Abstract
Background
Vulnerability to COVID-19 hospitalization has been linked to behavioral risk factors, including combustible psychoactive substance use (e.g., tobacco smoking). Paralleling the COVID-19 pandemic crisis have been increasingly permissive laws for recreational cannabis use. Cannabis use disorder (CUD) is a psychiatric disorder that is heritable and genetically correlated with respiratory disease, independent of tobacco smoking. We examined the genetic relationship between CUD and COVID-19 hospitalization.
Methods
We estimated the genetic correlation between CUD (case: n = 14,080; control: n = 343,726) and COVID-19 hospitalization (case: n = 9373; control: n = 1,197,256) using summary statistics from genome-wide association studies. Using independent genome-wide association studies conducted before the pandemic, we controlled for several covariates (i.e., tobacco use phenotypes, problematic alcohol use, body mass index, fasting glucose, forced expiratory volume, education attainment, risk taking, attention-deficit/hyperactivity disorder, Townsend deprivation index, chronic obstructive pulmonary disease, hypertension, and type 2 diabetes) using genomic structural equation modeling. Genetic causality between CUD and COVID-19 hospitalization was estimated using latent causal variable models.
Results
Genetic vulnerability to COVID-19 was correlated with genetic liability to CUD (rG = 0.423 [SE = 0.0965], p = 1.33 × 10−6); this association remained when accounting for genetic liability to related risk factors and covariates (b = 0.381–0.539, p = .012–.049). Latent causal variable analysis revealed causal effect estimates that were not statistically significant.
Conclusions
Problematic cannabis use and vulnerability to serious COVID-19 complications share genetic underpinnings that are unique from common correlates. While CUD may plausibly contribute to severe COVID-19 presentations, causal inference models yielded no evidence of putative causation. Curbing excessive cannabis use may mitigate the impact of COVID-19.
Keywords: Cannabis use disorder, COVID-19, Genetic correlations, Genome-wide association statistics, Tobacco use
Paralleling the COVID-19 pandemic has been an increase in substance use (1) and a continuation of increasingly permissive laws surrounding cannabis. On February 5, 2021, the state of Virginia voted to join 15 states (and the District of Columbia) that have already legalized recreational cannabis use. Legalization is associated with increased use, and 20% of individuals who have tried cannabis develop cannabis use disorder (CUD) (2), a moderately heritable (50%–60%) psychiatric syndrome (3) that shares risk with respiratory disease (4). Because the heterogeneous presentation of COVID-19 is partially attributable to host genomic background and respiratory symptoms are the primary reason for hospitalization and death (5), CUD may contribute to severe COVID-19 presentations.
The COVID-19 Host Genetics Initiative represents a collaboration of international investigators with data examining various aspects of COVID-19 illness via genome-wide association studies (GWASs). This effort includes laboratory-confirmed, physician-designated, and severe COVID-19 hospitalization, with comparisons to assessed and general population controls (5). Consistent with evidence that CUD is associated with heightened COVID-19 risk and lifetime substance use disorder is associated with COVID-19 hospitalization and death (6), phenome-wide association studies have begun to demonstrate genetic associations between these COVID-19 disease definitions and a host of substance use behaviors (7). Mendelian randomization (MR), an approach aimed at estimating causality using genetic instruments [e.g., genome-wide significant variants (8)], has also elucidated putative causal effects of correlated conditions, such as body mass index (BMI), on COVID-19 (9).
Here, we leveraged GWAS summary statistics to estimate whether genetic liability to CUD [case: n = 14,080; control: n = 343,726; (4)] may plausibly influence COVID-19 hospitalization [case: n = 9373; control: n = 1,197,256; (5)]. First, we estimated their genetic correlation using linkage disequilibrium score regression (LDSC) (10). Then, we used genomic structural equation modeling (gSEM) (11) to test whether the genetic correlation between CUD and COVID-19 is independent of potential confounders (i.e., tobacco-smoking phenotypes, problematic alcohol use, cannabis use, pulmonary function, metabolic traits, socioeconomic status, and impulsivity, as well as chronic conditions, such as chronic obstructive pulmonary disease [COPD], hypertension, and type 2 diabetes in supplemental analyses). Finally, we employed latent causal variable (LCV) (12) analyses to test for putative causal relationships between CUD and COVID-19 hospitalization. We hypothesized that genetic liability to CUD would be correlated with COVID-19 hospitalization and that we would find evidence of potential causal influence (i.e., association that is independent of confounders and evidence of putative causation in genetic causal modeling analyses).
Methods and Materials
The following sections describe the GWAS summary statistics that were used in analyses.
