Abstract
Background
Dynamics and persistence of neutralizing and non-neutralizing antibodies can give us the knowledge required for serodiagnosis, disease management, and successful vaccine design and development. The disappearance of antibodies, absence of humoral immunity activation, and sporadic reinfection cases emphasize the importance of longitudinal antibody dynamics against variable structural antigens.
Methods
In this study, twenty-five healthy subjects working in a SARS-COV-2 serodiagnostic assay development project were enrolled, and their sign and symptoms were followed up to six months. Three subjects showed COVID-19-like symptoms, and three subjects’ antibody dynamics were followed over 120 days by analyzing 516 samples. We have developed 12 different types of in-house ELISAs to observe the kinetics of IgG, IgM, and IgA against four SARS-CoV-2 proteins, namely nucleocapsid, RBD, S1, and whole spike (S1+S2). For the development of these assays, 30–104 pre-pandemic samples were taken as negative controls and 83 RT-qPCR positive samples as positive ones.
Results
All three subjects presented COVID-19-like symptoms twice, with mild symptoms in the first episode were severe in the second, and RT-qPCR confirmed the latter. The initial episode did not culminate with any significant antibody development, while a multifold increase in IgG antibodies characterized the second episode. Interestingly, IgG antibody development concurrent with IgM and IgA and persisted, whereas the latter two weans off rather quickly if appeared.
Conclusion
Antibody kinetics observed in this study can provide a pathway to the successful development of sero-diagnostics and epidemiologists to predict the fate of vaccination currently in place.
Keywords: antibody dynamics, COVID-19, SARS-CoV-2, reinfection, vaccination
Introduction
Worldwide spreading of SARS-CoV-2 caused by the novel coronavirus has a high infectious rate and has already claimed more than 3.5 million deaths till 30 May 2021.1 The previous two severe coronavirus infections in humans, ie, SARS-CoV and MERS-CoV, were epidemic in nature and geographically isolated.2,3
Symptoms associated with prevailing coronavirus infections that cause seasonal colds in humans include sore throat, cough, feverishness, congestion, wheezing, sputum, hoarseness, chills, dyspnea, diarrhea, rhinorrhea, sleep disturbance, muscle pain, fatigue, and joint pain.4–7 Similar to other ordinary human coronavirus infections, SARS-CoV-2 cases can be asymptomatic or symptomatic.8 COVID-19 symptomatic cases show symptoms similar to but often more severe than those presented by other common human coronaviruses. Additional symptoms include ageusia and anosmia, blood pressure fluctuation, myalgia, and severe respiratory complications.9,10 In extreme cases, patients may experience septic shock, metabolic acidosis, coagulation dysfunction, bleeding, organ failures, and even death.11,12
SARS-CoV-2 employs multiple tactics that enhance its prevalence rate. Escape mutation by the virus reduces the immunoglobulin-binding capacity, which can render certain vaccines less effective by reducing the efficacy of neutralizing antibodies, resulting in reinfection.13–17 Additionally, its virulence is enhanced by anchoring non-structural proteins (nsp) in double-membrane vesicles and capping mRNA, respectively, increasing its persistence capability and protecting the genome from intracellular viral host innate immune response.18–22 More than 4000 mutations have been reported for SARS-CoV-2, and the recent variants reported in the UK show cluster mutations in spike with escape mutation from South African variants, challenging long-term efficacies of spike-based vaccines.14,15,23,24
Patients generally develop antibody and memory T-helper cells against that particular virus, there has been a report of the decline of those cells along with Treg cells in severe cases of Covid-19.25,26 Additionally, observations of quick disappearances of neutralizing antibodies and activation of T-cell mediated immunity to eliminate SARS-CoV-2 without involving B-cell mediated immunity in multiple cases have also been reported.27,28 In line with these observations, reinfection/relapse with SARS-CoV-2 has been invoked to explain the recurring presence of SARS-CoV-2 RNA after testing negative by RT-qPCR, a gold standard test for COVID-19 diagnosis.29,30
Recently there has been a few reports of reinfection, there is still a raging debate on its overall frequency of occurrence.14,16,31–43 This raises questions about the efficacy of an effective vaccine. Understanding the behavioral pattern of antibody dynamics against SARS-CoV-2 antigens in a longitudinal study can shine some light on the veracity, or otherwise, of reinfection. Moreover, the success of serodiagnostic relies on proper antibody dynamic studies against proteins under consideration.
This article analyzed the kinetics of antibodies against four structural proteins of SARS-CoV-2 in three RT-qPCR positive patients. Our four monthly observations started 60 days before being RT-qPCR positive, during which time they could have been exposed to coronaviruses and showed coronavirus disease-like symptoms.
