of receptors on the T cell surface, such as Lymphocyte Function-associated Antigen 1 (LFA-1) receptor which binds to Intercellular Adhesion Molecule 1 (ICAM-1) [36, 37], revealing remarkable mechanical complexity of the IS. Additional complexity is found in considering TCR recognition of peptide-loaded Major Histocompatibility Complex (pMHC), the native ligand for this receptor. Engagement of the T cell through this interaction, which is bypassed using anti-CD3 antibodies, enhances substrate mechanosensing and can also exhibit a catch bond behavior [32, 36, 38]. Perhaps surprisingly, CD28 does not appear to exhibit mechanosensing, augmenting TCR signaling when engaged by either substrate-bound in in-solution ligands [9, 33]. Given the role of T cell expansion for cellular immunotherapy, this report focuses on model substrates presenting activating antibodies to CD3 and CD28, which are sufficient for initiating this process.
Most important to the design of biomaterials is our demonstration that T cell expansion initiated using such systems can show a biphasic response to material stiffness. This result can help reconcile earlier reports that mouse and human cells show contrasting responses and allowing studies in one system to better inform the other. The phenomenon of biphasic cell response to the elastic modulus of a substrate is an area of growing interest for a wide range of cell types [39–44]. Notably, increasing the density of anti-CD3 on the substrate surface shifts the T cell response of IL-2 secretion towards substrates of higher elastic modulus (Fig. 3B). This was surprising as increasing the mechanical resistance of a ligand to cell-generated forces is often considered to enhance receptor signaling; a higher ligand density would result in more signaling and a shift of the mechanosensing peak to materials of lower stiffness. A potential explanation for the shift shown in Fig. 3B is that T cells generate a certain level of force over a characteristic area, which is then split across the receptor-ligand interactions in that region. Increasing ligand density would then decrease the force applied per receptor, shifting the peak of response to higher stiffness. An additional unexpected result in this report is that while expansion is biphasic over the included range of substrate stiffness and a specific ligand density, late cell spreading and gene transcription (Figs. 4–6) show monotonic dependencies. This raised the prospect that biphasic cellular responses can result from the interaction of multiple, intersecting processes. That is, cellular functions showing a biphasic response (including IL-2 secretion and transcription of gene groups such as mTOR) might require processes that increase with elastic modulus and others that decrease; minimization of either process in response to elastic modulus can produce a biphasic response, supporting a more complex view of cell mechanosensing.
It is recognized that the choice of PA gels for this study limits the range of Young’s modulus that were examined. Indeed, polydimethylsiloxane (PDMS) elastomer can be much stiffer and is better suited for use in a manufacturable system for T cell activation [7, 8]. However, modulation of PDMS modulus over an extended range is also problematic, requiring changes between proprietary formulations [11] that might not be fully addressed and/or challenges associated with incomplete crosslinking of siloxane chains. Differences in ligand immobilization methods and presentation makes comparisons between PDMS and PA further problematic [45]. While there is the potential that this report misses additional complexity of T cell response at higher rigidity, we focus on PA gels as a single, well-recognized system capable of realizing stiffness that is closer to physiological range than PDMS for studies into the cellular-level mechanisms of mechanosensing.
Finally, this new knowledge of T cell sensitivity to substrate stiffness may enhance understanding of their interaction with APCs. The dendritic cell (DC) actin cytoskeleton has been shown to undergo maturation-associated changes, restraining the mobility of ligands such as ICAM-1, and changing cortical stiffness [46]. This functions to retain forces on receptors of interacting T cells and modulating mechanosensing at the IS; inhibiting the DC actin cytoskeleton disrupts T cell activation [47]. Furthermore, APCs have been shown to change their mechanical properties based on the state of inflammatory conditions [48]. The concept that T cells can exhibit complex, biphasic responses to DC mechanics, and that these could be regulated by converging processes that each respond monotonically to forces, sheds new insight into the language of T cell – APC communication and strategies for modulating this interaction.
