Abstract
Lung cancer is the most common cause of cancer deaths. The expression of the transcription factor C/EBPα (CCAAT/enhancer binding protein α) is frequently lost in non–small cell lung cancer, but the mechanisms by which C/EBPα suppresses tumor formation are not fully understood. In addition, no pharmacological therapy is available to specifically target C/EBPα expression. We discovered a subset of pulmonary adenocarcinoma patients in whom negative/low C/EBPα expression and positive expression of the oncogenic protein BMI1 (B lymphoma Mo-MLV insertion region 1 homolog) have prognostic value. We also generated a lung-specific mouse model of C/EBPα deletion that develops lung adenocarcinomas, which are prevented by Bmi1 haploinsufficiency. BMI1 activity is required for both tumor initiation and maintenance in the C/EBPα-null background, and pharmacological inhibition of BMI1 exhibits antitumor effects in both murine and human adenocarcinoma lines. Overall, we show that C/EBPα is a tumor suppressor in lung cancer and that BMI1 is required for the oncogenic process downstream of C/EBPα loss. Therefore, anti-BMI1 pharmacological inhibition may offer a therapeutic benefit for lung cancer patients with low expression of C/EBPα and high BMI1.
INTRODUCTION
Lung cancer accounts for 30% of tumor-related deaths (1). Most patients are refractory to current treatments; therefore, understanding the mechanisms that control lung tumorigenesis is essential to design new therapies. One event that frequently occurs in primary non–small cell lung cancer (NSCLC) is the lack of expression of the transcription factor CCAAT/enhancer binding protein α (C/EBPα) by either hypermethylation (2) or genetic loss (3). C/EBPα controls tissue-specific gene expression and promotes proliferation arrest in terminally differentiated cells from several tissues, including pulmonary cells (4). C/EBPα acts as a tumor suppressor in acute myeloid leukemia (4–7), and it is down-regulated in hepatic carcinogenesis (8), squamous cell skin carcinomas (9), and squamous cell cancers of the head and neck (10) and is dysregulated in prostate cancer (11).
C/EBPα knockout mice display perinatal lethality with severe hypoglycemia and respiratory distress (12, 13). Lung-specific in utero loss of C/EBPα results in delayed maturation of the lung, respiratory arrest, and lethality after birth, caused by epithelial cell expansion and loss of airspace (14, 15). C/EBPα loss causes lack of differentiation, hyperproliferation, and increased survival of type II alveolar cells, with an overall down-regulation of genes involved in differentiation, and up-regulation of proliferation, tumor progression, and cell survival genes (14). In addition, induction of C/EBPα expression in human lung cancer cells resulted in differentiation and growth reduction, attributable to proliferation arrest and apoptosis (16).
The mechanisms by which C/EBPα suppresses tumor formation are unclear. No pharmacological therapy is available to specifically target the large subsets of patients with loss of C/EBPα expression.
We hypothesized that C/EBPα could be performing its protective function by helping to restrain the activity of critical proto-oncogenes. Similar to what we observed in C/EBPα−/− hematopoietic stem cells (17), reciprocal expression between C/EBPα and BMI1 (B lymphoma Mo-MLV insertion region 1 homolog) also occurs in lung cancer. Here, we report that more than 80% of C/EBPα-negative/low adenocarcinoma patients are positive for the expression of the oncogenic protein BMI1 and that this expression pattern has prognostic value. By using short hairpin RNA (shRNA) against Bmi1 and a compound that inhibits it, we demonstrated that BMI1 has a critical in vivo role in supporting and maintaining the transformed phenotype of C/EBPα-null cells, pointing to the therapeutic potential of targeting BMI1 in C/EBPα-negative/low adenocarcinoma patients.
RESULTS
A subset of C/EBPα-negative/low and BMI1-positive human adenocarcinomas is amenable to BMI1 pharmacological inhibition
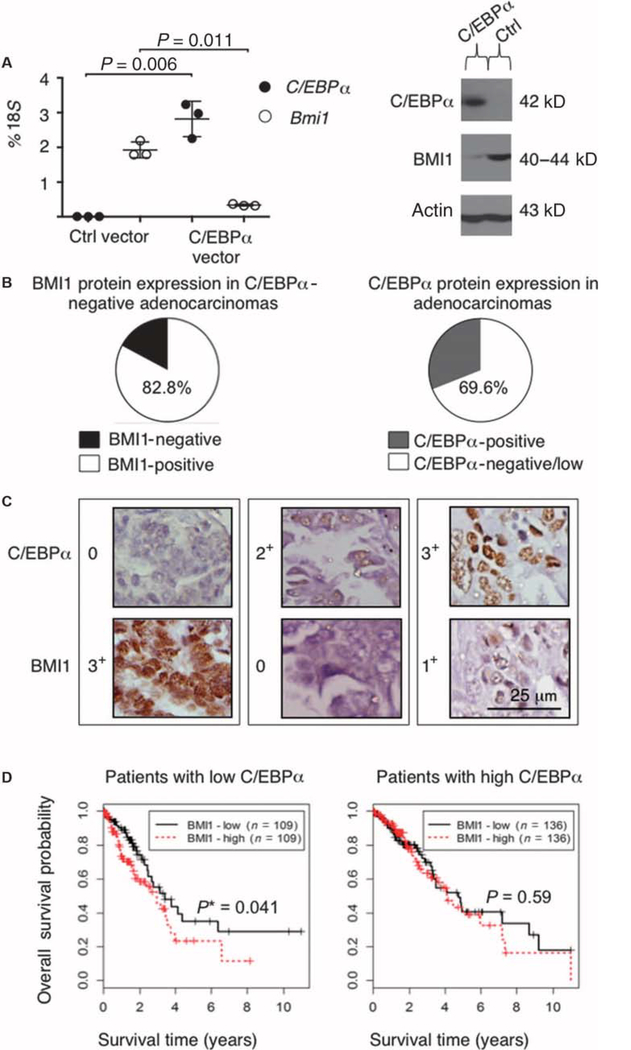
To address the molecular mechanisms by which C/EBPα suppresses transformation, we analyzed gene expression profile changes of a human adenocarcinoma cell line (H358) after overexpression of an inducible form of C/EBPα (16). Upon C/EBPα up-regulation, both BMI1 mRNA and protein were markedly reduced (Fig. 1A). To investigate whether C/EBPα performs its tumor-suppressing function by restraining BMI1 expression, we examined the correlation between C/EBPα and BMI1 protein amounts in primary lung cancer patient specimens through immunohistochemistry (IHC). The clinicopathological information available for the tissue microarray, including diagnosis, tumor stage, cancer stage, and lymph node involvement, is displayed in Table 1 (for adenocarcinoma cases) and table S1 (for all NSCLC cases, including adenocarcinomas). IHC analysis performed on 261 NSCLC patient tissues demonstrated a range of BMI1 protein expression (0 to 3+) in different NSCLC subtypes analyzed, as shown in fig. S1 (A to G). The adenocarcinoma cases (69.6%) were negative/low for C/EBPα expression (0, 1+), as previously described (16), and 82.8% of them were positive for BMI1 expression (1+ to 3+) (Fig. 1B). Examples of adenocarcinoma staining are presented in Fig. 1C. To investigate whether BMI1 expression correlates with prognosis in lung cancer for patients with negative/low C/EBPα expression, we compared overall survival in 490 lung adenocarcinoma samples available in The Cancer Genome Atlas [TCGA; lung adenocarcinoma (LUAD) data set at https://tcga-data.nci.nih.gov/tcga/], as well as three different publicly available microarray expression databases (18–20), containing data from additional 490 NSCLC patients. When the 490 TCGA adenocarcinoma patients were stratified according to absent/low C/EBPα expression, the survival difference between BMI1low and BMI1high lung cancers was in favor of BMI1low patients (P = 0.041) (Fig. 1D). All NSCLC microarray cohorts [fig. S2, A (n = 133, P = 0.044) and B (n = 181, P = 0.038)] showed a difference in survival between C/EBPα-negative/low BMI1low and BMI1high subgroups, except for the Tang et al. study (n = 176) (fig. S2C) (20).