Method and Materials
Case History and Clinical Characteristics
A cohort comprising twenty-five people, 15 male, and ten female, was selected for the study. The emergence of COVID-19 in China and its designation as a pandemic in 2020, this team started developing diagnostic kits for SARS-CoV-2 from March 2020 onwards. The subjects were regularly checked for any signs and symptoms, and were under serosurveillance to ensure the quality of the working environment. Among study subjects the three male who were actively involved in developing coronavirus diagnostics and frequently examined for suspected and confirmed COVID-19 through blood and nasopharyngeal, samples, showed COVID-19 like symptoms, were selected for the study. The studied subjects were healthy with no history of chronic disease or administration of any immunosuppressive drugs. Apart from direct exposure to infectious samples, Subject 01 (S01) was enrolled because he had exposure to the 2003 SARS-CoV outbreak and had an accidental exposure to SARS-CoV-2 positive serum parenterally, during inactivation of patient serum at the beginning of May. This led to the development of mild fibrosis around the inoculation site. Subject 02 (S02) and Subject 03 (S03) presented symptoms associated with coronavirus infection at the end of May 2020. This includes high- and low-grade fever, diarrhea, asthenia, sore throat, shortness of breath, and dry cough (Supplementary Table 1). Amid the ongoing pandemic, S02 and S03 took unproven treatment regimens, including ivermectin, doxycycline, paracetamol, normal saline, and vitamin-C and zinc.
Between Mid-June to the first week of July 2020, none of the subjects showed any symptoms associated with COVID-19 except for S02, who experienced sporadic rash and allergic reactions. However, after about 50 days from first exposure or symptoms, all three presented COVID-19-like symptoms, such as high-grade fever, blood pressure fluctuation, ageusia, anosmia, nausea, severe weakness, confusion, and muscle pain followed by sore throat, dry cough, slight respiratory distress, diarrhea, insomnia, and increase in urination (Supplementary Table 1). They tested positive for COVID-19 by RT-qPCR at the end of July.
Reverse Transcriptase Quantitative Real-Time PCR
RT-qPCR was done using patients’ oropharyngeal swabs. Two SARS-CoV-2 specific genes (N1 and N2) for the conserved nucleocapsid (N) protein region were targeted and simultaneously amplified. For total RNA extraction QIAamp Viral RNA Mini Kit was used, and purity of RNA was confirmed spectrophotometrically (Thermo Scientific, NanoDrop 2000c). From total RNA RT-qPCR was carried out using QIAGEN OneStep RT-PCR Kit with CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel.
Blood Sample Preparation to Investigate Antibody Dynamics
The patient’s blood was collected at various time intervals before the primary infection until 120 days. Blood was collected by venipuncture in tubes containing clotting factors. Later, serum was separated by centrifugation at 3500 rpm for 15 minutes at room temperature. The serum samples were stored at −80°C until further investigation. For negative control, two years old 30–104 serum samples from healthy subjects were applied. Moreover, as the positive control, 83 COVID-19 RT-qPCR positive serum samples were used.
Antibody Dynamics Analysis by in-House ELISA
The antibody profile of subjects was determined using in-house ELISA. We primarily investigated both the neutralizing (anti-receptor binding domain (RBD-IgG) and anti-Spike (S1-IgG) and non-neutralizing antibodies (anti-N-IgG) in patients’ serum using our in-house ELISA method.44,45 Briefly, commercially obtained RBD, S1, and N proteins (Sino Biological, China) were coated on ELISA plates (ExtraGene, USA). After blocking, diluted (1:100) serum samples were applied, and SARS-CoV-2 specific human IgG, IgM, and IgA were detected using HRP tagged goat anti-human IgG (The Native Antigen, UK), goat anti-human IgM (Abcam, USA), and goat anti-human polyclonal IgA (Sigma-Aldrich, USA). Furthermore, we investigated antibody dynamics against S1+S2 (Sino Biological, China) using the similar method. Results were obtained by a microplate reader (Thermo Scientific, USA) at 450 nm.
Statistical Methods
Data were presented as either mean, median, and standard deviation. Spearman rank correlation was used to evaluate the bivariate association between different in-house ELISA techniques. An independent sample t-test was used to see the difference in fold change of OD/cut-off value between the first and second episodes of infection. Paired sample t-test was used to see the change in OD/cut-off values of antibodies between the participants. All analyses were performed with Stata 13 (StataCorp, LP, College Station, Texas, USA). The graphical presentation was made using GraphPad Prism 8.3 or MS. Excel. A p-value <0.05 was considered significant.
Results
Twelve different types of ELISA were developed for this study, where IgA, IgM, and IgG antibody fold increased were measured against SARS-CoV-2 N, RBD, S1, and S1+S2 proteins. The sensitivity and specificity of each ELISA were measured separately (Supplementary Tables 2–5), and ROC curves were prepared for OD to Cut-off ratio for 0.95 and 1.0 for each assay (Supplementary Figures 1–4).
Antibodies developed in the three test subjects were measured using an in-house ELISA against SARS-CoV-2 specific antigens, ie, N, S1, whole spike (S1+S2 ECD), and RBD. Serum samples were taken at 14, 16, and 13-time points from S01, S02, and S03, respectively, over four months while observing the dynamics of IgA, IgM, and IgG antibodies against four SARS-CoV-2 specific proteins mentioned above. Graphs were plotted, and correlations were assessed using a total number of 516 observations (Figures 1–3; Supplementary Table 1; Supplementary Figures 4–7).
