Abstract
The deadly global outbreak of coronavirus disease-2019 (COVID-19) has forged an unrivaled threat to human civilization. Contemplating its profuse impact, initial risk management and therapies are needed, as well as rapid detection strategies alongside treatments with existing drugs or traditional treatments to provide better clinical support for critical patients. Conventional detection techniques have been considered but do not sufficiently meet the current challenges of effective COVID-19 diagnosis. Therefore, several modern techniques including point-of-care diagnosis with a biosensor, clustered regularly interspaced short palindromic repeats (CRISPR)-associated proteins that function as nuclease (Cas) technology, next-generation sequencing, serological, digital, and imaging approaches have delivered improved and noteworthy success compared to that using traditional strategies. Conventional drug treatment, plasma therapy, and vaccine development are also ongoing. However, alternative medicines including Ayurveda, herbal drugs, homeopathy, and Unani have also been enlisted as prominent treatment strategies for developing herd immunity and physical defenses against COVID-19. All considered, this review can help develop rapid and simplified diagnostic strategies, as well as advanced evidence-based modern therapeutic approaches that will aid in combating the global pandemic.
Keywords: COVID-19, Ayurveda, Herd immunity, Homeopathy
Graphical abstract
Overcoming the COVID-19 situation requires a balance between diagnosis and treatments.
Highlights
-
•
Novel coronavirus disease or COVID-19 is a pandemic outbreak and has initiated a global emergency.
-
•
The rapid diagnostic tool with immense sensitivity is essential for COVID-19 detection.
-
•
Novel treatment strategies along with vaccine development are urgently needed for treatment against COVID-19.
1. Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has resulted in a human disease called coronavirus disease-2019 (COVID-19), which has been currently evidenced in more than 200 countries worldwide, with the rapid spread of the disease being reported [1]. From its initial emergence in late 2019 until December 2020, the number of active cases rose to 74.8 million worldwide with 1.7 million casualties [2]. In this situation, accurate detection of suspected cases, proper isolation of infected patients, and active treatment procedures are required to arrest the pandemic.
Conventional nucleic acid-based detection techniques used in virological diagnosis have established themselves as a concrete technology. Detection of viral nucleic acids using real-time reverse transcriptase-polymerase chain reaction (rRT-PCR) is a well-known method owing to its high specificity and sensitivity [3]. In contrast to rRT-PCR, other modified PCR-based methods are also available. However, these diagnostic methods have various disadvantages to them such as a high rate of false-negative results [4,5], longer detection time, and compromises in the accurate detection of COVID-19 owing to the presence of inadequate genetic material in the throat or nasopharyngeal swab (NPS). Therefore, the development of novel detection methods and treatment strategies is imperative to supplement the current diagnosis.
Recent advancements in emerging techniques have led to the development of potential toolkits with advanced molecular biology and digital technology. The developed method includes utilization of clustered regularly interspaced short palindromic repeats (CRISPR)-associated proteins, which function as nuclease (Cas) for the detection of COVID-19 [6,7]. In addition, based on the CRISPR-Cas system technology against SARS-CoV-2, a group of Indian scientists have established an efficient, rapid, highly sensitive, and economical FnCas9 Editor Linked Uniform Detection Assay (FELUDA) kit for the successful detection of COVID-19, which provides results within 1 h [8]. Apart from the CRISPR-Cas system, aptamer-based technology has also gained attention for its sensitive detection of SARS-CoV-2 [9]. Additionally, isothermal amplification, rolling circle amplification, microarrays, biosensors such as field-effect transistors (FETs), plasmonics-based technologies [10], ultrafast advanced strand exchange amplification (ASEA) [11], immunoassays, next-generation sequencing (NGS) [12], and digital technology have also been used in COVID-19 detection.
In addition to developing diagnostic strategies, it is equally necessary to find treatments for the novel coronavirus (nCoV). Various treatment strategies, such as conventional drug treatment, plasma therapy, and vaccine development, have been proposed for COVID-19 treatment [13,14]. In this context, complementary and alternative medicines, such as Ayurveda, have also gained immense interest because of adequate clinical evidence [15] of their beneficial actions such as generation of herd immunity, anticancer and antimicrobial activities [16,17]. Recent reports have suggested that Ayurvedic treatment, involving an in-depth therapeutic strategy, can efficiently treat COVID-19 [18]. From past history, it has been deciphered that a combination of herbal formulations such as Shadanga Paniya and Ayurvedic medicine Agastya Harityaki is effective in treating respiratory turbulence and, recently, has been prescribed by the Ministry of Ayush, Government of India, for treatment against COVID-19 [19]. The inclusion of homeopathy, Unani, and herbal medicine has also been proven efficient in battling against COVID-19 [20,21].
In this review, we abridged the overview of the current updates on the diagnosis of COVID-19 and therapeutic strategies that are using traditional medicine as an immunity booster to help fight the COVID-19 global pandemic.
2. nCoV disease and clinical manifestations
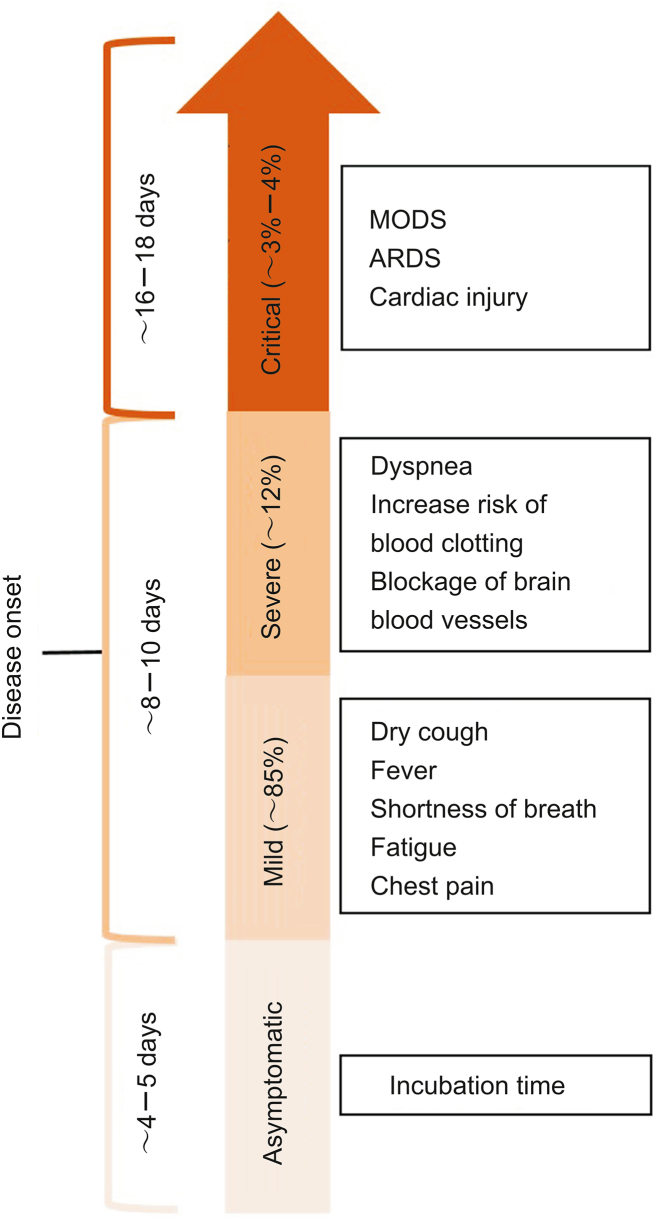
Coronaviruses (nCoVs) are large positive-sense single-stranded RNA viruses with lengths of approximately 26–32 kb [22,23] and with a spike glycoprotein on the envelope, which gives them the crown-like appearance. nCoVs are beta-coronaviruses of the subgenus Sarbecovirus and family Coronaviridae. nCoVs mainly originate from the B-lineage of the beta-coronaviruses and are closely related to their earlier strain SARS-CoV [24]. SARS-CoV-2 is a highly contagious pathogen that is transmitted by aerosols, mucous membranes, and respiratory droplets. The broad clinical spectrum of the illness ranges from asymptomatic or mildly symptomatic to severely symptomatic, leading to multi-organ dysfunction syndrome. The most common symptoms include dry cough, fever, shortness of breath, fatigue, tiredness, chest pain, increased risk of blood clotting, and blockage of brain blood vessels [25,26] (Fig. 1).
Fig. 1.
Different stages of clinical manifestations of COVID-19. Symptoms of COVID-19 mainly include fever, fatigue, dry cough, dyspnea, acute respiratory distress syndrome (ARDS), multi-organ dysfunction syndrome (MODS), and cardiac injury, with varied incubation time and disease onset.
3. Advance towards detection methods
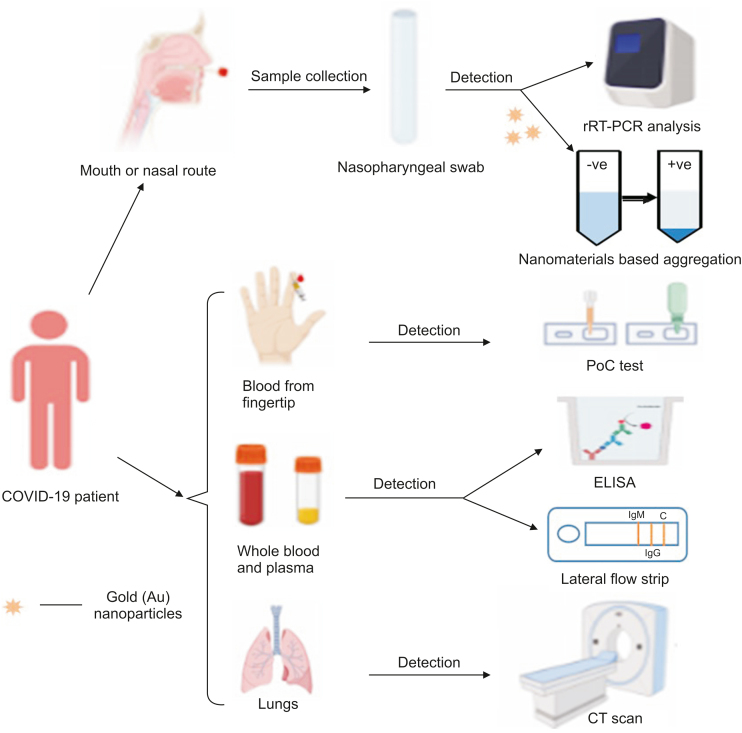
Recent critical events have reinforced the need for a global sensitive surveillance system that can detect the coronavirus during its early phase of outbreak and prevent its spread and amplification. Conventional methods are most often used for the identification of COVID-19, but several drawbacks to these methods have resulted in failure to effectively control the spread of the virus. Rigorous assessment using advanced high-end techniques during COVID-19 pandemic came into existence and thereby gained attention for their rapid and sensitive detection. Fig. 2 provides a diagrammatic representation of SARS-CoV-2 detection methods.