Supplementary Material
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Biotinylated Goat-anti-mouse | BioLegend | 405303 |
| Mouse-anti-human CD3 (OKT3) | BioXCell | BE0001-2 |
| Mouse-anti-human CD28 (9.3) | BioXCell | BE0248 |
| Biological Samples | ||
| Leukopack | New York Blood Center | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Acrylamide (40%) | Sigma | 01697 |
| Bis-acrylamide (2%) | Fisher | BP1404-250 |
| Ammonium persulfate (APS) | Sigma | A3678 |
| Tetramethylethylenediamine (TEMED) | Sigma | T7024 |
| Streptavidin-acrylamide | Fisher | S21379 |
| (3-Aminopropyl)triethoxysilane (APTES) | Sigma | 440140 |
| Glutaraldehyde | Sigma | G7776 |
| Alexa-fluor 488 | Thermo | A20000 |
| Blebbistatin | Sigma | 203391 |
| Prolong Diamond | ThermoFisher | P36961 |
| Qiazol | Qiagen | 79306 |
| Critical Commercial Assays | ||
| Human IL-2 Secretion kit (APC) | Miltenyi Biotec | 130-090-763 |
| RosetteSep Human T cell Enrichment Kit | Stemcell Technologies | 15061 |
| CellTrace CFSE Dilution Assay | ThermoFisher | C34554 |
| CellVue Jade | ThermoFisher | 88-0876-16 |
| Software and Algorithms | ||
| FlowJo 7.6 | FlowJo LLC | |
| FCS Express V6 | De Novo Software | |
| Graphpad Prism 7 | Graphpad Software | |
| kallisto | Bray et al. Nature Biotechnology. (2016) | |
| DESeq2 | Love et al. Genome Biology. (2014) | |
| fgsea | Sergushichev A. bioRxiv (2016) | |
| EnhancedVolcano | Blighe K. (2018) | |
| Other | ||
| RPMI 1640 | Thermo | 21870092 |
| HEPES Buffer | Gibco | 15630 |
| L-glutamine | Gibco | 25030 |
| Fetal Bovine Serum (FBS) | Gibco | Gibco-Performance 26140-079 |
| β-mercaptoethanol | Sigma-Aldrich | M3148 |
| Penicillin-streptomicin | Gibco | 15140122 |
| Scepter cell counter | Millipore | PHCC40500 |
Acknowledgements
RNA extraction for this work was performed in the Molecular Pathology Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University, supported by NIH grant P30 CA013696. RNA-sequencing for this work was performed in the Genomics and High Throughput Screening Shared Resource at the Columbia JP Sulzberger Genome Center, funded in part through the NIH/NCI Cancer Center Support Grant P30CA013696.
Funding
This study was supported by the NIH (R01AI10593 to LCK and TL1TR18753 to DJY) and the NSF (CMMI 1743420 to LCK and GRFP to LS).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data Availability
The raw/processed data required to reproduce these findings cannot be shared at this time due to technical or time limitations.
The data is in the process of submission to https://academiccommons.columbia.edu/.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Restifo NP, Dudley ME, Rosenberg SA, Adoptive immunotherapy for cancer: harnessing the T cell response, Nature Reviews Immunology 12 (2012) 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fesnak AD, June CH, Levine BL, Engineered T cells: the promise and challenges of cancer immunotherapy, Nature Reviews Cancer 16 (2016) 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dwarshuis NJ, Parratt K, Santiago-Miranda A, Roy K, Cells as advanced therapeutics: State-of-the-art, challenges, and opportunities in large scale biomanufacturing of high-quality cells for adoptive immunotherapies, Advanced Drug Delivery Reviews 114 (2017) 222–239. [DOI] [PubMed] [Google Scholar]
- [4].Ruella M, Kalos M, Adoptive immunotherapy for cancer, Immunological Reviews 257(1) (2014) 14–38. [DOI] [PubMed] [Google Scholar]
- [5].Levine BL, Mosca JD, Riley JL, Carroll RG, Vahey MT, Jagodzinski LL, Wagner KF, Mayers DL, Burke DS, Weislow OS, St DC. Louis CH. June, Antiviral Effect and Ex Vivo CD4+T Cell Proliferation in HIV-Positive Patients as a Result of CD28 Costimulation, Science 272(5270) (1996) 1939. [DOI] [PubMed] [Google Scholar]
- [6].Riches JC, Davies JK, McClanahan F, Fatah R, Iqbal S, Agrawal S, Ramsay AG, Gribben JG, T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production, Blood 121(9) (2013) 1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lambert LH, Goebrecht GKE, De Leo SE, O’Connor RS, Nunez-Cruz S, Li T-D, Yuan J, Milone MC, Kam LC, Improving T Cell Expansion with a Soft Touch, Nano Letters 17(2) (2017) 821–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dang AP, De Leo S, Bogdanowicz DR, Yuan DJ, Fernandes SM, Brown JR, Lu HH, Kam LC, Enhanced Activation and Expansion of T Cells Using Mechanically Soft Elastomer Fibers, Advanced Biosystems 2(2) (2018) 