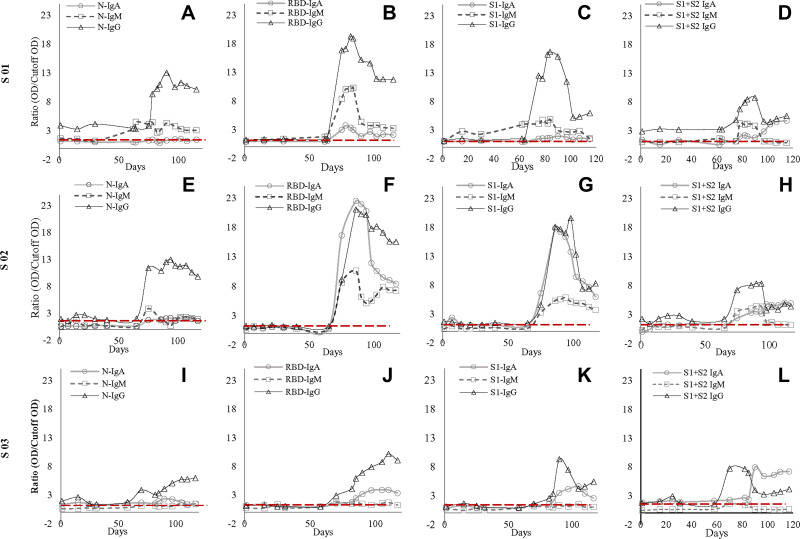
Figure 1.
Antibody dynamic of IgA, IgM, and IgG of the three subjects (S01, S02, S03) against SARS-CoV-2 nucleocapsid (N), receptor-binding domain (RBD), N-terminal of spike protein (S1), and spike protein (S1+S2) (A–L). Figure (A, E and I) represents antibody dynamics against nucleocapsid; (B, G, and J) against RBD; (C, G, and K) against S1; whereas (D, H, and L) against S1+S2 in S01, S02, and S03, respectively. IgG’s increase was consistent in all three subjects though there were some dissimilarities in the IgM and IgA antibody titer among them. However, the second exposure led to a multifold increase of IgG against all four SARS-CoV-2 proteins. In contrast, S01 failed to elicit an increase in IgA against SARS-CoV-2 proteins except for a four-fold increase against whole spike protein. Similarly, S03 failed to elicit IgM response against all four antigens.
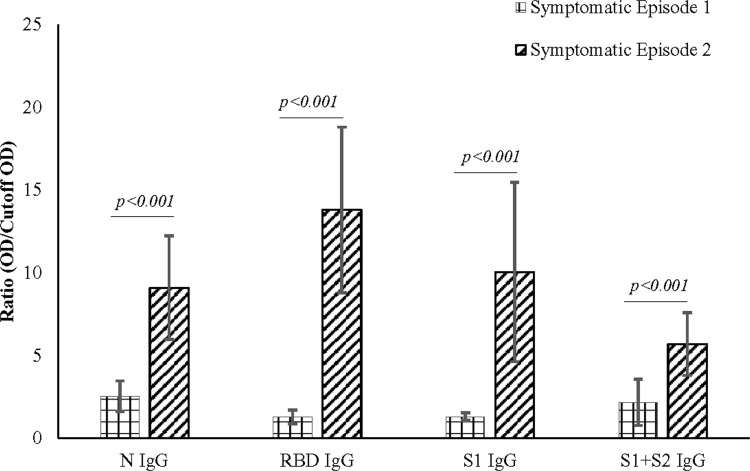
Figure 2.
Antibody expression during symptomatic episode 1 and symptomatic episode 2. IgG expression during symptomatic episode 2 was higher when compared to symptomatic episode 1 in all the subjects studied. The higher levels of antibodies correlate with the severity of the symptoms of the subjects.
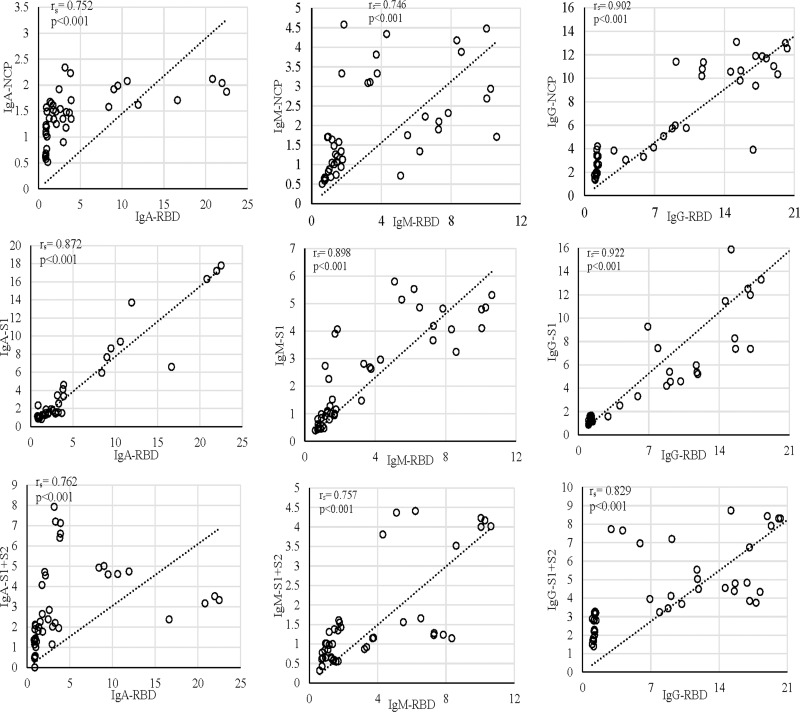
Figure 3.