Fig. 1. BMI1 expression is inversely associated with C/EBPα expression in human adenocarcinoma cells.
(A) Left: Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed on experimental samples (n=3) from the human lung adenocarcinoma cell line H358 transfected with a rapamycin-inducible C/EBPα-expressing vector or a rapamycin-inducible empty vector (as control), and induced with rapamycin (for 36 hours). The mean expression is presented as a percentage of 18S RNA. The P values for each sample are indicated on the basis of a two-sided Welch’s t test. Right: Western blot analysis was carried out in H358 cells transfected with the rapamycin-inducible C/EBPα-expressing vector or the rapamycin-inducible empty vector by using anti-C/EBPα and anti-BMI1 antibodies. Loading was assessed, after complete stripping of the membrane, with an anti-actin antibody. The expected size in kilodaltons is indicated. (B) Left: Pie chart demonstrating BMI1 protein expression in patient-derived adenocarcinomas that are negative or low for C/EBPα (staining intensity, 0 or 1+). BMI1 was considered positive when staining was scored as 1+ to 3+. Right: Pie chart showing C/EBPα protein expression in patient-derived adenocarcinomas, subdivided as negative/low (staining intensity, 0or 1+) or positive (staining intensity, 2+ or 3+). (C) Representative examples of IHC data from three patients’ tissues (each enclosed in a box), which were independently and blindly scored by two pathologists. Staining intensity was scored as follows: 0 (no staining); 1+ (mild staining); 2+ (moderate staining); and 3+ (strong staining). Scale bar, 25 μm. (D) Overall survival curves for 490 patients from TCGA lung adenocarcinoma cohort (https://tcga-data.nci.nih.gov/tcga/). Only 490 of the 521 TCGA samples have both RNA-sequencing and survival data. Patients are stratified according to low or high C/EBPα expression, defined as log2 expression < 9.0 or not, respectively. Survival probability is higher (P = 0.041) in BMI1low patients, as compared to BMI1high patients in the C/EBPαlow subgroup. The median expression of BMI1 used to define BMI1low and BMI1high patient subgroups was 10.2 in the C/EBPαlow group and 10.1 in the C/EBPαhigh group. The method of Kaplan and Meier was used for graphical displays of overall survival. The log-rank test was used to assess differences in overall survival. A similar approach was also applied for C/EBPαhigh samples. The P value and the sample size (n) for each subgroup are indicated on each plot. Among C/EBPαlow patients, 30 death events/109 patients presented with low BMI1 expression, and 39 death events/ 109patientspresentedwithhighBMI1expression. Among C/EBPαhigh patients, 40 death events/136 patients presented with low BMI1 expression, and 39 death events/136 patients presented with high BMI1 expression. Asterisk indicates statistical significance.
Table 1. Patient demographic and clinicopathological parameters for adenocarcinoma cases.
n (number of patients) = 92, unless otherwise indicated in parentheses.
| n | (%) | |
|---|---|---|
| Age (years) | ||
| Median | 59 | |
| Range | 32–77 | |
| Gender | ||
| Male | 53 | 57.6 |
| Female | 39 | 42.4 |
| Cancer stage | ||
| I | 35 | 38.1 |
| II | 29 | 31.5 |
| III | 28 | 30.4 |
| Tumor grade (n = 89) | ||
| 1 | 12 | 13.5 |
| 2 | 61 | 68.5 |
| 3 | 16 | 18.0 |
| Tumor stage* | ||
| T1 | 12 | 13.0 |
| T2 | 60 | 65.2 |
| T3 | 18 | 19.6 |
| T4 | 2 | 2.2 |
| Nodal stage* | ||
| N0 | 41 | 44.6 |
| N1 | 41 | 44.6 |
| N2 | 7 | 7.5 |
| N3 | 3 | 3.3 |
T and N are determined according to the TNM (tumor, lymph nodes, metastases) classification (American Joint Committee on Cancer, 7th edition).
Overall, our data show that more than 80% of C/EBPα-negative/low adenocarcinomas display positive BMI1 expression and that this expression pattern has prognostic value. We have thus identified a molecular subtype of human adenocarcinomas, which may be amenable to BMI1 inhibition therapy.
C/EBPα is a tumor suppressor in lung cancer, and its loss correlates with increased BMI1 expression
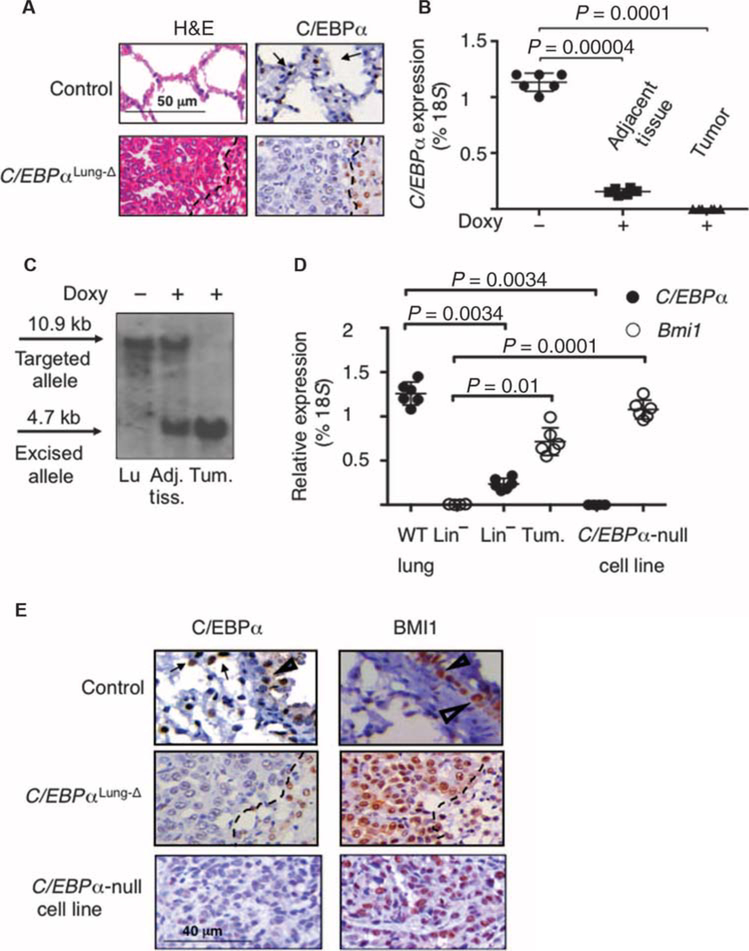
The observation that loss of C/EBPα is a common event in NSCLC (16) prompted us to study the in vivo consequences of C/EBPα deletion in adult lungs. We used the Clara Cell Secretory Protein (CCSP) promoter to drive conditional Cre recombinase–induced deletion of C/EBPα in pulmonary epithelial cells. Doxycycline-treated mice were referred to as C/EBPαLung-Δ mice, to indicate their targeted C/EBPα deletion in lung cells (fig. S3A). Histopathologic analysis showed that 6 to 8 months after doxycycline treatment, ~33% of the C/EBPαLung-Δ mice (n = 102) developed pulmonary adenocarcinomas (Fig. 2A, left panel), whereas no tumorigenesis could be detected in heterozygous mice (n = 58) (fig. S3B). Although most of the C/EBPαLung-Δ mice with tumors showed one lesion per lung, 12% of them displayed multiple foci per lung (fig. S3C). Pulmonary cells depleted of the hematopoietic and endothelial component [lineage-negative (Lin−) cells] obtained from tumors arising in C/EBPαLung-Δ mice did not express detectable amounts of C/EBPα RNA (Fig. 2B). Similarly, IHC staining confirmed the absence of C/EBPα protein (Fig. 2A, right panel), and Southern blot assays demonstrated complete excision of the C/EBPα gene in tumors (Fig. 2C). In summary, lung-specific deletion of C/EBPα in adult lung epithelium facilitates adenocarcinoma formation, validating the hypothesis that C/EBPα acts as a pulmonary tumor suppressor in vivo. Murine primary adenocarcinomas displayed reciprocal expression between C/EBPα and BMI1 (Fig. 2, D and E), similarly to human adenocarcinomas. Such a reciprocal gene expression pattern was recapitulated in a primary C/EBPα-null tumor cell line derived from the C/EBPαLung-Δ pulmonary tumors (Fig. 2, D and E), which was able to grow tumors upon subcutaneous injection into immunocompromised NSG (nonobese diabetic/severe combined immuno-deficient/interleukin-2 receptor γ null) mice (fig. S3D) and to form multiple pulmonary metastatic foci upon bloodstream injection (fig. S3E).