Fig. 2.
Schematic of several diagnostic methods for COVID-19 detection. All detection methods have been illustrated schematically. The methods include real-time reverse transcriptase-polymerase chain reaction (rRT-PCR), nanomaterials based aggregation assay, point-of-care (PoC), enzyme-linked immunosorbent assay (ELISA), and computed tomography (CT).
3.1. Conventional RT-PCR and its pitfalls
The standard reference test for coronavirus detection is reverse transcriptase polymerase chain reaction (RT-PCR), which is highly sensitive, specific, time-consuming, simple, and predominantly used for the detection of all types of CoVs [27,28]. Conventional RT-PCR detects viral RNA using fluorescent primers and thermal cycling steps. The process is sensitive, specific, and affordable; however, an error in the amplification process results in an adverse outcome, because of sample mishandling or the rapidly mutating nature of nCoV. A few reports on RT-PCR-based detection include targeting of the envelope (E) and RNA-dependent RNA polymerase (RdRp) genes, exhibiting 100% specificity [27]. Another RT-PCR-based detection by Nalla et al. [29] targeted the E gene and potentially detected samples with 100% specificity and sensitivity. To this end, Konrad et al. [30] also developed an approach that targets the E gene, rendering the present approach more sensitive than the earlier two RdRp gene assays; however, at later stages of the RT-PCR cycle, unspecified signals were obtained. In addition, the commercially available conventional kit for COVID-19 detection targeting the goal gene and direct saliva test is also advantageous over NPS, owing to easy and less contagious methods of detection, and is in the line towards routine diagnostics of COVID-19 [31,32].
3.2. Loop-mediated isothermal amplification (LAMP)
LAMP is a nucleic acid amplification technique that enables effective diagnosis and isolation strategies with high sensitivity, specificity, and efficiency under isothermal conditions at a single point temperature for pathogen detection. It is a significantly rapid, affordable, and highly reliable technique as compared to the conventional RT-PCR, and it establishes itself as a strong contender for direct use in diagnosis [33]. The method utilizes a single tube for DNA and RNA amplification, four primers specific to the experiment, and a DNA polymerase capable of strand displacement enabling the synthesis of 109 copies of target DNA in less than an hour at 65 °C [34]. Recently, Song's group [35] elucidated the design of a two-stage LAMP strategy (COVID-19 Penn-RAMP) that could be performed in closed tubes by means of colorimetric or fluorescence detection. These assays exhibited 10-fold higher sensitivity than conventional RT-PCR when tested with purified targets. Interestingly, Lamb et al. [36] demonstrated rapid SARS-CoV-2 RNA detection within 30 min. However, both these experimental evidences were executed in simulated patient samples with artificially spiked swabs and blood samples. Moreover, Zhang et al. [37] successfully performed LAMP-based experimentation to identify SARS-CoV-2 RNA from a pool of purified RNA or patient cell lysis by visual, colorimetric-based detection. For further validation, the experimental results were verified using RNA samples obtained from respiratory swabs of patient samples in Wuhan, China. Furthermore, the obtained results may be compared with spiked RNA samples, as well as patient samples. In addition, an innovative LAMP-based COVID-19 diagnostic kit was developed by the Sree Chitra Tirunal Institute for Medical Sciences and Technology, Thiruvananthapuram, India, named Chitra-Gene LAMP-N [38]. This test is regarded as a confirmatory sensitive diagnostic test for the SARS-CoV-2 nucleocapsid (N) gene. The detection time was determined to be 10 min, whereas the sampling to test time was highly remarkable at less than 2 h.
3.3. Rolling circle amplification-based method
Another time-saving method for the detection of target nucleic acids is rolling circle amplification (RCA), which can undergo signal amplification of 109-fold/circle within 90 min [39]. This method has been used for SARS-CoV detection. The setup was accomplished in solid and liquid, and presented preliminary results with a few clinical respiratory samples [40]. The foremost advantage of this molecular technique is the minimal reagent requirement and low false-positive results compared to those with traditional PCR techniques. However, the method is yet to be deployed for SARS-CoV-2 detection.
3.4. Microarray-based method
Microarray is a rapid, efficient, and high-tech SARS-CoV nucleic acid detection method. The process mainly relies on the generation of cDNA labeled with specific probes using reverse transcription. Before loading in each well for hybridization with solid-phase oligonucleotides fixed on the microarray, numerous washing steps were carried out to remove free DNA, thereby resulting in the occurrence of a signal due to the presence of specific viral nucleic acids [41]. Later, Shi et al. [42] designed an oligonucleotide microarray of 60-mer based on the TOR2 sequence and used it for SARS-CoV detection of clinical samples. Owing to the rapid mutation phenomenon of SARS-CoV, Guo et al. [43] designed a microarray for the detection of 24 single nucleotide polymorphism (SNP) mutations in the spike (S) gene of SARS-CoV with 100% sample detection accuracy. In addition, immunoglobulin profiling of IgG and IgM in response to SARS-CoV-2, resulting in a SARS-CoV-2 proteome microarray, has been constructed with approximately 18 of the 28 predicted proteins [44]. The results were simultaneously analyzed using a dual color strategy, proving that S1 and N proteins are suitable for COVID-19 diagnostics.
3.5. Immunodiagnostic tests
3.5.1. Lateral flow immunoassay (LFIA)
Recently, LFIA or immunochromatographic tests have drawn special attention in contrast to the available contemporary technologies. LFIA potentially offers economical and rapid diagnostic tests, which can be easily distributed and self-administered or performed by competent healthcare workers to test for SARS-CoV-2 antibodies. The LFIA test can detect the SARS-CoV-2 antibody and efficiently differentiate between IgG and IgM using whole blood, serum, and plasma [45]. The technique mainly follows the principle of the receptor-binding domain of the spike as well as nucleocapsid proteins. According to the study by Ragnesola et al. [46], LFIA often works as a cassette to determine the binding of SARS-CoV-2 antibodies found in convalescent plasma. The developed Clungene® SARS-CoV-2 IgG/IgM rapid test cassettes arbitrate the presence of antibodies in convalescent plasma of donors with a previous history of COVID-19. Bendavid et al. [47] illustrated a test by LFIA in approximately 3330 adults and children, and the results exhibited 82% sensitivity and 99.5% specificity. Another rapid LFIA test introduced by Premier Biotech (Minneapolis, MN, USA) demonstrated a clinical sensitivity of 80.3%, specificity as high as 99.5%, and an assay time as short as between 12 and 20 min [48]. Other LFIA assays with outstanding analytical performance for SARS-CoV-2 detection have been reported by various diagnostic companies such as Shanghai Kinbio Inc. (Shanghai, China), Auto Bio Diagnostics (Zhengzhou, China), and Artron Laboratories (Burnaby, Canada) [49,50]. Additionally, a rapid (10 min) and sensitive lanthanide-doped polystyrene nanoparticle has also been developed to detect anti-SARS-CoV-2 IgG [51]. Similarly, antibody-coated lateral flow strip-based detection has shown excellent specificity for rapid COVID-19 detection, and in most cases, IgG and IgM-class antibodies are primarily used. IgG and IgM expressions together exhibited positive results, while IgG or IgM alone displayed negative results for COVID-19 detection [52].
3.5.2. Enzyme-linked immunosorbent assay (ELISA)
ELISA is a qualitative or quantitative laboratory-based test with high sensitivity and selectivity. Typically, ELISA detects the presence of antibodies such as IgG and IgM in whole blood, plasma, or serum samples from patients [53]. Briefly, the patient samples were incubated on a plate-like platform coated with targeted viral proteins such as S-protein to achieve antibodies from patients and viral protein complexes. Following this, the bound protein-antibody complex is detected with another wash of antibodies, which produces different detection modalities such as colorimetric, electrochemical, and fluorescent readout. In the context of COVID-19, the analytical sensitivity of ELISA was reported to be as low as in the picomolar (pM) range, and the time of the assay was 2–5 h [54,55]. Interestingly, Krammer's group [56] described a two-staged ELISA protocol for the measurement of human antibody in response to that of the recombinant receptor binding domain or S-protein of nCoV. The first stage of the technique includes cutting-edge sample screening in a single-serum dilution with a receptor-binding domain (RBD). In contrast, the next step consists of samples with positive results obtained from the earlier step, and at the last step a thorough confirmatory ELISA test was performed for total S-protein. Another study by Liu et al. [57], conducted from the sera of 214 COVID-19 patients, involved detecting IgG and IgM using N and S ELISA. The study corroborated that the S ELISA showed higher sensitivity than N ELISA. Moreover, a comparative analysis between ELISA-based and gold-immunochromatographic assay (GICA) has also been performed [58]. The high sensitivity and short testing time suggest that immunoassays are a superior alternative to conventional detection methods. Moreover, Li et al. [59] recently reported a point-of-care diagnostic device with combined IgG–IgM testing platform, and the testing time was as short as 15 min. This over-reliance and effective management of pathogens may necessitate the rigorous development of antibody-based assays to help stop the uncontrollable spread of COVID-19.
3.5.3. Antigen detection test
Antigen detection test mainly detects viral antigens using the immune-capture method. Viral antigens can be detected during the active replication process of the virus, thereby establishing a highly specific test. The potential use of the antigen detection test can act as a triage test for the rapid identification of COVID-19 patients. Interestingly, recent reports have shown that monoclonal antibodies can be used against the N protein of SARS-CoV-2, and numerous simple, non-complex, and rapid test kits are under gradual development [60].
