Summary
Pseudoalteromonas luteoviolacea is a globally distributed marine bacterium that stimulates the metamorphosis of marine animal larvae, an important bacteria–animal interaction that can promote the recruitment of animals to benthic ecosystems. Recently, different P. luteoviolacea isolates have been shown to produce two stimulatory factors that can induce tubeworm and coral metamorphosis; Metamorphosis-Associated Contractile structures (MACs) and tetrabromopyrrole (TBP) respectively. However, it remains unclear what proportion of P. luteoviolacea isolates possess the genes encoding MACs, and what phenotypic effect MACs and TBP have on other larval species. Here, we show that 9 of 19 sequenced P. luteoviolacea genomes genetically encode both MACs and TBP. While P. luteoviolacea biofilms producing MACs stimulate the metamorphosis of the tubeworm Hydroides elegans, TBP biosynthesis genes had no effect under the conditions tested. Although MACs are lethal to larvae of the cnidarian Hydractinia symbiologicarpus, P. luteoviolacea mutants unable to produce MACs are capable of stimulating metamorphosis. Our findings reveal a hidden complexity of interactions between a single bacterial species, the factors it produces and two species of larvae belonging to different phyla.
Introduction
The free-swimming larvae of many marine invertebrates must settle and undergo metamorphosis to continue their life cycle as adults on the seafloor. Certain bacteria coating submerged surfaces may serve as important environmental cues that indicate a suitable habitat for larvae to settle and undergo metamorphosis (Hadfield, 2011; Cavalcanti et al., 2020). The larvae of diverse marine invertebrate animals undergo metamorphosis in response to stimulatory bacteria, including cnidarians, annelids, crustaceans, urchins and tunicates (Hadfield and Paul, 2001). This phenomenon of bacteria-induced metamorphosis is critical for the biofouling of ship hulls (Schultz et al., 2011), aquaculture of marine invertebrates like oysters (Yu et al., 2010), and the restoration of ecosystems such as coral reefs (Negri et al., 2001; Webster et al., 2004; Sneed et al., 2014). However, we have only just begun to understand broader mechanisms through which this beneficial microbe–animal interaction occurs.
Many larvae of marine invertebrates undergo metamorphosis in response to specific components of bacteria or products from bacteria, we call here ‘factors’, that are diverse in their chemical composition, physical properties, biological nature and ecological roles (Cavalcanti et al., 2020). Several bacterial factors responsible for inducing metamorphosis have been identified in diverse bacteria and include a small molecule (Tebben et al., 2011; Sneed et al., 2014), a protein (Huang et al., 2012; Shikuma et al., 2014; Ericson et al., 2019), polysaccharides, lipopolysaccharides (Freckelton et al., 2019; Guo et al., 2019) and (lyso)phospholipids (Leitz and Wagner, 1993; Guo et al., 2019). However, only two factors have been both characterized and the genes that encode their biosynthesis described; the small molecule 2,3,4,5-tetrabromopyrrole (TBP) (Tebben et al., 2011; Agarwal et al., 2014; Gamal et al., 2016) and the metamorphosis-inducing factor 1 (Mif1) protein that is carried by Metamorphosis Associated Contractile structures (MACs) (Shikuma et al., 2014; Ericson et al., 2019).
TBP stimulates the metamorphosis of several coral species (Tebben et al., 2011, 2015; Sneed et al., 2014) and was first identified as a metamorphosis-inducing compound through bioactivity-guided fractionation of bacterial extracts (Tebben et al., 2011). Isolated single species biofilms and organic extracts of Pseudoalteromonas sp. A3 (Negri et al., 2001), J010 (Tebben et al., 2011) and PS5 (Sneed et al., 2014) can induce the metamorphosis of coral larvae. However, when the coral larvae were exposed to either the individual strains or TBP extract, metamorphosis often occurred without attachment (Negri et al., 2001; Tebben et al., 2011, 2015; Sneed et al., 2014). Although TBP was proposed as a compound of importance for coral re-seeding and aquaculture (Tebben et al., 2011; Sneed et al., 2014), its ecological significance has been questioned since its discovery (Negri et al., 2001; Tebben et al., 2015) due to an intermediate phenotype where the larvae would metamorphose but remain unattached to the substrate. Additionally, the low abundance of pseudoalteromonads on encrusting algae may indicate its inability to provide a substantial signal for coral induction in situ (Tebben et al., 2015). Interestingly, TBP and other cyclic halogenated moieties were lethal to several species of phytoplankton (Whalen et al., 2018), demonstrating that TBP can elicit either negative or positive responses depending on the organism.
In contrast to the bioactivity-guided fractionation method used to identify TBP, MACs were discovered in P. luteoviolacea using bacterial genetics and functional mutants (Huang et al., 2012; Shikuma et al., 2014). MACs are a syringe-like complex thatisevolutionarily related to type 6 secretion systems and tailed bacteriophage. MACs are composed of conserved structural components including a rigid inner tube surrounded by a baseplate complex and contractile sheath. Contraction of the sheath propels the inner tube through cell membranes often delivering protein effectors to target cells (Basler et al., 2012; Shikuma et al., 2014). We recently identified an effector of MACs called Mif1 that is sufficient for inducing the metamorphosis of a tubeworm called Hydroides elegans (hereafter Hydroides) (Ericson et al., 2019). Instead of a small cyclic molecule like TBP, Mif1 is a 943 amino acid protein loaded within a macromolecular contractile injection system. While the discovery that MACs induce Hydroides metamorphosis brings us a step closer to determining one way that bacteria stimulate metamorphosis, it remains unclear what proportion of P. luteoviolacea isolates have this capability, and if other marine larvae respond to MACs by undergoing metamorphosis.
Although many marine bacteria from diverse phyla have been shown to induce the metamorphosis of marine invertebrates (Unabia and Hadfield, 1999; Tran and Hadfield, 2011; Freckelton et al., 2017; Guo et al., 2017) only a handful have been studied to identify and characterize their metamorphosis-inducing properties. One of these bacteria is Pseudoalteromonas luteoviolacea, which is a prodigious producer of bioactive compounds (Gauthier and Flatau, 1976; Laatsch and Pudleiner, 1989) and was shown to induce the metamorphosis of corals, sea urchins and tubeworms. P. luteoviolacea strain 2ta16 was isolated from the surface of corals (Rypien et al., 2010) and produces halogenated compounds, such as pentabromopseudilin and TBP (Agarwal et al., 2014). Strains H2 and A316 induce coral (Tran and Hadfield, 2011) and sea urchin metamorphosis (Huggett et al., 2006) respectively by a yet uncharacterized bacterial factor. P. luteoviolacea strain HI1 is genetically tractable, and is the subject of several studies showing that it is capable of inducing the metamorphosis of Hydroides by producing MACs (Huang and Hadfield, 2003; Huang et al., 2012; Shikuma et al., 2014, 2016; Ericson et al., 2019). These examples demonstrate the highly inductive nature of this bacterial species across diverse animal types. Nineteen genomes of P. luteoviolacea strains have been isolated and sequenced from oceans around the world (Rypien et al., 2010; Tran and Hadfield, 2011; Asahina and Hadfield, 2015; Maansson et al., 2016; Thøgersen et al., 2016) and display a significant diversity in gene content (Maansson et al., 2016; Busch et al., 2019). The chemical activity, stimulatory nature, genetic tractability and genomic diversity of P. luteoviolacea make this bacterium a particularly well-suited model for studying distinct metamorphosis-inducing factors in the laboratory.
The larvae of diverse marine invertebrates have been studied in the laboratory to investigate metamorphosis in response to bacteria (Hadfield, 2011; Cavalcanti et al., 2020). Two prominent animals used to study this process are the spirailian tubeworm, Hydroides, and the cnidarian hydroid, Hydractinia symbiolongicarpus (hereafter Hydractinia). Hydroides has been used as a model organism to study bacteria-stimulated metamorphosis because it is easily propagated in the laboratory (Hadfield et al., 1994; Nedved and Hadfield, 2008) and its larvae settle and undergo metamorphosis in response to biofilms composed of a natural consortia (Huang and Hadfield, 2003) or single strains of bacteria (Unabia and Hadfield, 1999). The colonial hydroid, Hydractinia has been used as an important model to study development, immunology, reproduction (Frank et al., 2001) and metamorphosis in response to Pseudoalteromonas species (Leitz and Wagner, 1993; Seipp et al., 2007; Guo et al., 2017, 2019). While the larvae of ecologically threatened animals, like stony corals that build coral reefs, are often difficult to obtain, Hydractinia serves as an accessible model cnidarian to investigate bacteria-stimulated metamorphosis.