Cannabis Use Disorder Summary Statistics
CUD summary statistics came from a GWAS (4) that meta-analyzed data from 20 datasets (18 datasets that were part of the Psychiatric Genomics Consortium Substance Use Disorders working group [European ancestry, 8277 cases and 23,497 controls], 1 dataset from The Lundbeck Foundation Initiative for Integrative Psychiatric Research [iPSYCH] [European ancestry, 2758 cases and 53,326 controls]; and 1 dataset from deCODE genetics [European ancestry, 6033 cases and 280,396 controls]). GWAS summary statistics were drawn from unrelated individuals of European ancestry (case: n = 14,080; control: n = 343,726). All cases were diagnosed with CUD; controls did not meet criteria for CUD. Psychiatric Genomics Consortium cases met criteria for a lifetime diagnosis of DSM-IV (or DSM-III-R) cannabis abuse or dependence via clinician ratings or semistructured interviews. Cases from the iPSYCH sample had ICD-10 codes of F12.1 (cannabis abuse), F12.2 (cannabis dependence), or both in the Danish Psychiatric Central Research Register. Cases in the deCODE sample met criteria for lifetime DSM-III-R or DSM-IV cannabis abuse or dependence or DSM-5 CUD according to diagnoses assigned at the National Center of Addiction Medicine in Iceland, with controls being derived from the general population of Iceland. The Psychiatric Genomics Consortium and iPSYCH samples used principal component analysis to account for additional population control, while deCODE used genomic control. Only summary data (i.e., single nucleotide polymorphism [SNP] identifier, effect size and standard error, p value, effect allele) were utilized (these files are available at https://www.med.unc.edu/pgc/download-results/). CUD has a heritability of 0.0522 on the liability scale (SE = 0.005), assuming a prevalence of 1.4% of the population developing CUD.
COVID-19 Hospitalization Summary Statistics
Several GWASs of COVID-19–related phenotypes were conducted by the COVID-19 Host Genetics Initiative (https://www.covid19hg.org) (5). Similar to CUD described above, only summary data were required for these analyses and were downloaded from https://www.covid19hg.org/results/. The round 54 release (original release in January 2021) was used. Cases were laboratory-confirmed COVID-19 hospitalized individuals (referred to as COVID “B2”), along with general population controls (n = 9373, excluding 613 cases from 23andMe that require additional permissions; control, n = 1,197,256). Multiple sites conducted GWASs of their individual data, and results were meta-analyzed. The COVID19 GWAS controlled for population structure using principal components and a genetic relatedness matrix for additional control. Under the Fort Lauderdale principles, publications using summary statistics may not present or discuss any genome-wide significant signals for the COVID-19 disease definitions. The full analytic plan (including control for ancestry) is available from the consortium here: https://docs.google.com/document/d/16ethjgi4MzlQeO0KAW_yDYyUHdB9kKbtfuGW4XYVKQg/edit. Heritability of the trait, which is likely underestimated due to unexposed controls, was estimated as 0.033 (0.009).
Additional GWAS Summary Statistics
To account for genetic confounders of the possible relationship between CUD and COVID-19, summary statistics were obtained from GWASs of the following traits.
Tobacco-Smoking Phenotypes
Tobacco-smoking phenotypes (13) included the following:
-
1.
Ever smoking tobacco (n = 632,802; h2 = 0.68 [SE = 0.0021]) is a binary phenotype that was coded positively for anyone who reported a lifetime history of smoking regularly, daily smoking for a month or longer, or smoking 100 or more cigarettes during their lifetime.
-
2.
Cigarettes smoked per day (n = 263,954; h2 = 0.072 [SE = 0.0068]) is a continuous phenotype representing how many cigarettes were smoked per day.
-
3.
Age at smoking initiation (n = 262,990; h2 = 0.047 [SE = 0.003]).
-
4.
Former versus current smoker, i.e., smoking cessation (n = 312,821; h2 = 0.032 [SE = 0.002]), was a binary phenotype in regular smokers reflecting whether they were smoking cigarettes at the time of data collection.
Problematic Alcohol Use
Problematic alcohol use (14) represented a meta-analysis of GWASs of alcohol dependence (binary), alcohol use disorder (binary), and the problems subscale of the Alcohol Use Disorder Identification Test (continuous) (n = 435,563; h2 = 0.068 [SE = 0.0032]).
Cannabis Use
Cannabis use (15) represented a binary measure reflecting whether an individual had ever used cannabis, even once, during their lifetime (n = 162,082; h2 = 0.068 [SE = 0.004]).
Metabolic Traits
Metabolic traits included the following:
-
1.
BMI, as a continuous measure of BMI (n = 795,640; h2 = 0.209 [SE = 0.006]) (16).
-
2.
