SUMMARY
During V(D)J recombination, RAG proteins introduce DNA double-strand breaks (DSBs) at recombination signal sequences (RSSs) that contain either 12- or 23-nt spacer regions. Coordinated 12/23 cleavage predicts that DSBs at variable (V) gene segments should equal the level of breakage at joining (J) segments. Contrary to this, here we report abundant RAG-dependent DSBs at multiple Vκ gene segments independent of V-J rearrangement. We find that a large fraction of Vκ gene segments are flanked not only by a bone-fide 12 spacer but also an overlapping, 23-spacer flipped RSS. These compatible pairs of RSSs mediate recombination and deletion inside the Vκ cluster even in the complete absence of Jκ gene segments and support a V(D)J recombination center (RC) independent of the conventional Jκ-centered RC. We propose an improved model of Vκ-Jκ repertoire formation by incorporating these surprisingly frequent, evolutionarily conserved intra-Vκ cluster recombination events.
In Brief
Shinoda et al. demonstrate previously unknown, frequent RAG-dependent recombination within the Ig Vκ gene cluster, which is mediated by evolutionarily conserved flipped RSS embedded in bona fide RSS. These intra-Vκ cluster recombination events alter the available Vκ gene pool by deleting Vκ segments and shape the Vκ-Jκ immune repertoire.
Graphical Abstract
INTRODUCTION
The immensely diverse repertoire of B and T cell antigen receptors (AgRs) are encoded in discrete variable (V), diversity (D), and joining (J) gene segments that undergo somatic recombination in lymphocyte precursors. V(D)J recombination is initiated by the lymphocyte-specific recombination activating gene (RAG-1 and ‒2) proteins that generate DNA double-strand breaks (DSBs) at recombination signal sequences (RSSs) flanking the V, D, and J gene segments (Teng and Schatz, 2015). The RSS is composed of a conserved heptamer and nonamer motif, separated by either a 12-bp (12RSS) or a 23-bp (23RSS) spacer. In every AgR locus (Sakano et al., 1979), efficient recombination occurs only between different types of gene segments flanked by two different types of RSS known as the 12/23 rule (Tonegawa, 1983). This rule is enforced by the RAG proteins that, in physiological conditions, introduce DSBs only in the context of a synaptic complex composed of a pair of 12/23 RSS (van Gent et al., 1996). After cleavage, the broken signal ends (SEs) and coding ends (CEs) are joined by the non-homologous end joining (NHEJ) pathway (Helmink and SleCκman, 2012). Recombination can occur by deletion of the intervening sequences or by intrachromosomal inversion. During deletional rerarrangement, joining of the two RSSs produce extrachromosomal excision circles (Shimizu and Yamagishi, 1992) that persist only in non-dividing precursors and are subsequently lost in mature lymphocytes (Livak and Schatz, 1996).
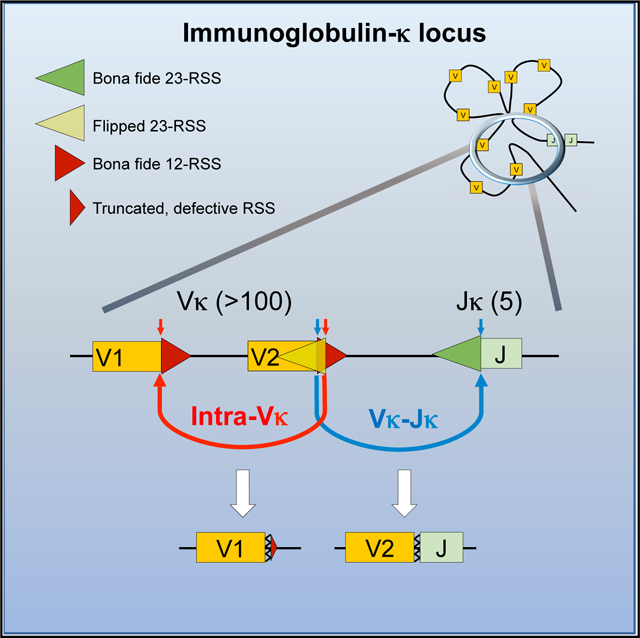
The mouse immunoglobulin κ (Igκ) locus contains more than 140 V gene segments and 4 functional J gene segments. In contrast to other AgR loci, there are many Vκ gene segments in either forward or reverse transcriptional orientation relative to the Jκ segments (Proudhon et al., 2015); thus, Vκ-to-Jκ joining can occur either deletionally or by inversion. In addition, the Igκ locus undergoes multiple rounds of rearrangements to generate a minimally self-reactive, tolerant Ig repertoire (Nemazee, 2017). Self-tolerance is also achieved through complete deletion of the Cκ gene segment mediated by the human Igκ deleting element (IGΚDE) (Siminovitch et al., 1987) or its murine equivalent sequences (recombining sequence or RS) (Durdik et al., 1984; Feddersen et al., 1990). Precursors with such unconventional V(D)J recombination events eventually develop into Igλ-expressing mature B cells. Ultimately, the final VκJκ repertoire is remarkably similar in bone marrow pre-B cells and mature splenic B cells (Aoki-Ota et al., 2012), indicating that, like in the IgH and T cell receptor (TCR)-β loci (Livak et al., 2000; Yu et al., 2002), the frequency of Vκ gene segment usage is predetermined largely by the gene rearrangement process rather than by cellular selection (Rubelt et al., 2016).
A major conundrum of V(D)J recombination is to understand how the simple, transposon-derived RAG recombinase controls the astonishingly finely tuned shape of the Ig and TCR repertoires. V(D)J recombination is initiated by the recruitment of RAG to trimethylated lysine 4 residue of histone 3 (H3K4me3), in a region termed recombination center (RC), located at the J loci of the AgRs (Desiderio, 2010; Ji et al., 2010; Matthews and Oettinger, 2009). The recently introduced RAG-scanning model proposes that within convergent CTCF-binding-element (CBE)-based chromatin loop domains, RAG scans directionally from an initiating RC for megabase (Mb) distances for compatible RSS pairs (Hu et al., 2015; Jain et al., 2018). How the RAG-scanning model could explain the widespread, balanced use of more than a 100 Vκ gene segments, some of them Mbs away from the Jκ-associated RC (Teng et al., 2015), is not fully understood.
Here, we report that a large fraction of RAG-mediated cleavage at Vκ gene segments occurs independent of the Jκ gene segments both in mouse and human and identify a second, autonomous RC within the Vκ cluster. This RC uses the bona fide Vκ 12RSS (hereafter referred to as bRSS) with overlapping, cryptic 23RSS and directs recombination between Vκ gene segments in competition with the Jκ RC. We provide a predictive model of Vκ gene segment use that incorporates intra-cluster Vκ rearrangements. This study highlights an evolutionarily conserved strategy that, uniquely among the AgR loci, shapes the Igκ repertoire in diverse species.
RESULTS
Extensive RAG-Mediated DSBs at Vκ Gene Segments Independent of Jκ
To evaluate the repertoire of RAG-mediated breakage in the Igκ locus, we applied END-seq, a method that maps DSBs at single-base-pair resolution (Canela et al., 2016) in Abelson virus (v-Abl)-transformed mouse pre-B cells. Here, RAG expression and Igκ locus recombination are induced following treatment with the v-Abl kinase inhibitor STI-571 (Muljo and Schlissel, 2003). DSBs were found reproducibly at 80% of annotated Vκ gene segments and at all four functional Jκ gene segments (Jκ1, Jκ2, Jκ4, and Jκ5) (Figures 1A and 1B). Rag1−/− pre-B cells did not show any breakage around the RSS (Figures 1A and 1B), confirming that these breaks are RAG mediated. END-seq analysis performed on ex-vivo-cultured primary mouse bone marrow pre-B cells showed a similar pattern of DSBs to v-Abl-transformed pre-B cells (Figure S1A).