Correlation of the anti-RBD antibody compared with anti-NCP, anti-S1, and anti-S1+S2 in S01, S02, and S03. In all the subjects, the increase in anti-RBD correlated with the increase in anti-S1. The first episode in all the graphs is signified with the cluster formed at the lower end of the x- and y-axis.
All three subjects showed mild elevation of IgG against N protein in their first episode, which continued to persist till the second episode (Figure 1). In the second episode, the sharp rise of these antibodies was observed, which continued to persist at a 7–12-fold increase compared to the first episode (p<0.001) for about 60 days till the end of our study (Figures 1 and 2). Initial elevation can confer cross-reaction against other coronaviruses, as observed for S01 (Figure 1).46 It is to be noted that the subjects did not undergo SARS-CoV-2 RT-qPCR during the first episode, though they did manifest antibody levels higher than two years old negative samples.
IgM dynamics against N, on the other hand, was variable, where S01 had a higher IgM fold compared to S02 (p<0.001) and S03 (p=0.001), while both S02 and S03 showed no significant difference (data not shown). The subject S03 continued to show a persisted elevation of 1.65-fold anti-N IgA throughout the study (Figure 1). Interestingly, when N-IgA, N-IgM, and N-IgG in all three subjects were compared with RBD antibodies against the respective class, N-IgG showed the highest correlation with RBD-IgG (rs=0.902; p<0.001) (Figure 3). Although, in the individual level, S01 and S03 showed insignificant correlation for IgA, rs=0.262; p=0.336 and rs=0.369; p=0.215, respectively (Supplementary Figure 5).
Concerning RBD and S1, none of the participants presented any increase in the first episode (Figure 1). In contrast, the second episode statistically significantly increased (p<0.001) almost 11-fold (first episode: 1.29±0.42; second episode: 13.80±5.00) in IgG against RBD (Figure 2). Similarly, an almost eightfold increase (first episode: 1.32±0.23; second episode: 10.05±5.42) in IgG against S1 (p<0.001) was also evident (Figure 2). The antibody was dynamic of RBD and S1 highly correlated (rs=0.923; p<0.001) (Figure 3). In all three subjects, after the sharp rise in IgG against RBD and S1 (8–20-fold), it plateaued to a 7–10-fold increase during the two months observation (Figure 1).
Like that of correlation between S1-IgG in all three subjects, S1-IgM also correlated significantly, rs= 0.898; p<0.001, with their respective RBD antibodies (Figure-3). Although anti-RBD and anti-S1 IgM titers in the three subjects varied at the individual level (Figure 1). While S01 and S02 presented more than a threefold increase, such increase was absent in S03 (Figure 1). Similar trend was observed for anti-N IgM (rs= 0.393; p=0.164) in S01 (Figure 1; Supplementary Figure 6) and anti-S1 IgM (rs= 0.632; p=0.021) for S03, compared to their respective RBD antibodies (Figure 1; Supplementary Figure 6).
However, in contrast to IgM and IgG, S02 presented the highest anti-RBD IgA (22.5-fold) and anti-S1 IgA (17.8-fold) increase compared to S01 (6.1-fold, 9.4-fold, respectively) and S03 (5.8-fold, fourfold, respectively) (Figure 1). Moreover, overall anti-S1 IgA (rs= 0.872; p<0.001) increase was comparable to anti-RBD (Figure 1; Figure 3). Although, anti-S1 IgA dynamics of S02 and S03 correlate highly, rs= 0.875; p<0.001 and rs= 0.927; p<0.001, respectively, compared to their RBD-IgA (Figure 1; Supplementary Figure 7), S01 did not present any increase in IgA for either of the SARS-CoV-2 proteins (Figure 1).
Whole spike proteins (S1+S2) have the propensity of expressing domains that may cross-react with other coronaviruses (Figure 1).46,47 In subject S01, the trend of IgM and IgA antibody dynamics against anti-whole spike closely mimics that of anti-N protein (Figure 1; Supplementary Figures 6 and 7). There was an approximately 3.5 and 4-fold increase of anti-whole spike IgM and IgA, respectively, in the second episode. Additionally, anti-whole spike IgG increased significantly (p<0.001) in the second episode (5.70±1.89) when compared with the first episode (2.71±1.40) (Figure 2). The IgM against whole spike proteins persisted for two weeks for subjects S01 and S02 but never increased for S03 (Figure 1). Surprisingly, all three subjects showed abrupt IgA rise against this protein when IgG started to wade off (Figure 1).