In this work, we aimed to determine what proportion of P. luteoviolacea isolates possess the genes encoding MACs, and what phenotypic effect MACs and TBP have on other larval species. We use comparative genomics to illustrate the distribution of the MACs biosynthesis gene clusters across diverse isolates of P. luteoviolacea and find that roughly half of the sequenced P. luteoviolacea strains encode the genes responsible for the production of both MACs and TBP. We construct P. luteoviolacea mutants lacking the ability to produce TBP or MACs, and directly compare the phenotypic responses of two model animals, Hydroides and Hydractinia. We show that P. luteoviolacea HI1 produces the two previously characterized factors, MACs and TBP, that have very different phenotypic effects on larvae from different phyla, including eliciting no apparent response, death or metamorphosis. Taken together, these results highlight the utility in studying P. luteoviolacea as a model bacterium to further characterize the effect of bacterial factors on diverse animals and their phenotypic responses, including metamorphosis.
Results
Many P. luteoviolacea strains encode both TBP and MACs genes
Pseudoalteromonas luteoviolacea is a globally distributed Gammaproteobacterium that exhibits a broad genetic diversity. A previous work has demonstrated that some P. luteoviolacea strains possess the biosynthesis genes and the ability to produce brominated natural products (Busch et al., 2019). However, a similar survey has not yet been performed for genes encoding MACs. To explore this, we identified several experimentally confirmed genes important for MACs production in strain HI1, including the baseplate (macB), tube (macT) and sheath (macS) structural genes (Shikuma et al., 2014) and the metamorphosis-inducing effector (mif1) gene (Ericson et al., 2019) (Fig. 1A). The MACs genes were blasted against all 19 complete and draft genomes of P. luteoviolacea available from GenBank (Table S1) including a Pseudoalteromonas sp. outgroup ATCC 29581 (Cress et al., 2013). We then identified the proportion of strains that encode MACs (macB, macS, macT and mif1) and/or TBP (bmp1–4) (Gamal et al., 2016) biosynthesis genes by blastn (Camacho et al., 2009). We also reconstructed a P. luteoviolacea phylogeny using 71 bacterial ribosomal genes (Eren et al., 2015; Delmont and Eren, 2018; Lee, 2019). As observed previously (Vynne et al., 2012; Busch et al., 2019), P. luteoviolacea strains fall within one of two major clades (Fig. 1B). All P. luteoviolacea strains queried possessed MACs gene homologues with significant homology to essential MACs genes from strain HI1 (represented by purple arrows in Fig. 1B and listed in Table S1). Furthermore, the genomic architecture of the bmp genes and production of pentabromopseudilin was confirmed in roughly half of the strains (Busch et al., 2019). The blue pentagons (Fig. 1B) show that brominated natural product biosynthesis is not restricted to the phylogenetic distribution. Our results show that all P. luteoviolacea strains examined have the genetic capacity to produce MACs and nearly half of the strains have the genetic capacity to produce both TBP and MACs.
Fig 1.
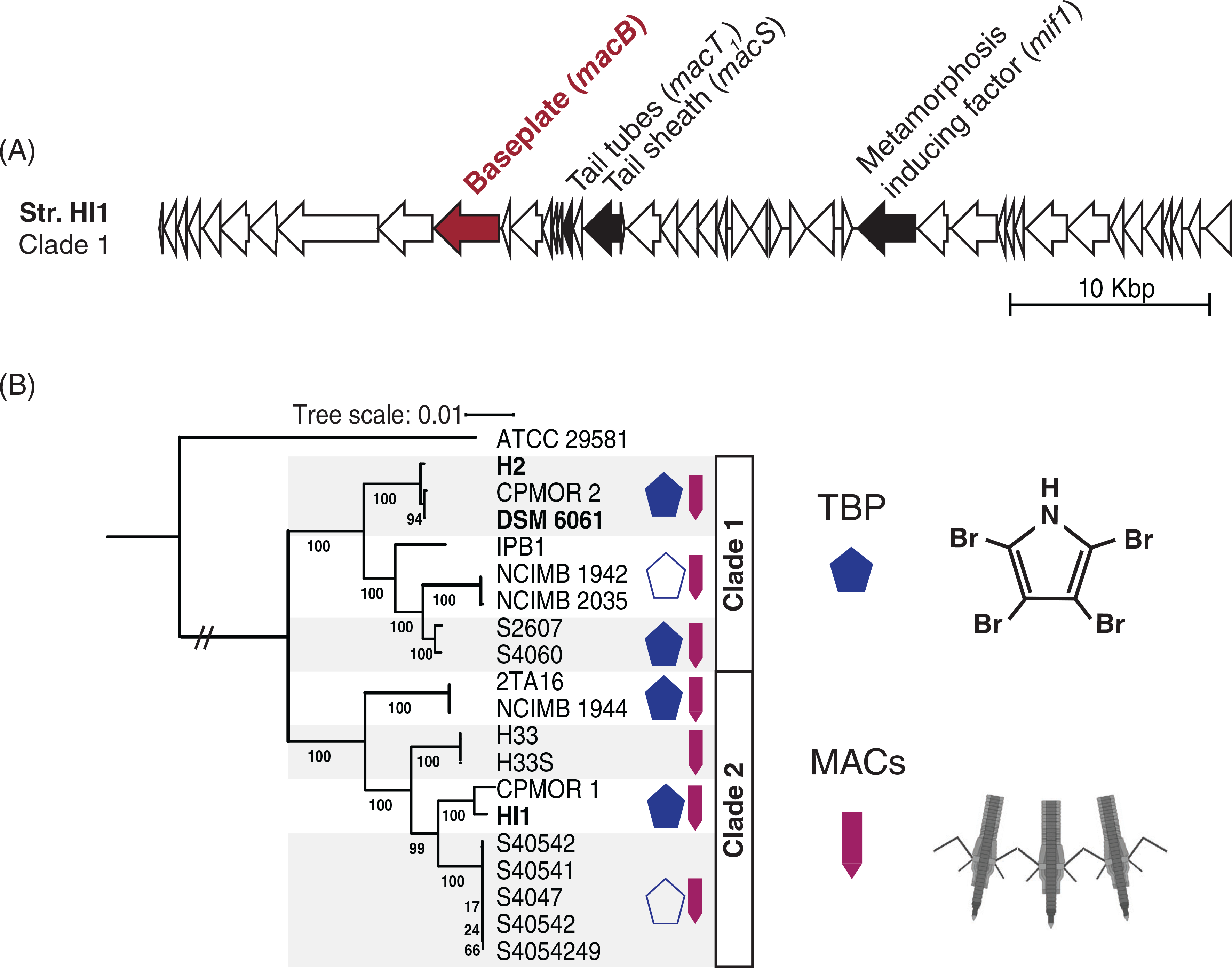
Diverse Pseudoalteromonas luteoviolacea strains encode the biosynthesis genes for TBP and MACs. (A) MACs gene cluster from strain HI1. The filled arrows denote the genes interrogated by the blast and are previously shown to be necessary for MACs function. The red filled arrow represents the macB gene, which is knocked out to create the nonfunctional MACs (Shikuma et al., 2014) strain used in the biofilm metamorphosis assays. (B) Maximum likelihood phylogeny of 19 sequenced P. luteoviolacea genomes including Pseudoalteromonas sp. ATCC 29581 as an outgroup. The bootstrap values represent 100 re-samples. The banded boxes indicate highly conserved subgroups for which symbol representations apply throughout the group. The blue pentagon denotes strains that produce TBP. Hollow pentagons represent strains that encode some genes in the bmp gene cluster, but do not experimentally produce halogenated compounds. Purple arrows indicate the presence of MACs genes (macB, macS, macT, and mif1).
P. luteoviolacea strain HI1 produces both TBP and MACs
We have shown previously that P. luteoviolacea strain HI1 produces MACs (Shikuma et al., 2014), and it was recently shown that this same strain also possesses the bmp gene cluster (Busch et al., 2019) (Fig. 2A). Although we have previously shown that a ΔmacB mutant is unable to produce functional MACs (Shikuma et al., 2014), the ability of P. luteoviolacea strain HI1 to produce TBP was unknown. We therefore tested whether strain HI1 is capable of producing TBP, and whether the brominase Bmp2 (Agarwal et al., 2014; Gamal et al., 2016) is required for production. Pseudoalteromonas luteoviolacea. mutant strains were constructed using double-homologous recombination (Shikuma et al., 2014; Ericson et al., 2019; Rocchi et al., 2019). The mutant strains contain in-frame deletions of the bmp2 gene (Fig. 2A; blue), shown previously to be required for TBP production (Agarwal et al., 2014; Gamal et al., 2016), the macB gene (Fig. 1B; purple), encoding an essential structural component of the MACs baseplate and a ΔmacBΔbmp2 mutant that is unable to produce both MACs and TBP (Table S2). Using QToF LC–MS/MS (Agilent 6530 Accurate Mass; CA, USA) we determined that P. luteoviolacea strain HI1 produces TBP (Fig. 2B and Fig. S1a), and a mutation in the bmp2 gene abrogated TBP biosynthesis (Fig. 2B). We complemented the bmp2 gene on a constitutively expressed plasmid, which enabled a small but detectable amount of TBP despite the absence of the gene at its native locus (Fig. S1b). This finding suggests that there are no other active brominases responsible for the production of TBP in P. luteoviolacea under the conditions tested.