3.5.4. Other immunoassays
Other rapid immunoassay-based diagnostic methods include a multiplexed single-molecule array (SIMOA) established by the renowned company Quanterix (MA, USA). It targets the responses of three important immunoglobulins, IgG, IgM, and IgA, to four viral proteins (spike protein, S1 subunit, receptor binding domain, and N protein). The method is executed in only one sample, aiming towards 12 antibodies for isotype-viral protein quantification. An ultrasensitive immunological response SIMOA with high specificity and sensitivity was further hypothesized compared to that with a single interaction in ELISA for COVID-19 detection. During the first week of infection, the method showed sensitivity and specificity of 86% and 100%, respectively, and after the first week classified as later stage, 100% sensitivity and specificity were observed [61].
The proteomic screening tool, a protein microarray method (PMM), simultaneously analyzes a mixture of proteins. Moreover, the PEPperCHIP® SARS-CoV-2 Proteome Microarray (PEPperPRINT) was developed based on the genomic construction of SARS-CoV-2 isolated from the virus Wuhan-Hu-1 (GenBank ID: MN908947.3), which has been used for serological screening of at least 5000 individual peptides present throughout the viral proteome. Since 2002, PMM has successfully diagnosed Chinese subjects as SARS-positive due to its adequate sensitivity to ELISA tests. Therefore, the method is considered to be more reliable than conventional immunoassays, owing to its determination of more protein antigens of the virus [62].
3.6. Next-generation sequencing (NGS)
NGS plays a decisive role in virus detection. NGS platform was recruited for the famous Human Genome Project and has been implemented to detect SARS-CoV-2. “Billion To One (BTO)”, professional cancer molecular diagnostic company, has designed a technique called the qSanger-COVID-19 test capable of a detection 30 times faster than that with conventional qPCR methods [63]. Numerous NGS-based COVID-19 toolkits have been used for the detection of viral mutations. Additionally, a UK-based company, Youseq, recently developed a highly sensitive COVID-19 NGS Library Prep Kit, which functions effectively in the presence of minimal viral titers [64]. Following this, BGI Biotechnology, a Chinese company, developed a metagenomic-based sequence detection kit, NGS COVID-19 Kit, for the rapid detection of SARS-CoV-2 [12]. The kit can also detect viral sequences and has been approved by the National Medical Products Administration, China. Additionally, in this alarming situation, the rapid spread of the variants of SARS-CoV-2, such as the UK B.1.1.7 strain, defined by multiple spike proteins such as deletion 69–70, deletion 144, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H, and the Brazilian B.1.1.28 strain with the first detected S protein mutation E484K in the sequence of a patient from Rio de Janeiro, Brazil [65], is gaining increased concern. This 484 K.V2 variant has also spread to several other countries, such as England, Singapore, and Canada. Therefore, the gradual emergence of these new strains and the increase in reinfection critically urge the sequencing of more SARS-CoV-2 samples. Public health agencies leverage sequencing technology to provide critical data for viral surveillance of SARS-CoV-2 variants. Moreover, single-cell RNA sequencing can also firmly address SARS-CoV-2 infection along with the strong guidance of previous literature study [66].
3.7. Point-of-care (PoC)-based detection for COVID-19
The PoC device can be a foremost option for rapid, highly sensitive detection of COVID-19, which can be used at home. PoC device can be an alternative and a much superior solution for widespread detection of SARS-CoV-2 among various populations.
PoC platforms, receiving emergency use authorization (EUA) from the US Food and Drug Administration (USFDA), are manufactured by renowned groups such as Abbott (IL, USA) and Cepheid (CA, USA) [67]. Abbott ID NOW COVID-19 test kits are also on the frontline by the fabrication of 6.6 lb, a small toaster-like lightweight box known as Abbott's ID NOW™. The Cepheid Xpert® Xpress SARS-CoV-2 system works in a random manner to achieve the result within 30 min. It is also noted that the Abbott technology missed one-third of the samples detected as positive by Cepheid Xpert Xpress [68]. Another reputed non-government company in India, the Fast Sense Diagnostics, has proposed a technology called CovE-Sens by developing two modules for early detection of COVID-19, within a few minutes [69]. ePlex® SARS-CoV-2 is a qualitative automated nucleic acid multiplex platform capable of automated extraction, amplification, and detection using a single-use cartridge. This entire setup is a very simple visualized readout similar to that of the pregnancy test kit. The developed PoC system is based on a specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) technology and is called SHERLOCK testing in one pot (STOP COVID) [70]. The system utilizes the AapCas12b protein, which is efficiently integrated with the guided RNA containing spacer sequence for reliable, specific, and sensitive detection of the nCoV N gene. This efficient technique may be further used at home using samples such as saliva. However, most of the aforementioned EUA-approved technologies need rapid testing to achieve their sensitivity, specificity, and reliability.
3.8. Biosensors
Due to the lack of clinical evaluation of PoC devices, other emerging techniques such as biosensors have been developed for rapid, ultrasensitive detection of COVID-19. The technology is successful in detecting the earliest point of infection from NPS within minutes, owing to smart exposure and a rationalized protocol for virus diagnosis.
Currently, a study has reported a photonics-technology that manipulates light; the ultrasensitive demonstrator could detect “day 1” infections in patients with low viral load, representing a breakthrough in tackling the coronavirus pandemic [71]. A plasmonic biosensor with dual attitude, such as the plasmonic photothermal (PPT) effect and localized surface plasmon resonance (LSPR) sensing transduction, has been demonstrated as a phenomenal approach towards the medical diagnosis of SARS-CoV-2 [10]. The detection procedure was initiated through nucleic acid hybridization using gold nano-islands with a two-dimensional architecture and was functionalized using complementary DNA receptors. Moreover, the fabricated biosensor precisely detected a distinct target from a multigene mixture with a detection limit as low as 0.22 pM. Another biosensor for sensitive S1 protein detection underwent steady and thorough monitoring of asymptomatic patients. Moreover, the bioelectric response change in membrane-engineered cell lines constituted with human chimeric spike S1 antibody was measured using the fabricated biosensor following the combined bioelectric recognition assay principle and the membrane engineering-based molecular identification technology [72,73]. The viral antigen was detected at a lower level of pg/ng under approximately 3 min using this fabricated biosensor, and no sample pre-processing was required [74]. In addition, immunological field-effect transistor (FET)-based sensing technology is a promising tool for the sensitive detection of viral antigens from clinical samples [75,76]. Recently, a graphene-based FET sensor was fabricated for the rapid detection of SARS-CoV-2 antigen in clinical samples using a specific antibody against the SARS-CoV-2 spike protein [77]. However, all these technologies need to be extensively explored for tackling the COVID-19 pandemic outbreak.
Single-stranded DNA or RNA oligonucleotides selected randomly from nucleic acid libraries called aptamers have formerly paved the way for the successful detection of biomolecules, such as proteins and neurotransmitters [78]. Moreover, detection of SARS-CoV N protein using nanoarray RNA aptamer chip at a concentration as low as 2 pg/mL deserves mention [9]. COVID-19 aptamer-linked immobilized sorbent assay (ALISA)-based detection of SARS-CoV-2 with 90% sensitivity and 99% specificity was first developed by an eminent Indian company THSTI [79]. With the gradual progression in the aptameric detection of SARS-CoV-2, another group of researchers from Pinpoint Science's Laboratory in Berkley, California, USA, developed an efficient handheld device that could potentially detect the targeted protein of SARS-CoV-2 within 30 s [80]. However, the reliability and sensitivity of this technology require further trials with a large sample size.
3.9. Chest computed tomography (CT)
A few recent reports have demonstrated the disadvantages of the conventional RT-PCR technique. An instance was evidenced where two patients infected with nCoV were tested negative with RT-PCR, which led the clinicians to prescribe chest CT scans as an important criterion for clinical diagnosis of nCoV [81]. Recurring issues with a similar rate of false-negative RT-PCR testing during disease progression and treatment have been separately reported [82]. Such observations have prompted practitioners to recommend CT technique as an effective parameter to be considered along with RT-PCR to make decisions on patient discharge and evaluation of treatment protocols. Indeed, a retrospective study has drawn a conclusion in favor of this technique, ascribing 97.2% sensitivity to CT compared to the 83.3% with RT-PCR [83]. At this juncture, chest CT scans could be a potential diagnostic tool to formulate a standard paradigm indicative of a nCoV infection.
3.10. Digital approach towards SARS-CoV-2 detection
A digital approach has been conceived for the detection of asymptomatic COVID-19 patients through the phenomenon of recording the forced-cough by using cell phones and thereby steadily combined with artificial intelligence (AI) [84]. A varied survey with 5,320 people was conducted by a popular MIT website with cough recordings of COVID-19 patients. A framework of AI speech processing with acoustic biomarker extractors has been developed, and the recordings were converted into a convolutional neural network (CNN) for early screening of patients. Furthermore, a saliency map was created for the real-time monitoring of patients, a non-invasive detection method with no cost. This advanced CNN-based method has a training subset of 4,256 samples and was validated on 1,064 samples. Apart from the CNN architecture, Quatieri et al. [85] from MIT proposed modeling of speech as well as signal-processing framework to be the estimated parameters for the detection of different states of COVID-19. The outcome of the framework was a dynamicity analysis of human behavior in real surroundings to potentially locate COVID-19 patients. Another remarkable study on asymptomatic COVID-19 patients based on speech recordings has elucidated vocal biomarkers as potent indicators for disease prediction. Biomarkers are developed as a consequence of infection due to respiratory, laryngeal, and articulatory muscular movements [86].
3.11. Looming techniques towards nCoV detection
3.11.1. CRISPR-Cas technology and reactive polymers
CRISPR mainly deals with repeated nucleotide sequences along with aced spacer sequences, and Cas is correlated with CRISPR-associated proteins depicting the nuclease enzymatic function. An RNA targeting CRISPR enzyme Cas 13 was recently developed for rapid and portable sensing of nucleic acids [87]. Zhang's group [88] reported that Cas 13 is generally programmed to target, thereby destroying the genomes of diverse mammalian single-stranded RNA viruses. Moreover, they developed a platform known as SHERLOCK that combined isothermal pre-amplification with Cas 13 for the detection of single molecules such as DNA or RNA. It could detect single-stranded RNA of the Zika virus or dengue and the presence of mutations in liquid biopsy samples of patients. Recently, the same group has developed a protocol for COVID-19 detection using CRISPR technology [89]. The protocol mainly includes the use of RNA fragments of synthetic SARS-CoV-2, followed by continuous detection of SARS-CoV-2 target sequences in a range between 20 and 200 aM, that is, 10–100 copies/μL of input by a dipstick within 60 min, without requiring detailed instrumentation. In addition to CRISPR-Cas 13, CRISPR-Cas 12 system DNA endonuclease-targeted CRISPR trans reporter (DETECTR) also plays an important role in the potential detection of SARS-CoV-2 targeting the E and N2 genes [6]. The all-in-one dual CRISPR-Cas 12a system (AIOD-CRISPR) also exhibited versatile roles in the ultrasensitive detection of COVID-19 to achieve a detection limit of up to 5–11 copies of viral RNA/microliter within 90 min [90]. CRISPR-Cas 9 gene editing protocol pens establishment of impressive, rapid, highly sensitive, and economical FELUDA kit developed by a group of Indian scientists [8]. The kit can detect SARS-CoV-2 genetic material within 60 min.