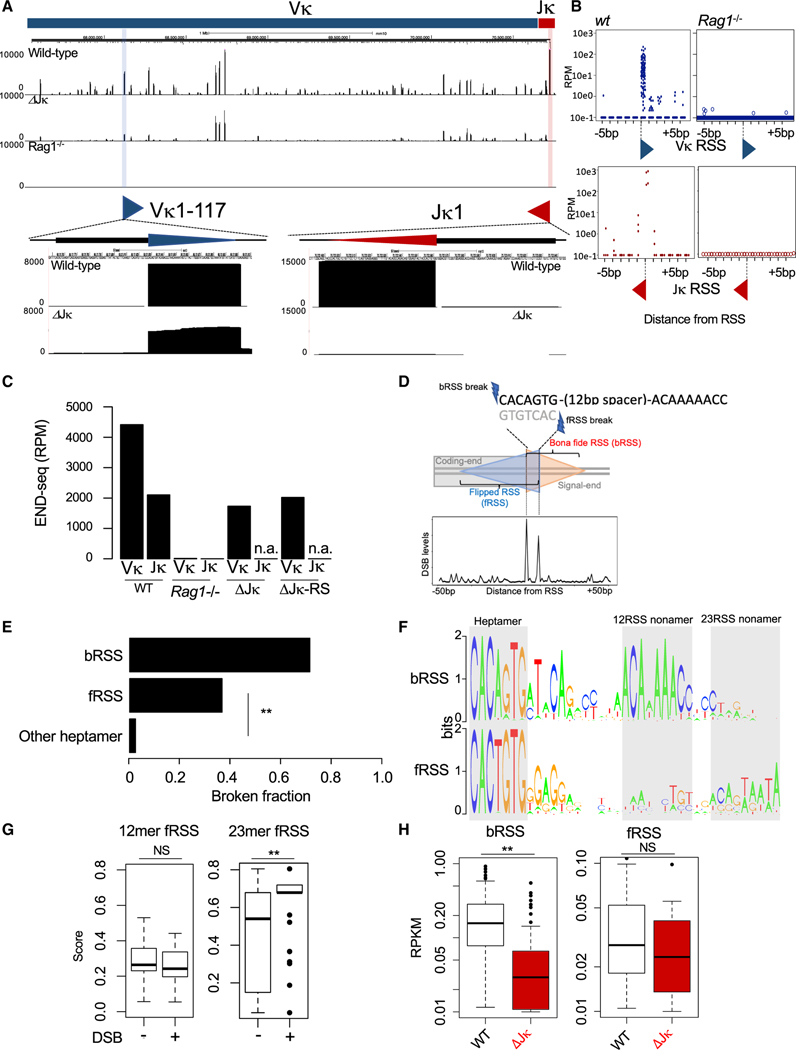
Figure 1. RAG-Dependent DSBs in the Vκ Cluster at Flipped RSS (fRSS) that Overlap with Bona Fide RSS (bRSS).
(A) Top panel: END-seq reads at the Igκ locus from WT (top track), ΔJκ (middle track), and Rag1−/− (bottom track) v-Abl-transformed pre-B cells. Positions of the Vκ1–117 (blue) and Jκ1 (red) genes are marked on the top. Bottom panel: magnification of the Vκ1–117 and Jκ1 gene segments. Vκ1–117 and Jκ1 breakages are highlighted in blue and red, respectively; blue and red triangles depict the corresponding 12RSS and 23RSS, respectively.
(B) Nucleotide resolution cumulative mapping of DSB sites by END-seq within ±5 bp surrounding Vκ RSS (top) or Jκ RSS (bottom) in WT (left) or Rag1−/− (right) v-Abl-transformed pre-B cells.
(C) Quantification of END-seq reads within the Vκ and Jκ gene clusters (represented as reads per milion [RPM]) in WT, ΔJκ, and ΔJκ-RS v-Abl-transformed pre-B cells. n.a., not applicable.
(D) Top panel: schematic diagram of bRSS and the partially overlapping fRSS. Double-strand breaks for bRSS occur at the 5′ end of the heptamer and for fRSS at the 3′ end of the heptamer. Orange triangle represents bRSS, and blue triangle represents fRSS. Bottom panel: aggregate plot for END-seq signal at Vκ RSS surrounding region (±50 bp).
(E) Fraction of broken bRSS, fRSS, and other heptamers within the Vκ cluster in v-Abl-transformed pre-B cells, as detected by END-seq. *p < 1e−10, Fisher’s test.
(F) Consensus sequence logo of Vκ bRSS (top) and fRSS (bottom). Heptamer and nonamer motifs at 12-bp and 23-bp spacer distances are highlighted in gray.
(G) Boxplots of average PWM score of 12-bp spacer RSS (left) or 23-bp spacer RSS (right) sequences of fRSS with (+) or without (−) DSBs, as detected by END-seq in WT v-Abl-transfomed pre-B cells. NS, p = 0.82; **p < 1e−7; t test.
(H) Boxplot quantification of END-seq reads present at Vκ bRSS (left) and fRSS (right) DSBs in WT (blaCκ) and ΔJκ (red) v-Abl-transfomed pre-B cells. See also Figure S1. NS, p = 0.11; **p < 1e−14; paired t test.
Coordinated RAG cleavage following the 12/23 rule of V(D)J recombination dictates that the total number of DSBs at Vκ 12RSS and Jκ 23RSS would be nearly equal. Surprisingly, the total number of DSBs detected at Vκ gene segments was more than 2-fold higher than that of Jκ gene segments (Figure 1C; for average of 7 replicates see Figure S1B).
To test whether all Vκ breaks are the result of attempted V-to-J rearrangement, we applied END-seq on a pre-B cell clone where all five Jκ gene segments were deleted (termed ΔJκ; see Figures S1C and S1D). Strikingly, Vκ breaks continued to be detected at substantial levels in the ΔJκ clone (Figures 1A and 1C). We considered the possibility that the previously described Cκ-deleting 23RSS elements (Durdik et al., 1984; Feddersen et al., 1990; Siminovitch et al., 1987) may contribute to the excess of Vκ breaks. However, the removal of the Jκ-Cκ intronic deleting elements (RS), in addition to Jκ (termed ΔJκ-RS; see Figures S1C and S1D) did not eliminate RAG-dependent Vκ breaks (Figure 1C). The above findings raised two alternatives: either DSBs at Vκ 12RSS are occurring in violation of the 12/23 rule or there are other, unidentified 23RSS in the Igκ locus.
Identification of a 23-mer RSS within the Vκ Locus
Taking advantage of the single-nucleotide resolution capacity of END-seq, we noticed a significant enrichment of breaks not only at the first nucleotide of the heptamer, where RAG normally introduces a precise DSB, but also at the last nucleotide of the heptamer of the bRSS (Figure 1D). Due to the inherent near-palindromic nature of the consensus heptamer sequence (5′-CACAGTG), this motif in the opposite direction on the other strand can be read as a flipped heptamer (5′-CACTGTG) and could be part of a flipped RSS (hereafter referred to as fRSS; see Figure 1D).
Although most bRSS and about one-third of fRSS showed DSBs precisely at the first position of their heptamers, virtually no other heptamer-like sequences were broken in the Vκ cluster (Figure 1E). We searched the DNA sequences following all heptamers for nonamer-like motifs by using a position weight matrix (PWM) approach (see STAR Methods). The heptamers of bRSS are followed by a strong nonamer at exactly a 12-bp distance, as expected for a well-defined 12RSS (Figure 1F). The flipped heptamers are also frequently followed by a nonamer-like motif but at a 23-bp distance (Figure 1F). We compared the sequences of fRSS with respect to their DSB status and found that broken fRSS had a higher 23RSS PWM score (see STAR Methods) than non-broken fRSS, whereas both cleaved and non-cleaved fRSS had equally low 12RSS PWM scores (Figure 1G; Figure S1E). Finally, although the number of bRSS breaks showed a significant reduction in the ΔJκ clone, the number of fRSS breaks remained similar between wild-type (WT) and ΔJκ cells (Figure 1H; Figure S1F), indicating that fRSS are used independently of V-to-Jκ rearrangement.
Collectively, these results identify a number of 23-mer RSS in the mouse Vκ locus, which overlap with the heptamer of the Vκ bRSS in a flipped orientation. A substantial fraction of these fRSSs, specficially those with higher quality 23RSS, is cleaved by RAG. We hypothesize that these fRSS embedded within the Vκ cluster could explain many of the widespread, Jκ-independent DSBs described above.