Fig. 2. Lung-specific C/EBPα knockout mice develop pulmonary adenocarcinoma.
(A) Left: Representative histological lung sections of C/EBPαLung-Δ mice and control littermates, stained with hematoxylin and eosin (H&E). “Control” indicates littermates that do not have at least one of the transgenic alleles (CCSP-rtTA, Cre, or C/EBPαloxP/loxP). Right: Representative IHC data showing C/EBPα protein expression (brown and indicated by arrows). The dashed line separates the C/EBPαLung-Δ tumor area (to the left of the dashed line) fromthe normal-appearing tissue, which surrounds the tumor. Scale bar, 50 μm. (B) qRT-PCR analysis was performed in duplicate using RNA from lineage-depleted (CD45.1−,CD45.2−,andCD31−) pulmonary cells from five doxycycline-untreated (Doxy −) (n = 5) and five doxycycline-treated (Doxy +) (n=5) C/EBPαloxP/loxP CCSP-rtTA+ Cre+ mice, as well as fluorescence-activated cell sorting–purified lineage-depleted pulmonary cells from five tumors (n = 5). Adjacent tissue represents the “normal-appearing” area in the vicinity of the adenocarcinoma, which does not display a transformed phenotype. The mean expression is presented as a percentage of 18S RNA amount. Two-sided Welch’s t test showed a significant statistical difference among uninduced tissue versus tumor (P = 0.0001) and induced normal-appearing tissue versus tumor (P = 0.00004). (C) Representative Southern blot analysis of lung genomic DNA from doxycycline-untreated (Doxy −) and doxycycline-treated (Doxy +) C/EBPαloxP/loxP CCSP-rtTA+ Cre+ mice. In doxycycline-treated mice, both tumor adjacent tissue and adenocarcinoma tissue were aluated, as shown, indicating nearly complete excision in tumor cells. No unexcised allele could be detected in tumors, as compared to uninduced pulmonary tissue, in which excision is undetectable. The sizes of the targeted (10.9-kb) and excised (4.7-kb) alleles are shown. Lu, lung; Adj. tiss., adjacent nontumor tissue; Tum., tumor tissue. (D) qRT-PCR was performed in duplicate using RNA extracted from three lineage-depleted (CD45.1−, CD45.2−, and CD31−) tumors (n = 3) and wild-type (WT) pulmonary cells (n = 3), as well as the C/EBPα-null cell line (n = 3). The mean expression is presented as a percentage of 18S RNA. P values for each sample are indicated on the basis of a two-tailed Welch’s t test. (E) IHC analysis performed on C/EBPαLung-Δ adenocarcinomas and the C/EBPα-null pulmonary tumor cell line confirmed a reciprocal relationship between C/EBPα and BMI1 protein expression. Representative C/EBPα (dark brown) and BMI1 (brown) IHC in lung sections from control, C/EBPαLung-Δ, and the C/EBPα-null tumor cell line derived from C/EBPαLung-Δ tumors are shown. Filled arrowheads indicate alveolar epithelial cells, and triangles indicate bronchioepithelial cells. Tumor tissue is to the left of the dashed line in the C/EBPαLung-Δ sections (middle panels). Scale bar, 40 μm.
C/EBPα restrains BMI1 expression
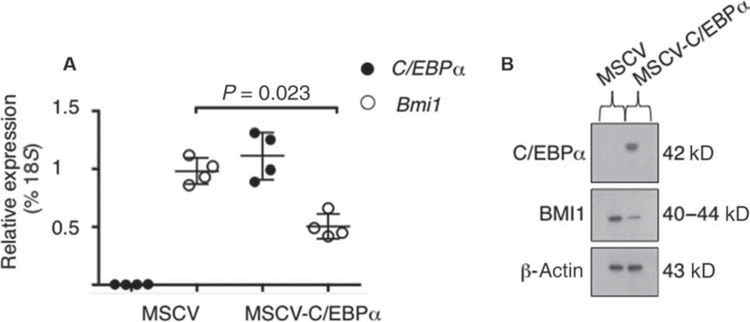
Retroviral introduction of C/EBPα in the murine C/EBPα-null pulmonary tumor cells was sufficient to decrease both Bmi1 mRNA (Fig. 3A) and protein expression (Fig. 3B), suggesting that C/EBPα is involved in Bmi1 gene regulation. Similarly, overexpression of an inducible form of C/EBPα (16) in the human adenocarcinoma cell line H358 was able to decrease BMI1 expression by 83%, as compared to the empty vector control (Fig. 1A), suggesting that repression of BMI1 by C/EBPα is conserved between murine and human pulmonary cells. Together, these data indicate that C/EBPα negatively affects BMI1 expression in lung cancer cells.
Fig. 3. C/EBPα negatively affects BMI1 expression.
(A) qRT-PCR of C/EBPα (black circles) and Bmi1 (white circles) was performed in C/EBPα-null cell lines transduced with either a retroviral construct to overexpress murine C/EBPα (MSCV-C/EBPα-IRES-GFP) or a control vector (MSCV-IRES-GFP). qRT-PCR was performed on GFP+ cells 3 days after the infection. Mean expression is presented as a percentage of 18S RNA. Assays were performed in duplicate on two independent biological replicates (n = 2). Data were compared by Welch’s t test, and the P value for Bmi1 expression is indicated. (B) Western blot analysis carried out in C/EBPα-null cell lines transduced with either a retroviral construct to overexpress murine C/EBPα (MSCV-C/EBPα-IRES-GFP) or a control vector (MSCV-IRES-GFP). Total protein lysates were immunoblotted with anti-C/EBPα and anti-BMI1 antibodies. Loading was assessed with an anti-actin antibody. The expected size in kilodaltons is indicated.