Apart from CRISPR-Cas technology, another non-conventional method that may serve as a potential platform for SARS-CoV-2 detection, is the use of reactive polymer-grafter devices that may utilize antibodies to detect dsRNA [91]. In this device, a polymer-coated surface-reactive poly (pentafluorophenyl acetate) was applied to immobilize the J2 antibodies on the platform surface. The main advantage of this type of graft device is that antibody binding is sequence-independent, and therefore, the platform may serve as a universal virus detection unit.
3.11.2. Exhaled breath condensate (EBC)
This study aimed to establish a potential diagnostic method for SARS-CoV-2 detection. nCoV is mainly spread via droplets, and in the case of bronchoalveolar lavage (BAL), the diagnosis sensitivity and specificity indexes are the highest. Recently, scientists have proposed that EBC could be used as an efficient diagnostic tool for SARS-CoV-2 detection. Moreover, the use of EBC for examination purposes is a novel approach for the assessment of inflammation within the respiratory tract. The collection of samples is non-invasive, easy, and can be repeated several times, and the process may be executed in severe COVID-19 patients and also in small children covered with special facial masks. During the examination process, patients were asked to breathe out normally for approximately 10–15 min into an apparatus supported by a cold system to accumulate the condensate. The probable utilization of this method could be justified in accordance with the perspective of public healthcare; but the number of false-positive results contributes to the continuous spread of COVID-19 worldwide [92,93].
4. Treatment strategies towards SARS-CoV-2
Initially, the generalized treatment of COVID-19 patients included complete bed rest along with an adequate diet and competent medications. In severe cases, a special care or organ support system is also needed to treat or prevent respiration-related complications, along with adequate therapeutic measurements. In this section, we focus on the various established and developed treatment paradigms recommended for proper treatment of the COVID-19 pandemic.
4.1. Respiratory therapy
Hypoxemia patients with COVID-19 with less than 90% oxygen saturation generally require mild to moderate work of breathing resulting from other causes, and the maintenance of greater than 90% oxygen saturation using high-flow oxygen is a much-needed support. In addition, patients with acute respiratory distress syndrome (ARDS), multi-organ dysfunction syndrome (MODS), hemodynamic instability, severe hypoxia, or multiple high-risk conditions such as hypertension, diabetes mellitus, cancer, cardiovascular disease, or chronic respiratory disorder should be immediately treated with mechanical ventilation and endotracheal intubation [94]. Furthermore, a robust circulatory system with improved microcirculation, vasoactive drugs, and regular monitoring of hemodynamics are also needed for active treatment support.
4.2. Conventional drug-based treatment
Drug-based treatment with well-known anti-malarial agents such as chloroquine and its derivative hydroxychloroquine deserves special mention, because it has elicited profound antiviral effects against several viruses such as hepatitis B, HCoV-229E, and human immunodeficiency virus (HIV) type 1. These potent anti-malarial agents block viral entry into cells by glycosylation inhibition of the host receptors and by elevating the endosomal pH, thereby suppressing the production as well as the release of factors that mediate the inflammatory complications of viral diseases [95]. These antiviral effector functions emphasize the rationale for using chloroquine to treat SARS-CoV-2 infection. In light of the advantages of chloroquine use, several drawbacks also have been gaining attention in recent times. Chloroquine causes tissue damage and several diseases such as retinopathy, neuropathy, cardiomyopathy, and myopathy [96]. A recent study of COVID-19 patients using chloroquine diphosphate in Brazil could not conclude any discrete conclusion and ended early, as most of the patients died due to severe arrhythmias. Moreover, chloroquine simultaneously inhibits proliferation and T-cell activation, inducing apoptosis, which may adversely affect antiviral immune surveillance [97]. It also inhibits antigen-antibody reactions, along with the inhibition of proteolysis, chemotaxis, and phagocytosis. In addition, chloroquine acts as an autophagic inhibitor, thereby preventing fusion between autophagosomes and lysosomes [98]. Thus, autophagy inhibition by chloroquine may lead to cellular death and secondary infection.
As early as in February 2020, hydroxychloroquine was identified as a potential inhibitor of SARS-CoV-2 in vitro. The in vitro activity of hydroxychloroquine in comparison to chloroquine after 24 h of growth resulted in a lower half-maximal effective concentration (EC50) for SARS-CoV-2 [99]. Recently, reports have shown that chloroquine and its analogue hydroxychloroquine have been successfully used to treat a series of COVID-19 cases, as observed by improved radiologic findings, enhanced viral clearance, and reduced disease progression [100]. Experimental evidence from putative randomized clinical trials (RCTs) and prospective open-label studies have claimed that hydroxychloroquine and azithromycin are potentially effective drugs against COVID-19 [101]. Another non-randomized, non-blinded, open-label trial study recruiting 80 patients was conducted. The patients were administered 600 mg of hydroxychloroquine for 10 days, followed by 250 mg of azithromycin to alleviate COVID-19 symptoms [102]. However, the aforementioned claims were offset by three studies, none of which conclude any significant benefit from this treatment [[103], [104], [105]]. However, these two drug candidates-hydroxychloroquine and chloroquine, other antiviral agents, corticosteroids, and inflammation inhibitor agents were found to affect few secondary endpoints, and future clinical trials may verify the current data to suggest a broad spectrum role against COVID-19. The details of various antiviral agents, corticosteroids, and inflammation inhibitors are shown in Table 1 [[106], [107], [108], [109], [110], [111], [112], [113], [114], [115], [116], [117], [118], [119], [120]] and Fig. 3.
Table 1.
Drug-based treatment for COVID-19.
| Drug types | Drug | Drug target | Trial outcome | Refs. |
|---|---|---|---|---|
| Antiviral agents (protease/spike glycoprotein inhibitor) | Lopinavir/ritonavir | Inhibiting 3CLpro | Showing no benefit compared to standard care | [106,107] |
| Nelfinavir | Inhibiting 3CLpro | Potential treatment option against SARS-CoV-2 | [108] | |
| Griffithsin | Inhibiting the spike mediated fusion with the membrane | Preventing >80% infection | [109] | |
| Nafamostat | Inhibiting the spike mediated fusion with the membrane | Preventing SARS-CoV-2 infection in human lung cells | [110,111] | |
| Antiviral agents (nucleoside analogues) | Remdesivir (GS- 5734) | Inhibitor of RNA polymerase, binding to viral RdRp and acting as an RNA chain terminator | Under clinical trial; showing promising results. | [112,113] |
| Favipiravir | Inhibitor of RNA polymerase | Showing efficacy against SARS-CoV-2 | [114,115] | |
| Galidesivir (BCX4430) | Inhibitor of RNA polymerase and premature termination of RNA transcription | Broad spectrum antiviral activity | [116] | |
| Corticosteroids | Methylprednisolone | Decreasing host inflammatory response | Reducing death by one-third of critical COVID-19 patients | [117] |
| Inflammation inhibitors | Tocilizumab | IgG1 monoclonal antibody with IL-6 receptor | Reduction in mortality | [118,119] |
| Anakinra | Monoclonal antibody with IL-1 receptor | Clinical improvement in 72% in patients | [119] | |
| Acalabrutinib | Tyrosine-kinase inhibitor | Improving oxygenation, effect on C reactive protein and IL-6 | [119] | |
| Interferons | α- interferon | Producing interferon response | Effective in clinical trial | [120] |
IL: Interleukin.
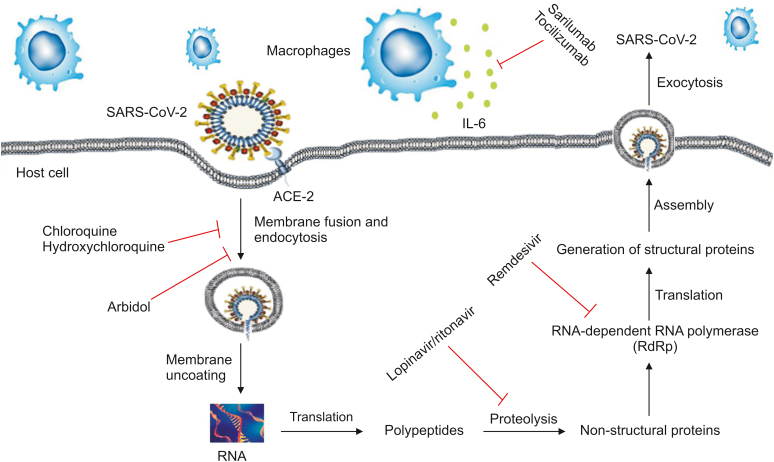
Fig. 3.
Simplified representation of SARS-CoV-2 life cycle along potential drug targets. Arbidol, chloroquine, and hydroxychloroquine block membrane fusion or endocytosis. Lopinavir/ritonavir inhibits the proteolysis of translated peptide, whereas remdesivir targets RNA dependent RNA polymerase (RdRp) to inhibit SARS-CoV-2 protein generation. Immunomodulatory agents, sarilumab, and tocilizumab, target IL-6 receptor limit cytokine release. ACE2: angiotensin-converting enzyme-2; IL-6: interleukin-6.