Extensive Intra-Vκ Cluster Rearrangements Mediated by 12/23 Pairs of bRSS and fRSS
To determine whether these 23-bp spacer fRSSs actually mediate rearrangements within the Igκ locus, we used high-throughput genome-wide translocation sequencing (HTGTS) (Lin et al., 2016). This method captures joining between a pre-defined site, termed the “bait,” with its fusion partner that originates elsewhere in the genome. We compared the number of HTGTS joints of V-Jκ genes with the number of intra-Vκ cluster bRSS/fRSS-mediated joining events. Consistent with the END-seq data, HTGTS performed with the Jκ1 CE as a bait revealed a large number of of Jκ1 joining events to Vκ gene segments using the Vκ bRSS (Figures 2A and 2B). In contrast, only one-half to two-thirds of the junctions from the highly used (Aoki-Ota et al., 2012) Vκ1–117 (Figures 2A and 2B) or Vκ8–34 RSSs (Figures S2A and S2B) were V-Jκ rearrangements, whereas a sizeable fraction involved recombination between bRSS and fRSS within the Vκ cluster in pre-B cells (Figures 2A and 2B; Figures S2A and S2B). These intra-Vκ cluster rearrangements were also entirely retained in ΔJκ pre-B cells (Figures 2A and 2B; Figures S2A and S2B). HTGTS performed with the Vκ1–117 CE bait on sorted, primary mouse bone marrow pre-B cells showed a similar pattern to the v-Abl pre-B cell clones (Figure S2C).
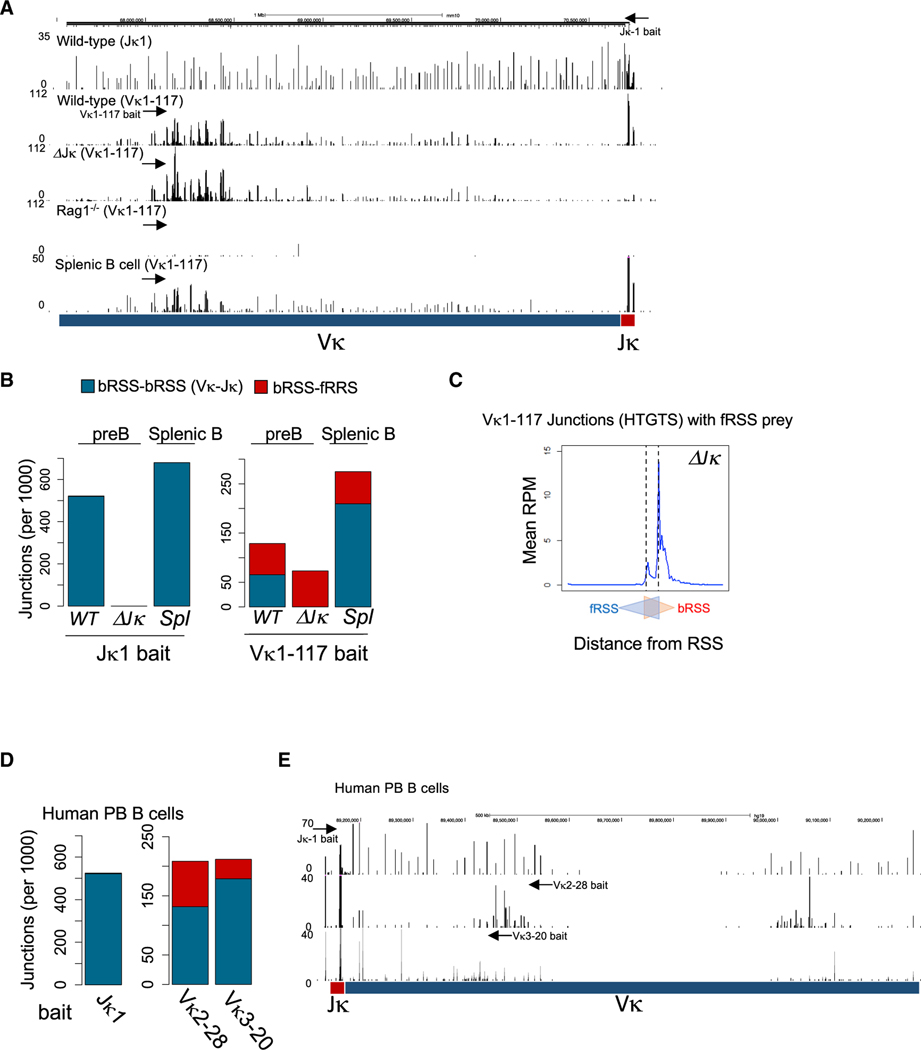
Figure 2. Frequent fRSS-Mediated Intra-Vκ Cluster Rearrangements in Mouse and Human B Cells.
(A) HTGTS junction profiles in WT (Jκ1 bait and Vκ1–117 bait), ΔJκ (Vκ1–117 bait), and Rag1−/− (Vκ1–117 bait) v-Abl-transformed pre-B cells and in WT splenic B cells within the Igκ locus. Arrows indicate the approximate positions of the Jκ1 and Vκ1–117 bait primers.
(B) The number of junctions (per 1,000 total junctions) of bRSS-bRSS (Vκ-Jκ in blue) and bRSS-fRSS (intra-Vκ in red) recombinations in v-Abl-transformed pre-B cells (WT and ΔJκ) or splenic B cells detected by the Jκ1 (left) or Vκ1–117 (right) bait.
(C) Aggregate Vκ1–117 junctions surrounding the Vκ RSSs in ΔJκ v-Abl-transformed pre-B cells. The first nucleotides of bRSS and fRSS are depicted as two overlapping triangles.
(D) The number of junctions of bRSS-bRSS (Vκ-Jκ in blue) and bRSS-fRSS (intra-Vκ in red) recombinations in human peripheral blood B cells detected by HTGTS with Jκ1 (left), Vκ2–28, and Vκ3–20 (right) baits.
(E) HTGTS junction profiles of human peripheral B cells within the Igκ locus. The arrows indicate the approximate positions of the Jκ1, Vκ2–28, and Vκ3–20 bait primers.
See also Figure S2.
The Jκ1 CE bait products included diverse V gene segments across the entire Vκ cluster, whereas the Vκ CE baits captured predominantly nearby Vκ cluster targets (Figure 2A; Figure S2A). When we analyzed Vκ1–117 CE bait products according to the published Vκ locus chromatin loop distribution (Karki et al., 2018) (Figure S2D), we found that more than 80% of Vκ cluster bRSS-fRSS joints resided within the same loop as the Vκ1–117 gene segment (Figure S2E). In contrast, a pre-B cell clone with a CRISPR-Cas9 DSB bait site inserted near the Vκ1–117 gene segment captured prey sequences across the entire Vκ locus. Only about 30% of these junctions resided within the same loop as the Cas9 bait site, which was similar to the calculated random frequency (Figure S2E). These data suggest that intra-Vκ cluster rearrangements occur in accordance with the RAG scanning model of canonical 12/23 V(D)J recombination.
We also examined the precise location of prey sequences to which the Vκ1–117CE bait joined. We found that the majority of bRSS-fRSS-mediated joining occurred between the Vκ1–117 CE sequences and sequences flanking the heptamer of the fRSS of other partner Vκ gene segments (Figure 2C with ΔJκ clone and Figure S2F with WT clone). HTGTS performed in DNA Ligase IV- or Artemis-deficient pre-B cells by using the Vκ1–117 CE bait confirmed that both V-Jκ and intra-Vκ cluster joints were NHEJ dependent (Figure S2G).
Strikingly, intra-Vκ cluster rearrangements mediated by bRSS-fRSS interactions were also detected in splenic, IgM+ mature B cells in similar pattern, albeit at somewhat reduced proportions, as in pre-B cells (Figures 2A and 2B). Furthermore, HTGTS analysis of the Igκ locus in purified, human peripheral blood B cells showed a similarly large number of intra-Vκ cluster junctions from the Vκ2–28 gene segment—orthologous to the mouse Vκ1–117—or the Vκ3–20 gene segment (Figures 2D and 2E). Overall, mature B cells exhibited a surprisingly high frequency of intra-Vκ cluster rearrangements using 12/23 bRSS-fRSS pairs both in mice (24% of Vκ1–117 joints) and humans (37% of Vκ2–28 joints), suggesting that the fRSSs are extensively used during Igκ locus rearrangements. These results from peripheral B cells also support the notion that bRSS-fRSS-mediated intra-Vκ cluster joints are primary rearrangements on the chromosome rather than byproducts of secondary rearrangements on extrachromosomal circles, which are mostly lost in mature B cells.