Discussion
Our study looks into antibody dynamics of COVID-19 and how they correlate with the disease outcome with symptoms. Fortunately, all three subjects survived the infection but with varying degrees of post-COVID-19 effects. In all three subjects, the first episode of symptoms cannot be conclusively defined as SARS-CoV-2 infection due to the lack of RT-qPCR testing results. However, the subjects presented a pool of symptoms that are common for coronavirus infection, indicating the possibility of being infected with SARS-CoV-2 or with other species of coronavirus.4
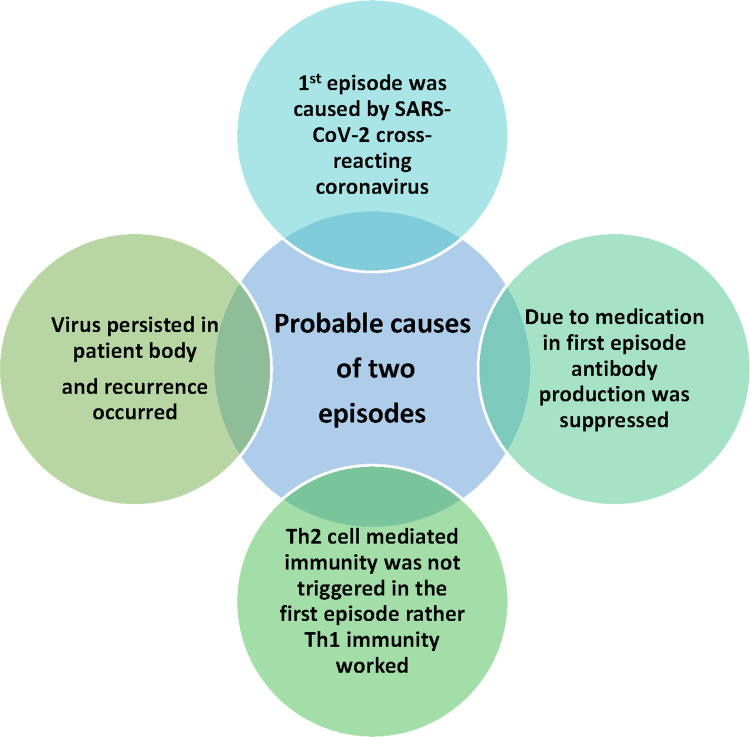
Upon investigating antibody dynamics, it was observed that in the first episode, all the subjects presented a slight rise in anti-N and anti-whole spike protein (S1+S2), while such development being absent for anti-RBD and anti-S1 antibodies. Both S02 and S03 peaked for the antibodies mentioned above after 20 days of symptom onset in the second phase. We suspect the first episode may have been due to infection with any of the common coronaviruses. This is mainly due to approximately 24.6–90.5% similarity of the nucleocapsid sequences shared amongst the coronaviruses.48 Furthermore, it has also been noted that the S2 region is conserved within members of the Coronaviridae family members while the S1 region varies.49,50 Henceforth, there is a possibility of infection with any other common coronaviruses (Figure 4).
Figure 4.
Probable causes for two episodes of symptoms. The first episode did not present a significant antibody increase. The symptoms presentation during the first episode may be due to cross-reacting coronavirus infection. Medication may have resulted in the suppression of antibody development during the first episode. In addition to it, lack of Th2 cell-mediated immunity activation in the first episode may cause the lack of antibody development. This, in turn, may lead to virus persistence or viral reinfection, resulting in a second episode of the presentation of the symptoms, which may have led to activation of Th2 cell-mediated immunity.
Another observation is that for S01, the anti-N and anti-(S1+S2) increase was maintained continuously until the second episode. This anomaly may be explained due to S01’s previous exposure to SARS-CoV during the 2003 outbreak. The exposure may have played a significant role in maintaining the antibody titer of anti-N and anti-(S1+S2), while S02 and S03 presented a sharp decline. Nevertheless, previous exposure did not cross-protect against SARS-CoV-2, as S01 suffered from symptomatic COVID-19 symptoms like others while the neutralizing antibodies formed 10–12 days after symptom onset (Figure 4).
The second episode is characterized by the sharp increase in antibody titer (Figures 1–3). The multi-fold rise in the antibody titer was observed approximately 12 days from the onset of symptoms mentioned in earlier reports.51,52 However, during the study period, the subjects did not lose the antibody titer, ultimately contributing to their immunity from subsequent infection with SARS-CoV-2. However, they all experienced from post-COVID-19 and again COVID-19-like symptoms, nevertheless, were repeatedly revealed negative for SARS-CoV-2 RNA at RT-qPCR tests. The antibody titer’s dynamic increases against the four viral proteins highly correlated with the symptoms’ severity in all subjects. Though an increase in IgG was consistent throughout the subjects, there was some dissimilarity in the IgM and IgA antibody titer.
IgA antibody protects from viruses by blocking its binding onto the mucosal membrane.53,54 Previous studies mention the development of IgA before the development of IgM.55 There also have been reports that the early development of IgA would provide better protection by neutralizing the viruses at the site of infection.53 Our study presented IgA antibodies’ development against the four proteins for S02, which coincide with the subject’s symptoms with epithelial lining, like severe diarrhea, although no respiratory involvement was observed. In the case of S01 and S03, IgA increased multifold against whole-spike protein only.