4.3. Passive antibody therapy
Passive antibody therapy generally involves the administration of antibodies against a given agent to a susceptible individual to provide immediate immunity to prevent or treat infectious diseases. Experiences from previous CoV outbreaks such as the SARS-CoV-1 have shown that convalescent sera contain neutralizing antibodies to the relevant virus provide immediate immunity [121]. In recent studies, five critical COVID-19 patients were treated with convalescent plasma for 10 to 22 days. Of these, three patients were discharged from the hospital after a little more than 50 days, and the remaining two were found to be in a stable condition after 37 days of transfusion [122]. In addition, an RCT was designed to properly investigate whether plasma can be used to prevent infectious COVID-19 [123]. With antibody therapy, anticoagulant therapy by enoxaparin reduces intensive care unit (ICU) mortality [124]. Moreover, according to recent reports, early convalescent plasma may slow down the progression of illness in older adults with COVID-19 [125,126]. Early infusion of this simple and inexpensive technique may help high-risk patients.
4.4. Vaccine development
The current SARS-CoV-2 outbreak demands an immense effort to develop vaccines which will help control and reduce disease transmission by creating herd immunity. Vaccines against COVID-19 are currently in progress involving numerous vaccine candidates. In the process of COVID-19 vaccine development, previously noteworthy bacille Calmette-Guerin (BCG) immunization is found to be beneficial against COVID-19 due to its non-specific immune response [127]. However, few clinical trials are underway to evaluate its efficacy among healthcare workers who are in direct contact with COVID-19 patients [128]. There are also other reported vaccines in the clinical stages against COVID-19 (Table 2) [[129], [130], [131], [132], [133], [134], [135], [136], [137], [138]].
Table 2.
List of vaccines undergoing clinical and preclinical trials.
| Candidate | Characteristics of vaccine | Company | Stage/approval along with dates | Efficiency rate (%) | Refs. |
|---|---|---|---|---|---|
| Sputnik V | Non-replicating viral vector | Gamaleya Research Institute, Acellena Contract Drug Research and Development | Approved in Russia (12 August 2020) | 91.6 | [129] |
| BNT 162 | mRNA vaccine | Pfizer, BioNTech, Fosun Pharma | Approved in UK, Canada (9 November 2020) | 95 | [130] |
| mRNA-1273 | mRNA vaccine | Moderna/NIAID | Approved in USA (23 December 2020) | 94.1 | [131] |
| ChAdOx1 | Attenuated adenoviral construct expressing SARS-CoV-2 protein | Oxford University/Astra Zeneca | Approved in UK (30 December 2020) | 63 | [132] |
| CoronaVac | Inactivated | Sinovac Biotech | Limited use in China (July 2020) | [132] | |
| Inactivated | Inactivated | Sinopharm/Wuhan Institute of Biological Products | Limited use in China, UAE (September 2020) | [132] | |
| BBIBP-CorV | Inactivated | Sinopharm/Beijing Institute of Biological Products | Approved in UAE (9 December 2020), China (31 December 2020) | 79.34 | [132] |
| Covaxin | Inactivated | Bharat Biotech | Approved in India (3 January 2021) | [132] | |
| Ad5-nCoV | Adenovirus type 5 vector that expresses SARS-CoV-2 spike protein | CanSino Biologicals | Phase 3 | 65.7 | [133] |
| NVX-CoV2373 | Nanoparticle vaccine | Novavax | Phase 3 | [133] | |
| Adeno-based (rAd26-S+rAd5-S) | Non-replicating viral vector | Gamaleya Research Institute | Phase 3 | [133] | |
| Bacillus Calmette-Guerin | Repurposed vaccine | Murdoch Children's Research Institute | Phase 2/3 | [133] | |
| GX-19 | DNA vaccine | Genexine | Phase 1/2 | [133] | |
| ARCoV | mRNA vaccine | Academy of Military Medical Sciences, Suzhou Abogen Biosciences and Walvax Biotechnology | Phase 1 | [133] | |
| ZyCoV-D | DNA vaccine | ZydusCadila | Phase 2 | [134] | |
| Inactivated | Inactivated | Institute of Medical Biology at the Chinese Academy of Medical Sciences | Phase 2 | [134] | |
| – | mRNA vaccine | CureVac | Phase 2 | [134] | |
| LNP-nCoVsaRNA | RNA vaccine | Imperial College London | Phase 1/2 | [134] | |
| Live attenuated virus | Codon deoptimized live attenuated vaccines | Codagenix/Serum Institute of India | Preclinical | [134] | |
| GRAd-COV2 | Adenovirus type | ReiThera | Phase 1 | [134] | |
| QazCovid | Inactivated | Research Institute for Biological Safety Problems | Phase 1 | [134] | |
| INO-4800 | DNA vaccine | Inovio Pharmaceuticals |
Phase 2/3 | [135] | |
| ‘Molecular clamp’ vaccine platform | Fast-response technology | University of Queensland | Phase 1 | [136] | |
| MVA-VLP | Modified vaccine Ankara-Virus like particles vaccine platform | GeoVax-BravoVax | Preclinical | [137] | |
| Anhui Zhifei Longcom's vaccine (NCT04466085) | Protein subunit vaccine | Institute of Microbiology and Anhui Zhifei Longcom Biopharmaceutical | Phase 2 | [138] |
4.5. Complementary and alternative medicines
The diverse field of medical practices encompassed by the National Center for Complementary and Integrative Health (NCCIH) of the National Institute of Health (NIH), USA, includes Ayurveda, homeopathy, Unani, and naturopathy, which can aid in accurate diagnosis of different diseases. This globally recognized medical system utilizes the potential of traditional medicines that have been used since primitive times for the treatment of diverse diseases and have been implemented in the treatment of influenza, Japanese encephalitis, and HIV [139]. To suppress the present scenario, the Department of Ayush, Government of India, recommended Indian herbal medicines as potential candidates to combat COVID-19 [15].
4.5.1. Ayurvedic treatment
Ancient Ayurvedic treatments have been applied in cases of various infectious diseases, and successful results have been obtained since ancient period. In the modern era of 2020, Ayurvedic treatment has been carried out in 43 patients to diagnose its efficacy against COVID-19 [18]. The treatments included systematic medicines, diet, and sleeping patterns. During the initial phase of 1–13 days of fever, patients were allowed to consume a diet of rice porridge, Yusha, Bhakta [140], Sudarsana Churna tablets [141], Dhanwantara Gutika tablets [142], and Talisadi Churna with honey [143], and patients were prevented from sleeping during the day as well as at night. After the fever subsided, the second phase of treatment, from days 14 to 30, included diets such as milk, ghee [143], and medicine Vidrayadi Ghritam [144], and the sleeping pattern remained the same as that for the initial phase. Further deterioration from the disease was prevented, and the patients did not reach critical condition [18]. Likewise, traditional Chinese medicine (TCM) has attracted great attention during the COVID-19 pandemic, and more than 60,000 cases have been successfully treated with TCM [145]. Additionally, Shadanga Paniyaan herbal formulation and Ayurvedic medicine Agastya Harityaki are being widely used to treat high fever, headache, shivering, and respiratory problems. Indeed, the Ministry of Ayush, Government of India, recommends a combination of these herbal drugs against SARS-CoV-2. Some of the recommended Ayurvedic formulations such as Pippali rasayana, Laghu Vasant Malati, Sanjeevani Vati, Tribhuvan Keerti rasa, Brihata Vata Chintamni rasa, Mrityunjaya rasa, and Siddha Makardhvaja rasa were also assumed to be effective against moderate to severe SARS-CoV-2 infections [146,147]. Other recommended medicinal formulations by the Department of Ayush include Tirakatu and Tulasi to curb the coronavirus infections in India [20].
4.5.2. Homeopathic treatment
After the emergence of SARS-CoV-2 in India, the Central Council for Research in Homeopathy (CCRH), Government of India, recommended consumption of Arsenicum album-30 for three consecutive days on an empty stomach, and repeated one month later if the infection prevailed [20]. However, reports regarding the success of the above treatment are still not available to form a concrete conclusion. Fifty patients in Italy with probable, suspected, and confirmed cases were treated with homeopathic medicines under isolated conditions, and no adverse conditions were recorded. Some ancient homeopathic medications that were prescribed against COVID-19 include Bryonia alba, Arsenicum album, Phosphorus flavus, Atropa belladonna, and Natrum muriaticum. Moreover, homeopathic treatment is considered as an individualized therapy for definite and specific pathological outcomes. Therefore, homeopathic ultra-high diluted solution products, such as Aconitum napellus, Arsenicum album, Eupatorium perfoliatum, Gelsemium, or Ipecacuana, could help treat early pathogenesis, while Antimuonium tartaricum, Baptisia, or Camphor officinalis would be beneficial in later stages for both respiratory and systematic treatment [[148], [149], [150], [151], [152]].
4.5.3. Unani treatment
Historically, Unani treatment has been used to combat numerous diseases. According to the eminent Persian scholar Najeebuddin Samarqandi, Unani medicine has a beneficial role against Humma-e-Wabaiya (epidemic fever) and Nazla-e-Wabaiya (epidemic influenza). The symptoms of COVID-19 resemble those diseases mentioned, which enhance anti-COVID possibilities of Unani medicine [153]. Unani medicinal treatment has also been recommended by the Government of India against the virus. Patients were advised to consume Sharbat Unnab (10–20 mL), Sharbat Nazla (10 mL), Tiryaq Arba (3–5 g), Ikseer Bukhar (2 pills), and Qurs e Suaal (2 tablets) twice daily along with lukewarm water. The Unani drugs Lactuca sativa (Tukhm and Kahu Mukashar) and Rosa damascene (Gule Surkh) are used to treat sore throat and can possibly curb infections [20]. Moreover, in terms of sanitation, air purification, and immune modulation, Unani medicine could also be effective in reducing SARS-CoV-2 infections [154].
4.5.4. Herbal treatment
Immunity status plays a crucial role in COVID-19 patients, and herbal medicine accounts for immunomodulatory activities exhibiting anti-COVID possibilities [155,156]. The herbal extracts of Anthemis hyaline, Nigella sativa, and Citrus sinensis have been reported to inhibit corona virus replication in vitro [157]. An extract from Eupatorium fortunei, a well-known herbal medicinal plant, can inhibit RNA viruses, including human coronavirus [158]. These probable herbal drugs can be tested against SARS-CoV-2 and may become an essential therapeutic agent against it. Additionally, the medicinal plant Artemisia annua, recommended by the World Health Organization, may act as a possible treatment against COVID-19 [159]. Interestingly, the extracts of 13 herbs, including Forsythia suspense, Ephedra sinica, Lonicera japonica, and Isatis indigotica, were tested in SARS-CoV-2 infected Vero E6 cells [21]. They were found to inhibit viral replication, affect the morphology of viral cells, and reduce the production of pro-inflammatory cytokines. Moreover, many countries have used curcumin for its effective antioxidant, anti-inflammatory, anti-mutagenic, anticancer, anti-Alzheimer's disease, and antibacterial activities, and owing to its broad range of activities, curcumin could also be used as an efficient anti-COVID therapy [[160], [161], [162]].