Vκ fRSSs Exploit Conserved Framework Region 3 (FR3) Codons as a 23RSS Nonamer
Evolutionary conservation of non-coding 12 and 23RSS motifs is a hallmark of V(D)J recombination (Tonegawa, 1983). In contrast to bRSS, the fRSS is largely localized within the 3′ end of the Vκ coding sequences that could exert conflicting evolutionary selection on an otherwise regulatory, non-coding element. We found, however, that the sequence composition of Vκ gene segments allows the co-existence of both selection forces. Specifically, the 23-bp spacer encodes for the complementarity-determining region 3 (CDR3) of the Vκ domain, whereas the nonamer motif corresponds to the highly conserved YY[F/H]C anchor residues of the FR3 (Figure 3A). Frequently, the nine nucleotides of the predominant three codons of YYC resemble the canonical nonamer sequence 5′-ACAATAGTA (Figure 3A). Thus, in the Vκ locus, a highly conserved nonamer-like motif of FR3 and the flipped heptamer overlapping with the bRSS, separated exactly by 23 nt of less conserved CDR3, can create a 23-mer fRSS to pair with the Vκ 12-mer bRSS (Figure 3A).
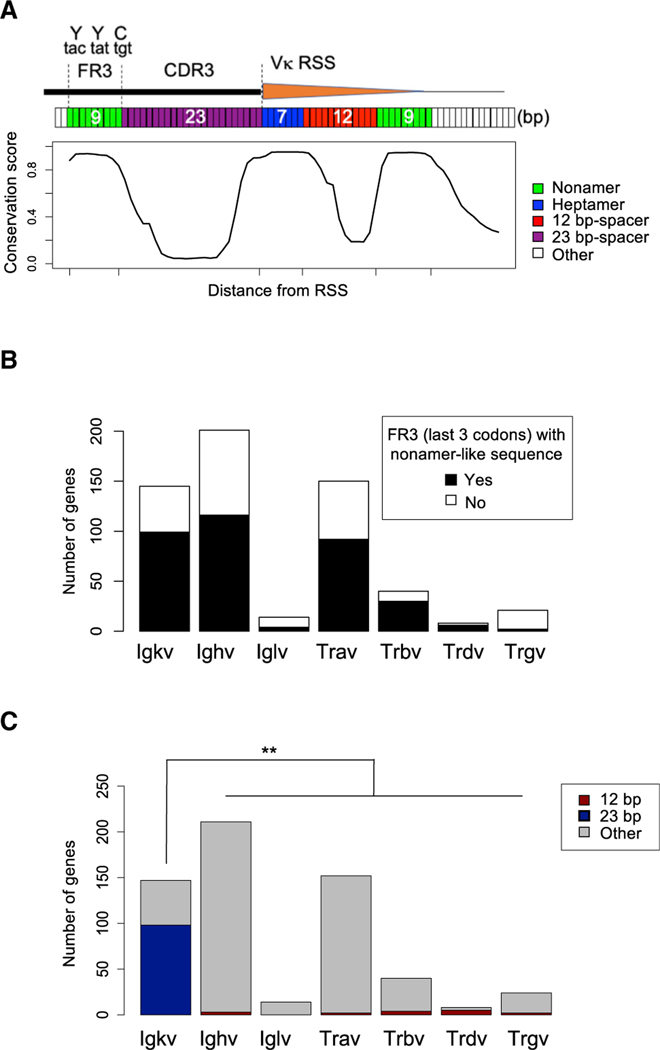
Figure 3. Vκ Gene Segments Harbor Unique, Evolutionarily Conserved Frss.
(A) Conservation score of the region surrounding the Vκ RSS region. Top diagram shows the position of FR3 and CDR3 at the 3′ end of Vκ coding region. Heptamer (blue), spacer (purple and red), and nonamer (green) regions are highlighted.
(B) Number of V gene segments with FR3 with the last 3 codons encoded by nonamer-like sequences in the Igκ, Igh, Igl, Tra, Trb, Trd, and Trg loci.
(C) Number of V gene segments with CDR3 with length of 12 bp (red), 23 bp (blue), or other number of nucleotides (gray) in the Igκ, Igh, Igl, Tra, Trb, Trd, and Trg loci.
See also Figure S3. **p < 1e−10, Fisher’s test.
Given the conservation of the YY[F/H]C motif in all Ig and TCR variable domains and the fact that this triplet can be frequently encoded by a nonamer-like motif (Figure 3B), we sought to determine whether the embedded fRSS could be a more general phenomenon extending beyond the Vκ gene segments. Because all other AgR loci have 23-mer bRSS flanking the V genes, we focused our search on 12-mer fRSS. In all AgR loci, except the Igκ and λ light chains, the V domains contain a shorter CDR3, typically 2–5 amino acids in length, following the conserved FR3 and could potentially encode a flipped 12RSS. However, we found only a few V gene segments that appear to contain a high scoring flipped 12RSS (see STAR Methods), primarily due to the frequent deviation of CDR3 sequences from the 12-nt length, which is absolutely necessary for a 12RSS spacer (Figure 3C).
To test directly whether RAG-mediated DSBs occur at putative fRSSs in another AgR locus, we performed END-seq on CD4/CD8 double-positive (DP) thymocytes that actively undergo TCRα locus recombination and compared Vκ and Vα loci with respect to their bRSS and fRSS sequences and breakage (Figures S3A and S3B). High level bRSS breakage is seen both in pre-B cells at the Vκ gene segments and in thymocytes at the Vα gene segments, as expected (Figures 1A and S3A). Both Vκ and Vα clusters contain a large number of palindromic heptamer motifs (Figure S3C, marked as fRSS); however, only the Vκ fRSSs contain frequent DSBs (Figure S3B), whereas none of the Vα palindromic heptamers showed breakage at the opposite end of the heptamer in thymocytes (Figure 3B). Thus, we conclude that other AgR loci besides Vκ do not have conserved, functional fRSSs with either 12-bp or 23-bp spacers and, at least in the TCRα locus, do not undergo RAG-mediated cleavage in thymocytes at predicted, opposite end positions of palindromic heptamers anywhere within their V clusters.
An Autonomous RC in the Vicinity of fRSSs Initiate Intra-Vκ Cluster Recombination
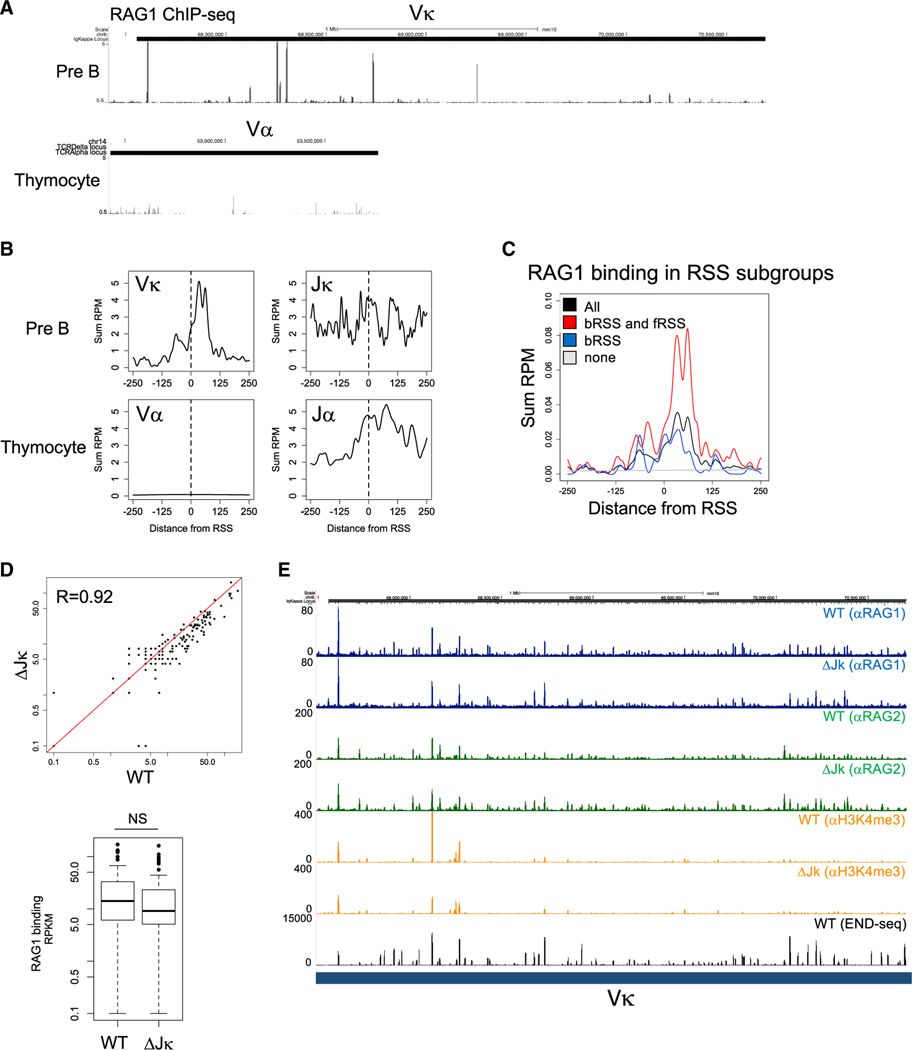
V(D)J recombination is initiated by the binding of RAG1/2 to active, H3K4me3-marked chromatin at the J region of the AgR loci, termed the nascent RC (Jain et al., 2018; Ji et al., 2010). The chromatin-bound RAG complex associated with the RSS of a J gene segment linearly tracks the chromatin for compatible RSS to form a paired complex with the V or D gene segment RSS (Hu et al., 2015; Lin et al., 2018; Zhao et al., 2016). Given that intra-Vκ cluster rearrangements are independent of the Jκ gene segments and of the natural Igκ locus RC (Figure 1C), we hypothesized that these rearrangements are initiated by autonomous RAG binding within the Vκ locus. To test this prediction, we examined RAG1 chromatin immunoprecipitation sequencing (ChIP-seq) data obtained from RAG1-D708A mice, where a catalytically inactive, mutant RAG1 retains its binding to RSS but is incapable of cleaving DNA (Teng et al., 2015). In these mice, robust RAG binding to both Jκ and Jα clusters from bone marrow pre-B cells and DP thymocytes, respectively, were documented previously (Ji et al., 2010). However, when we compared RAG ChIP-seq signals in the V clusters, Vκ gene segments in bone marrow pre-B cells, but not Vα in DP thymocytes, also showed substantial RAG1 binding (Figures 4A and 4B). Strikingly, the highest level of RAG1 binding in the Vκ cluster is detected at bRSS with overlapping, cleaved fRSSs (Figure 4C). Further ChIP-seq analysis of v-Abl-transformed pre-B cells revealed that RAG1 and RAG2 binding, as well as the levels of H3K4me3 throughout the Vκ locus, are nearly completely retained in the ΔJk clone at the level observed in WT pre-B cells (Figures 4D and 4E).