Fig 2.
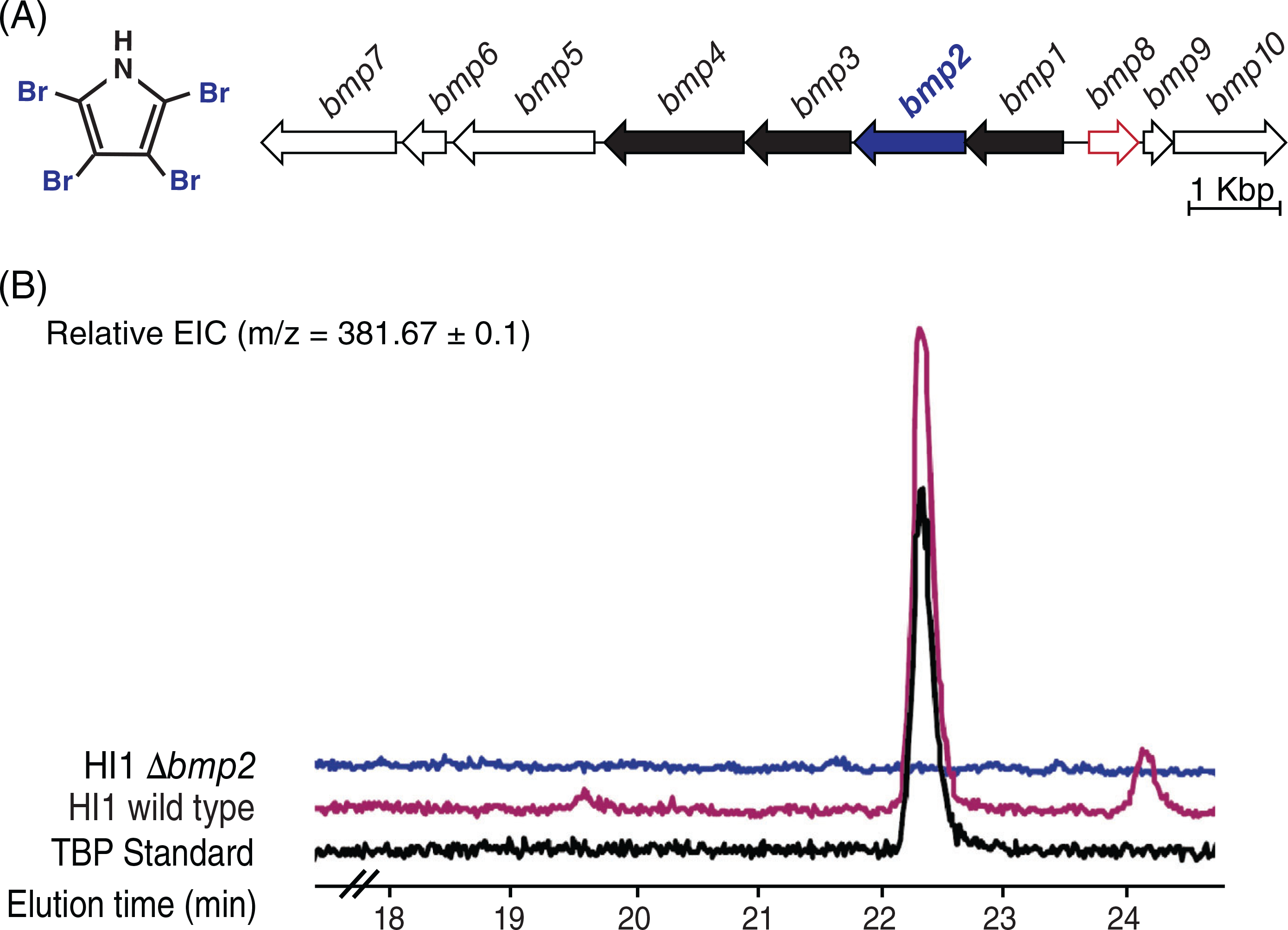
Pseudoalteromonas luteoviolacea HI1 wild type produces TBP, while the Δbmp2 strain does not. (A) Genomic arrangement of the bmp gene cluster in strain HI1. Bolded genes bmp1–4 have previously been validated to code for TBP biosynthesis. The blue bolded gene bmp2 was deleted to create a nonfunctional TBP mutant in P. luteoviolacea. The gene outlined in red bmp8 is a pseudogene. (B) Representative ion chromatogram (EIC = 381.67) overlaid the comparison of an organic extraction of wild type and Δbmp2 strains to the TBP standard. Cultures were grown in MB and SWT media overnight in triplicate to quantify TBP production in P. luteoviolacea (see Fig. S1a).
Pseudoalteromonas luteoviolacea stimulates Hydractinia metamorphosis in the absence of MACs
TBP has been implicated as an inducer of coral metamorphosis by testing the effect of fractionated and purified TBP on coral larvae (Tebben et al., 2011, 2015; Sneed et al., 2014). To determine whether larvae of Hydractinia respond to purified TBP in a similar manner, we investigated whether synthesized, exogenously added TBP (Zheng et al., 2018) at similar concentrations to those tested for corals (Sneed et al., 2014; Tebben et al., 2015) stimulates Hydractinia metamorphosis. The development of Hydractinia larvae after exposure to TBP or bacteria was quantified after 72 h and scored positively for metamorphosis if they developed stolons and tentacles (Fig. 3A). Upon exposure to a range of TBP concentrations, Hydractinia did not undergo metamorphosis, and at the highest tested concentrations (1000 and 750 nM) TBP was lethal (Fig. 3B; Table S3). These results indicate that Hydractinia larvae do not undergo metamorphosis in response to TBP under the conditions tested.
Fig 3.
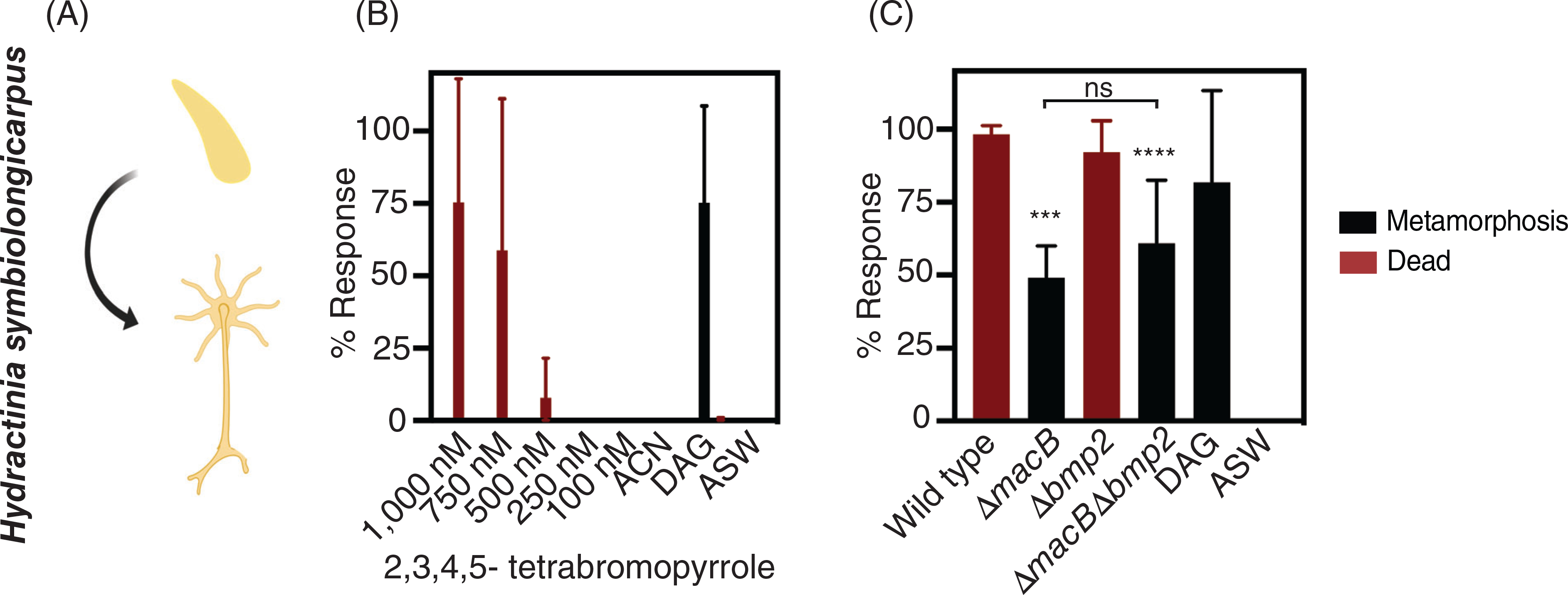
Pseudoalteromonas luteoviolacea stimulates Hydractinia metamorphosis in the absence of MACs. (A) Schematic of larval and metamorphosis phenotypes scored for Hydractinia larvae. All metamorphosis assays are performed in 96-well plates with either (B) the addition of synthesized TBP or (C) monoculture biofilms of P. luteoviolacea wild type or mutant strains. Phenotypic response of Hydractinia larvae to treatments containing (B) increasing concentrations of purified TBP and (C) P. luteoviolacea wild type or mutant biofilms. The bars represent the average of three biological replicates (n = 3). Values for the biological replicates were determined by averaging four technical replicates per treatment. The biological replicates were performed on different batches of larvae on different days. Error bars denote standard deviation. Asterisks above the bars denote statistical significance compared to the (B) ACN and (C) ASW controls. ACN is a 2% (v/v) acetonitrile solvent control. ASW is a filtered artificial seawater and is used as the negative control in all assays. DAG (1,2-Diocanoyl-sn-glycerol) is a chemical stimulant of metamorphosis and is used as a control for the competency of Hydractinia larvae at a concentration of 100 μM. (c) Statistical analyses were performed with a one-way ANOVA corrected for multiple comparisons by false discovery rate (FDR) using the two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli (GraphPad Prism), where ***p = 0.0029, and ****p = 0.0006. No statistical difference was found between treatments ΔmacB and ΔmacBΔbmp2.