Fasting glucose, a continuous measure derived from whole blood, plasma, or serum, using assays specific for each cohort (n = 58,074; h2 = 0.100 [SE = 0.016]) (16,17).
-
3.
Type 2 diabetes was a binary diagnosis from a meta-analysis (supplemental analyses: n = 659,316; h2 = 0.140 [SE = 0.008]) (18).
-
4.
Hypertension from the UK Biobank (field code: 131287; n = 361,141; h2 = 0.005 [SE = 0.001]) (19); reference group were subjects in the UK Biobank who did not have a hypertension diagnosis.
Respiration
-
1.
One-second forced expiratory volume (FEV1) was a continuous spirometry measure derived from the UK Biobank (field code: 3036; n = 272,338; h2 = 0.199 [SE = 0.007]) (19).
-
2.
COPD from the UK Biobank (field code: 20002; n = 361,141; h2 = 0.117 [SE = 0.004]) (19); reference group were subjects in the UK Biobank who did not have a COPD diagnosis.
Sociodemographic
-
1.
Townsend deprivation index (TDI) is a continuously distributed census-based measure of social deprivation. This was taken in the UK Biobank (field code: 189; n = 336,798; h2 = 0.038 [SE = 0.002]) (19).
-
2.
Educational attainment is a continuous measure of number of years of schooling completed (n = 766,345; h2 = 0.107 [SE = 0.0026]) (20).
Indices of Impulsivity
-
1.
Risk taking, which included a meta-analysis of various indices of an individual’s willingness or interest in taking risks in general (n = 466,571; h2 = 0.047 [SE = 0.002]) (21).
-
2.
Attention-deficit/hyperactivity disorder (ADHD), representing binary clinical diagnoses (n = 55,374; h2 = 0.232 [SE = 0.014]) (22).
Along with the CUD GWAS, all GWASs of confounding traits accounted for population stratification using principal component analysis, were conducted with data collected before the COVID-19 pandemic, and are free of confounding by COVID diagnosis. Data accession URLs or dbGAP Study Accession identifiers are provided below.
-
1.
Smoking phenotypes: https://conservancy.umn.edu/handle/11299/201564
-
2.
Problematic alcohol use: phs001672.v3.p1
- 3.
-
4.
BMI: https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files
-
5.
Fasting glucose: https://www.magicinvestigators.org/downloads/
-
6.
Type 2 diabetes: https://cnsgenomics.com/content/data
-
7.
FEV1, COPD, TDI, hypertension: http://www.nealelab.is/uk-biobank
-
8.
Educational attainment: https://www.thessgac.org/data
-
9.
Risk taking: https://www.thessgac.org/data
- 10.
Statistical Analyses
LD Score
LDSC (10) was used to estimate genetic correlations between traits using SNPs with minor allele frequencies > 0.01 and INFO > 0.90. LDSC estimates the genetic correlation between two traits based on GWAS summary statistics (i.e., effect sizes). Both traits do not need to be assessed in the same individuals or even the same GWAS. LDSC also separates any potential sample overlap or other sources of confounding (e.g., population stratification) from SNP heritability (i.e., when examining one trait) and SNP-genetic correlation (SNP-rG; i.e., when examining a pair of traits) estimates.
We first applied standard filters to the summary statistics of each trait (only SNPs with minor allele frequencies > 0.01 and INFO scores > 0.9). Palindromic and multiallelic SNPs and insertion/deletion polymorphisms were excluded before estimating genetic correlations. SNPs with <1000 individuals were removed (this was partially done to remove SNPs specific to any minority ancestral populations). All summary statistics were aligned with reference genome data from the Haplotype Reference Consortium (23) and normalized to a standard z statistic, which placed all summary statistics on the same scale for analyses. LDSC beta weights and LD scores were pregenerated from 1000 Genomes phase 3 European GWAS data included in the LDSC software download. LDSC estimates were subsequently also used for gSEM analyses, and the LDSC fourth-order moments were used as inputs for LCV analyses.
One advantage of LDSC is the ability to account for spurious sample overlap in GWASs contributing to the genetic correlation. Sample overlap can induce population stratification (e.g., overlapping controls may be more related to each other) and, when unaccounted for, can upwardly bias the genetic correlation estimate. However, LDSC parses overlap from genetic correlation into the model intercept (i.e., intercept not at origin), while the slope, which is used to calculate the bivariate SNP-rG, remains unbiased. Assuming the intercept is not constrained at 0, this allows the model to remain robust to sample overlap when estimating the rG (10). This remains true even when the heritability estimates themselves are poorly estimated (24). Therefore, the LDSC intercept was freed from 0 to account for population stratification caused by either sample overlap or latent population structure.