BMI1 is required for lung tumor initiation and maintenance
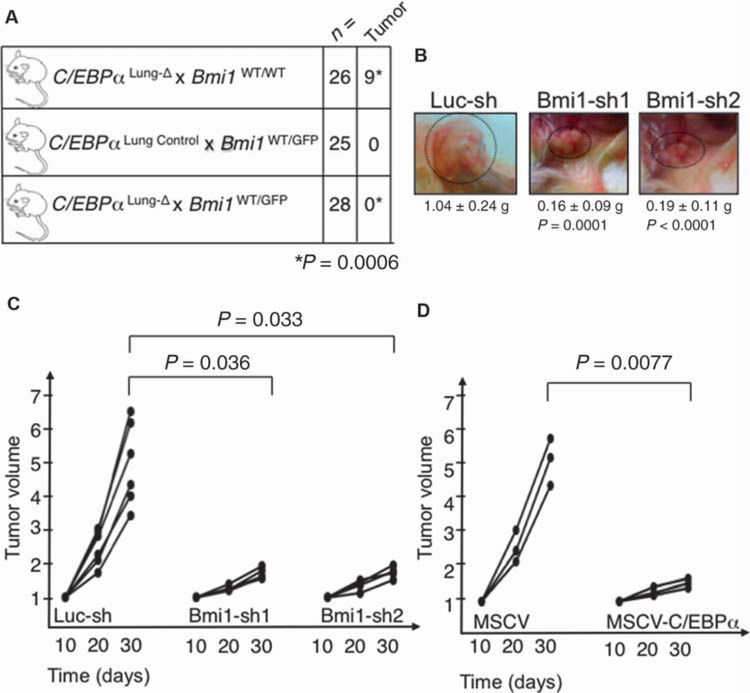
Next, we asked whether tumor initiation was affected in C/EBPα conditional lung knockout mice in the context of Bmi1 haploinsufficiency (Bmi1WT/GFP) (fig. S4A) (21). These mice display a ~50% reduction in Bmi1 RNA expression in Lin− lung cells, as compared to Bmi1WT/WT mice (fig. S4B). Upon doxycycline treatment, C/EBPαLung-Δ × Bmi1WT/WT mice (n = 26) developed tumors at the expected incidence (34.6%), whereas C/EBPαLung-Δ animals with reduced BMI1 expression (n = 28), as well as control mice (n = 25), did not develop any tumors (Fig. 4A), demonstrating that decreased BMI1 expression was sufficient to inhibit tumor formation. To test whether Bmi1 knockdown was sufficient to also affect tumor growth in already transformed cells, we performed subcutaneous transplantation assays in NSG mice. After transduction of the murine C/EBPα-null pulmonary tumor cells with green fluorescent protein (GFP)–coupled lentiviral shRNAs against Bmi1 (sh1 and sh2) or a control shRNA (sh-Luciferase), we verified that Bmi1 RNA (fig. S4C, left panel) and protein expression (fig. S4C, right panel) was silenced in GFP+ cells. Injection of cells displaying Bmi1 knockdown resulted in the formation of smaller tumors (Bmi1-sh1 versus control, P = 0.0001; Bmi1-sh2 versus control, P < 0.0001; Fig. 4B), as well as a decreased percentage of mice with tumors (83.3% of Bmi1-sh1–infected and 66.6% of Bmi1-sh2–infected mice developed tumors, as compared to 100% of the Luc-sh-control–infected mice). At day 30, tumor growth was specifically affected in mice injected with Bmi1-sh1–infected (P = 0.036) or Bmi1-sh2–infected cells (P = 0.033), as compared to sh-Luciferase–infected cells (Fig. 4C). At the time of harvest, GFP+ tumor cells from both Bmi1-sh1– and Bmi1-sh2–infected cells maintained reduced Bmi1 expression (fig. S4D). At harvest, tumors derived from Bmi1-sh1– and Bmi1-sh2–infected cells were mainly GFP−, as compared to Luc-sh-control–derived tumors (fig. S4, E, lower panel, and F), suggesting that the tumors found in Bmi1-sh1 and Bmi1-sh2 grafts were derived from the small percentage (less than 3%, as shown in fig. S4, E, upper panel, and F) of nontransduced (GFP−) cells that have outgrown the GFP+ population when co-injected into the NSG mice. Murine C/EBPα-null pulmonary tumor cells transduced with a GFP-coupled retroviral construct to overexpress C/EBPα (MSCV-C/EBPα) demonstrated a 50% decrease in BMI1 expression as compared to control vector (MSCV)–transduced cell (Fig. 3A). Therefore, we assessed their ability to grow tumors in subcutaneous transplantation assays in NSG mice. Injection of GFP+ MSCV-C/EBPα–transduced cells resulted in the formation of smaller tumors at day 30 (P = 0.0077), as compared to GFP+ MSCV-transduced cells (Fig. 4D). Overall, these data indicate that BMI1 is required to maintain the transformed phenotype of C/EBPα-null pulmonary cancer cells and allow their growth.
Fig. 4. BMI1 is required for lung tumor formation in C/EBPαLung-Δ mice.
(A) Detection of tumorigenic foci in lungs of mice with reduced Bmi1 expression. Pups were treated with doxycycline, and the presence of tumors was scored between 7.5 and 9.5 months after treatment termination, when mice were 9 to 11 months old. The difference in tumorigenesis was estimated to be statistically significant by the two-sided Fisher’s exact test (P = 0.0006). (B) Representative pictures of tumors growing subcutaneously in NSG mice after injection of cells infected with control shRNA lentivirus (Luc-sh), Bmi1-sh1, and Bmi1-sh2. Three mice were injected on each flank with each lentiviral construct (n = 6 measurements, two per mouse). The mean weight and P values, analyzed by a two-sided Welch’s test, are indicated. There was no difference between Bmi1-sh1 and Bmi1-sh2 (P = 0.681). (C) Tumor size was measured with calipers at 10, 20, and 30 days after injection of cells infected with control shRNA lentivirus (Luc-sh), Bmi1-sh1, or Bmi1-sh2 into immunocompromised mice (three mice per lentiviral construct; mice were injected on both flanks, n = 6). The figure indicates the tumor burden volume versus time since injection (in days). Data normalized to tumor size at day 10 are shown for each time point. At day 30, each of the Bmi1-sh constructs was significantly different from controls, using a two-sided Welch’s t test. In the control animals, tumor burden was 4.95 times that of day 10, compared to 1.69 times that of day 10 for Bmi1-sh1 (P = 0.0036), and 1.77 times that of day 10 for Bmi1-sh2 (P = 0.033). There was no difference between Bmi1-sh1 and Bmi1-sh2 (P = 0.453). (D) Tumor size was measured with calipers at 10, 20, and 30 days after injection of GFP+-sorted cells infected with control retrovirus (MSCV) or C/EBPα-expressing retrovirus (MSCV-C/EBPα) into immunocompromised mice (n = 3 mice per retroviral construct). The figure indicates tumor burden versus time since injection (in days). Data normalized to tumor size at day 10 are shown for each time point. The P value is indicated in the figure, as calculated by the Welch’s two-sided t test.
BMI1 inhibition therapy reduces tumor growth in lung tumorigenesis
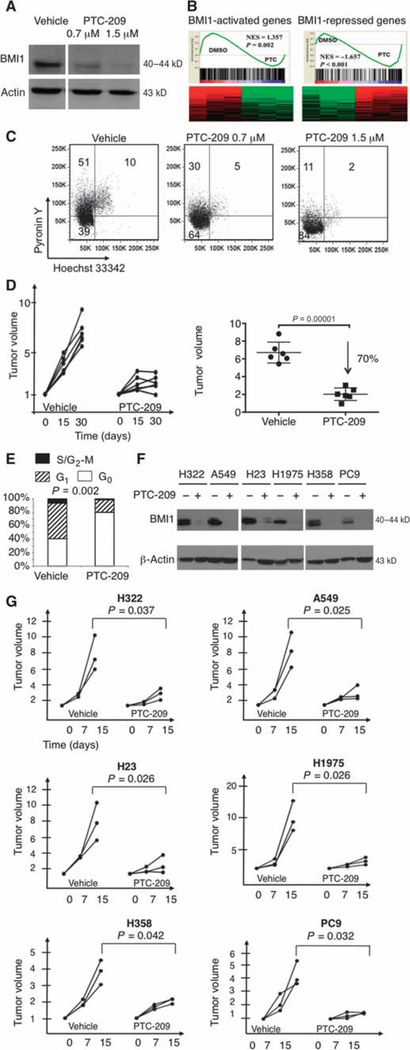
We next investigated whether modulation of BMI1 expression was sufficient to reduce tumor burden. To devise a targeted therapy to antagonize the oncogenic effect of BMI1, we used a compound (PTC-209) (fig. S5A) capable of reducing BMI1 translation (22). PTC-209 is a low–molecular weight molecule identified by high-throughput screening of compounds using gene expression modulation by small molecules (GEMS) technology, in which the GEMS reporter vector contains the luciferase open reading frame flanked by and under post-transcriptional control of the BMI1 5′ and 3′ untranslated regions (22). Western blots of the C/EBPα-null line exposed to increasing concentration of PTC-209 (0.7 and 1.5 μM) or 0.5% dimethyl sulfoxide (DMSO) control showed that BMI1 protein was efficiently decreased in a dose-dependent manner (Fig. 5A), to 35 and 5% that of DMSO-treated cells, respectively (fig. S5B). BMI1 is a key component of the epigenetic complex PRC1 (Polycomb repressive complex 1), and other components of the PRC1/2 complex (RING1, EZH1, EZH2, and EED) were unaffected (fig. S5C, upper panel) or just mildly affected (SUZ12) by PTC-209 treatment (fig. S5C, lower panel). PTC-209 treatment was also accompanied by a slightly increased histone 3 with trimethylated lysine 27 and by abrogation of histone 2A lysine 119 ubiquitination, as expected, because BMI1 catalyzes the H2AK119Ub modification (fig. S5C, lower panel). Gene expression profiling and gene set enrichment analysis (GSEA) of C/EBPα-null cells exposed to PTC-209 (1.5 μM) or DMSO control showed enrichment of BMI1 target genes (Fig. 5B). Specifically, most BMI1-activated targets (23) were down-regulated in PTC-treated cells (P = 0.002), and most genes repressed by BMI1 (23) were up-regulated with PTC treatment (P < 0.001). These results corroborate the observation that PTC-209 effectively inhibits BMI1 protein expression (Fig. 5A) and that Bmi1 mRNA is also down-regulated upon PTC-209 treatment (fig. S5D).