Figure 4. RAG1/2 Complex Forms an Autonomous RC at the Vκ Cluster.
(A) Publically accessible ChIP-seq data (RPM mapped reads) for RAG1 binding in the Vκ cluster in primary bone marrow pre-B cells (top) or in the Vα cluster in DP thymocytes (bottom) from RAG1(D708A) mutant mice (Teng et al., 2015).
(B) Aggregate plot of RAG1 ChIP-seq signal around bRSS for Vκ, Jκ, Va, and Jα loci (±250 bp).
(C) Aggregate plot of RAG1 ChIP-seq signal around Vκ RSS, classified as all RSS (blaCκ), cleaved bRSS with overlapping cleaved fRSS (red), cleaved bRSS with overlapping non-cleaved fRSS (blue), or at RSS with no cleavage at all (gray) (±250 bp).
(D) Scatterplot correlation of RAG1 ChIP-seq signal at each Vκ RSS in WT and ΔJκ v-Abl-transformed pre-B cells (top). Boxplot of total RAG1 ChIP-seq signal at Vκ RSS in WT and ΔJκ v-Abl-transformed pre-B cells (bottom).
(E) ChIP-seq (RPM mapped reads) for RAG1 (blue), RAG2 (green), and H3K4me3 (orange) in WT and ΔJκ v-Abl-transformed pre-B cells across the Igκ locus. END-seq tracks (a replicate sample different from the one shown in Figure 1A) are displayed in the bottom.
These results support the hypothesis that RAG1/RAG2 are recruited locally to the Vκ locus to form an autonomous Vκ RC. This Vκ RC can initiate intra-Vκ cluster rearrangement by using the Vκ-associated bRSS and fRSS pairs, independent of the Jκ RC.
Predictive Model of the Igκ Light Chain Repertoire
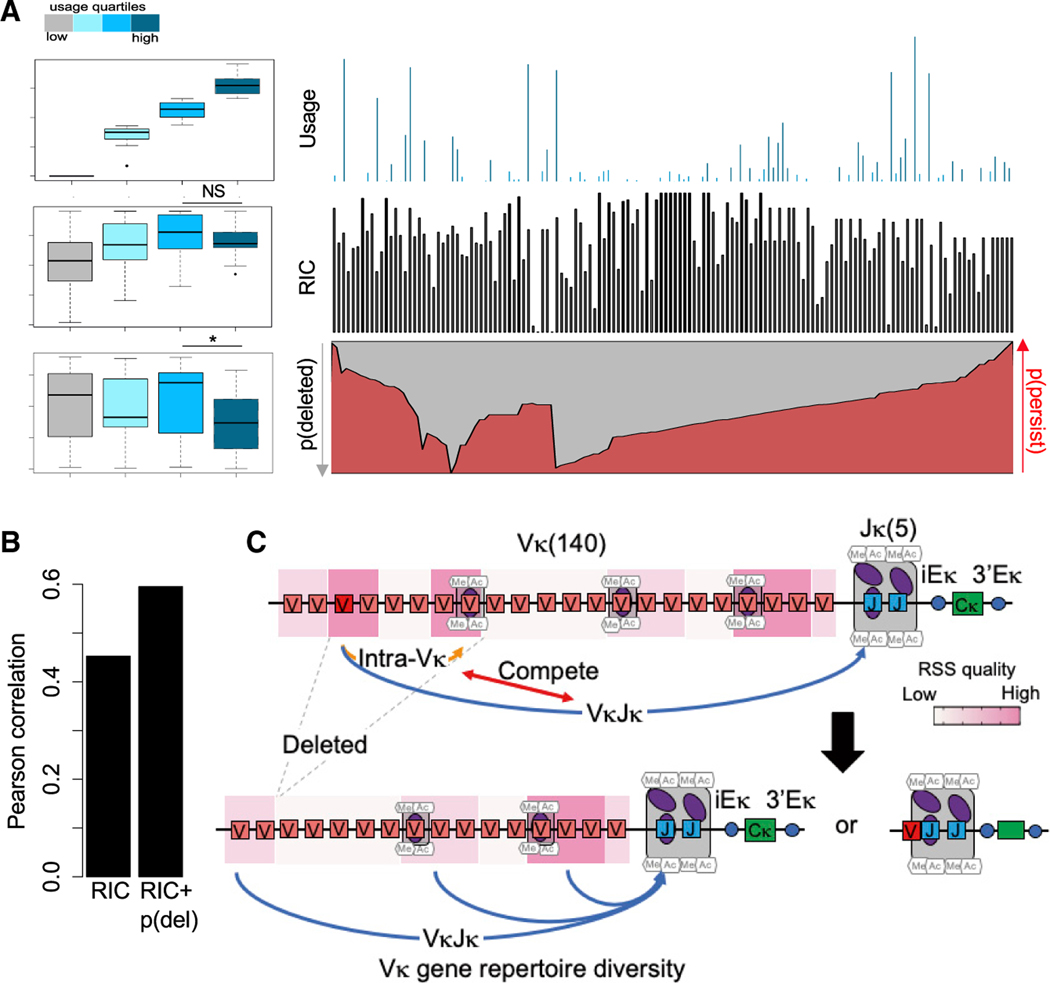
Numerous studies have shown that the biochemical properties of RAG-RSS interactions significantly shape the developing combinatorial AgR repertoire and that this repertoire is minimally altered by subsequent cellular selection for the TCRβ (Livak et al., 2000; Wu et al., 2003), IgH (Yu et al., 2002), or Igκ loci (Aoki-Ota et al., 2012). Several algorithms have been developed to measure the functionality of the RSS, mostly based on sequence conservation, but these measures, such as the Recombination Information Content (RIC) score (Cowell et al., 2002), predict gene segment use or DSBs with modest success in AgR loci, including the mouse Igκ locus (Figures 5A and 5B). The majority of Vκ gene segments are flanked by high PWM nonamer score bRSS (Figure S4A), whereas RSS cleavage and gene segment use vary significantly across the locus (Figure S4A). Recombination mediated by bRSS-fRSS pairs would destroy the bRSS and render both participating Vκ gene segments non-functional. In addition, deletional bRSS-fRSS recombination would also remove all intervening Vκ gene segments from the chromosome. We hypothesized that bRSS-fRSS-mediated recombination and deletion within the Vκ cluster could significantly influence V gene segment availability for V-Jκ rearrangements.
Figure 5. Predictive Model for the Generation of the Igκ Repertoire.