Similarly, IgM development before IgG or vice versa may decide the outcome and severity of the disease.51 In S01, IgM development was absent during both the first and second episodes, which the previous exposure hypothesis can explain. Furthermore, the subject did recover early after a short bout of the severity of symptoms. In previous reports, the development of IgG before the development of IgM signified less severity of the symptoms in subjects.51 One interesting observation in our study was for both the S01 and S02, and there was an increase in IgM for whole-spike protein but not for RBD and S1 proteins in the second episode. This would signify that the IgM increase was for the S2 part of the whole spike protein rather than the S1 part. Our observation coincides with SARS-CoV’s previous findings where IgG developed simultaneously with IgM and IgA.56 Moreover, IgM and IgA disappeared rapidly in all three cases, indicating a unique class-switching phenomenon, which needs to be confirmed with a large population study.
At the end of the study, the IgG antibody titers against all SARS-CoV-2 proteins, essentially N, RBD, and spike, remained high. However, further, observation is required to identify the overall titer of antibodies over two to three years. It is noted that SARS-CoV and MERS-CoV are known to protect for 2–3 years approximately.57,58
One of our study’s main limitations is that it presents only three subjects and the same gender. The lack of diversity may result in the skewness of our conclusion. However, though our study is limited to three subjects, the significance lies in the difference of antibody development in different individuals due to underlying various physiological conditions. Additionally, it also provides direct evidence of the correlation between the severity of disease and antibody development.59 This information would provide the researchers with a starting point to study antibody development dynamics after vaccination. It would also provide insights into the persistence of antibodies after being exposed to the SARS-CoV-2 virus. Moreover, our study would help researchers design effective IgM/IgG/IgA serodiagnostic kits by selecting the SARS-CoV-2 specific proteins.
Recommendations
On the event of SARS-CoV-2 infection, it is essential to observe the antibody levels.
The early presentation of IgG or IgM would provide a better idea about the probability of the severity of the infection.
Further investigation is to be carried out to observe the correlation of IgA with the outcome of the infection.
Antibody levels are to be observed after vaccination with SARS-CoV-2.
Observation of antibody levels is necessary to analyze the probability of reinfection with SARS-CoV-2.
Further studies with larger sample size and more extended periods are required to establish the facts provided in this study.
Acknowledgments
The authors express their sincere thanks to all the participants who were willing to participate in the study. They also thank their team members for their support work during the current pandemic situation. The authors are grateful to Prof. Mohammed S. Razzaque, MBBS, Ph.D. of Lake Erie College of Osteopathic Medicine (Pennsylvania, USA), for reading the manuscript and providing valuable suggestions. A portion of this study was published in pre-print server https://ssrn.com/abstract=3704264.
Funding Statement
There is no funding to report.
Article Highlights
The antibody dynamics of COVID-19 patients have been evaluated before the infection period and continued until 120 days.
Antibody dynamics against four different viral proteins, ie, nucleocapsid (N), receptor-binding domain (RBD), and spikes (S1 and S1+S2), have been investigated.
Dynamics of three type’s antibodies, ie, IgA, IgM, and IgG, have been identified.
SARS-CoV-2 specific IgG develops simultaneously with IgM and IgA.
IgM and IgA antibodies specific against SARS-CoV-2 weans off rapidly than IgG.
Data Sharing Statement
All data underlying the findings in our study are freely available in the manuscript. For additional information, please refer to http://www.grblbd.com.
Ethical Approval
All procedures performed in this study involving human participants were in accordance with the ethical standards of the national research committee as well as the 1964 Helsinki Declaration. Henceforth, the study was approved by the National Research Ethics Committee of Bangladesh [Reference No.: BMRC/NREC/2019-2022/697]. All the subjects of the study provided their written informed consent. The history of all subjects was noted in the questionnaire prior to collection of blood samples. The privacy of all participants in this study is ensured as all the information provided by them remains undisclosed.
Consent for Publication
All authors reviewed and approved the final version and have agreed to be accountable for all aspects of the work, including any issues related to accuracy or integrity.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agreed to be accountable for all aspects of the work.
Disclosure
Dr. Mohd Raeed Jamiruddin, Mr. Md. Ahsanul Haq, Prof. Dr. Mohib Ullah Khondoker, Prof. Dr. Bijon Kumar Sil, and Dr. Nihad Adnan report a patent 10202006327W pending/licensed to Intellectual Property Office of Singapore. The authors report no other conflicts interest in this work.