Fig. 5. BMI1 pharmacological inhibition reduces tumorigenicity of C/EBPα-null tumor cells.
(A) Western blot assay with an anti-BMI1 antibody was carried out in the C/EBPα-null tumor cell line treated for 48 hours with the BMI1 inhibitor (0.7 and 1.5 μM) or 0.5% DMSO vehicle as a control. Loading was assessed, after complete stripping of the membrane, with an anti-actin antibody. The expected size in kilodaltons is indicated. (B) GSEA shows enrichment of BMI1-activated targets (P = 0.002) in vehicle (DMSO)–treated cells, as compared to PTC-209–treated cells (left panel). GSEA also shows enrichment in BMI1-repressed targets in PTC-209–treated cells, as compared to vehicle (DMSO)–treated cells (right panel, P < 0.001). Normalized enrichment score (NES) is indicated in each panel. Fold change cutoff value of 2 and false discovery rate–adjusted P value cutoff of 0.05 were used for heat presentation. (C) Cell cycle analysis of the C/EBPα-null tumor cell line after treatment for 48 hours with 0.7 and 1.5 μM BMI1 inhibitor and vehicle as control. Cell cycle status was determined by pyronin Y/Hoechst 33342 staining (Hoechst 33342 stains DNA and pyronin Y stains RNA). Percentages of cells in G0 (double-negative population), G1 (pyronin Y+ population), and S/G2-M phases (double-positive population) are indicated. Specifically, the average percentage of the population in cell cycle arrest is 61 ± 4.3% (cells treated with 0.7 μM PTC-209, P = 0.05), 76.4 ± 7.6% (cells treated with 1.5 μM PTC-209, P = 0.01), and 46 ± 7% (DMSO-treated cells). Assays were performed as triplicate and analyzed by running three two-sample one-sided t tests, corrected by a Bonferroni adjustment. (D) Tumor size was measured with calipers at the beginning of the treatment and every 15 days from initiation of treatment until termination. Mice were treated daily for 1 month with PTC-209 (50 mg/kg) or vehicle control. The left panel indicates tumor burden versus time of treatment (in days). Growth curves of six vehicle-treated mice are shown on the left, and those of six PTC-209–treated mice are on the right. Data (n = 6 per group) are normalized to the tumor burden detected at the beginning of treatment and are shown for each time point. The normalized tumor volume at day 30 is indicated in the histogram (right panel). The difference in tumor growth is statistically significant (P = 0.00001), as calculated by the two-sided Welch’s t test. (E) The graph indicates the percentages of cells in G0 (white), G1 (striped), and S/G2-M phases (black). Cell cycle analysis was performed by pyronin Y/Hoechst 33342 staining of subcutaneously transplanted tumors treated with the PTC-209 compound (n = 4) or the vehicle only (n = 4) as control. Welch’s two-sided t test was used to calculate differences in G0 (P = 0.002) between PTC-209– and vehicle-treated cells. (F) Western blot analysis of the indicated human adenocarcinoma cell lines treated for 48 hours with the BMI1 inhibitor (1.5 μM, +) or 0.5% DMSO vehicle as control (−). Protein lysates were immunoblotted with an anti-BMI1 antibody, and loading was assessed with an anti-actin antibody. The expected size in kilodaltons is indicated. Treatment with PTC-209 at 1.5 μM reduces the expression of BMI1 to 4.2% (H322), 1.87% (A549), 6.33% (H23), 9.08% (H1975), 6.1% (H358), and 5.4% (PC9), as compared to the expression with 0.5% DMSO. (G) NSG mice were subcutaneously injected with different human adenocarcinoma cell lines, as indicated on each plot. Mice were treated daily for 2 weeks with PTC-209 (50 mg/kg) or vehicle control. Tumor burden was measured with calipers at the beginning of the pharmacological treatment and after 7 and 15 days from initiation of treatment. The figure indicates tumor volume versus time of treatment (in days). Each graph shows growth curves of three vehicle-treated mice on the left and curves from three PTC-209–treated mice on the right. Data (n = 3 per group) are normalized to the tumor volume measured at the beginning of treatment and are shown per time point. The difference in tumor size at day 15 was statistically significant (P values indicated in the figure), as calculated by Welch’s two-sided t test, in every xenograft model.
Further, we observed that BMI1 in vitro inhibition resulted in an increased percentage of C/EBPα-null lung cancer cells arrested in a nondividing state (Fig. 5C), which prompted us to test PTC-209 effects on in vivo tumor maintenance. After developing measurable tumors, NSG mice were treated daily with PTC-209 or vehicle control for 1 month. Tumor growth was specifically affected in PTC-209–treated animals (n = 6), in which the tumor burden was decreased by 70% (P = 0.00001) compared to that observed in vehicle-treated mice (n = 6) (Fig. 5D). PTC-209 treatment altered the cell cycle profile, with BMI1 in vivo inhibition consistently showing an increase in cells in G0 phase (average of 76.9 ± 2.3% in PTC-209–treated compared to 39.1 ± 8.7% in vehicle-treated cells, P = 0.002), as shown in Fig. 5E.
A panel of human NSCLC cell lines (adenocarcinoma cell lines H322, A549, H23, H1975, H358, PC9, Calu-3, H1755, and H1650; adenosquamous carcinoma cell line H647; bronchial carcinoid cell line H727; and carcinoma cell lines Calu-1 and H1299) expressed almost undetectable amounts of C/EBPα and medium/high amounts of BMI1 (fig. S5E). We could not identify any NSCLC cell line with substantial expression of C/EBPα. In contrast, C/EBPα was expressed in medium/high amounts in the breast cancer cell line MDA-231 and in the ovarian cancer cell lines Ovcar-3, Skov3, and 36M2, which all had medium to low amounts of BMI1 expression (fig. S5E).
Upon treatment of the adenocarcinoma cell lines H322, A549, H23, H1975, H358, and PC9 with PTC-209 (1.5 μM) or DMSO control for 48 hours, the amount of BMI1 protein decreased across the six cell lines to a mean of 5.49% of the expression in the absence of the inhibitor (SD, 2.39) (Fig. 5F), and PTC-209 treatment increased the percentage of cells arrested in a nondividing state in all the human adenocarcinoma cell lines analyzed (fig. S5F). Consistently, when mice xenografted with the human adenocarcinoma cell lines were treated with daily injections of PTC-209, they all responded to BMI1 in vivo inhibition treatment, showing a significant tumor growth reduction (P = 0.037 for H322 cells, P = 0.025 for A549 cells, P = 0.026 for H23 cells, P = 0.026 for H1975 cells, P = 0.042 for H358 cells, and P = 0.032 for PC9 cells), as compared to vehicle-treated mice (Fig. 5G). Overall, our data suggest that BMI1 is a critical and druggable target, because PTC-209 effectively reduced tumor growth and maintenance in C/EBPα-null/low tumors positive for BMI1 expression.