1700167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Judokusumo E, Tabdanov E, Kumari S, Dustin Michael L., Kam Lance C, Mechanosensing in T Lymphocyte Activation, Biophysical Journal 102(2) (2012) L5–L7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Saitakis M, Dogniaux S, Goudot C, Bufi N, Asnacios S, Maurin M, Randriamampita C, Asnacios A, Hivroz C, Different TCR-induced T lymphocyte responses are potentiated by stiffness with variable sensitivity, eLife 6 (2017) e23190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wahl A, Dinet C, Dillard P, Nassereddine A, Puech P-H, Limozin L, Sengupta K, Biphasic mechanosensitivity of T cell receptor-mediated spreading of lymphocytes, Proceedings of the National Academy of Sciences (2019) 201811516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gioia L, Siddique A, Head SR, Salomon DR, Su AI, A genome-wide survey of mutations in the Jurkat cell line, BMC genomics 19(1) (2018) 334–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kandow CE, Georges PC, Janmey PA, Beningo KA, Polyacrylamide Hydrogels for Cell Mechanics: Steps Toward Optimization and Alternative Uses, Methods in Cell Biology, Academic Press; 2007, pp. 29–46. [DOI] [PubMed] [Google Scholar]
- [14].Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification, Nature Biotechnology 34 (2016) 525. [DOI] [PubMed] [Google Scholar]
- [15].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol 15(12) (2014) 550–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Blighe K, Enhancedvolcano: Publication-ready volcano plots with enhanced colouring and labeling, 2018. [Google Scholar]
- [17].Yu G, Wang L-G, Han Y, He Q-Y, clusterProfiler: an R package for comparing biological themes among gene clusters, OMICS 16(5) (2012) 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sergushichev AA, An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation, bioRxiv (2016) 060012. [Google Scholar]
- [19].Hulsen T, de Vlieg J, Alkema W, BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams, BMC Genomics 9(1) (2008) 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shen K, Thomas VK, Dustin ML, Kam LC, Micropatterning of costimulatory ligands enhances CD4+ T cell function, Proceedings of the National Academy of Sciences 105(22) (2008) 7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sims TN, Soos TJ, Xenias HS, Dubin-Thaler B, Hofman JM, Waite JC, Cameron TO, Thomas VK, Varma R, Wiggins CH, Sheetz MP, Littman DR, Dustin ML, Opposing Effects of PKCtheta and WASp on Symmetry Breaking and Relocation of the Immunological Synapse, Cell 129(4) (2007) 773–785. [DOI] [PubMed] [Google Scholar]
- [22].O’Connor RS, Hao X, Shen K, Bashour K, Akimova T, Hancock WW, Kam LC, Milone MC, Substrate Rigidity Regulates Human T Cell Activation and Proliferation, The Journal of Immunology 189(3) (2012) 1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Deeg J, Axmann M, Matic J, Liapis A, Depoil D, Afrose J, Curado S, Dustin ML, Spatz JP, T Cell Activation is Determined by the Number of Presented Antigens, Nano Letters 13(11) (2013) 5619–5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles, Proceedings of the National Academy of Sciences 102(43) (2005) 15545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hu J, Gondarenko AA, Dang AP, Bashour KT, O’Connor RS, Lee S, Liapis A, Ghassemi S, Milone MC, Sheetz MP, Dustin ML, Kam LC, Hone JC, High-Throughput Mechanobiology Screening Platform Using Micro- and Nanotopography, Nano Letters 16(4) (2016) 2198–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tamzalit F, Wang MS, Jin W, Tello-Lafoz M, Boyko V, Heddleston JM, Black CT, Kam LC, Huse M, Interfacial actin protrusions mechanically enhance killing by cytotoxic T cells, Science Immunology 4(33) (2019) eaav5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Basu R, Whitlock BM, Husson J, Le Floc’h A, Jin W, Oyler-Yaniv A, Dotiwala F, Giannone G, Hivroz C, Biais N, Lieberman J, Kam LC, Huse M, Cytotoxic T Cells Use Mechanical Force to Potentiate Target Cell Killing, Cell 165(1) (2016) 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Stroka KM, Aranda-Espinoza H, Neutrophils display biphasic relationship between migration and substrate stiffness, Cell Motility 66(6) (2009) 328–341. [DOI] [PubMed] [Google Scholar]