Modeling Genetic Covariances Using gSEM
gSEM (11) is a form of classical structural equation modeling where, in place of manifest phenotypes, genomic liability to those phenotypes are used to test specific hypotheses. gSEM is performed using GWAS summary statistics and a genetic correlation matrix (created using LDSC) as input for the multivariable models. Genetic effects of CUD predicting COVID-19 hospitalization vulnerability above and beyond other variables of interest (i.e., other substance use, cardiometabolic, respiratory, and sociodemographic measures) were modeled as a multiple regression in gSEM. Substance use phenotypes were tested in separate models to avoid multicollinearity among them. Within a multiple regression framework, the standardized beta coefficient approximates the partial rG when accounting for covariates.
For both respiratory difficulties and metabolic syndromes, we elected to focus our primary analyses on covariates representing continuous variation in genetic susceptibility to these syndromes (i.e., covarying for FEV1 and fasting glucose values). In supplemental sensitivity analyses, we examined whether genetic risk for chronic conditions related to respiration (i.e., COPD) and metabolic syndrome (i.e., type 2 diabetes and hypertension) were included instead of FEV1 and fasting glucose. Similarly, we also examined whether the exclusion of TDI as a covariate modified our findings. While TDI is a good index of genetic liability to neighborhood deprivation exposure, it is specific to the census characteristics of the UK Biobank sample.
LCV Analysis
LCV (12) is related to MR (8), which is a type of instrumental variable analysis where the instrument is genetic liability to an exposure (here, CUD). Broadly, genetic causality models are a form of instrumental variable analysis that treat genomic risk for a given phenotype, in our case CUD, as proxy measures for assignment to elevated exposure on these phenotypes at the group level [for additional information, see (25)]. As such, these models theoretically test whether one phenotype may plausibly cause another. Here, our genomic causal inference models stipulate that individuals are randomly assigned to polygenic liability to CUD, which can be interpreted to reflect a group-level likelihood of CUD expression (which would include noise from those with high genomic liability who do not have a CUD diagnosis). As such, if genomic liability to CUD is associated with COVID-19 hospitalization, then it is plausible that CUD may cause COVID-19 hospitalization. Importantly, LCV analysis builds on more traditional MR approaches by accounting for pleiotropy (i.e., genomic risk that is shared between CUD and COVID-19 hospitalization) using fourth-order moments from LDSC. Furthermore, LCV includes genetic variants across the entire genome as “instruments,” unlike traditional MR methods that select the most strongly associated variants (typically with p < 5 × 10−8).
The LCV model is premised on estimating a latent variable L that represents the consistency of effects of trait 1 on trait 2; i.e., if there are causal effects of trait 1 (CUD) on trait 2 (COVID-19 hospitalization), genetic associations with trait 1 (CUD) would correlate with the latent causal variable (L). The extent to which the latent causal variable L causes CUD (qCUD) versus COVID-19 (qCOVID) is expressed as a ratio between 0 and 1 (i.e., 0.3 for 30%). This ratio is further distilled from the genetic correlation between CUD and COVID-19 (ρg) to obtain the extent to which CUD and COVID-19 might be causally related. As qCUD and qCOVID are simultaneously estimated, no additional multiple testing is required to account for reciprocal effects of outcome on exposure. The extent to which the latent causal variable mediates the genetic correlation between the two traits is quantified by the genetic causality proportion, an estimate of the degree to which each trait is correlated with the latent genetic variable (ranging from 0, reflecting no genetic causality, to |1|, indicating full genetic causality). For example, a genetic causality proportion of 0.50 suggests that 50% of SNP effect sizes are consistent with trait A (i.e., CUD) causing trait B (i.e., COVID-19 hospitalization); however, potential residual mechanisms (pleiotropy and error) may remain.
Results
Genetic Correlations
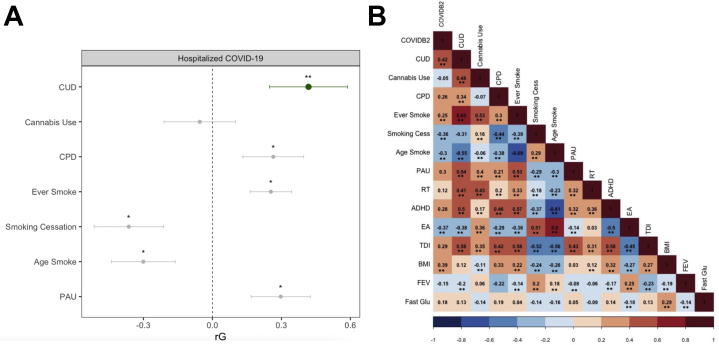
CUD and COVID-19 hospitalization were genetically correlated, even after Bonferroni correction for all traits examined (SNP-rG = 0.418 [SE = 0.097], p = 1.34 × 10−6) (Figure 1). COVID-19 hospitalization was also genetically correlated with other variables, most notably BMI, smoking cessation, and educational attainment (rG > 0.3). As has been previously demonstrated (4), CUD was most highly correlated with cannabis use (rG = 0.48 [SE = 0.042]) and other substance use measures (rG > 0.31–0.48).