References
- 1.World Health Organization. WHO Coronavirus (COVID-19) Dashboard. 2021. Available at: https://covid19.who.int/. Accessed May 31, 2021. [Google Scholar]
- 2.De Wit E, Van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523. doi: 10.1038/nrmicro.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrosillo N, Viceconte G, Ergonul O, Ippolito G, Petersen E. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect. 2020;26(6):729–734. doi: 10.1016/j.cmi.2020.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh EE, Shin JH, Falsey AR. Clinical impact of human coronaviruses 229E and OC43 infection in diverse adult populations. J Infect Dis. 2013;208(10):1634–1642. doi: 10.1093/infdis/jit393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu R, Zhang L, Tan W, et al. Characterization of human coronavirus 229E infection among patients with respiratory symptom in Beijing, Oct-Dec, 2007. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi= Zhonghua Shiyan He Linchuang Bingduxue Zazhi= Chinese Journal of Experimental and Clinical Virology. 2009;23(5):367–370. [PubMed] [Google Scholar]
- 6.Kaiser L, Regamey N, Roiha H, Deffernez C, Frey U. Human coronavirus NL63 associated with lower respiratory tract symptoms in early life. Pediatr Infect Dis J. 2005;24(11):1015–1017. doi: 10.1097/01.inf.0000183773.80217.12 [DOI] [PubMed] [Google Scholar]
- 7.Friedman N, Alter H, Hindiyeh M, Mendelson E, Shemer Avni Y, Mandelboim M. Human coronavirus infections in Israel: epidemiology, clinical symptoms and summer seasonality of HCoV-HKU1. Viruses. 2018;10(10):515. doi: 10.3390/v10100515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Xu X, Ruan J, Lin S, Jiang J, Ye H. Quadruple therapy for asymptomatic COVID-19 infection patients. Expert Rev Anti-Infect Ther. 2020;18:617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long Q-X, Tang X-J, Shi Q-L, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26:1200–1204. [DOI] [PubMed] [Google Scholar]
- 10.Heidari F, Karimi E, Firouzifar M, et al. Anosmia as a prominent symptom of COVID-19 infection. Rhinology. 2020;58(3):302–303. doi: 10.4193/Rhin20.140 [DOI] [PubMed] [Google Scholar]
- 11.Carfì A, Bernabei R, Landi F. Persistent symptoms in patients after acute COVID-19. JAMA. 2020;324(6):603. doi: 10.1001/jama.2020.12603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020;71(16):2027–2034. doi: 10.1093/cid/ciaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziegler K, Steininger P, Ziegler R, Steinmann J, Korn K, Ensser A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill. 2020;25(39). doi: 10.2807/1560-7917.ES.2020.25.39.2001650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv. 2021. [DOI] [PubMed] [Google Scholar]
- 15.Collier DA, De Marco A, Ferreira IATM, et al. SARS-CoV-2 B.1.1.7 escape from mRNA vaccine-elicited neutralizing antibodies. MedRxiv. 2021. doi: 10.1101/2021.01.19.21249840 [DOI] [Google Scholar]
- 16.Nonaka CKV, Franco MM, Graf T, et al. Genomic evidence of SARS-CoV-2 reinfection involving E484K spike mutation, Brazil. Emerg Infect Dis. 2021;27(5):1522–1524. doi: 10.3201/eid2705.210191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jangra S, Ye C, Rathnasinghe R, et al. The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera. medRxiv. 2021. [Google Scholar]
- 18.Chaturvedi R, Naidu R, Sheth S, Chakravarthy K. Efficacy of serology testing in predicting reinfection in patients with SARS-CoV-2. Disaster Med Public Health Prep. 2020;1–7. doi: 10.1017/dmp.2020.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020;112(5):3588–3596. doi: 10.1016/j.ygeno.2020.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shahmohamadnejad S, Nabavi SF, Habtemariam S, et al. May we target double-membrane vesicles and oxysterol-binding protein to combat SARS-CoV-2 infection? Cell Biol Int. 2020;44(9):1770–1772. doi: 10.1002/cbin.11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein S, Cortese M, Winter SL, et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. BioRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viswanathan T, Arya S, Chan S-H, et al. Structural basis of RNA cap modification by SARS-CoV-2. Nat Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-17496-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rambaut A, Loman N, Pybus O et al.; on behalf of COVID-19 Genomics Consortium UK (CoG-UK). Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. 2020. [Google Scholar]
- 24.Adnan N, Khondoker MU, Rahman MS, et al. Coding-complete genome sequences and mutation profiles of nine SARS-CoV-2 strains detected from COVID-19 patients in Bangladesh. Microbiol Resour Announc. 2021;10(10):e00124–21. doi: 10.1128/MRA.00124-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day CL, Lauer GM, Robbins GK, et al. Broad specificity of virus-specific CD4+ T-helper-cell responses in resolved hepatitis C virus infection. J Virol. 2002;76(24):12584–12595. doi: 10.1128/JVI.76.24.12584-12595.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ibarrondo FJ, Fulcher JA, Goodman-Meza D, et al. Rapid decay of anti–SARS-CoV-2 antibodies in persons with mild Covid-19. N Engl J Med. 2020;383(11):1085–1087. doi: 10.1056/NEJMc2025179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med. 2007;357(19):1903–1915. doi: 10.1056/NEJMoa066092 [DOI] [PubMed] [Google Scholar]