(A) The Vκ RSSs are divided into four quartiles based on published use data in splenic B cells (Aoki-Ota et al., 2012). RIC score and probablility of deletion are calculated for each classified RSS. Boxplot summary (left) and distribution (right) of the Vκ gene segment use (top), RIC score (middle), and probability of deletion/persistence (gray/red, respectively) (bottom) are shown (for calculations see STAR Methods). NS, p = 0.16; *p = 0.006; t test.
(B) Pearson correlation coefficient between Vκ use against RIC score and combined RIC score and probability of deletion.
(C) Schematic model displaying competition between intra-Vκ cluster rearrangement and Vκ-Jκ rearrangement. Intra-Vκ cluster rearrangement results in the deletion of various Vκ gene segments, which allows for diverse V-Jκ rearrangements to fine tune the Vκ gene repertoire. See also Figure S4.
We, therefore, sought to incorporate the probability of loss of Vκ gene segments by bRSS-fRSS-mediated recombination into the prediction of Vκ gene segment use. By considering all possible bRSS/fRSS pairs, we assigned a probability of deletion [p(del)] score to each Vκ gene segment that reflects the probability that a given Vκ gene gets deleted through intra-Vκ cluster rearrangement. We assigned each RSS pair a weight proportional to the RIC score of its bRSS and fRSS component, their position relative to each other on chromosomal loop domains, and the binding of RAG1 measured by ChIP-seq (Figure S4B; see STAR Methods). We then binned the Vκ gene segments into four quartiles with respect to their usage in V-Jκ rearrangements (group 1–4 from low to high use, respectively) and compared these groups based on their bRSS RIC score and deletion probability. We noticed that the bRSS RIC score correctly predicted the ranking of use in three groups (3 > 2 > 1). However, RIC score values did not increase for the highest used group 4 RSS, i.e., the RIC score failed to place this group at the top of the use ranking (Figure 5A). Importantly, group 4 also has the lowest probability of being deleted. By incorporating the p(del) score together with the RIC score, the predictive power of Vκ gene segment use was significantly improved (Figure 5B). We, therefore, propose that the frequency at which a given Vκ gene segment is used is determined both by the strength of its bRSS and its probability to be deleted by intra-Vκ cluster rearrangement (see model in Figure 5C). Notably, stronger Vκ bRSSs tend to rearrange more not only with Jκ RSS but also with fRSS flanking other Vκ genes; therefore, the stronger a bRSS is, the more likely it is to undergo intra-Vκ cluster deletion. These two competing rerrangement pathways reduce the dominance of V gene segments flanked by the best bRSS and allow for the generation of a more balance Igκ repertoire.
DISCUSSION
The 12/23 rule is the cornerstone of ordered rearrangement of V, D, and J gene segments in vertebrate lymphocyte precursors. Indeed, it may have arisen very early at the time of the domestication of the RAG transposon (Zhang et al., 2019). It was, therefore, very surprising to observe extensive RAG-mediated DNA breaks and recombination events within the Vκ cluster in the complete absence of Jκ gene segments and Jκ RC. Although cryptic RSSs (Lieberman et al., 2009) and atypical rearrangement events in the Igκ locus had been reported (Li et al., 2016; Vinocur et al., 2009), their use appears to be relatively infrequent, as determined by HTGTS (Lin et al., 2016) and END-seq (Canela et al., 2016). In contrast, we observed RAG-dependent intra-Vκ cluster recombination frequencies for commonly used Vκ gene segments, in both mouse and human, that occur at comparable levels to the bona fide V-Jκ rearrangement frequencies of the same V gene segment. Many of these products are retained in mature, selected splenic B cells and should, therefore, reside on the chromosome rather than on extrachrosmosomal circles.
Sequence conservation of the fRSS is unique because it is embedded within the 3′ end of the V gene segment where it encodes the last 11 residues of the V domain. The nonamer encodes the highly conserved YYC motif that is present, with minor variations, in the V domains of all AgRs (Figure 3B). This raised the possibility that V gene segment clusters of other AgR loci also might contain fRSS, but further experiments showed that the Vκ cluster is unique in that the majority of its CDR3-encoding regions are 23 bp long and, therefore, can serve as a compatible 23RSS for its bona fide 12RSS. The germline-encoded CDR3 in Vκ is uniquely long (7–8 amino acids) and functions to span the gap that in other loci is filled by non-templated nucleotide addition catalyzed by terminal deoxynucleotidyl transferase (TdT), which is not expressed in light-chain-rearranging pre-B cells (Victor et al., 1994). In summary, the unique organization of the Igκ locus with 12-bp spacer bRSSs flanking the V gene segments, the near perfect palindrome structure of the consensus heptamer, the laCκ of TdT activity in pre-B cells compensated by unusually long germline-encoded CDR3, and the nonamercoding potential of the highly conserved YYC residues in FR3 all contributed to the evolutionarily conserved emergence of a flipped 23RSS to mediate intra-Vκ cluster recombination.
Even if structural requirements for a functional 12/23 RRS pair are met, recruitment of the RAG proteins mediated by RAG2-H3K4me3 interactions is critical to allow initiation of rearrangement. The discovery of exclusive J gene segment-associated RCs in all AgR loci (Ji et al., 2010) offered an elegant explanation for the spatially and temporarily ordered rearrangement process seen in lymphocyte precursors (Schatz and Ji, 2011). It was unexpected, therefore, that we found an exception to this rule and detected significant RAG protein binding to several Vκ gene segments even in the complete absence of Jκ RSS and Jκ RC. Indeed, the widespread cleavage and recombination activity at the Vκ gene segments independent of Jκ gene segments are fully consistent with the idea that an autonomous, Vκ-associated RC would also be present in pre-B cells. The presence and activity of the Vκ RC in ΔJκ pre-B cells clearly indicate that the two RCs can function independently of each other. We propose that the Vκ RC controls intra-Vκ cluster, whereas the Jκ RC controls V-to-J recombination. We note that an analogous situation exists in the TCRα/δ locus where the embedded TCRδ J gene segments form a RC in the middle of the larger TCRα locus. Early deletional TCRδ rearrangements have been shown to be important for subsequent balanced Vα-to-Jα recombinations to take place in developing thymocytes (Carico et al., 2017). Perhaps the reason why evolution did not maintain 12-mer fRSS and autonomous RC in the Vα cluster is because the embedded TCRδ locus provides an internal RC proximal to the Vα genes.
The sheer magnitude of intra-Vκ cluster recombination and its evolutionary conservation imply an important physiological role in Igκ repertoire formation. Rearrangements mediated by bRRS-fRSS pairs could occur either by deletion or inversion, depending on the orientation of the participating RSS. Both reactions would result in a loss of the participating Vκ gene segments from the final V-Jκ repertoire due to irreversible damage to the bRSS. In addition, deletion would remove multiple additional Vκ gene segments contained within the intervening sequences (Figure 5C).
The presence and activity of the Vκ RC in ΔJκ pre-B cells clearly indicate that the two RCs can function independently of each other. We have found that the probability of intra-Vκ cluster recombination was highest among V gene segments flanked by high RIC score bRSS. This would lead to a more balanced use of Vκ gene segments in the final V-Jκ repertoire. Our model adds a fundamentally different layer to the complex regulation of Vκ gene segment use, which includes the dynamic regulation of the 3D conformation of the Igκ locus (Lin et al., 2012) and the developmental activation of intra-Vκ enhancer elements, such as E88 (Barajas-Mora et al., 2019). Generally, intra-Vκ cluster recombination events occur in close proximity to the bait Vκ gene segment (Figure 2A), in accordance with the RAG scanning model of V(D)J recombination (Hu et al., 2015; Jain et al., 2018), which restricts recombination to gene segments located within the same chromatin loop domain. Based on the RAG scanning model, Vκ gene segments near to convergent CBE sites are predicted to be the most frequent targets for rearrangement controlled by the Jκ RC. The presence of an additional, possibly competing RC in the Vκ cluster could further increase the probability of other, CBE-distal V gene segments to gain access to RAG proteins anchored at the Jκ RC and undergo productive V-to-Jκ rearrangements.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Andre Nussenzweig (andre_nussenzweig@nih.gov). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture and Mice
Abelson-transformed pre-B (v-Abl) cell lines WT, Rag1−/−, Lig4−/−, and Artemis−/− (Clone1 and Clone2) were obtained from Barry Sleckman (Bredemeyer et al., 2006). In all experiments, pre-B cells were arrested in G1 with 3 μM STI-571 (imatinib) for 48 hr (Bredemeyer et al., 2006). ΔJk and ΔJk-RS clones were generated by CRISPR/Cas9-mediated gene deletion. Ten million v-Abl cells were electroporated with hCas9 plasmid (Addgene 41815), expression plasmids for two guide RNA (gRNAs) targeting sequences that flank the region to be deleted, and a plasmid encoding hCD4 (pcDNA3.1-Hygro-delta-hCD4). hCD4+ cells were purified 24 hour post-transfection using magnetic beads (STEMCELL Technologies 18052), passaged for ~7 days, subcloned by single cell sorting, and screened for deletions using multiple independent primer pairs outside and inside of the gRNA target sites. The gRNA sequences are shown in Table S1. gRNAs were cloned into pKLV-U6gRNA(BbsI)-PGKpuro2ABFP (Addgene 62348). PCR primers for screening the deletions are provided in Table S1. PCR products spanning deletion sites were purified and confirmed with Sanger sequencing.