5. Conclusion and future perspectives
In the current scenario, steps such as countrywide lockdown, self-quarantine, social distancing, and control of infection sources have been initiated to ensure the safety of the people. These restrictions may have beneficial effects in the short-term, but partially fail in the long-term owing to economic issues. Finding ways to live with this virus is the best option using preventive care, which can reduce the financial crisis. However, the exact mechanism of this viral infection is still unclear and needs to be validated by adequate clinical trials, advanced diagnosis, and therapies to combat this devastating situation.
The optimization of diagnostics and treatment strategies is a dynamically developing field during this pandemic outbreak, supported by contemporary medicine and compatible healthcare protocols. Scientists and clinicians have focused on implementing the most sensitive, specific, rapid, and reliable diagnostic toolkit; however, the COVID-19 lacks sufficient data to determine the standards for interpretation by PoC devices. Advocation of a treatment strategy using a conventional drug, respiratory therapy, plasma therapy, vaccine development, complementary and alternative medicine may pave the way towards a successful treatment paradigm. Nevertheless, efforts of thorough treatment development might be a gamble because of safety issues. In this context, commercialized vaccines promise the hope to delay or stop disease progression, especially in older adults with a higher risk of being affected by COVID-19.
In conclusion, this review helps collate knowledge about the current diagnosis and therapeutic scenario, including rapid molecular, serological, digital-based detection, traditional drug treatments, molecular therapies, complementary and alternative drug treatments against SARS-CoV-2; these techniques may help stabilize the clinical crisis faced by the world.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Footnotes
Peer review under responsibility of Xi’an Jiaotong University.
References
- 1.He S., Zhou C., Lu D. Relationship between chest CT manifestations and immune response in COVID-19 patients. Int. J. Infect. Dis. 2020;98:125–129. doi: 10.1016/j.ijid.2020.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO coronavirus COVID-19, https://covid19.who.int/?gclid=Cj0KCQiAifzBRDjARIsAEElyGLzLAGDf_LYNk44vviUC34Z2_lnvxwy_IO3GXoxznnOaGjGZ60mOi8aAk1SEALw_wcB. (Accessed 24 March 2020).
- 3.Mullis K., Faloona F., Scharf S. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 1986;51(Pt 1):263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 4.Chan J.F.W., Yuan S., Kok K.H. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zu Z.Y., Jiang M.D., Xu P.P. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296:E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broughton J.P., Deng X., Yu G. CRISPR-Cas12- based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gootenberg J.S., Abudayyeh O.O., Lee J.W. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azhar Md, Phutela R., Ansari A.H. Rapid, field-deployable nucleobase detection and identification using FnCas9. bioRxiv. 2020 doi: 10.1101/2020.04.07.028167. [DOI] [Google Scholar]
- 9.Ahn D.G., Jeon I.J., Kim J.D. RNA aptamer-based sensitive detection of SARS coronavirus nucleocapsid protein. Analyst. 2009;134:1896–1901. doi: 10.1039/b906788d. [DOI] [PubMed] [Google Scholar]
- 10.Qiu G., Gai Z., Tao Y. Dual functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 11.Ma C., Jing H., Zhang P. An ultrafast one-step assay for the visual detection of RNA virus. Chem. Commun. (Camb) 2018;54:3118–3121. doi: 10.1039/c8cc00150b. [DOI] [PubMed] [Google Scholar]
- 12.Kumar R., Nagpal S., Kaushik S. COVID-19 diagnostic approaches: different roads to the same destination. Virus Dis. 2020;31:97–105. doi: 10.1007/s13337-020-00599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alanagreh L., Alzoughool F., Atoum M. The human coronavirus disease COVID-19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020;9:E331. doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le T.T., Andreadakis Z., Kumar A. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 15.Homoeopathy for prevention of corona virus Infections. Unani medicines useful in symptomatic management of corona virus infection. Compilation by PIB Delhi Advisory for corona virus, https://pib.gov.in/PressReleasePage.aspx?PRID¼1600895. (Accessed 29 January 2020).
- 16.Ruidas B., Chaudhury S.S., Pal K. A novel herbometallic nanodrug has the potential for antibacterial and anticancer activity through oxidative damage. Nanomedicine (Lond) 2019;14:1173–1189. doi: 10.2217/nnm-2018-0187. [DOI] [PubMed] [Google Scholar]
- 17.Ruidas B., Sur T.K., Pal K. Herbometallic nano-drug inducing metastatic growth inhibition in breast cancer through intracellular energy depletion. Mol. Biol. Rep. 2020;47:3745–3763. doi: 10.1007/s11033-020-05467-7. [DOI] [PubMed] [Google Scholar]
- 18.Girija P.L.T., Sivan N. Ayurvedic treatment of COVID-19/SARS-CoV-2: a case report. J. Ayurveda Integr. Med. 2020;20:30042–30045. doi: 10.1016/j.jaim.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Government of India, Ministry of Ayush Guidelines of Ayurveda practitioners for COVID-19. https://www.ayush.gov.in/docs/ayurved-guidlines.pdf
- 20.Maurya V.K., Kumar S., Bhatt M.L.B. In: Coronavirus Disease 2019 (COVID-19). Medical Virology: From Pathogenesis to Disease Control. Saxena S., editor. Springer; Singapore: 2020. Therapeutic Development and Drugs for the treatment of COVID-19. [Google Scholar]
- 21.Li R., Hou Y., Huang J. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol. Res. 2020;156:104761. doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu R., Zhao X., Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;588:E6. doi: 10.1038/s41586-020-2951-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Wang Y., Chen Y. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J. Med. Virol. 2020;92:568–576. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corman V.M., Landt O., Kaiser M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noh J.Y., Yoon S.W., Kim D.J. Simultaneous detection of severe acute respiratory syndrome, middle east respiratory syndrome, and related bat coronaviruses by real-time reverse transcription PCR. Arch. Virol. 2017;162:1617–1623. doi: 10.1007/s00705-017-3281-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nalla A.K., Casto A.M., Huang M.W. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020;58:e00557–e00620. doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konrad R., Eberle U., Dangel A. Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Euro Surveill. 2020;25:2000173. doi: 10.2807/1560-7917.ES.2020.25.9.2000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Kasteren P.B., van der Veer B., van den Brink S. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128:104412. doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams E., Bond K., Zhang B. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00776-20. e00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 34.Francois P., Tangomo M., Hibbs J. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. 2011;62:41–48. doi: 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- 35.El-Tholoth M., Bau H.H., Song J. A single and two-stage, closed-tube, molecular test for the 2019 novel coronavirus (COVID-19) at home, clinic, and points of entry. ChemRxiv. 2020 doi: 10.26434/chemrxiv.11860137.v1. [DOI] [Google Scholar]
- 36.Lamb L.E., Bartolone S.N., Ward E. Rapid detection of novel coronavirus/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS One. 2020;15 doi: 10.1371/journal.pone.0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Odiwuor N., Xiong J. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. 2020 doi: 10.1101/2020.02.26.20028373. [DOI] [Google Scholar]
- 38.Indian scientists develop low cost diagnostic test kit for COVID-19, http://www.ddnews.gov.in/health/indian-scientists-develop-low-cost-diagnostic-test-kit-covid-19 (Accessed 17 April 2020).
- 39.Chapin S.C., Doyle P.S. Ultrasensitive multiplexed microRNA quantification on encoded gel microparticles using rolling circle amplification. Anal. Chem. 2011;83:7179–7185. doi: 10.1021/ac201618k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W.K., Fang C.T., Chen H.L. Detection of severe acute respiratory syndrome coronavirus RNA in plasma during the course of infection. J. Clin. Microbiol. 2005;43:962–965. doi: 10.1128/JCM.43.2.962-965.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Q., Li J., Deng Z. Comprehensive detection and identification of seven animal coronaviruses and human respiratory coronavirus 229E with a microarray hybridization assay. Intervirology. 2010;53:95–104. doi: 10.1159/000264199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi R., Ma W., Wu Q. Design and application of 60mer oligonucleotide microarray in SARS coronavirus detection. Chin. Sci. Bull. 2003;48:1165–1169. doi: 10.1007/BF03183928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo X., Geng P., Wang Q. Development of a single nucleotide polymorphism DNA microarray for the detection and genotyping of the SARS coronavirus. J. Microbiol. Biotechnol. 2014;24:1445–1454. doi: 10.4014/jmb.1404.04024. [DOI] [PubMed] [Google Scholar]
- 44.Jiang H.W., Li Y., Zhang H.N. SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat. Commun. 2020;11:3581. doi: 10.1038/s41467-020-17488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osher G., Lamb C.C., Ibarra Y. Observational study of SARS-CoV-2 antibody immune response in a cohort of patients at a North Suburban Chicago, Illinois, in a physician’s practice. LymphoSign J. 2020;7:104–108. [Google Scholar]
- 46.Ragnesola B., Jin D., Lamb C.C. COVID19 antibody detection using lateral flow assay tests in a cohort of convalescent plasma donors. BMC Res. Notes. 2020;13:372. doi: 10.1186/s13104-020-05212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bendavid E., Mulaney B., Sood N. COVID-19 antibody seroprevalence in Santa Clara county, California. Int. J. Epidemiol. 2021;50:410–419. doi: 10.1093/ije/dyab010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Interim guidelines for collecting and handling of clinical specimens for COVID-19 testing, Compilation by Centres for Disease Control and Prevention, https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html. (Accessed 26 February 2021).