We next queried whether Hydractinia responds to intact P. luteoviolacea cells within biofilms, capable of producing both TBP and MACs, TBP alone, MACs alone, or neither. To this end, we exposed Hydractinia larvae to biofilms of P. luteoviolacea wild type, ΔmacB, Δbmp2, or ΔmacBΔbmp2 mutant strains. Exposure to wild type biofilms of P. luteoviolacea was lethal after they appeared to initiate the settlement process (Fig. 3C; Table S3). Similarly, the biofilms of Δbmp2 resulted in the mortality of larvae. Interestingly, both mutants that lacked functional MACs (ΔmacB and ΔmacbΔbmp2) stimulated the metamorphosis of Hydractinia larvae (Fig. 3C; Table S3). Our results show that MACs are lethal to Hydractinia larvae, deletion of the bmp2 gene in P. luteoviolacea has no effect under the conditions tested and P. luteoviolacea stimulates metamorphosis in the absence of MACs.
Pseudoalteromonas luteoviolacea biofilms stimulate Hydroides metamorphosis via MACs, not TBP
We next questioned how the larvae of a different animal, Hydroides, respond to P. luteoviolacea and its metamorphosis factors. To this end, we exposed Hydroides larvae to purified TBP and subsequently the same panel of mutant P. luteoviolacea strains with and without MACs and TBP. Hydroides larvae were scored after 24 h of exposure to TBP or bacterial biofilms and assessed as metamorphosed if they developed branchial radioles and a primary proteinaceous tube (Fig. 4A). At a 500 nM concentration, TBP alone resulted in up to 30% of Hydroides metamorphosis compared to the acetonitrile solvent control (Fig. 4B; Table S3; p = 0.0054). Notably, all Hydroides larvae that metamorphosed were attached to the well. Higher TBP concentrations tested (1000, 750 and 500 nM), resulted in the death of a fraction of Hydroides larvae.
Fig 4.
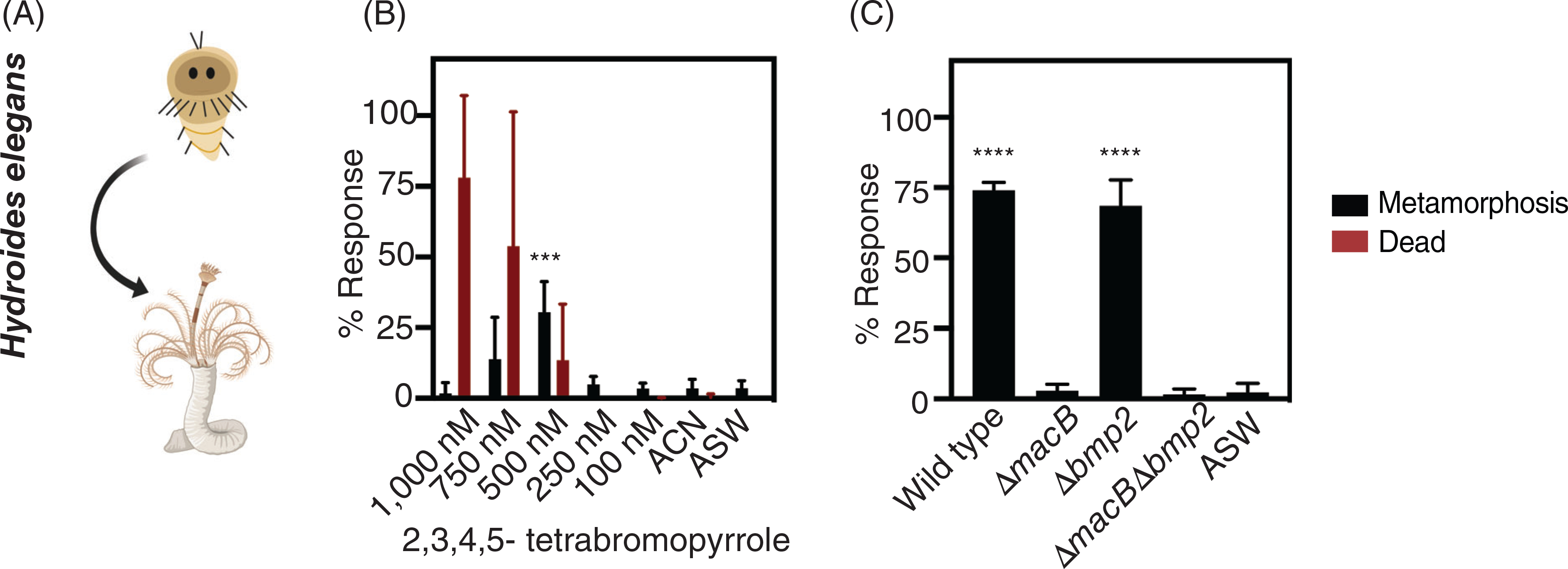
Pseudoalteromonas luteoviolacea MACs stimulate the metamorphosis of Hydroides larvae.
(A) Schematic of larval and metamorphosis phenotypes scored for Hydroides. Metamorphosis assays are performed as described previously (Shikuma et al., 2014). Phenotypic response of Hydroides larvae to treatments containing (B) increasing concentrations of purified TBP and (C) mutant biofilms. The bars represent the average of (B) five biological replicates (n = 5) and (C) three biological replicates (n = 3). A statistical power analysis aided with the determination for the appropriate number of biological replicates. Values for the biological replicates were found by averaging four technical replicates per treatment. The biological replicates were performed on different batches of larvae on different days. Error bars denote standard deviation. Asterisks above the bars denote statistical significance compared to the (B) ACN and (C) ASW controls. Statistical analyses were performed with a (B) Kruskal–Wallis ANOVA and (C) one-way ANOVA both corrected for multiple comparisons by FDR using the two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli (GraphPad Prism), where ***p = 0.0054, and ****p < 0.0001.
We next tested whether biofilms of P. luteoviolacea wild type and each macB or bmp2 mutant elicited a phenotypic response in Hydroides larvae. While biofilms of wild type P. luteoviolacea stimulated the metamorphosis of Hydroides, the ΔmacB mutant abrogated metamorphosis (Fig. 4C; Table S3), consistent with previous findings (Shikuma et al., 2014). In contrast, Hydroides larvae were stimulated to metamorphose by the Δbmp2 mutant, producing a similar response to the wild type. Like the ΔmacB mutant, the ΔmacBΔbmp2 mutant did not induce metamorphosis. Our results show that Hydroides larvae undergo metamorphosis in response to MACs and are unaffected by mutation of the bmp2 gene under the conditions tested.
Discussion
Pseudoalteromonas luteoviolacea produces both TBP and MACs, which are factors that have been shown to stimulate the metamorphosis of corals and tubeworms respectively. However, the effect of MACs and TBP on different animal types and the distribution of MACs genes in P. luteoviolacea strains have not yet been determined.
Purified TBP stimulates Hydroides metamorphosis but has no effect on Hydractinia larvae
We found that purified TBP induces a moderate level of Hydroides metamorphosis at intermediate (500 nM) concentrations and resulted in death at higher (750 and 1000 nM) concentrations. Coral larvae were found to undergo metamorphosis, many without attaching to the substrate (Negri et al., 2001; Tebben et al., 2011, 2015; Sneed et al., 2014), when exposed to similar TBP concentration ranges used in previous studies (Sneed et al., 2014; Tebben et al., 2015). Interestingly, we found that all Hydroides larvae metamorphosed with attachment. It is currently unknown whether the cellular processes that control attachment and metamorphosis are different between corals and Hydroides. Although Hydractinia belongs to the same phylum as stony corals that undergo metamorphosis in response to TBP, Hydractinia larvae did not undergo metamorphosis in response to TBP and at the highest concentrations tested, TBP was lethal.