DISCUSSION
A better understanding of the heterogeneity of lung cancer may produce innovations in treatment strategies to potentially overcome resistance, relapse, and progression of the cancer. Previously, we have shown that C/EBPα expression is frequently decreased/abrogated in human lung cancers (4, 16). We have also suggested that the loss of C/EBPα could be an important event in the multistep program of transformation (14). However, the key molecular events required for C/EBPα-deficient cells to trigger lung tumorigenesis were still unclear. Here, we identified a subset of C/EBPα-negative/low adenocarcinoma patients, in which we observed an inverse correlation of C/EBPα and BMI1 expression. We highlighted the therapeutic potential of targeting BMI1 in this subset of patients by describing a compound that inhibits BMI1 and blocks its oncogenic role. We generated mice with specific targeted deletion of C/EBPα in adult lung epithelium, which developed pulmonary adenocarcinomas with 33% penetrance, thus establishing C/EBPα as a bona fide lung tumor suppressor in vivo. Tumors growing in the murine model recapitulated the abnormal expression of the C/EBPα/BMI1 axis observed in more than 80% of C/EBPα-negative/low adenocarcinoma patients, confirming that they are an important preclinical tool. We showed that C/EBPα-null tumor formation is dependent on the presence of two functional Bmi1 alleles, implying that Bmi1 gene dosage is a critical checkpoint that lung cells must overcome to achieve transformation.
BMI1 is a key component of the epigenetic complex PRC1, which is frequently overexpressed in human cancers, including NSCLCs, lymphomas, leukemias, neuroblastoma, skin tumors, and breast and colorectal carcinomas (24–32). BMI1 inhibition was recently found to reduce tumor burden in human colorectal xenograft models (22), similarly to what we observed in lung cancer. Specifically, by knocking down Bmi1, a gene that lies at the heart of stem cells’ self-renewal machinery (33), Kreso et al. observed a decrease in human colon cancer tumor mass in mouse xenografts, as well as a decrease in the total number of tumors initiated, suggestive of BMI1’s role in regulating cancer-initiating cells (22). Thus, identification of factors modulating BMI1 expression has generated major clinical interest. Among the mechanisms proposed to affect its expression, BMI1 copy number was analyzed in NSCLCs but found to be unchanged (32). Chromosomal aberrations that may result in up-regulation of BMI1 expression were shown in a case of T cell acute lymphoblastic leukemia (34). Increased gene transcription of BMI1 has been described in Hodgkin’s lymphoma cell lines, in which nuclear factor κB signaling contributes to its up-regulation (35). In breast and gastric cancers, BMI1 expression is repressed by MEL-18, a Polycomb gene with tumor suppressor activity (36). Nonetheless, the exact mechanisms controlling BMI1 up-regulation in lung cancer and other epithelial tumors remain unidentified. Because our data indicate that C/EBPα contributes to inhibit BMI1 expression, we propose that reducing C/EBPα protein relieves its repressive activity on BMI1 transcription. We showed that inhibition of BMI1 expression by shRNA in tumorigenic C/EBPα-null cells impairs their tumor-propagating ability. Similarly, pharmacological inhibition of BMI1 protein impairs lung tumor growth in C/EBPα-deficient/low cells by efficiently decreasing cell cycle progression, demonstrating the role of BMI1 in the transformed phenotype of C/EBPα-null/low tumor cells. In addition, we observed that BMI1 expression correlates with prognosis among CEBPα-null/low lung adenocarcinoma patients.
Further studies will be needed to elucidate the molecular mechanism through which C/EBPα loss contributes to pulmonary tumorigenesis. C/EBPα may repress BMI1 through down-regulation of c-MYC. C/EBPα directly represses c-MYC in human myeloid cells (37). Conversely, stable overexpression of c-MYC results in an up-regulation of BMI1 expression, whereas knockdown of c-MYC results in a substantial down-regulation of the endogenous BMI1 expression in the human fibrocystic breast epithelial cell line MCF10A (36). MEL-18 down-regulates BMI1 expression in breast and gastric cancers via c-MYC repression (36) and may be involved in this regulatory loop.
Down-regulation/inactivation of C/EBPα is a required step in the development of several tumor types in addition to NSCLC, making its downstream effectors potential markers and therapeutic targets. Loss of C/EBPα in keratinocytes contributes through yet unknown mechanisms to the accumulation of ultraviolet light B–induced mutations and accelerates skin cancer progression (38). In hematopoietic malignancies, C/EBPα modulates cell growth through repression of oncogenes c-Myc, N-Myc, Bmi1, and Sox4 (17, 37, 39, 40). We hypothesize that the C/EBPα/BMI1 axis will be central to cancer cell biology in other malignancies as well, given the numerous reports underlining both decreased activity of C/EBPα and increased expression of BMI1 in many cancer types (4, 7, 41).
In conclusion, our studies show that C/EBPα is a tumor suppressor in lung cancer and that BMI1 is a downstream mediator of the oncogenic process. This suggests that the lung cancer subtype defined by the loss of C/EBPα expression might specifically benefit from BMI1 inhibitory therapy, which is being evaluated in a clinical trial (clinical trial identifier NCT02404480 at ClinicalTrials.gov). Because BMI1 plays a substantial role in many solid tumors, including one of the most aggressive models of lung cancer, defined by inducible expression of mutated K-Ras (42), and its expression is positively correlated with tumor growth, invasion, metastasis, prognosis, and recurrence (43), our findings are of interest to help design better therapeutics for oncologic cancer patients displaying a positive BMI1 signature.
MATERIALS AND METHODS
Detailed protocols are provided in Supplementary Materials and Methods.
Study design
The objective of this study was to assess the physiological consequences of the genetic deletion of C/EBPα in lung epithelial cells. A murine pulmonary-specific C/EBPα conditional knockout model was generated to study the tumor suppressor function of C/EBPα.
The numbers of mice required for the experiments depicted in Fig. 4 and fig. S3 were not based on differences identified a priori, nor were formal power calculations done before conducting those experiments. In the experiment using 28 Bmi1-haploinsufficient C/EBPαLung-Δ mice and 26 C/EBPαLung-Δ mice with normal Bmi1 expression (Fig. 4A), there would have been 86% power by Fisher’s exact test to distinguish between a 30% rate of tumor in the 26 C/EBPαLung-Δ mice with normal Bmi1 expression and a 1% rate of tumor in the 28 Bmi1-haploinsufficient C/EBPαLung-Δ mice, testing at the 0.05 two-sided level.
In the experiment that uses 102 C/EBPαLung-Δ mice and 58 C/EBPαLung-Δ (heterozygous) mice (Fig. 3B), there would have been greater than 99% power to detect a difference in the rate of tumor formation of 30% compared to 1%, using Fisher’s exact test and testing at the 0.05 two-sided level of significance.
shRNA-mediated or pharmacological inhibition of BMI1 was also evaluated to determine BMI1’s role as a therapeutic target in C/EBPα-negative/low adenocarcinoma xenografts. Replicate experiments were performed two or three times. For drug treatments, animals were randomly assigned to treatment. Mice were randomized into various groups, but the experimenter was not blinded to the identities of individual groups. All collected data were used in statistical analyses. Exact numbers for each experiment are included in the figure legends. All of the mouse studies were approved by ethical committees of the Beth Israel Deaconess Medical Center. IHC data were independently and blindly scored by two pathologists.
Supplementary Material
Materials and Methods
Fig. S1. IHC data show BMI1 and C/EBPα expression in NSCLC patient subtypes.
Fig. S2. C/EBPα determines BMI1-related overall survival probability in NSCLC patients.
Fig. S3. C/EBPαLung-Δ mice develop pulmonary adenocarcinomas.
Fig. S4. Schematic representation shows generation of Bmi1-haploinsufficient C/EBPαLung-Δ mice and subcutaneous transplantation models.