- 49.Lin D., Liu L., Zhang M. Evaluations of the serological test in the diagnosis of 2019 novel coronavirus (SARS-CoV-2) infections during the COVID-19 outbreak. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:2271–2277. doi: 10.1007/s10096-020-03978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lassaunière R., Frische A., Harboe Z.B. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv. 2020 doi: 10.1101/2020.04.09.20056325. [DOI] [Google Scholar]
- 51.Chen Z., Zhang Z., Zhai X. Rapid and sensitive detection of anti-SARS-CoV-2 IgG using lanthanide doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 52.Yüce M., Filiztekin E., Özkaya K.G. COVID-19 diagnosis - a review of current methods. Biosens. Bioelectron. 2021;172:112752. doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao S.Y., Wu Y., Liu H. Evolving status of the 2019 novel coronavirus infection: proposal of conventional serologic assays for disease diagnosis and infection monitoring. J. Med. Virol. 2020;92:464–467. doi: 10.1002/jmv.25702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weissleder R., Lee H., Ko J. COVID-19 diagnostics in context. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc1931. [DOI] [PubMed] [Google Scholar]
- 55.Younes N., Al-Sadeq D.W., AL-Jighefee H. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses. 2020;12:E582. doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stadlbauer D., Amanat F., Chromikova V. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr. Protoc. Microbiol. 2020;57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu L., Liu W., Zheng Y. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. Microb. Infect. 2020;22:206–211. doi: 10.1016/j.micinf.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiang J., Yan M., Li H. Enzyme-linked immunoassay and colloidal gold immunochromatographic assay kit for detection of novel coronavirus (SARS-Cov-2) causing an outbreak of pneumonia (COVID-19) medRxiv. 2020 doi: 10.1101/2020.02.27.20028787. [DOI] [Google Scholar]
- 59.Li Z., Yi Y., Luo X. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng M.P., Papenburg J., Desjardins M. Diagnostic testing for severe acute respiratory syndrome-related coronavirus 2: a narrative review. Ann. Intern. Med. 2020;172:726–734. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Norman M., Gilboa T., Ogata A.F. Ultra-sensitive high-resolution profiling of anti-SARS-CoV-2 antibodies for detecting early seroconversion in COVID-19 patients. Nat. Biomed. Eng. 2020;4:1180–1187. doi: 10.1038/s41551-020-00611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu H., Hu S., Jona G. Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4011–4016. doi: 10.1073/pnas.0510921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Next-generation sequencing delivers 30x faster COVID-19 testing than qPCR. Compilation by Rapid microbiology. 2020 https://www.rapidmicrobiology.com/news/next-generation-sequencing-delivers-30x-faster-covid-19-testing-than-qpcr (Accessed 22 April 2020) [Google Scholar]
- 64.SARS-COV-2 coronavirus NGS library prep kit, Compilation by Youseq, https://youseq.com/product/sars-cov-2-coronavirus-ngs-library-prep-kit/15.
- 65.Voloch C.M., da Silva F. Jr R., de Almeida L.G.P. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021;95:e00119–e00121. doi: 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Speranza E., Williamson B.N., Feldmann F. Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abe8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giri B., Pandey S., Shrestha R. Review of analytical performance of COVID-19 detection methods. Anal. Bioanal. Chem. 2021;413:35–48. doi: 10.1007/s00216-020-02889-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Basu A., Zinger T., Inglima K. Performance of Abbott ID NOW COVID-19 rapid nucleic acid amplification test in nasopharyngeal swabs transported in viral media and dry nasal swabs, in a New York City academic institution. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01136-20. e01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pune based fast sense develops rapid coronavirus test kits, https://www.deccanherald.com/national/west/pune-based-fastsense-develops-rapid-coronavirus-test-kits-822805.html. (Accessed 26 March 2021).
- 70.Joung J., Ladha A., Saito M. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv. 2020 doi: 10.1101/2020.05.04.20091231. [DOI] [Google Scholar]
- 71.Photonics-21 to develop saliva test to detect COVID-19 with lasers. Compilation by Science Buisness, https://sciencebusiness.net/network-updates/photonics21-develop-saliva-test-detect-covid-19-lasers. (Accessed 7 April 2020).
- 72.Kintzios S., Pistola E., Panagiotopoulos P. Bioelectric recognition assay (BERA) Biosens. Bioelectron. 2001;16:325–336. doi: 10.1016/s0956-5663(01)00127-0. [DOI] [PubMed] [Google Scholar]
- 73.Kokla A., Blouchos P., Livaniou E. Visualization of the membrane engineering concept: evidence for the specific orientation of electro inserted antibodies and selective binding of target analytes. J. Mol. Recogn. 2013;26:627–632. doi: 10.1002/jmr.2304. [DOI] [PubMed] [Google Scholar]
- 74.Mavrikou S., Moschopoulou G., Tsekouras V. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors (Basel) 2020;20:E3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Janissen R., Sahoo P.K., Santos C.A. InP nanowire biosensor with tailored biofunctionalization: ultrasensitive and highly selective disease biomarker detection. Nano Lett. 2017;17:5938–5949. doi: 10.1021/acs.nanolett.7b01803. [DOI] [PubMed] [Google Scholar]
- 76.Liu J., Chen X., Wang Q. Ultrasensitive monolayer MoS2 field-effect transistor based DNA sensors for screening of Down syndrome. Nano Lett. 2019;19:1437–1444. doi: 10.1021/acs.nanolett.8b03818. [DOI] [PubMed] [Google Scholar]
- 77.Seo G., Lee G., Kim M.J. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 78.Sinha K., Mukhopadhyay C.D. Quantitative detection of neurotransmitter using aptamer: from diagnosis to therapeutics. J. Biosci. 2020;45:44. [PubMed] [Google Scholar]
- 79.Aptamer-based assay developed for coronavirus detection, Compilation by Science Chronicle, https://journosdiary.com/2020/07/12/aptamer-based-assay-developed-for-coronavirus-detection/. (Accessed 12 July 2020).
- 80.Pinpoint's low-cost handheld Covid-19 Aptamer-based diagnostic device in development, Compilation by Rapid Microbiology, https://www.rapidmicrobiology.com/news/pinpoint39s-low-cost-handheld-covid-19-aptamer-based-diagnostic-device-in-development. (Accessed 9 March 2020).
- 81.Li D., Wang D., Dong J. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep learning-based CT diagnosis and insights from two cases. Korean J. Radiol. 2020;21:505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Y., Yao L., Li J. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020;92:903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Long C., Xu H., Shen Q. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur. J. Radiol. 2020;126:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laguarta J., Hueto F., Subirana B. COVID-19 artificial intelligence diagnosis using only cough recordings. IEEE Open J. Eng. Med. Biol. 2020;1:275–281. doi: 10.1109/OJEMB.2020.3026928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quatieri T.F., Talkar T., Palmer J.S. A framework for biomarkers of COVID-19 based on coordination of speech-production subsystems. IEEE Open J. Eng. Med. Biol. 2020;1:203–206. doi: 10.1109/OJEMB.2020.2998051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Signs of COVID-19 may be hidden in speech signals, Compilation by MIT news, https://medicalxpress.com/news/2020-07-covid-hidden-speech.html. (Accessed 9 July 2020).
- 87.Wright A.V., Nuñez J.K., Doudna J.A. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 88.Freije C.A., Myhrvold C., Boehm C.K. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell. 2019;76:826–837. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.A protocol for detection of COVID-19 using CRISPR diagnostics, Compilation by Broad Institute, https://www.broadinstitute.org/files/publications/special/COVID19%20detection%20(updated).pdf.
- 90.Ding X., Yin K., Li Z. All-in-one dual CRISPR-Cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. bioRxiv. 2020 doi: 10.1101/2020.03.19.998724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ku J., Kim S., Park J. Reactive polymer targeting dsRNA as universal virus detection platform with enhanced sensitivity. Biomacromolecules. 2020;21:2440–2454. doi: 10.1021/acs.biomac.0c00379. [DOI] [PubMed] [Google Scholar]
- 92.Khoubnasabjafari M., Jouyban-Gharamaleki V., Ghanbari R. Exhaled breath condensate as a potential specimen for diagnosing COVID-19. Bioanalysis. 2020;12:1195–1197. doi: 10.4155/bio-2020-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jerzyńska J., Brzozowska A., Bobrowska-Korzeniowska M. Usefulness of exhaled breath condensate for evaluation of markers of airway inflammation in children with asthma. Pediatr. Pol. 2009;84:437–445. [Google Scholar]
- 94.Whittle J.S., Pavlov I., Sacchetti A.D. Respiratory support for adult patients with COVID-19. J. Am. Coll. Emerg. Physicians Open. 2020;1:95–101. doi: 10.1002/emp2.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Keyaerts E., Vijgen L., Maes P. In-vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004;323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Al-Bari M.A.A. Chloroquine analogues in drug discovery: new directions of uses, mechanisms of actions and toxic manifestations from malaria to multifarious diseases. J. Antimicrob. Chemother. 2015;70:1608–1621. doi: 10.1093/jac/dkv018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Savarino A., Shytaj I.L. Chloroquine and beyond: exploring anti-rheumatic drugs to reduce immune hyperactivation in HIV/AIDS. Retrovirology. 2015;12:51. doi: 10.1186/s12977-015-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Klionsky D.J., Abdelmohsen K., Abe A. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12(3rd edition):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yao X., Ye F., Zhang M. In-vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;71:732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 101.Gautret P., Lagier J.C., Parola P. Clinical and microbiological effect of a combination of hydroxychloroquine and azithromycin in 80 COVID-19 patients with at least a six-day follow up: a pilot observational study. Trav. Med. Infect. Dis. 2020;34:101663. doi: 10.1016/j.tmaid.2020.101663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Molina J.M., Delaugerre C., Goff J.L. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med. Maladies Infect. 2020;50:384. doi: 10.1016/j.medmal.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Borba M.G.S., Val F.F.A., Sampaio V.S. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (sars-cov-2) infection: a randomized clinical trial. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.8857. [DOI] [PubMed] [Google Scholar]