Mature resting B cells were obtained from spleens of 6–12 week old wild-type C57BL/6 mice using magnetic depletion with anti-CD43 MicroBeads (Miltenyi Biotech). Primary mouse bone marrow pre-B cells for END-Seq analyses were obtained from 6–12 week old Vavp-Bcl2-transgenic mice (Ortega-Molina et al., 2015) and cultured in the presence of recombinant IL-7 (ThermoFisher) for 7–10 days at 2 × 106 cells/ml in vitro. To induce cell cycle arrest, cells were resuspended in media without IL-7 and maintained at 2 × 106 cells/ml for 48 hours and harvested for END-seq. Primary mouse bone marrow pre-B cells for HTGTS analyses were obtained from 6–12 week old wild-type, C57BL/6 mice with flow cytometry cell sorting on a FACSAria II cell sorter (BD Biosciences) using a combination of anti-B220, anti-CD43 (BioLegend), anti-CD24 (ThermoFisher/eBioscience) and anti-IgM antibodies (BD Biosciences) (Lin et al., 2016). Both male and female mice were indiscriminately used for these studies. Human peripheral blood B cells were isolated from deidentified, donated PBMC using human B cell isolation kit (STEMCELL Technologies 17954). All animal experimentation was approved by the NCI Animal Care and Use Committee.
METHOD DETAILS
END-seq
Single cell suspension of v-Abl cells (30 million), primary pre B cells (10 million) or thymocytes (70 million) were embedded in a single agarose plug (1% agarose final). Embeded cells were lysed and digested using Proteinase κ (50°C, 1 hour then 37°C for 7 hours). Plugs were rinsed in TE buffer and treated with RNase A at 37°C for 1 hour and DNA ends were blunted. DNA was retained in agarose plugs to prevent shearing throughout the ssDNA blunting reactions. The first blunting reaction was performed using ExoT (NEB, M0265S) for 45 min, 24°C. After blunting, two washes were performed in NEB Buffer 2 (1x), followed by A-tailing to attach dA to the free 3′-OH (Klenow 3′- > 5′exo-, NEB, M0212S). The A-tailed product was ligated to “END-seq hairpin adaptor 1,” listed in reagents section, using NEB QuiCκ Ligation Kit (NEB, M2200S).
Agarose plugs were melted and dissolved, and DNA was sonicated to a median shear length of 170bp using a Covaris S220 sonicator for 4 min at 10% duty cycle, peak incident power 175, 200 cycles per burst at 4°C. DNA was ethanol-precipitated and dissolved in 70 μL TE buffer. 35 μL of Dynabeads were washed twice with 1 mL Binding and Wash Buffer (1xBWB) (10 mM Tris-HCl pH8.0, 1 mM EDTA, 1 M NaCl, 0.1% Tween20). Beads were recovered using a DynaMag-2 magnetic separator (12321D, Invitrogen). Supernatants were discarded. Washed beads were resuspended in 70 μL 2xBWB (10 mM Tris-HCl pH8.0, 2 mM EDTA, 2 M NaCl) combined with the 70 μL of sonicated DNA and incubated at 24°C for 30 min in a ThermoMixer C at 400 rpm.
Following 30 min mixing, the supernatant was removed and the bead-bound biotinylated DNA was washed 3 times with 1 mL 1xBWB, twice with 1 mL EB buffer, once with 1 mL T4 ligase reaction buffer (NEB) and then resuspended in 50 mL of end-repair reaction mix (0.4 mM of dNTPs, 2.7 U of T4 DNA polymerase (NEB), 9 U of T4 Polynucleotide Kinase (NEB) and 1 U of Klenow fragment (NEB). The end-repair reaction was incubated at 24°C for 30 min in a ThermoMixer C at 400 rpm (tubes were vortexed every 10 min). The supernatant was removed using a magnetic separator and beads were then washed once with 1 mL 1xBWB, twice with 1 mL EB buffer, once with 1 mL NEBNext dA-Tailing reaction buffer (NEB) and then resuspended in 50 mL ofA-tailing reaction with NEBNext dA-Tailing reaction buffer (NEB) and 20 U of Klenow fragment exo- (NEB). The A-tailing reaction was incubated at 37°C for 30 min in a ThermoMixer C at 400 rpm (tubes were vortexed every 10 min). The supernatant was removed using a magnetic separator and washed once with 1 mL NEBuffer 2 and then resuspended in 115 mL of Ligation reaction with QuiCκ Ligase buffer (NEB), 6,000 U of QuiCκ Ligase (NEB) and ligated to “END-seq hairpin adaptor 2” and incubated at 25°C for 30 min in a ThermoMixer C at 400 rpm. Ligation reactions were stopped by adding 50 mM of EDTA, then beads were washed 3 times with BWB and 3 times with EB, and eluted in 8 mL of EB. Hairpin adaptors were digested using USER enzyme (NEB, M5505S) at 37°C, 30 minutes. PCR amplification was performed in 50 mL reaction with 10 mM primers 5′-CAAGCAGAAGACGGCATACGA-GATNNNNNNGTGACTG GAGTTCAGACGTGTGCTCTTCCGATC*T-3′ and 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTT CCGATC*T-3′, and 2X Kapa HiFi HotStart Ready mix (Kapa Biosciences), where * represents a phosphothiorate bond and NNNNNN a Truseq index sequence. PCR amplification was performed at 98°C, 45 s; 15 cycles [98°C, 15 s; 63°C, 30 s; 72°C, 30 s]; 72°C, 5 min. PCR reactions were cleaned with AMPure XP beads, and 200–500 bp fragments were isolated after running on 2% agarose gel. Libraries were purified using QIA-quiCk Gel Extraction Kit (QIAGEN). Library concentration was determined with KAPA Library Quantification Kit for Illumina Platforms (Kapa Biosystems). Sequencing was performed on using Illumina NextSeq 500 or 550 (75bp single end reads). A more detailed END-seq protocol can be found in Canela et al. (2016), (2017).