It is currently unclear whether TBP is an ecologically relevant stimulant of metamorphosis or how TBP stimulates metamorphosis in marine larvae. Studies of TBP exposure to phytoplankton reveals that TBP induces the release of intracellular calcium stores (Whalen et al., 2018). TBP exposure to mammal microsomes triggers Ca2+ efflux by activating the Ryanodine receptor, RyR1, and inhibiting microsomal sarcoplasmic/endoplasmic reticulum Ca2+ ATPase, SERCA1a (Zheng et al., 2018). Calcium signalling and membrane potential depolarization have been linked to the induction of Hydroides metamorphosis (Carpizo-Ituarte and Hadfield, 1998; Holm et al., 1998; Chen et al., 2012). Furthermore, exposure of a calcium ionophore to the larvae of the sea urchin, Strongylocentrotus purpuratus, resulted in a similar percentage of metamorphosis as compared to TBP-stimulated metamorphosis in Hydroides (Amador-Cano et al., 2006). Taken together, these studies provide a potential link between TBP and calcium signalling mediating invertebrate larvae metamorphosis.
Mutation of bmp2 in P. luteoviolacea HI1 has no apparent effect on Hydroides or Hydractinia larvae
We found that mutation of the bmp2 brominase in P. luteoviolacea had no effect on Hydroides or Hydractinia larvae under the conditions tested in this work. Although we observed Hydroides metamorphosis in response to purified TBP, mutation of bmp2 had no effect on Hydroides metamorphosis in response to P. luteoviolacea biofilms under the conditions tested. These results suggest that MACs from P. luteoviolacea biofilms are the primary stimulant of Hydroides metamorphosis while TBP from P. luteoviolacea biofilms has no effect. Our results show that phenotypic responses can be very different when comparing exposure to purified factors versus live bacteria where genetic interrogation is possible.
While our investigations show that P. luteoviolacea can produce TBP, we found that TBP production by P. luteoviolacea biofilms does not impact Hydroides metamorphosis. One possible explanation is that TBP is not produced at the same concentration as the coral metamorphosis-inducing Pseudoalteromonas under the conditions tested (Tebben et al., 2011; Sneed et al., 2014). We found that P. luteoviolacea grown in different media [seawater tryptone (SWT) and Marine Broth (MB)] produced significantly different concentrations of TBP in our study (Fig. S1a). Differences in concentrations of available bromine may account for the measured differences. SWT contains a 0.056 g/L concentration of potassium bromide while MB contains at higher concentration of 0.080 g/L. Furthermore, many strains of P. luteoviolacea contain the bmp gene cluster (bmp1–10) and produce a suite of polybrominated natural compounds including the antibacterial compound, pentabromopseudilin (Laatsch and Pudleiner, 1989; Busch et al., 2019), and its associated monomeric molecules, such as TBP (Agarwal et al., 2014). Importantly, strains of Pseudoalteromonas capable of inducing coral larvae from TBP extract possess a truncated version of the bmp gene cluster (bmp1–4,9,10) (Gamal et al., 2016) that produce TBP almost exclusively. Future experiments with P. luteoviolacea and a truncated bmp gene cluster (bmp1–4, 9 and 10 only) could elucidate the function of TBP-producing pseudoalteromonads and their potential effect on the larvae of different animals.
MACs are a double-edged sword, depending on the animal
Here, we show that P. luteoviolacea MACs stimulate Hydroides metamorphosis while they are lethal to Hydractinia larvae. MACs carry two characterized effector proteins; one effector called Mif1 that stimulates Hydroides metamorphosis (Ericson et al., 2019), and another effector called Pne1 that is toxic to insect and mouse cell lines ex vivo (Rocchi et al., 2019). Interestingly, MACs were also observed to be lethal in Hydroides at high biofilm densities and crude extract concentrations (Shikuma et al., 2014). While the ecological role of MACs has not yet been determined, the range of phenotypes (i.e., metamorphosis and death) in response MACs demonstrates that they can elicit positive or negative phenotypes depending on the concentration tested and animal type. Future work into the function of Mif1, Pne1 and other putative MACs effectors could help to explain how each effector elicits a specific phenotypic response in different animals or cells.
Our finding that P. luteoviolacea mutants lacking macB induce Hydractinia metamorphosis suggests that P. luteoviolacea produces one or more additional uncharacterized factor(s) that stimulate Hydractinia metamorphosis. This finding may not be surprising as other Pseudoalteromonas and Alteromonas species have been previously isolated from Hydractinia echinata and induce their metamorphosis (Leitz and Wagner, 1993; Klassen et al., 2015a; Klassen et al., 2015b; Guo et al., 2017). Furthermore, a recent study found that purified (lyso) phospholipids and polysaccharides are strong inducers of Hydractinia metamorphosis (Guo et al., 2019). Other recent studies have implicated outer membrane vesicles and lipopolysaccharides as bacterial stimulants of metamorphosis (Freckelton et al., 2017, 2019). Hydractinia is found in temperate oceans, while P. luteoviolacea was isolated from a tropical environment. Although P. luteoviolacea HI1 and Hydractinia were not isolated from the same environment, both partners of this model interaction are genetically tractable (Huang et al., 2012; Sanders et al., 2018) and could serve as a strong platform for determining mechanisms underlying bacteria-induced metamorphosis in cnidarians.
Genes encoding MACs are part of the core P. luteoviolacea pangenome
In this work, we found that many P. luteoviolacea strains possess genes required for the biosynthesis of two known metamorphosis-inducing factors (TBP and MACs). We show that MACs genes are found in all sequenced P. luteoviolacea strains, suggesting that MACs biosynthesis genes may be a conserved feature of the P. luteoviolacea pangenome. Extracellular contractile injection systems may be a common mechanism of host–microbe interactions, as genes encoding structures related to MACs have been found in diverse bacteria and archaea (Böck et al., 2017; Chen et al., 2019), including Bacteroidales bacteria from the human gut (Rojas et al., 2020). Although the 19 strains of P. luteoviolacea we analysed possess genes encoding MACs, it remains to be tested whether they are capable of producing a functional contractile injection system that deploys effectors into target cells. A previous study tested the type strain DSM 6061/ATCC 33492/NCIMB 1893 (Fig. 1B) and found the strain was unable to induce metamorphosis in Hydroides (Huang et al., 2012). We find that the DSM 6061 strain possesses the known genetic components necessary to produce MACs (Fig. 1A; bolded genes: macB, macS, macT, and mif1) with high nucleotide identity (Table S1). Interestingly, strain H2 (Fig. 1B; clade 1, bold), which is within the same lineage as DSM 6061, induces the metamorphosis of the coral Pocillopora damicornis through a yet undetermined mechanism (Tran and Hadfield, 2011). These results raise the possibility that P. luteoviolacea could elicit a positive or negative interaction with marine larvae depending on the expression of its various factor arsenal. Future studies into the regulation of MACs and TBP expression under laboratory growth conditions could explain differences between the metamorphosis inducing capabilities among P. luteoviolacea strains.
Future directions and challenges
The role that bioactive products play in the ecology of Pseudoalteromonas species still requires significant investigation. In the environment, pseudoalteromonads have been found associated with diverse marine plants and animals (Holmström and Kjelleberg, 1999; Bowman, 2007) and possess diverse antagonistic properties; for example, P. luteoviolacea was shown to inhibit the growth of other marine bacteria, algae (Holmström et al., 2002; Rypien et al., 2010), fungi (Holmström et al., 2002; Atencio et al., 2018) and here we show that P. luteoviolacea is lethal to Hydractinia larvae. At the same time, P. luteoviolacea was shown to stimulate the metamorphosis of corals (Tran and Hadfield, 2011), urchins (Huggett et al., 2006) and tubeworms (Shikuma et al., 2014) in independent laboratory studies. The factors of P. luteoviolacea that facilitate or inhibit metamorphosis beyond the organisms investigated within this study remain unknown and are interesting targets for future work. For example, it has been shown that Histamine derived from algae or its associated microbes induces metamorphosis in sea urchins (Swanson et al., 2004), but Huggett et al. show that P. luteoviolacea can induce metamorphosis as well (Huggett et al., 2006). It is currently not clear whether P. luteoviolacea produces histamine which induces metamorphosis, or a potential other bacterial factor may be capable of inducing metamorphosis in sea urchins.
The capability of P. luteoviolacea to facilitate or inhibit settlement and metamorphosis leads us to question which underlying molecular processes enable bacterial factors to influence some animals to metamorphose, but not others. These results suggest that selective inductive capabilities elicited by bacteria could have an influence on animal recruitment in the environment. Future investigations into the molecular targets and regulation of these bioactive factors in natural biofilm assemblages may help shed light on their ecological role. However, before these implications can be fully interpreted, we must consider ecologically relevant concentrations of the bacteria and their factors, their distribution in varying environments, and the potential role of the bacteria and their associated factors in natural assemblages among other organisms.