- 104.Magagnoli J., Narendran S., Pereira F. Outcomes of hydroxychloroquine usage in United States veterans hospitalized with Covid-19. medRxiv. 2020 doi: 10.1101/2020.04.16.20065920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mahevas M., Tran V.T., Roumier M. No evidence of clinical efficacy of hydroxychloroquine in patients hospitalized for COVID-19 infection with oxygen requirement: results of a study using routinely collected data to emulate a target trial. medRxiv. 2020 doi: 10.1101/2020.04.10.20060699. [DOI] [Google Scholar]
- 106.Cao B., Wang Y., Wen D. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Recovery Collaborative Group Lopinavir–ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2020;396:1345–1352. doi: 10.1016/S0140-6736(20)32013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu Z., Peng C., Shi Y. Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. BioRxiv. 2020 doi: 10.1101/2020.01.27.921627. [DOI] [Google Scholar]
- 109.Cai Y., Xu W., Gu C. Griffithsin with a broad-spectrum antiviral activity by binding glycans in viral glycoprotein exhibits strong synergistic effect in combination with a pan-coronavirus fusion inhibitor targeting SARS-CoV-2 spike S2 subunit. Virol. Sin. 2020;35:857–860. doi: 10.1007/s12250-020-00305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hoffmann M., Schroeder S., Kleine-Weber H. Nafamostatmesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00754-20. e00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Drug Target Review. Nafamostat Inhibits SARS-CoV-2 Infection, Preventing COVID-19 Transmission. NEWS; 2020. [Google Scholar]
- 112.Williamson B.N., Feldmann F., Schwarz B. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585:273–276. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang M., Cao R., Zhang L. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li G., Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 115.Cai Q., Yang M., Liu D. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering (Beijing) 2020;6:1192–1198. doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Elfiky A.A. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ledford H. Coronavirus breakthrough: dexamethasone is first drug shown to save lives. Nature. 2020;582:469. doi: 10.1038/d41586-020-01824-5. [DOI] [PubMed] [Google Scholar]
- 118.Biran N., Ip A., Ahn J. Tocilizumab among patients with COVID-19 in the intensive care unit: a multicentre observational study. Lancet Rheumatol. 2020;2:e603–e612. doi: 10.1016/S2665-9913(20)30277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Salama C., Han J., Yau L. Tocilizumab in patients hospitalized with COVID-19 pneumonia. N. Engl. J. Med. 2021;384:20–30. doi: 10.1056/NEJMoa2030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cavalli G., De Luca G., Campochiaro C. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;2:e325–e331. doi: 10.1016/S2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roschewski M., Lionakis M.S., Sharman J.P. Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cascella M., Rajnik M., Cuomo A. StatPearls. StatPearls Publishing; Treasure Island (FL): 2020. Features, evaluation, and treatment of coronavirus (COVID-19) [PubMed] [Google Scholar]
- 123.Zhang J.S., Chen J.T., Liu Y.X. A serological survey on neutralizing antibody titer of SARS convalescent sera. J. Med. Virol. 2005;77:147–150. doi: 10.1002/jmv.20431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shen C., Wang Z., Zhao F. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. J. Am. Med. Assoc. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kollias A., Kyriakoulis K.G., Dimakakos E. Thromboembolic risk and anticoagulant therapy in COVID-19 patients: emerging evidence and call for action. Br. J. Haematol. 2020;189:846–847. doi: 10.1111/bjh.16727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Early convalescent plasma may lower risk of severe COVID in seniors, Compilation prepared by Centre for infectious disease research and policy, https://www.cidrap.umn.edu/news-perspective/2021/01/early-convalescent-plasma-may-lower-risk-severe-covid-seniors#:∼:text=Selection%20of%20donor%20plasma%20with,within%20the%20next%202%20days. (Accessed 6 January 2021).
- 127.Arts R.J.W., Moorlag S.J.F.C.M., Novakovic B. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe. 2018;23:89–100. doi: 10.1016/j.chom.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 128.BCG Clinical Trials Compilation by U.S. National library of medicine. https://clinicaltrials.gov/ct2/results?cond=COVID&term=BCG&cntry=&state=&city=&dist=
- 129.Jones I., Roy P. Sputnik V COVID-19 vaccine candidate appears safe and effective. Lancet. 2021;397:642–643. doi: 10.1016/S0140-6736(21)00191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Polack F.P., Thomas S.J., Kitchin N. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2021;384:1576–1578. [Google Scholar]
- 131.Baden L.R., El Sahly H.M., Essink B. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kyriakidis N.C., López-Cortés A., González E.V. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccines. 2021;6:28. doi: 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Draft landscape and tracker of COVID-19 candidate vaccines. Compilation by WHO, https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines. (Accessed 26 March 2021).
- 134.Coronavirus vaccine tracker, Compilation by the New York Times, https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html. (Accessed 25 March 2021).
- 135.Inovio selected by Cepi to develop vaccine against new coronavirus inovio, Compilation by Inovio Pharmaceuticals Inc, http://ir.inovio.com/news-and-media/news/press-release-details/2020/Inovio-selected-byCEPI-to-develop-vaccine-against-new-coronavirus/default.aspx. (Accessed 23 January 2020).
- 136.Race to develop coronavirus vaccine, Compilation by University of Queensland Australia, https://www.uq.edu.au/news/article/2020/01/race-develop-coronavirus-vaccine.
- 137.Geovax and Bravovax (Wuhan, China) to collaborate on development of coronavirus vaccine, Compilation by GeoVax, https://www.geovax.com/news/geovax-and-bravovax-wuhan-china-to-collaborate-ondevelopment-of-coronavirus-vaccine.
- 138.Li Y.D., Chi W.Y., Su J.H. Coronavirus vaccine development: from SARS and MERS to COVID-19. J. Biomed. Sci. 2020;42:104. doi: 10.1186/s12929-020-00695-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Saxena S.K., Haikerwal A., Gadugu S. Complementary and alternative medicine in alliance with conventional medicine for dengue therapeutics and prevention. Futur. Virol. 2017;12:399–402. [Google Scholar]
- 140.Chandramurthy P.H. first ed. Varanasi; 2001. Chowkhamba Sanskrit Series, Sharngadharasamhita of Sharngadharacharya, Madhyamakhanda, Kwatha Kalpana; pp. 169–170. Chapter 2, Verses 154-156. [Google Scholar]
- 141.Shastri P. Sharngadharasamhita of sharngadhara, madhyamakhanda, snehakalpana. Verse. 2002;14 [Google Scholar]
- 142.Krishnan K.V., Pillai V.S.G. sixteenth ed. Vol. 29. Vidyarambam Publishers; Alleppey: 1989. (Sahasrayogam, Commentary of Sujanapriya, Gutikayogam). [Google Scholar]
- 143.Sharma R.K., Dash V.B., editors. Chowkhambha Sanskrit Series, Charakasamhita of agnivesha, Varanasi. 2003. [Google Scholar]
- 144.Srikantha Murthy K.R. Krishnadas Academy; Varanasi: 2003. Ashtanga Hrdayam of Vagbhatacharya. [Google Scholar]
- 145.Rastogi S., Pandey D.N., Singh R.H. COVID-19 pandemic: a pragmatic plan for Ayurveda intervention. J. Ayurveda Integr. Med. 2020 doi: 10.1016/j.jaim.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bisht D., Sharma Y., Mehra B. A clinical study to evaluate the efficacy of pippalirasayana in certain respiratory disorders. AYU. 2009;30:337–341. [Google Scholar]
- 147.Panigrahi H.K. Efficacy of ayurvedic medicine in the treatment of uncomplicated chronic sinusitis. Ancient Sci. Life. 2006;26:6–11. [PMC free article] [PubMed] [Google Scholar]
- 148.Italian MDs Study Results on Homeopathic Treatment of 50 COVID-19 Patients None of Whom Needed Hospitalization, Compilation prepared by Dana Ullman, MPH, CCH. (Accessed 7 April 2020).
- 149.Vithoulkas G. International Academy of Classical Homeopathy; Alonissos, Greece: 1997. Materia Medica Viva. [Google Scholar]
- 150.Ghegas V. Homeo-Study VZW; Genk, Belgium: 2000. The Classical Homeopathic Lectures. [Google Scholar]
- 151.Schroyens F. 5. Homeopathic Book Publishers; London: 1993. Synthesis Repertorium Homeopathicum Syntheticum. [Google Scholar]
- 152.Adler U.C., Adler M.S., Hotta L.M. Homeopathy for COVID-19 in primary care: a structured summary of a study protocol for a randomized controlled trial. Trials. 2021;22:109. doi: 10.1186/s13063-021-05071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Islam A. Origin and development of unani medicine: an analytical study. Intellect. Discourse. 2018;26:23–49. [Google Scholar]
- 154.Khan S.H., Rahman A.K.M. Prophylactic and therapeutic approach in unani medicine to counter the COVID-19: a review. Med. Clin. Rev. 2020;6:110. [Google Scholar]
- 155.Zhang L., Liu Y. Potential interventions for novel coronavirus in China: a systematic review. J. Med. Virol. 2020;92:479–490. doi: 10.1002/jmv.25707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sharma M., Anderson S.A., Schoop R. Induction of multiple pro-inflammatory cytokines by respiratory viruses and reversal by standardized Echinacea, a potent antiviral herbal extract. Antivir. Res. 2009;83:165–170. doi: 10.1016/j.antiviral.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 157.Ulasli M., Gurses S.A., Bayraktar R. The effects of Nigella sativa (Ns), Anthemishyalina (Ah) and Citrus sinensis (Cs) extracts on the replication of coronavirus and the expression of TRP genes family. Mol. Biol. Rep. 2014;41:1703–1711. doi: 10.1007/s11033-014-3019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Choi J.G., Lee H., Hwang Y.H. Eupatorium fortunei and its components increase antiviral immune responses against RNA viruses. Front. Pharmacol. 2017;8:511. doi: 10.3389/fphar.2017.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.WHO supports scientifically-proven traditional medicine, https://www.afro.who.int/news/who-supports-scientifically-proven-traditional-medicine. (Accessed 4 May 2020).
- 160.Kocaadam W.B., Şanlier N. Curcumin, an active component of turmeric (Curcuma longa), and its effects on health. Crit. Rev. Food Sci. Nutr. 2017;13:2889–2895. doi: 10.1080/10408398.2015.1077195. [DOI] [PubMed] [Google Scholar]
- 161.Mukhopadhyay C.D., Ruidas B., Chaudhury S.S. Role of curcumin in treatment of alzheimer's disease. Int. J. Neurorehabil. 2017;4:274. [Google Scholar]
- 162.Manoharan Y., Haridas V., Vasanthakumar K.C. Curcumin; a wonder drug as a preventive measure for COVID-19 management. Indian J. Clin. Biochem. 2020;35:373–375. doi: 10.1007/s12291-020-00902-9. [DOI] [PMC free article] [PubMed] [Google Scholar]