Figure 1.
Genetic associations between substance use phenotypes and COVID-19 hospitalization. (A) Genetic correlations between COVID-19 hospitalization and substance use phenotypes. Cannabis use disorder (CUD) is highlighted in green. ∗Indicates nominally significant. ∗∗Indicates Bonferroni significant after accounting for all genetic correlations tested. Age of smoking initiation was reverse coded, such that younger age was more associated with other negative life outcomes and would be easier to interpret. (B) Heat map of genetic correlations between all phenotypes included in the study. COVID-19 hospitalization [Variable COVIDB2 from the COVID Host Genomics Initiative (4)]. ∗∗Indicates Bonferroni significant for all tests run (number of tests = 120). ADHD, attention-deficit/hyperactivity disorder; Age Smoke, age of smoking initiation; BMI, body mass index; Cannabis Use, any lifetime cannabis use; CPD, cigarettes/day; EA, education attainment; Ever Smoke, ever smoking tobacco; Fast Gluc, fasting glucose; FEV1, forced expiratory volume for 1 second; PAU, problematic alcohol use; RT, risk taking; Smoking Cess, smoking cessation; TDI, Townsend deprivation index.
Genetic Association Accounting for Other Factors
Despite genetic correlations between COVID-19 hospitalization and other related phenotypes (e.g., tobacco use phenotypes, BMI) (Figure 1), a series of gSEM models revealed an independent association between CUD and COVID-19 hospitalization (Table 1) when accounting for genomic liability to the following covariates: 1) tobacco phenotypes (i.e., cigarettes per day, age of smoking initiation, ever smoking tobacco, smoking cessation), 2) other substance use phenotypes (i.e., problematic alcohol use, lifetime cannabis use), 3) cardiometabolic traits (i.e., BMI, fasting glucose), 4) respiration (i.e., FEV1), 5) indices of impulsivity (i.e., risk taking, ADHD), and 6) socioeconomic status (i.e., educational attainment, TDI1) (Table S1). Interestingly, despite prior reports of genetic associations between tobacco-smoking measures and COVID-19 disease definitions, none of the tobacco-smoking phenotypes remained significantly associated with COVID-19 hospitalization in the multivariable gSEM (Table 1). In addition to the significant association with CUD, only BMI and educational attainment (except when adjusting for cannabis use or smoking cessation) remained genetically correlated with genetic vulnerability to COVID-19 hospitalization in the multivariable models (Table 1).
Table 1.
COVID-19 Genetic Correlations When Accounting for Potential Confounding Variables
| Substance Use Phenotype | CUD | β Substance Use Phenotypea | EA | TDI | BMI | FEV1 | Fast Gluc | β Risk | β ADHD |
|---|---|---|---|---|---|---|---|---|---|
| Cannabis Use | 0.539b | −0.300 | 0.015 | 0.057 | 0.312b | 0.025 | −0.024 | −0.007 | −0.058 |
| CPD | 0.381b | 0.042 | −0.193b | −0.052 | 0.327b | 0.011 | 0.006 | −0.026 | −0.092 |
| Age Smoke | 0.398b | 0.067 | −0.215b | −0.037 | 0.336b | 0.009 | 0.010 | −0.025 | −0.064 |
| Smoking Cessation | 0.402b | −0.205 | −0.114 | −0.122 | 0.329b | 0.014 | −0.0003 | −0.054 | −0.069 |
| Ever Smoke | 0.439b | −0.123 | −0.191b | −0.021 | 0.344b | 0.011 | −0.003 | −0.029 | −0.052 |
| PAU | 0.315 | 0.162 | −0.211b | −0.082 | 0.345b | 0.004 | 0.009 | −0.036 | −0.088 |
Standardized beta estimates for CUD and substance use phenotypes were taken from a multiple regression parameterized in gSEM. When all of the above covariates were included in the model simultaneously with PAU and CUD, the partial rG between CUD and COVID-19 was no longer significant (rG = 0.315, p = .08). This was largely due to the number of covariates; when only PAU was included in the model, the partial r effect size for CUD was similar in magnitude and significant (r = 0.364, p = .004).