Conclusion
Pseudoalteromonas luteoviolacea’s ability to produce diverse bioactive compounds and distinct metamorphosis-inducing factors makes this bacterium an interesting model to study bacteria-induced metamorphosis in animals from different phyla. Our results emphasize that there is a complex set of interactions between bacteria, the factors they produce and animal responses, even when studied under controlled laboratory conditions. Using approaches like those used in this work to compare and identify the effects of different bacterial factors on metamorphosis may aid in unravelling this complexity and could provide a deeper understanding of the molecular underpinnings of bacteria-induced metamorphosis in divergent animals.
Experimental procedures
Construction of Δbmp2 and ΔmacBΔbmp2 mutants
Using a double-homologous recombination technique as described previously (Shikuma et al., 2014; Ericson et al., 2019; Rocchi et al., 2019), we created P. luteoviolacea HI1 in-frame deletion strains of the bmp2 (brominase) gene, shown previously to be required for TBP production (Agarwal et al., 2014; Gamal et al., 2016), the macB gene, encoding an essential structural component of the MACs baseplate and a ΔmacbΔbmp2 mutant that is unable to produce both MACs and TBP. We complemented the bmp2 mutant by constitutively expressing bmp2 from a plasmid in trans (Lee et al., 1998). A list of strains, plasmids, and primers constructed and used in this study can be found in Tables S2 and S4.
Detection of TBP production by P. luteoviolacea HI1
Pseudoalteromonas luteoviolacea was grown in two growth medias, SWT (35.9 g/L Instant Ocean, 2.5 g/L Bacto Tryptone, 1.5 g/L Bacto Yeast and 1.5 ml/L glycerol) and MB (BD 2216) to address the differences in growth conditions previously used to describe MACs (Huang et al., 2012; Shikuma et al., 2014) and TBP (Agarwal et al., 2014; Sneed et al., 2014). Single colonies of wild type and Δbmp2 were inoculated in triplicate, grown in 5 ml SWT and MB media, and incubated shaking (200 rpm) at 28°C for 16 h. The cultures were extracted twice with an equal volume of ethyl acetate (EtOAc) and concentrated under a stream of nitrogen. The samples were re-suspended in 100 μl methanol, filtered in a 0.2 μm column and 10 μl of the sample was injected into a Luna C18 reversed-phase analytical HPLC column (5 μm, 250 mm × 4.6 mm, Phenomenex; CA, USA). TBP was measured on an Accurate Mass QtoF LC–MS/MS (Agilent 6530 Accurate Mass), run at 0.5 ml/min in negative mode. Eluent was detected using electrospray ionization-mass spectrometry monitoring m/z 150–2200 in negative mode with a speed of 32 500 m/z per second. A solvent system of acetonitrile and water both containing 0.1% formic acid (v/v) was used. Samples were eluted over a 30 min method with a gradient from 10% to 70% acetonitrile over 15 min, 70%–80% over the next 10 min, and then immediately to 100% for 5 min before returning back to 10% acetonitrile. Quantification of TBP was based on the relative intensities of 2,3,4,5-TBP to synthetic standards. 2,3,4,5-TBP was synthesized following the procedure as previously described (Chekan et al., 2019).
Culture of Hydroides and Hydractinia
Hydroides elegans adults were collected from Quivira Basin, San Diego, CA, USA. The larvae were spawned and fed living Isochrysis cultures daily (Carolina Cat# 153180, NC, USA) as previously described (Nedved and Hadfield, 2008; Shikuma et al., 2014). Larvae were maintained in beakers containing filtered artificial seawater (ASW) (Instant Ocean Cat # SS15–10, VA, USA) at 35 PSU water until competent (between 6 and 8 days).
Colonies of H. symbiolongicarpus were maintained, attached to microscope slides in ASW at 29 PSU at 20°C, a light:dark cycle of 14:10 with constant aeration, fed 4 days a week with 3–5 days old brine shrimp (artemia) (Carolina, Cat# 142242), and twice a week with frozen blended oyster. ASW was changed 5 days a week. Embryos were collected 2 h after the onset of light and maintained in the above-mentioned conditions inside of 100 mm plastic Petri dishes with ASW 29 PSU supplemented with ampicillin 100 μg ml−1 and kanamycin 5 μg ml−1 for 7 days. Prior to the metamorphosis assay, competent larvae were transferred to autoclaved ASW 29 PSU to reduce the load of bacteria and antibiotics.
Biofilm metamorphosis assay
Single species biofilms of P. luteoviolacea were produced and tested for their ability to induce metamorphosis as previously described (Huang and Hadfield, 2003; Shikuma et al., 2014). Briefly, bacterial strains were struck onto SWT and MB agar plates and incubated overnight at 28°C. Single colonies were inoculated into 5 ml of SWT and MB broth and incubated overnight for 14–16 h with agitation (200 rpm). Cells were removed from the culture, pelleted at 4000 g, and washed twice and re-suspended with ASW. Cell density was adjusted to OD600 of 1 (approximately 107–108 cells ml−1) for Hydractinia and was further diluted to OD 0.5 for Hydroides (1:1 with ASW) to evoke optimal metamorphic responses. Aliquots of 100 μl were added to 96-well plates to form biofilms over a 2-h incubation period at room temperature. The excess culture and unattached cells were removed from each well. Each well contained 20–40 competent larvae (6–8 days old) and filtered ASW with a final volume of 100 μl for Hydroides and 200 μl for Hydractinia. The percentage of metamorphosis was scored after 24 h (Hydroides) and 72 h (Hydractinia). While the complete metamorphosis of Hydractinia can occur 24 h post-induction (Seipp et al., 2007), the death phenotype produced similar phenotypes during early differentiation. This influenced qualitative scoring daily and quantitative scoring after 72 h. Four technical replicates of each treatment and three biological replicates were performed on separate occasions. Both media conditions produced similar metamorphosis outcomes for Hydroides and Hydractinia. Graphs displayed for metamorphosis assays represent the media that produced the most prominent results. We display biofilm metamorphosis assays with Hydroides grown in MB media and Hydractinia grown in SWT media.
Exogenous TBP metamorphosis assays
The pure 2,3,4,5-TBP standard was synthesized as previously described (Zheng et al., 2018). Eleven milligrams of TBP was re-suspended in acetonitrile (0.0059 M) and was diluted into ASW (35 PSU for Hydroides and 29 PSU for Hydractinia), which contained a final acetonitrile concentration of 2% (v/v). The concentrations tested were: 1000, 750, 500, 250, 100, 10, 1 and 0.1 nM. We saw no phenotype for the lowest concentrations tested and therefore did not include them in the final analysis. The diluted TBP was aliquoted (100 μl) into 96-well plates. Approximately 20–40 competent larvae (Hydroides and Hydractinia) were added to each well. Four technical replicates were performed for each treatment with Hydractinia and eight technical replicates were performed with Hydroides. Three and five biological replicates were repeated on different occasions for Hydractinia and Hydroides respectively.
Comparative genomics
Pseudoalteromonas luteoviolacea draft and complete genomes were downloaded from NCBI (Table S1). The representations of the MACs and TBP gene clusters in P. luteoviolacea were created using Easyfig (Sullivan et al., 2011) (v2.2.2_OS). Fasta files containing the gene clusters for MACs (Shikuma et al., 2014) and TBP (Gamal et al., 2016) were used to perform a tblastx on the P. luteoviolacea HI1 strain. MACs genes, macB, macS, macT1, and mif1 were selected to perform a blastn (Camacho et al., 2009) against all of the other sequenced strains for P. luteoviolacea. The sequenced genomes were formatted into a blast database through the galaxy server (Cock et al., 2015).
All P. luteoviolacea genomes were then analysed through the Anvi’o pangenomics (v6) pipeline (Eren et al., 2015; Delmont and Eren, 2018) using their publicly available tutorials. The pangenome was generated using the parameter values (Van Dongen and Abreu-Goodger,−2012; Benedict et al., 2014): –minbit 0.5; mcl-inflation −10; −use-ncbi-blast. Once the pangenome was generated, the sequences for bacterial hmm hits were concatenated. A total of 71 ribosomal bacterial genes (Lee, 2019) (modified as noted here: https://github.com/merenlab/anvio/tree/master/anvio/data/hmm/Bacteria_71) were used to create the phylogeny. The concatenated sequences were aligned using MAFFT and the alignment algorithm G-ins-I (Katoh et al., 2019). A maximum likelihood phylogeny was constructed using the LG + G + I + F substitution model (Guindon et al., 2010) with the Smart Model Selection (Lefort et al., 2017) feature for PhyML from the ATGC bioinformatics webserver. Bootstrap values (100 resamples) were calculated to ensure tree robustness. The tree was manipulated and viewed in iTOL (Letunic and Bork, 2016).