Fig. S5. The BMI1 inhibitor molecule PTC-209 inhibits BMI1 and induces cell cycle arrest in vitro.
Table S1. Patient demographic and clinicopathological parameters for all NSCLC cases.
Acknowledgments:
We thank the Dana-Farber Cancer Institute and Beth Israel Deaconess Medical Center Flow Cytometry facilities; M. Loh, M. E. Nga, and Y. H. Pang for help with patients’ IHC analysis, L. Pastorino and members of the Tenen laboratory for helpful discussions; and PTC Therapeutics (South Plainfield, NJ) for donating the BMI1 inhibitor PTC-209.
Funding:
This work was funded by NIH/NCI (National Cancer Institute) P50 CA90578 Project 2 grant (to D.G.T. and B.H.); the NCIS (National University Cancer Institute of Singapore) Yong Siew Yoon Research grant through donations from the Yong Loo Lin Trust, the Singapore Ministry of Health’s National Medical Research Council under its Singapore Translational Research (STaR) Investigator Award, and the National Research Foundation of Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative and PO1 CA66996 Project 3 and 1R35CA197697 grants (to D.G.T.); FAMRI (Flight Attendant Medical Research Institute) YCSA (Young Clinical Scientist Award) 052409 and FAMRI CIA (Clinical Innovator Awards) 103063 (to E.L.); FAMRI YCSA 072165 (to D.S.B.); Doctors Cancer Foundation Award (to E.L.); IASLC (International Association for the Study of Lung Cancer) Award (to E.L.); MIUR (Ministry of Education, University and Research) Flagship InterOmics Project (to E.L. and M.C.M.); and MSMT Navrat grant LK21307 (to M.-A.J.). R.S.W. was supported by a José Carreras fellowship, FIJC-10. C.B. was supported by an NCI T32/K12/R25 award. W.C.Z. and B.L.’s work was supported by Agency for Science, Technology and Research (A*STAR), Singapore. R.A.S. was supported by the National Medical Research Council of Singapore (NMRC/CG/NCIS/2010). D.N. was supported by Dana-Farber/Harvard Cancer Center support grant 5P30 CA006516.
Footnotes
Competing interests: Y.-C.M., N.S., L.C., and T.W.D. are employees of PTC Therapeutics, which has a patent for PTC-209. All other authors declare that they have no competing interests.
Data and materials availability: The array data have been deposited in the Genomic Spatial Event (GSE) database with accession no. GSE56935. The BMI1 inhibitor clinical trial identifier is NCT02404480 (ClinicalTrials.gov).
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/8/350/350ra104/DC1
REFERENCES AND NOTES
- 1.Siegel R, Naishadham D, Jemal A, Cancer statistics, 2012. CA Cancer J. Clin 62, 10–29 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Tada Y, Brena RM, Hackanson B, Morrison C, Otterson GA, Plass C, Epigenetic modulation of tumor suppressor CCAAT/enhancer binding protein a activity in lung cancer. J. Natl. Cancer Inst 98, 396–406 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Girard L, Zöchbauer-Müller S, Virmani AK, Gazdar AF, Minna JD, Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 60, 4894–4906 (2000). [PubMed] [Google Scholar]
- 4.Koschmieder S, Halmos B, Levantini E, Tenen DG, Dysregulation of the C/EBPα differentiation pathway in human cancer. J. Clin. Oncol 27, 619–628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, Behre G, Hiddemann W, Tenen DG, Dominant-negative mutations of CEBPΑ, encoding CCAAT/enhancer binding protein-α (C/EBPα), in acute myeloid leukemia. Nat. Genet 27, 263–270 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Koschmieder S, D’Alò F, Radomska H, Schöneich C, Chang JS, Konopleva M, Kobayashi S, Levantini E, Suh N, Di Ruscio A, Voso MT, Watt JC, Santhanam R, Sargin B, Kantarjian H, Andreeff M, Sporn MB, Perrotti D, Berdel WE, Müller-Tidow C, Serve H, Tenen DG, CDDO induces granulocytic differentiation of myeloid leukemic blasts through translational up-regulation of p42 CCAAT enhancer–binding protein alpha. Blood 110, 3695–3705 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tenen DG, Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer 3, 89–101 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Tan EH, Hooi SC, Laban M, Wong E, Ponniah S, Wee A, Wang N.-d., CCAAT/enhancer binding protein a knock-in mice exhibit early liver glycogen storage and reduced susceptibility to hepatocellular carcinoma. Cancer Res. 65, 10330–10337 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Oh H-S, Smart RC, Expression of CCAAT/enhancer binding proteins (C/EBP) is associated with squamous differentiation in epidermis and isolated primary keratinocytes and is altered in skin neoplasms. J. Invest. Dermatol 110, 939–945 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Bennett KL, Hackanson B, Smith LT, Morrison CD, Lang JC, Schuller DE, Weber F, Eng C, Plass C, Tumor suppressor activity of CCAAT/enhancer binding protein a is epigenetically down-regulated in head and neck squamous cell carcinoma. Cancer Res. 67, 4657–4664 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Yin H, Radomska HS, Tenen DG, Glass J, Down regulation of PSA by C/EBPα is associated with loss of AR expression and inhibition of PSA promoter activity in the LNCaP cell line. BMC Cancer 6, 158 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flodby P, Barlow C, Kylefjord H, Åhrlund-Richter L, Xanthopoulos KG, Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein a. J. Biol. Chem 271, 24753–24760 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ, Impaired energy homeostasis in C/EBP alpha knockout mice. Science 269, 1108–1112 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Bassères DS, Levantini E, Ji H, Monti S, Elf S, Dayaram T, Fenyus M, Kocher O, Golub T, K.-k. Wong, B. Halmos, D. G. Tenen, Respiratory failure due to differentiation arrest and expansion of alveolar cells following lung-specific loss of the transcription factor C/EBPα in mice. Mol. Cell. Biol 26, 1109–1123 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martis PC, Whitsett JA, Xu Y, Perl A-K, Wan H, Ikegami M, C/EBPα is required for lung maturation at birth. Development 133, 1155–1164 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Halmos B, Huettner CS, Kocher O, Ferenczi K, Karp DD, Tenen DG, Down-regulation and antiproliferative role of C/EBPα in lung cancer. Cancer Res. 62, 528–534 (2002). [PubMed] [Google Scholar]
- 17.Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM, Shigematsu H, Levantini E, Huettner CS, Lekstrom-Himes JA, Akashi K, Tenen DG, Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBPα. Immunity 21, 853–863 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Zhu C-Q, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, Thomas RK, Naoki K, Ladd-Acosta C, Liu N, Pintilie M, Der S, Seymour L, Jurisica I, Shepherd FA, Tsao M-S, Prognostic and predictive gene signature for adjuvant chemotherapy in resected non–small-cell lung cancer. J. Clin. Oncol 28, 4417–4424 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Der SD, Sykes J, Pintilie M, Zhu C-Q, Strumpf D, Liu N, Jurisica I, Shepherd FA, Tsao M-S, Validation of a histology-independent prognostic gene signature for early-stage, non–small-cell lung cancer including stage IA patients. J. Thorac. Oncol 9, 59–64 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Tang H, Xiao G, Behrens C, Schiller J, Allen J, Chow C-W, Suraokar M, Corvalan A, Mao J, White MA, Wistuba II, Minna JD, Xie Y, A 12-gene set predicts survival benefits from adjuvant chemotherapy in non–small cell lung cancer patients. Clin. Cancer Res 19, 1577–1586 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosen N, Yamane T, Muijtjens M, Pham K, Clarke MF, Weissman IL, Bmi-1-green fluorescent protein-knock-in mice reveal the dynamic regulation of Bmi-1 expression in normal and leukemic hematopoietic cells. Stem Cells 25, 1635–1644 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Kreso A, van Galen P, Pedley NM, Lima-Fernandes E, Frelin C, Davis T, Cao L, Baiazitov R, Du W, Sydorenko N, Moon Y-C, Gibson L, Wang Y, Leung C, N Iscove N, Arrowsmith CH, Szentgyorgyi E, Gallinger S, Dick JE, O’Brien CA, Self-renewal as a therapeutic target in human colorectal cancer. Nat. Med 20, 29–36 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Douglas D, Hsu JH-R, Hung L, Cooper A, Abdueva D, van Doorninck J, Peng G, Shimada H, Triche TJ, Lawlor ER, BMI-1 promotes Ewing sarcoma tumorigenicity independent of CDKN2A repression. Cancer Res. 68, 6507–6515 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM, Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in Eμ-myc transgenic mice. Cell 65, 753–763 (1991). [DOI] [PubMed] [Google Scholar]