- [29].Kim ST, Takeuchi K, Sun Z-YJ, Touma M, Castro CE, Fahmy A, Lang MJ, Wagner G, Reinherz EL, The αβ T Cell Receptor Is an Anisotropic Mechanosensor, Journal of Biological Chemistry 284(45) (2009) 31028–31037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li Y-C, Chen B-M, Wu P-C, Cheng T-L, Kao L-S, Tao M-H, Lieber A, Roffler SR, Cutting Edge: Mechanical Forces Acting on T Cells Immobilized via the TCR Complex Can Trigger TCR Signaling, The Journal of Immunology 184(11) (2010) 5959. [DOI] [PubMed] [Google Scholar]
- [31].Husson J, Chemin K, Bohineust A, Hivroz C, Henry N, Force Generation upon T Cell Receptor Engagement, PLOS ONE 6(5) (2011) e19680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Das DK, Feng Y, Mallis RJ, Li X, Keskin DB, Hussey RE, Brady SK, Wang J-H, Wagner G, Reinherz EL, Lang MJ, Force-dependent transition in the T-cell receptor β-subunit allosterically regulates peptide discrimination and pMHC bond lifetime, Proceedings of the National Academy of Sciences 112(5) (2015) 1517–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bashour KT, Tsai J, Shen K, Lee J-H, Sun E, Milone MC, Dustin ML, Kam LC, Cross Talk between CD3 and CD28 Is Spatially Modulated by Protein Lateral Mobility, Molecular and Cellular Biology 34(6) (2014) 955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hui KL, Balagopalan L, Samelson LE, Upadhyaya A, Cytoskeletal forces during signaling activation in Jurkat T-cells, Molecular Biology of the Cell 26(4) (2014) 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jin W, Tamzalit F, Chaudhuri PK, Black CT, Huse M, Kam LC, T cell activation and immune synapse organization respond to the microscale mechanics of structured surfaces, Proceedings of the National Academy of Sciences 116(40) (2019) 19835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Blumenthal D, Chandra V, Avery L, Burkhardt JK, Mouse T cell priming is enhanced by maturation-dependent stiffening of the dendritic cell cortex, eLife 9 (2020) e55995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hu KH, Butte MJ, T cell activation requires force generation, Journal of Cell Biology 213(5) (2016) 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu B, Chen W, Brian D. Zhu Evavold, C, Accumulation of Dynamic Catch Bonds between TCR and Agonist Peptide-MHC Triggers T Cell Signaling, Cell 157(2) (2014) 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Engler AJ, Sen S, Sweeney HL, Discher DE, Matrix Elasticity Directs Stem Cell Lineage Specification, Cell 126(4) (2006) 677–689. [DOI] [PubMed] [Google Scholar]
- [40].Ulrich TA, de Juan Pardo EM, Kumar S, The Mechanical Rigidity of the Extracellular Matrix Regulates the Structure, Motility, and Proliferation of Glioma Cells, Cancer Research 69(10) (2009) 4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ambrosi D, Duperray A, Peschetola V, Verdier C, Traction patterns of tumor cells, Journal of Mathematical Biology 58(1) (2008) 163. [DOI] [PubMed] [Google Scholar]
- [42].Isenberg BC, DiMilla PA, Walker M, Kim S, Wong JY, Vascular Smooth Muscle Cell Durotaxis Depends on Substrate Stiffness Gradient Strength, Biophysical Journal 97(5) (2009) 1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lo C-M, Wang H-B, Dembo M, Wang Y.-l., Cell Movement Is Guided by the Rigidity of the Substrate, Biophysical Journal 79(1) (2000) 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Oakes PW, Patel DC, Morin NA, Zitterbart DP, Fabry B, Reichner JS, Tang JX, Neutrophil morphology and migration are affected by substrate elasticity, Blood 114(7) (2009) 1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Trappmann B, Gautrot JE, Connelly JT, Strange DG, Li Y, Oyen ML, Cohen Stuart MA, Boehm H, Li B, Vogel V, Spatz JP, Watt FM, Huck WT, Extracellular-matrix tethering regulates stem-cell fate, Nat Mater 11(8) (2012) 642–9. [DOI] [PubMed] [Google Scholar]
- [46].Comrie WA, Li S, Boyle S, Burkhardt JK, The dendritic cell cytoskeleton promotes T cell adhesion and activation by constraining ICAM-1 mobility, The Journal of Cell Biology 208(4) (2015) 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Al-Alwan MM, Rowden G, Lee TDG, West KA, Cutting Edge: The Dendritic Cell Cytoskeleton Is Critical for the Formation of the Immunological Synapse, The Journal of Immunology 166(3) (2001) 1452. [DOI] [PubMed] [Google Scholar]
- [48].Bufi N, Saitakis M, Dogniaux S, Buschinger O, Bohineust A, Richert A, Maurin M, Hivroz C, Asnacios A, Human Primary Immune Cells Exhibit Distinct Mechanical Properties that Are Modified by Inflammation, Biophysical Journal 108(9) (2015) 2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.