Supplementary Material
Fig. S1. Pseudoalteromonas luteoviolacea HI1 wild type produces TBP, while the Δbmp2 strain does not. (A) Wild type cultures were grown in triplicate in SWT and MB media overnight. The cultures were extracted with two volumes of ethyl acetate and concentrated under a stream of nitrogen gas. Extracts were resuspended in methanol, filtered and injected into a C18 reversed-phase analytical HPLC column. TBP was measured on an Accurate Mass QToF LC–MS/MS, run at 0.5 ml/min in negative mode. Quantification of TBP was determined by integrating the 381.67 mass (m + 4) peak from the isotope distribution of the mass spectrum to enhance the signal to noise ratio. Gaussian statistics were calculated using an unpaired t-test with Welch’s correction, where *p ≦ 0.0325. Error bars represent the standard deviation. (B) The bmp2 complemented cultures were grown overnight in MB media in duplicate and the same extraction and analysis procedures from (A) were used to determine the presence of TBP in the complemented strains. Horizontal lines represent the mean in both (A) and (B).
Table S1. List of Pseudoalteromonas luteoviolacea strains used for Phylogeny and MACs genes used for blastn
Table S2. List of strains and plasmids used in this study.
Table S3. Summary table of Hydractinia and Hydroides metamorphosis assay results.
Table S4. List of Primers used in this study
Acknowledgements
We would like to thank Dr. Giselle Cavalcanti, Kyle Malter, Dr. Linda Wegley-Kelly, and our anonymous reviewers for their valuable feedback on the manuscript. We thank Dr. Matthew Nicotra from the University of Pittsburgh for generously gifting Hydractinia symbiologicarpus colonies and providing us with significant technical support. The schematics were created with Biorender.com. This work was supported by the National Science Foundation (2017232404, A.T.A.; 1942251, N.J.S. and OCE-1837116, B.S.M.), the Office of Naval Research (N00014-20-1-2120, N.J.S.; N00014-17-1-2677, N.J.S. and N00014-16-1-2135, N.J.S.), the National Institutes of Health (R01-ES030316, B.S.M. and T32-GM067550, T.N.P.), and the Alfred P. Sloan Foundation, Sloan Research Fellowship (N.J.S.).
Data Availability Statement
The P. luteoviolacea genomes were downloaded from NCBI GenBank, and Table S1 provides a list of accessions.
References
- Agarwal V, El Gamal AA, Yamanaka K, Poth D, Kersten RD, Schorn M, et al. (2014) Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat Chem Biol 10: 640–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador-Cano G, Carpizo-Ituarte E, and Cristino-Jorge D ( 2006) Role of protein kinase C, G-protein coupled receptors, and calcium flux during metamorphosis of the sea urchin Strongylocentrotus purpuratus. Biol Bull 210: 121–131. [DOI] [PubMed] [Google Scholar]
- Asahina AY, and Hadfield MG (2015) Draft genome sequence of Pseudoalteromonas luteoviolacea HI1, determined using Roche 454 and PacBio single-molecule real-time hybrid sequencing. Genome Announc 3: e01590–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atencio LA, Dal Grande F, Young GO, Gavilán R, Guzmán HM, Schmitt I, et al. (2018) Antimicrobial-producing Pseudoalteromonas from the marine environment of Panama shows a high phylogenetic diversity and clonal structure. J Basic Microbiol 58: 747–769. [DOI] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, and Mekalanos JJ (2012) Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483: 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict MN, Henriksen JR, Metcalf WW, Whitaker RJ, and Price ND (2014) ITEP: an integrated toolkit for exploration of microbial pan-genomes. BMC Genomics 15: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böck D, Medeiros JM, Tsao HF, Penz T, Weiss GL, Aistleitner K, et al. (2017) In situ architecture, function, and evolution of a contractile injection system. Science 357: 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JP (2007) Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar Drugs 5: 220–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch J, Agarwal V, Schorn M, Machado H, Moore BS, Rouse GW, et al. (2019) Diversity and distribution of the bmp gene cluster and its Polybrominated products in the genus Pseudoalteromonas. Environ Microbiol 21: 1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, and Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpizo-Ituarte E, and Hadfield MG (1998) Stimulation of metamorphosis in the polychaete Hydroides elegans Haswell (Serpulidae). Biol Bull 194: 14–24. [DOI] [PubMed] [Google Scholar]
- Cavalcanti GS, Alker AT, Delherbe N, Malter KE, and Shikuma NJ (2020) The influence of bacteria on animal metamorphosis. Annu Rev Microbiol 74: 137–158. [DOI] [PubMed] [Google Scholar]
- Chekan JR, Lee GY, El Gamal A, Purdy TN, Houk KN, and Moore BS (2019) Bacterial tetrabromopyrrole debrominase shares a reductive dehalogenation strategy with human thyroid deiodinase. Biochemistry, Vol. 58(52), pp. 5329–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Song N, Liu B, Zhang N, Alikhan NF, Zhou Z, et al. (2019) Genome-wide identification and characterization of a superfamily of bacterial extracellular contractile injection systems. Cell Rep 29: 511–521.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZF, Wang H, and Qian PY (2012) Characterization and expression of calmodulin gene during larval settlement and metamorphosis of the polychaete Hydroides elegans. Comp Biochem Physiol - B Biochem Mol Biol 162: 113–119. [DOI] [PubMed] [Google Scholar]
- Cock PJA, Chilton JM, Grüning B, Johnson JE, and Soranzo N (2015) NCBI BLAST+ integrated into galaxy. Gigascience 4: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress BF, Erkert KA, Barquera B, and G Koffas MA (2013) Draft genome sequence of Pseudoalteromonas luteoviolacea strain B (ATCC 29581). Genome Announc 1: 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmont TO, and Eren AM (2018) Linking pangenomes and metagenomes: the Prochlorococcus meta-pangenome. PeerJ 6: e4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, and Delmont TO (2015) Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3: e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson CF, Eisenstein F, Medeiros JM, Malter KE, Cavalcanti GS, Zeller RW, et al. (2019) A contractile injection system stimulates tubeworm metamorphosis by translocating a proteinaceous effector. Elife 8: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank U, Leitz T, and Müller WA (2001) The hydroid Hydractinia: a versatile, informative cnidarian representative. Bioessays 23: 963–971. [DOI] [PubMed] [Google Scholar]
- Freckelton ML, Nedved BT, Cai Y-S, Cao S, Turano H, Alegado RA, and Hadfield MG (2019) Bacterial lipopolysaccharide induces settlement and metamorphosis in a marine larva. bioRxiv 851519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freckelton ML, Nedved BT, and Hadfield MG (2017) Induction of invertebrate larval settlement; different bacteria, different mechanisms? Sci Rep 7: 42557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamal A, Agarwal V, Diethelm S, Rahman I, Schorn MA, Sneed JM, et al. (2016) Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair. Proc Natl Acad Sci U S A 113: 3797–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier MJ, and Flatau GN (1976) Antibacterial activity of marine violet-pigmented Alteromonas with special reference to the production of brominated compounds. Can J Microbiol 22: 1612–1619. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, and Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Guo H, Rischer M, Sperfeld M, Weigel C, Menzel KD, Clardy J, and Beemelmanns C (2017) Natural products and morphogenic activity of γ-Proteobacteria associated with the marine hydroid polyp Hydractinia echinata. Bioorganic Med Chem 25: 6088–6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Rischer M, Westermann M, and Beemelmanns C (2019) Two distinct bacterial biofilm components trigger metamorphosis in the colonial hydrozoan Hydractinia echinata. bioRxiv 2019.12.23.887182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield M, and Paul VJ (2001) Natural chemical cues for settlement and metamorphosis of marine-invertebrate larvae. In Marine Chemical Ecology, 13: 431–461. Boca Raton: CRC Press. [Google Scholar]
- Hadfield M, Unabia C, Smith C, and Michael TM (1994) Settlement preferences of the ubiquitous fouler Hydroides elegans. Recent Developments in Biofouling Control: 65–74. Rotterdam, Netherlands: Taylor & Francis. [Google Scholar]
- Hadfield MG (2011) Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Ann Rev Mar Sci 3: 453–470. [DOI] [PubMed] [Google Scholar]
- Holm ER, Nedved BT, Carpizo-Ituarte E, and Hadfield MG (1998) Metamorphic-signal transduction in Hydroides elegans (Polychaeta: Serpulidae) is not mediated by a G protein. Biol Bull 195: 21–29. [DOI] [PubMed] [Google Scholar]
- Holmström C, Egan S, Franks A, McCloy S, and Kjelleberg S (2002) Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiol Ecol 41: 47–58. [DOI] [PubMed] [Google Scholar]
- Holmström C, and Kjelleberg S (1999) Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol Ecol 30: 285–293. [DOI] [PubMed] [Google Scholar]