- 29.Yang JR, Deng DT, Wu N, Yang B, Li HJ, Pan XB. Persistent viral RNA positivity during the recovery period of a patient with SARS‐CoV‐2 infection. J Med Virol. 2020;92(9):1681–1683. doi: 10.1002/jmv.25940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oishee MJ, Ali T, Jahan N, et al. COVID-19 Pandemic: Review of Contemporary and Forthcoming Detection Tools. Infect Drug Resist. 2021;14:1049–1082. doi: 10.2147/IDR.S289629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.To KK, Hung IF, Ip JD, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nainu F, Abidin RS, Bahar MA, et al. SARS-CoV-2 reinfection and implications for vaccine development. Hum Vaccin Immunother. 2020;16(12):3061–3073. doi: 10.1080/21645515.2020.1830683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prado-Vivar B, Becerra-Wong M, Guadalupe JJ, et al. A case of SARS-CoV-2 reinfection in ecuador. Lancet Infect Dis. 2020. doi: 10.1016/S1473-3099(20)30910-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan PKS, Lui G, Hachim A, et al. Serologic responses in healthy adult with SARS-CoV-2 reinfection, Hong Kong, August 2020. Emerg Infect Dis. 2020;26(12):3076–3078. doi: 10.3201/eid2612.203833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulder M, van der Vegt D, Oude Munnink BB, et al. Reinfection of SARS-CoV-2 in an immunocompromised patient: a case report. Clin Infect Dis. 2020. doi: 10.1093/cid/ciaa1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldman JD, Wang K, Roltgen K, et al. Reinfection with SARS-CoV-2 and failure of humoral immunity: a case report. medRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Elslande J, Vermeersch P, Vandervoort K, et al. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis. 2020. doi: 10.1093/cid/ciaa1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zucman N, Uhel F, Descamps D, Roux D, Ricard JD. Severe reinfection with South African SARS-CoV-2 variant 501Y.V2: a case report. Clin Infect Dis. 2021. doi: 10.1093/cid/ciab129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salzer HJF. Emerging COVID-19 reinfection four months after primary SARS-CoV-2 infection. Wien Med Wochenschr. 2021. doi: 10.1007/s10354-021-00813-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sicsic I, Chacon AR, Zaw M, Ascher K, Abreu A, Chediak A. A case of SARS-CoV-2 reinfection in a patient with obstructive sleep apnea managed with telemedicine. BMJ Case Rep. 2021;14(2):e240496. doi: 10.1136/bcr-2020-240496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novoa W, Miller H, Mattar S, Faccini-Martinez AA, Rivero R, Serrano-Coll H. A first probable case of SARS-CoV-2 reinfection in Colombia. Ann Clin Microbiol Antimicrob. 2021;20(1):7. doi: 10.1186/s12941-020-00413-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrington D, Kele B, Pereira S, et al. Confirmed reinfection with SARS-CoV-2 variant VOC-202012/01. Clin Infect Dis. 2021. doi: 10.1093/cid/ciab014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tillett RL, Sevinsky JR, Hartley PD, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2021;21(1):52–58. doi: 10.1016/S1473-3099(20)30764-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sil BK, Adnan N, Oishee MJ, et al. Development and evaluation of two rapid indigenous IgG-ELISA immobilized with ACE-2 binding peptides for detection neutralizing antibodies against SARS-CoV-2. medRxiv. 2020. [Google Scholar]
- 45.Sil BK, Jahan N, Haq MA, et al. Development and performance evaluation of a rapid in-house ELISA for retrospective serosurveillance of SARS-CoV-2. PLoS One. 2021;16(2):e0246346. doi: 10.1371/journal.pone.0246346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uddin MB, Hasan M, Harun-Al-Rashid A, Ahsan MI, Imran MAS, Ahmed SSU. Ancestral origin, antigenic resemblance and epidemiological insights of novel coronavirus (SARS-CoV-2): global burden and Bangladesh perspective. Infect Genet Evol. 2020;84:104440. doi: 10.1016/j.meegid.2020.104440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tilocca B, Soggiu A, Sanguinetti M, et al. Comparative computational analysis of SARS-CoV-2 nucleocapsid protein epitopes in taxonomically related coronaviruses. Microbes Infect. 2020;22(4–5):188–194. doi: 10.1016/j.micinf.2020.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madu IG, Roth SL, Belouzard S, Whittaker GR. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J Virol. 2009;83(15):7411–7421. doi: 10.1128/JVI.00079-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Long QX, Liu B-Z, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845–848. [DOI] [PubMed] [Google Scholar]
- 52.Pan Y, Guan H, Zhou S, et al. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur Radiol. 2020;30:3306–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Renegar KB, Small PA, Boykins LG, Wright PF. Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. J Immunol. 2004;173(3):1978–1986. doi: 10.4049/jimmunol.173.3.1978 [DOI] [PubMed] [Google Scholar]
- 54.Béné MC, de Carvalho Bittencourt M, Eveillard M, Le Bris Y. Good IgA bad IgG in SARS-CoV-2 infection? Clin Infect Dis. 2020;71(15):897–898. doi: 10.1093/cid/ciaa426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boes M. Role of natural and immune IgM antibodies in immune responses. Mol Immunol. 2000;37(18):1141–1149. doi: 10.1016/S0161-5890(01)00025-6 [DOI] [PubMed] [Google Scholar]
- 56.Hsueh PR, Huang LM, Chen PJ, Kao CL, Yang PC. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin Microbiol Infect. 2004;10(12):1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mo H, Zeng G, Ren X, et al. CHAN‐YEUNG M: longitudinal profile of antibodies against SARS‐coronavirus in SARS patients and their clinical significance. Respirology. 2006;11(1):49–53. doi: 10.1111/j.1440-1843.2006.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Payne DC, Iblan I, Rha B, et al. Persistence of antibodies against Middle East respiratory syndrome coronavirus. Emerg Infect Dis. 2016;22(10):1824. doi: 10.3201/eid2210.160706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roltgen K, Powell AE, Wirz OF, et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol. 2020;5(54):eabe0240. doi: 10.1126/sciimmunol.abe0240 [DOI] [PMC free article] [PubMed] [Google Scholar]