- 25.Kim JH, Yoon SY, Jeong S-H, Kim SY, Moon SK, Joo JH, Lee Y, Choe IS, Kim JW, Overexpression of Bmi-1 oncoprotein correlates with axillary lymph node metastases in invasive ductal breast cancer. Breast 13, 383–388 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Kim JH, Yoon SY, Kim C-N, Joo JH, Moon SK, Choe IS, Choe Y-K, Kim JW, TheBmi-1 oncoprotein is overexpressed in human colorectal cancer and correlates with the reduced p16INK4a/p14ARF proteins. Cancer Lett. 203, 217–224 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Cui H, Hu B, Li T, Ma J, Alam G, Gunning WT, Ding H-F, Bmi-1 is essential for the tumorigenicity of neuroblastoma cells. Am. J. Pathol 170, 1370–1378 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merkerova M, Bruchova H, Kracmarova A, Klamova H, Brdicka R, Bmi-1 over-expression plays a secondary role in chronic myeloid leukemia transformation. Leuk. Lymphoma 48, 793–801 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Lee K, Adhikary G, Balasubramanian S, Gopalakrishnan R, McCormick T, Dimri GP, Eckert RL, Rorke EA, Expression of Bmi-1 in epidermis enhances cell survival by altering cell cycle regulatory protein expression and inhibiting apoptosis. J. Invest. Dermatol 128, 9–17 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vrzalikova K, Skarda J, Ehrmann J, Murray PG, Fridman E, Kopolovic J, Knizetova P, Hajduch M, Klein J, Kolek V, Radova L, Kolar Z, Prognostic value of Bmi-1 oncoprotein expression in NSCLC patients: A tissue microarray study. J. Cancer Res. Clin. Oncol 134, 1037–1042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koch L-K, Zhou H, Ellinger J, Biermann K, Höller T, von Rücker A, Büttner R, Gütgemann I, Stem cell marker expression in small cell lung carcinoma and developing lung tissue. Hum. Pathol 39, 1597–1605 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Vonlanthen S, Heighway J, Altermatt HJ, Gugger M, Kappeler A, Borner MM, van Lohuizen M, Betticher DC, The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br. J. Cancer 84, 1372–1376 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wicha MS, Targeting self-renewal, an Achilles’ heel of cancer stem cells. Nat. Med 20, 14–15 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Larmonie NSD, Dik WA, Beverloo HB, van Wering ER, van Dongen JJM, Langerak AW, BMI1 as oncogenic candidate in a novel TCRB-associated chromosomal aberration in a patient with TCRgd+ T-cell acute lymphoblastic leukemia. Leukemia 22, 1266–1267 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Dutton A, Woodman CB, Chukwuma MB, Last JIK, Wei W, Vockerodt M, Baumforth KRN, Flavell JR, Rowe M, Taylor AMR, Young LS, Murray PG, Bmi-1 is induced by the Epstein-Barr virus oncogene LMP1 and regulates the expression of viral target genes in Hodgkin lymphoma cells. Blood 109, 2597–2603 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Guo W-J, Datta S, Band V, Dimri GP, Mel-18, a polycomb group protein, regulates cell proliferation and senescence via transcriptional repression of Bmi-1 and c-Myc oncoproteins. Mol. Biol. Cell 18, 536–546 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansen LM, Iwama A, Lodie TA, Sasaki K, Felsher DW, Golub TR, Tenen DG, C-Myc isa critical target for C/EBPα in granulopoiesis. Mol. Cell. Biol 21, 3789–3806 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson EA, Zhu S, Hall JR, House JS, Ranjan R, Burr JA, He Y-Y, Owens DM, Smart RC, C/EBPα expression is downregulated in human nonmelanoma skin cancers and inactivation of C/EBPα confers susceptibility to UVB-induced skin squamous cell carcinomas. J. Invest.Dermatol 131, 1339–1346 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye M, Zhang H, Amabile G, Yang H, Staber PB, Zhang P, Levantini E, Alberich-Jordà M, Zhang J, Kawasaki A, Tenen DG, C/EBPα controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat. Cell Biol 15, 385–394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Alberich-Jorda M, Amabile G, Yang H, Staber PB, Di Ruscio A, Welner RS, Ebralidze A, Zhang J, Levantini E, Lefebvre V, Valk PJM, Delwel R, Hoogenkamp M, Nerlov C, Cammenga J, Saez B, Scadden DT, Bonifer C, Ye M, Tenen DG, Sox4 is a key oncogenic target in C/EBPα mutant acute myeloid leukemia. Cancer Cell 24, 575–588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao L, Bombard J, Cintron K, Sheedy J, Weetall ML, Davis TW, BMI1 as a novel target for drug discovery in cancer. J. Cell. Biochem 112, 2729–2741 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Dovey JS, Zacharek SJ, Kim CF, Lees JA, Bmi1 is critical for lung tumorigenesis and bronchioalveolar stem cell expansion. Proc. Natl. Acad. Sci. U.S.A 105, 11857–11862 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng X, Wang Y, Zheng X, Liu C, Su B, Nie H, Zhao B, Zhao X, Yang H, ShRNA-mediated knockdown of Bmi-1 inhibit lung adenocarcinoma cell migration and metastasis. Lung Cancer 77, 24–30 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Oguro H, Iwama A, Morita Y, Kamijo T, van Lohuizen M, Nakauchi H, Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J. Exp. Med 203, 2247–2253 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Euhus DM, Hudd C, LaRegina MC, Johnson FE, Tumor measurement in the nude mouse. J. Surg. Oncol 31, 229–234 (1986). [DOI] [PubMed] [Google Scholar]
- 46.Chua S-W, Vijayakumar P, Nissom PM, Yam C-Y, Wong VVT, Yang H, A novel normalization method for effective removal of systematic variation in microarray data. Nucleic Acids Res. 34, e38 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teschendorff AE, Naderi A, Barbosa-Morais NL, Caldas C, PACK: Profile analysis using clustering and kurtosis to find molecular classifiers in cancer. Bioinformatics 22, 2269–2275 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Wang J, Wen S, Symmans WF, Pusztai L, Coombes KR, The bimodality index: A criterion for discovering and ranking bimodal signatures from cancer gene expression profiling data. Cancer Inform. 7, 199–216 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peltz SW, Welch EM, Trotta CR, Davis T, Jacobson A, Targeting post-transcriptional control for drug discovery. RNA Biol. 6, 329–334 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Bhattacharyya A, Trotta CR, Peltz SW, Mining the gems – a novel platform technology targeting post-transcriptional control mechanisms. Drug Discov. Today 12, 553–560 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Fig. S1. IHC data show BMI1 and C/EBPα expression in NSCLC patient subtypes.
Fig. S2. C/EBPα determines BMI1-related overall survival probability in NSCLC patients.
Fig. S3. C/EBPαLung-Δ mice develop pulmonary adenocarcinomas.
Fig. S4. Schematic representation shows generation of Bmi1-haploinsufficient C/EBPαLung-Δ mice and subcutaneous transplantation models.
Fig. S5. The BMI1 inhibitor molecule PTC-209 inhibits BMI1 and induces cell cycle arrest in vitro.
Table S1. Patient demographic and clinicopathological parameters for all NSCLC cases.