- Huang S, and Hadfield MG (2003) Composition and density of bacterial biofilms determine larval settlement of the polychaete Hydroides elegans. Mar Ecol Prog Ser 260: 161–172. [Google Scholar]
- Huang Y, Callahan S, and Hadfield MG (2012) Recruitment in the sea: bacterial genes required for inducing larval settlement in a polychaete worm. Sci Rep 2: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett MJ, Williamson JE, De Nys R, Kjelleberg S, and Steinberg PD (2006) Larval settlement of the common Australian sea urchin Heliocidaris erythrogramma in response to bacteria from the surface of coralline algae. Oecologia 149: 604–619. [DOI] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, and Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20: 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen JL, Rischer M, Wolf T, Guo H, Shelest E, and Clardy J (2015a) Genome sequences of three Pseudoalteromonas strains (P1–8, P1–11, and P1–30), isolated from the marine hydroid Hydractinia echinata. Genome Announc 3: 2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen JL, Wolf T, Rischer M, Guo H, Shelest E, Clardy J, and Beemelmanns C (2015b) Draft genome sequences of six Pseudoalteromonas Strains, P1–7a, P1–9, P1–13-1a, P1–16-1b, P1–25, and P1–26, which induce larval settlement and metamorphosis in Hydractinia echinata. Genome Announc 3: 1477–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laatsch H, and Pudleiner H (1989) Marine Bakterien, I. Synthese von Pentabrompseudilin, einem cytotoxischen Phenylpyrrol aus Alteromonas luteo-violaceus. Liebigs Ann. Chem. 1989: 863–881. [Google Scholar]
- Lee AK, and Falkow S (1998) Constitutive and inducible green fluorescent protein expression in Bartonella henselae. Infect Immun 66: 3964–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MD (2019) GToTree: a user-friendly workflow for phylogenomics. Bioinformatics 35: 4162–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort V, Longueville J-E, and Gascuel O (2017) SMS: smart model selection in PhyML. Mol Biol Evol 34: 2422–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitz T, and Wagner T (1993) The marine bacterium Alteromonas espejiana induces metamorphosis of the hydroid Hydractinia echinata. Mar Biol 115: 173–178. [Google Scholar]
- Letunic I, and Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maansson M, Vynne NG, Klitgaard A, Nybo JL, Melchiorsen J, Nguyen DD, et al. (2016) An integrated metabolomic and genomic mining workflow to uncover the biosynthetic potential of bacteria. mSystems 1: e00028–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedved BT, and Hadfield MG (2008) Hydroides elegans (Annelida: Polychaeta): a model for biofouling research. In Marine and Industrial Biofouling. Berlin, Heidelberg: Springer, pp. 203–217. [Google Scholar]
- Negri AP, Webster NS, Hill RT, and Heyward AJ (2001) Metamorphosis of broadcast spawning corals in response to bacteria isolated from crustose algae. Mar Ecol Prog Ser 223: 121–131. [Google Scholar]
- Rocchi I, Ericson CF, Malter KE, Zargar S, Eisenstein F, Pilhofer M, et al. (2019) A bacterial phage tail-like structure kills eukaryotic cells by injecting a nuclease effector. Cell Rep 28: 295–301.e4. [DOI] [PubMed] [Google Scholar]
- Rojas MI, Cavalcanti GS, McNair K, Benler S, Alker AT, Cobián-Güemes AG, et al. (2020) A distinct contractile injection system gene cluster found in a majority of healthy adult human microbiomes. mSystems 5(4): e00648–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rypien KL, Ward JR, and Azam F (2010) Antagonistic interactions among coral-associated bacteria. Environ Microbiol 12: 28–39. [DOI] [PubMed] [Google Scholar]
- Sanders SM, Ma Z, Hughes JM, Riscoe BM, Gibson GA, Watson AM, et al. (2018) CRISPR/Cas9-mediated gene knockin in the hydroid Hydractinia symbiolongicarpus. BMC Genomics 19: 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MP, Bendick JA, Holm ER, and Hertel WM (2011) Economic impact of biofouling on a naval surface ship. Biofouling 27: 87–98. [DOI] [PubMed] [Google Scholar]
- Seipp S, Schmich J, Kehrwald T, and Leitz T (2007) Metamorphosis of Hydractinia echinata - natural versus artificial induction and developmental plasticity. Dev Genes Evol 217: 385–394. [DOI] [PubMed] [Google Scholar]
- Shikuma NJ, Antoshechkin I, Pilhofer M, and Newman DK (2016) Stepwise metamorphosis of the tubeworm Hydroides elegans is mediated by a bacterial inducer and MAPK signaling. Proc Natl Acad Sci U S A 113: 10097–10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikuma NJ, Pilhofer M, Weiss GL, Hadfield MG, Jensen GJ, and Newman DK (2014) Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 343: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneed JM, Sharp KH, Ritchie KB, and Paul VJ (2014) The chemical cue tetrabromopyrrole from a biofilm bacterium induces settlement of multiple Caribbean corals. Proc R Soc B Biol Sci 281: 20133086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MJ, Petty NK, and Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27: 1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson RL, Williamson JE, De Nys R, Kumar N, Bucknall MP, and Steinberg PD (2004) Induction of settlement of larvae of the sea urchin Holopneustes purpurascens by histamine from a host alga. Biol Bull 206: 161–172. [DOI] [PubMed] [Google Scholar]
- Tebben J, Motti CA, Siboni N, Tapiolas DM, Negri AP, Schupp PJ, et al. (2015) Chemical mediation of coral larval settlement by crustose coralline algae. Sci Rep 5: 10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebben J, Tapiolas DM, Motti CA, Abrego D, Negri AP, Blackall LL, et al. (2011) Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas bacterium. PLoS One 6: e19082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thøgersen MS, Delpin MW, Melchiorsen J, Kilstrup M, Månsson M, Bunk B, et al. (2016) Production of the bioactive compounds violacein and indolmycin is conditional in a maeA mutant of Pseudoalteromonas luteoviolacea S4054 lacking the malic enzyme. Front Microbiol 7: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran C, and Hadfield MG (2011) Larvae of Pocillopora damicornis (Anthozoa) settle and metamorphose in response to surface-biofilm bacteria. Mar Ecol Prog Ser 433: 85–96. [Google Scholar]
- Unabia CRC, and Hadfield MG (1999) Role of bacteria in larval settlement and metamorphosis of the polychaete Hydroides elegans. Mar Biol 133: 55–64. [Google Scholar]
- Van Dongen S, and Abreu-Goodger C (2012) Using MCL to extract clusters from networks. Methods Mol Biol 804: 281–295. [DOI] [PubMed] [Google Scholar]
- Vynne NG, Mansson M, and Gram L (2012) Gene sequence based clustering assists in dereplication of Pseudoalteromonas luteoviolacea strains with identical inhibitory activity and antibiotic production. Mar Drugs 10: 1729–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Smith LD, Heyward AJ, Watts JEM, Webb RI, Blackall LL, and Negri AP (2004) Metamorphosis of a Scleractinian coral in response to microbial biofilms. Appl Environ Microbiol 70: 1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen KE, Kirby C, Nicholson RM, O’Reilly M, Moore BS, and Harvey EL (2018) The chemical cue tetrabromopyrrole induces rapid cellular stress and mortality in phytoplankton. Sci Rep 8: 15498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, He W, Li H, Yan Y, and Lin C (2010) Larval settlement and metamorphosis of the pearl oyster Pinctada fucata in response to biofilms. Aquaculture 306: 334–337. [Google Scholar]
- Zheng J, Mckinnie SMK, El Gamal A, Feng W, Dong Y, Agarwal V, et al. (2018) Organohalogens naturally biosynthesized in marine environments and produced as disinfection byproducts Alter Sarco/endoplasmic reticulum Ca2+ dynamics. Environ Sci Technol 52: 5469–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Pseudoalteromonas luteoviolacea HI1 wild type produces TBP, while the Δbmp2 strain does not. (A) Wild type cultures were grown in triplicate in SWT and MB media overnight. The cultures were extracted with two volumes of ethyl acetate and concentrated under a stream of nitrogen gas. Extracts were resuspended in methanol, filtered and injected into a C18 reversed-phase analytical HPLC column. TBP was measured on an Accurate Mass QToF LC–MS/MS, run at 0.5 ml/min in negative mode. Quantification of TBP was determined by integrating the 381.67 mass (m + 4) peak from the isotope distribution of the mass spectrum to enhance the signal to noise ratio. Gaussian statistics were calculated using an unpaired t-test with Welch’s correction, where *p ≦ 0.0325. Error bars represent the standard deviation. (B) The bmp2 complemented cultures were grown overnight in MB media in duplicate and the same extraction and analysis procedures from (A) were used to determine the presence of TBP in the complemented strains. Horizontal lines represent the mean in both (A) and (B).
Table S1. List of Pseudoalteromonas luteoviolacea strains used for Phylogeny and MACs genes used for blastn
Table S2. List of strains and plasmids used in this study.
Table S3. Summary table of Hydractinia and Hydroides metamorphosis assay results.
Table S4. List of Primers used in this study
Data Availability Statement
The P. luteoviolacea genomes were downloaded from NCBI GenBank, and Table S1 provides a list of accessions.