ADHD, attention-deficit/hyperactivity disorder; Age Smoke, age of smoking initiation; BMI, body mass index; Cannabis Use, any lifetime cannabis use; CPD, cigarettes/day; CUD, cannabis use disorder; EA, education attainment; Ever Smoke, ever smoking tobacco; Fast Gluc, fasting glucose; FEV1, forced expiratory volume for 1 second; gSEM, genomic structural equation modeling; GWAS, genome-wide association study; PAU, problematic alcohol use; Risk, risk taking; TDI, Townsend deprivation index.
Substance use phenotypes were entered and tested separately to avoid multicollinearity among them. Each row represents genetic correlations with COVID-19 hospitalization from one model. The multiple rows indicate the separate models run substituting each substance use phenotype (listed in the first column). The model was COVID-19 hospitalization = CUD + substance use phenotype + BMI + TDI + EA + FEV1 + Fasting Gluc + ADHD + Risk taking + error. The original GWASs accounted for standard GWAS covariates (age, sex, genetic principal components, etc.).
p < .05.
In secondary sensitivity analyses, we examined whether including GWASs of chronic diagnosable health conditions, including COPD (rather than FEV1) and type 2 diabetes (rather than fasting glucose), as well as hypertension as putative genetic covariates influenced our findings. While the GWAS of these disorder states represents greater severity and potential COVID-19 risk, they do not capture the range of variability in the underlying predisposing symptoms (i.e., respiration and metabolic dysregulation). Indeed, the genetic correlation between FEV1 and COPD was 0.190 (SE = 0.020); fasting glucose was genetically correlated with type 2 diabetes (rG = 0.564 [SE = 0.055]) and hypertension (rG = 0.256 [SE = 0.055]). CUD was genetically correlated with type 2 diabetes (rG = 0.149 [SE = 0.041]) and hypertension (rG = 0.223 [SE = 0.070]) and weakly with COPD (rG = 0.098 [SE = 0.037]). Likewise, COVID-19 hospitalization was genetically correlated with type 2 diabetes (rG = 0.363 [SE = 0.060]), hypertension (rG = 0.419 [SE = 0.140]), and COPD (rG = 0.231 [SE = 0.050]). When FEV1 and fasting glucose were replaced by COPD, type 2 diabetes, and hypertension, results remained largely unchanged and CUD remained genetically correlated with COVID-19 hospitalization, except when controlling for marijuana use, in which CUD was reduced to marginally significant but effect sizes were similar (Table S2; see Figure S1 for full genetic correlation matrix).
Genetic Causality
LCV analyses did not provide significant evidence that liability to CUD may be genetically causal for COVID-19 hospitalization. Even though the effect size of the genetic causality proportion was substantial, it was not statistically significant (genetic causality proportion estimate = 0.38, SE = 0.36, p = .58); that is, after accounting for pleiotropy between CUD and COVID-19, there was no statistically significant evidence of a potentially causal effect of genetic liability to CUD on COVID-19 hospitalization risk.
Discussion
Our findings suggest that genetic liability to CUD is correlated with risk for COVID-19 hospitalization. The genetic correlation between CUD and COVID-19 hospitalization was independent of potential confounding variables (e.g., other substance inhalation phenotypes, cardiometabolic traits, respiration, socioeconomic status indicators). In contrast, genetic liability to lifetime cannabis ever-use showed a nonsignificant, but negative, genetic correlation with COVID-19 hospitalization (Figure 1 and Table 1); divergent associations of cannabis use and CUD in their genetic association have been observed for other phenotypes (e.g., BMI, which is also positively genetically correlated with CUD and negatively with cannabis use) (4). Furthermore, when genomic liability to lifetime cannabis use was included in models, the association between CUD and COVID-19 hospitalization became stronger (Figure 1 and Table 1). Collectively, these data suggest that genetic liability to CUD and COVID-19 hospitalization is shared and unique from many potentially confounding variables. While such findings suggest that it is plausible that CUD may have an independent causal impact on severe COVID-19 outcomes, genomic causal models (i.e., LCV analyses) did not support a causal role of CUD on COVID-19 hospitalization.
Putative mechanisms underlying the genetic association between CUD and COVID-19 hospitalization warrant attention in future research. Our gSEM analyses indicated that BMI, tobacco smoking (as well as other substance use), and socioeconomic factors did not account for the association between CUD and COVID-19 hospitalization. Many immunologic processes may be involved, because endogenous cannabinoid receptors, especially CB2 receptors, are ubiquitously expressed in immune tissues and likely participate in immune signaling (26). In addition, studies have also begun to explore the relationship between ACE2 expression and cannabinoid compounds (27). Studies examining potential biological mechanisms underpinning the genetic correlation between CUD and COVID-19 hospitalization are needed (e.g., potential shared effects on ACE2, inflammation).
LCV analyses estimated that 38% of the genetic effects were consistent with CUD causing COVID-19. This result should be re-examined when larger GWASs of CUD and COVID-19 become available. Prior analyses, conducted with an earlier data release (release September 4, 2020), found that 60% of the genetic effects were consistent with causal effects of CUD on COVID-19, a statistically significant finding (genetic causality proportion estimate = 0.63, SE = 0.21, p = 4.0 × 10−6) (28). This reduction in causal effects might reflect greater precision in the genetic causality proportion estimate or an increase in heterogeneity across the samples in the latest COVID-19 GWASs (e.g., of cases with varying degrees of illness severity). Nonetheless, the current nonsignificant causal results are consistent with another preprint that utilized traditional MR methods (29).
Some limitations are noteworthy. First, the use of population controls (i.e., individuals who may or may not have had COVID-19) in the COVID-19 hospitalization GWAS and the resulting case-control imbalance may have affected heritability estimate precision. Second, predominant composition of European ancestry in constituent GWASs may limit generalizability to other populations. Third, while there is convergent phenotypic evidence consistent with our findings (6), our findings await replication attempts; unfortunately, we are unaware of any currently available datasets. Furthermore, much like any other instrumental variable analysis, third variable confounding is possible. We accounted for several plausible confounders of this genetic causal pathway between CUD and COVID-19 using gSEM, but other unmeasured factors cannot be excluded. Cannabis use could also reflect a mixture of lighter, casual use and heavy use, including current frequent use. Unfortunately, we still lack a large GWAS of cannabis frequency that could be used to test casual use versus frequency versus problematic use hypotheses. Finally, as described above, we assume that genomic liability to CUD is a good instrument for manifest CUD (likewise, that genomic liability to COVID-19 indices manifest COVID-19 hospitalization).
As the world continues to endure surges in COVID-19, identifying putative risk factors associated with severe presentations may mitigate the worldwide impact of this disease. In contrast to anecdotal evidence and media reports (30) that cannabis may attenuate COVID-19, these data urge caution in light of the continued wave of cannabis legalization during the COVID-19 pandemic.
Acknowledgments and Disclosures
This work was supported by the National Institute on Drug Abuse (NIDA) (Grant No. DA007261-17 [to ASH]), National Institute of Mental Health (NIMH) (Grant No. MH016880 [to CLM]), NIDA (Grant No. HD007289-30 [to EAW]), National Institute on Alcohol Abuse and Alcoholism (NIAAA) (Grant No. F32AA027435 [to ECJ]), NIMH and NIDA (Grant Nos. MH109532 and K02DA032573 [to AA]), and National Institute on Aging, NIAAA, and NIDA (Grant Nos. AG052564, AA027827, and DA046224 [to RB]).
ASH, ECJ, AA, and RB developed the research questions. ASH, SMCC, CLM, and EAW conducted analyses. ASH, AA, and RB drafted the manuscript. ASH, ECJ, AA, and RB provided critical revision of the manuscript for important intellectual content. ASH had full access to all data in the study and takes responsibility for the integrity of the data and accuracy of analyses.
We also would like to acknowledge the COVID-19 Host Genetics Initiative for access to summary statistics (https://www.covid19hg.org/acknowledgements/). Links to publicly available GWAS summary data used in analyses are provided in the methods.
The authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
As the Townsend deprivation index is a geographic index from the UK Biobank that may reflect a multitude of factors (e.g., migration), we repeated analyses excluding this as a covariate. As depicted in Table S1, conclusions are not altered by doing so.
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.bpsgos.2021.06.005.
Supplementary Material
References
- 1.Farhoudian A., Ramin Radfar S., Mohaddes Ardabili H., Rafei P., Ebrahimi M., Khojasteh Zonoozi A., et al. A global survey on changes in the supply, price, and use of illicit drugs and alcohol, and related complications during the 2020 COVID-19 pandemic [published online ahead of print Aug 6] Front Psychiatry. 2021 doi: 10.3389/fpsyt.2021.646206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hasin D.S. US epidemiology of cannabis use and associated problems. Neuropsychopharmacology. 2018;43:195–212. doi: 10.1038/npp.2017.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verweij K.J.H., Zietsch B.P., Lynskey M.T., Medland S.E., Neale M.C., Martin N.G., et al. Genetic and environmental influences on cannabis use initiation and problematic use: A meta-analysis of twin studies. Addiction. 2010;105:417–430. doi: 10.1111/j.1360-0443.2009.02831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson E.C., Demontis D., Thorgeirsson T.E., Walters R.K., Polimanti R., Hatoum A.S., et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry. 2020;7:1032–1045. doi: 10.1016/S2215-0366(20)30339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.COVID-19 Host Genetics Initiative The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet. 2020;28:715–718. doi: 10.1038/s41431-020-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q.Q., Kaelber D.C., Xu R., Volkow N.D. COVID-19 risk and outcomes in patients with substance use disorders: Analyses from electronic health records in the United States. Mol Psychiatry. 2021;26:30–39. doi: 10.1038/s41380-020-00880-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wendt F.R., de Lillo A., Pathak G.A., de Angelis F., Polimanti R. Host genetic liability for severe COVID-19 overlaps with alcohol drinking behavior and diabetic outcomes in over 1 million participants. medRxiv. 2020 doi: 10.1101/2020.11.08.20227884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith G.D., Ebrahim S. What can Mendelian randomisation tell us about modifiable behavioural and environmental exposures? BMJ. 2005;330:1076–1079. doi: 10.1136/bmj.330.7499.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leong A., Cole J.B., Brenner L.N., Meigs J.B., Florez J.C., Mercader J.M. Cardiometabolic risk factors for COVID-19 susceptibility and severity: A Mendelian randomization analysis. PLoS Med. 2021;18 doi: 10.1371/journal.pmed.1003553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics Consortium, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grotzinger A.D., Rhemtulla M., de Vlaming R., Ritchie S.J., Mallard T.T., Hill W.D., et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav. 2019;3:513–525. doi: 10.1038/s41562-019-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Connor L.J., Price A.L. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat Genet. 2018;50:1728–1734. doi: 10.1038/s41588-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu M., Jiang Y., Wedow R., Li Y., Brazel D.M., Chen F., et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou H., Sealock J.M., Sanchez-Roige S., Clarke T.K., Levey D.F., Cheng Z., et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci. 2020;23:809–818. doi: 10.1038/s41593-020-0643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasman J.A., Verweij K.J.H., Gerring Z., Stringer S., Sanchez-Roige S., Treur J.L., et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci. 2018;21:1161–1170. doi: 10.1038/s41593-018-0206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yengo L., Sidorenko J., Kemper K.E., Zheng Z., Wood A.R., Weedon M.N., et al. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manning A.K., Hivert M.F., Scott R.A., Grimsby J.L., Bouatia-Naji N., Chen H., et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue A., Wu Y., Zhu Z., Zhang F., Kemper K.E., Zheng Z., et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat Commun. 2018;9:2941. doi: 10.1038/s41467-018-04951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abbott L., Bryant S., Churchouse C., Ganna A., Howrigan D., Palmer D., et al. Results of an updated "round 2" version of GWAS for the UK Biobank. 2018. http://www.nealelab.is/uk-biobank Available at: Accessed October 18, 2020.
- 20.Lee J.J., Wedow R., Okbay A., Kong E., Maghzian O., Zacher M., et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlsson Linnér R., Biroli P., Kong E., Meddens S.F.W., Wedow R., Fontana M.A., et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51:245–257. doi: 10.1038/s41588-018-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demontis D., Walters R.K., Martin J., Mattheisen M., Als T.D., Agerbo E., et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boef A.G.C., Dekkers O.M., le Cessie S. Mendelian randomization studies: A review of the approaches used and the quality of reporting. Int J Epidemiol. 2015;44:496–511. doi: 10.1093/ije/dyv071. [DOI] [PubMed] [Google Scholar]
- 26.Lu H.C., MacKie K. An introduction to the endogenous cannabinoid system. Biol Psychiatry. 2016;79:516–525. doi: 10.1016/j.biopsych.2015.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang B., Kovalchuk A., Li D., Rodriguez-Juarez R., Ilnytskyy Y., Kovalchuk I., Kovalchuk O. In search of preventative strategies: Novel high-CBD Cannabis sativa extracts modulate ACE2 expression in COVID-19 gateway tissues. Aging (Albany NY) 2020;12:22425–22444. doi: 10.18632/aging.202225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatoum A.S., Morrison C.L., Winiger E.A., Johnson E.C., Agrawal A., Bogdan R. Genetic liability to cannabis use disorder and COVID-19 hospitalization. medRxiv. 2020 doi: 10.1101/2020.11.15.20229971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosoff D.B., Yoo J., Lohoff F.W. A genetically-informed study disentangling the relationships between tobacco smoking, cannabis use, alcohol consumption, substance use disorders and respiratory infections, including COVID-19. medRxiv. 2021 doi: 10.1101/2021.02.11.21251581. [DOI] [Google Scholar]
- 30.Earlenbaugh E. Cannabis shows potential to help and harm in coronavirus cases: Experts explain why. 2020. https://www.forbes.com/sites/emilyearlenbaugh/2020/08/11/cannabis-shows-potential-to-help-and-harm-in-coronavirus-cases-experts-explain-why/#2da9a29b33ea Available at: Accessed September 23, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.