ChIP-Seq
ChIP-seq was performed as described previously (Canela et al., 2017). Rabbit monoclonal antibodies for RAG1 (mAb 23) and RAG2 (mAb 39) (Coster et al., 2012) and a polyclonal rabbit antibody for H3K4me3 (Millipore) were used. Cells were fixed by adding 37% formaldehyde (F1635, Sigma) to a final concentration of 1% and incubated at 37°C for 10 min. Fixation was quenched by addition of 1M glycine (Sigma) in PBS at a final concentration of 125 mM. Twenty million fixed cells were washed twice with cold PBS and pellets were snap frozen in dry ice and stored at 80°C. Fixed cell pellets of 20 million cells were thawed on ice and resuspended in 2 mL of cold RIPA buffer (10 mM TrisHCl pH 7.5, 1 mM EDTA, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 1 Complete Mini EDTA free proteinase inhibitor (Roche)). Sonication was performed using the Covaris S220 sonicator at duty cycle 20%, peak incident power 175, cycle/burst 200 for 30 min at 4°C or using the Branson sonifier at amplitude 35%, 12 cycles of 20 s sonication and 30 s of pause at 4°C. Chromatin were clarified by centrifugation at 21,000 g at 4°C for 10 min and precleared with 80 ul prewashed Dynabeads protein A (ThermoFisher) for 30 min at 4°C. 40 μL prewashed Dynabeads protein A were incubated with 10 μg of each respective antibody in 100 μL of PBS for 10 min at room temperature in continuous mixing, washed twice in PBS for 5 min and added to 1 mL of chromatin followed by overnight incubation at 4°C on a rotator. Beads were then collected in a magnetic separator (DynaMag-2 Invitrogen), washed twice with cold RIPA buffer, twice with RIPA buffer containing 0.3M NaCl, twice with LiCl buffer (0.25 M LiCl, 0.5% Igepal-630, 0.5% sodium deoxycholate), once with TE (10 mM Tris pH 8.0, 1mM EDTA) plus 0.2% Triton X-100, and once with TE. Crosslinking was reversed by incubating the beads at 65°C for 4 hr in the presence of 0.3% SDS and 1mg/mL of Proteinase κ (Ambion). DNA was purified using Zymo ChIP DNA clean and concentrator kit (Zymo Research) and eluted in 20 μl. The entire ChIP DNA was used to prepare Illumina sequencing libraries. End-repair was performed in 75 μL of T4 ligase reaction buffer, 0.4 mM of dNTPs, 4 U of T4 DNA polymerase (NEB), 13.5 U of T4 Polynucleotide Kinase (NEB) and 1.5 U of Klenow fragment (NEB) at 24°C for 30 min in a ThermoMixer C at 400 rpm. End-repair reaction was cleaned using 2X Agencourt AMPure XP beads and eluted in 15 μL of EB that was used for A-tailing reaction in 30 μL of NEBNext dA-Tailing reaction buffer (NEB) with 7.5 U of Klenow fragment exo- (NEB) at 37°C for 30 min. The 30 μL of the A-tailing reaction was mixed with QuiCκ Ligase buffer 2X (NEB), 3,000 U of Quick ligase and 5 nM of annealed adaptor (Illumina truncated adaptor) in a volume of 75 μL and incubated at 25°C for 20 min. The adaptor was prepared by annealing the following HPLC oligos: 5′-phosphate/GATCGGAAGAGCACACGTCT-3′ and 5′-ACACTCTTTCCCTACACGACGCT CTTCCGATC*T-3′ (*phosphorothioate bond). Ligation was stopped by adding 50mM of EDTA and cleaned with 1.8X Agencourt AM- Pure XP beads and eluted in 15ul of EB that was used for PCR amplification in a 50 uL reaction with 1 μM primers TruSeq barcoded primer p5, AATGATACGGCGACCACCGAGATCTACACNNNNNNNNACACTCTTTCCCTACACGACGCTCTTCCGATC*T, TruSeq barcoded primer p7, CAAGCAGAAGACGGCATACGAGANNNNNNNGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC*T, (NNNNNNNN represents barcode and * a phosphothiorate bond), and 2X Kapa HiFi HotStart Ready mix (Kapa Biosciences). The temperature settings during the PCR amplification were 45 s at 98°C followed by 15 cycles of 15 s at 98°C, 30 s at 63°C, 30 s at 72°C and a final 5 min extension at 72°C. PCR reactions were cleaned with Agencourt AMPure XP beads (BeCκman Coulter), run on a 2% agarose gel and a smear of 200–500bp was cut and gel purified using QIAquiCk Gel Extraction Kit (QIAGEN). Library concentration was determined with KAPA Library Quantification Kit for Illumina Platforms (Kapa Biosystems). Sequencing was performed on the Illumina Nextseq500 (75bp single-end reads).
HTGTS
HTGTS was performed as described (Hu et al., 2015). Genomic DNA was extracted from splenic B cells, isolated B cells from human PBMC, or v-Abl cells arrested in G1 for 3 days by treatment with 3 μM of STI-571. Briefly, 20–50 ug of DNA was fragmented via sonication on a. Diagenode bioruptor and subjected to linear PCR amplification with a biotinylated primer. Single-stranded PCR products were purified via Dynabeads MyONE C1 streptavidin beads (Life Technologies, 65002) and ligated to bridge adaptors. Adaptor-ligated products were amplified via nested PCR with indexed locus-specific primers and primer annealed to adaptor. The PCR products were further tagged with Illumina sequencing adaptor sequences, size-selected (fragment size 500 – 1,000 bp) via gel extraction and loaded to Illumina MiSeq for paired-end 250 bp sequencing. For translocation analysis, the standard LAM-HTGTS bioinformatic pipeline was used (Hu et al., 2015). Primer information can be found in Table S1.
Generation of Cas9/gRNA DSBs for HTGTS Libraries
Cas9/gRNA was design to generate DSBs 654 bp downstream of the Vκ1–117 bRSS breakage site. HTGTS primer was design that allowed 5′ broken ends of these DSBs to be used as a bait. WT v-Abl cells were treated with 3 μM imatinib at a concentration of 5×105 cells/ml for 30 hours followed by nucleofection of pX330-Cas9-Vκ1–117-CRSPR plasmid (15 μg) into 20×106 G1-arrested cells (2 reactions; 10×106 cells each reaction) using X-001 program of Amaxa Nucleofector II (Lonza) with Human B cell Nucleofector kit (Lonza). Transfected cells were cultured in DMEM medium with 15% (v/v) FBS plus 3 μM imatinib for 3 more days before harvested for genomic DNA. HTGTS libraries were prepared and processed as described above. Primer information can be found in Table S1.
Genome Alignment
For END-seq and ChIP-seq, tags were aligned to the mouse (GRCm38p2/mm10) or human (GRCm37/hg19) genomes using Bowtie (version 1.1.2) (Langmead et al., 2009) with the options–best–all –strata–l 50. For ChIP-seq, we allow 2 mismatches and discarded tags with multiple alignments (-n 2 -m 1). For END-seq, we allowed 3 mismatches and kept the best strata for tags with multiple alignments (-n 3 –k 1). HTGTS reads, and the identification of sites that underwent chromosomal translocations to the bait site, were done using HTGTS pipeline, as described earlier (Hu et al., 2016)
Genomic annotations
bRSSs were identified at the flanks of each Ig/TCR segment as defined by GENCODE version M22 (Ensmbl 97) (Hunt et al., 2018). Ig/TCR CDR3 and FR3 were obtained from IMGT data base. In cases where more than one allele existed for a gene segment, all alleles were included. Vκ loop annotation was taken from published data (Karki et al., 2018).
Sequence Conservation
Sequence conservation around RSSs was calculated as the averaged PhastCons (60-vertebrates) (Siepel et al., 2005) conservation score over all sequences at a specific position.
Calculating RAG mediated breakage from END-seq data
Total RAG mediated breakage in a locus was calculated as the total breakage within the locus that occurs precisely at the 5′ of the minimal RAG target sequence – CAC/GTG. Signal was calculated as reads-per-million (RPM).
Heptamer/Nonamer/RSS scores
To estimate the strength of individual heptamer/nonamer, a position-weight-matrix (PWM) was first built based on Vκ bRSSs. Using this PWM, heptamer/nonamer score was defined as , where PWMi,j is the probability to for nucleotide i at position j in Vκ RSS, Si,j is 1 if heptamer/nonamer S has nucleotide i at position j, and 0 otherwise, and Ej is the entropy at position j.
RSS strength quantification is also obtained by using the published online algorithm (Merelli et al., 2010), which calculates RIC scores (Cowell et al., 2002).
Estimating Probability of Deletion of Vκ segment
The probability of a Vκ segment to get deleted is obtained by combination of three factors.
The first is the sum of probabilities of all the possible 12/23 (bfRSS/flRSS) pairs (see Figure S4B for illustration). This probability, p(pair) was calculated as the product of the probability of the two RSS to be functional, which in turn is estimated as MinMax function of the RSS RIC score (p12 and p23 for 12 and 23RSS, respectively):
The second is a binary factor, r, indicating whether RAG binds to at least one of the RSS pair mates based on the RAG ChIP seq data (Figures 4A and 4E).
The third factor, l, is a loop coefficient, giving higher pairing probability for RSS pairs that share the same loop (see Genomic Annotations section above).
Using the above factors, p(del) can be described as:
where k ≥ min(i,j) and k ≤ max(i,j).
Data Visualization
Aligned-reads bed files were first converted to bedgraph files using bedtools genomecov (Quinlan and Hall, 2010) following by bedGraphToBigWig to make a bigwig file (Kent et al., 2010). Visualization of genomic profiles was done by the UCSC browser (Kent et al., 2002). Genome browser profiles were normalized to present RPM.
Heatmaps were produced using the R package pheatmap. For Seq logo representation, position-weight-matrices (PWMs) was first produced from the array of sequences. These PWMs were used as an input for seq logo using R package seqLogo.
QUANTIFICATION AND STATISTICAL ANALYSIS
Fisher test has been used for comparison of fraction of breakage between fRSS and other Vκ heptamers (Figure 1C), the CDR3 size of the different AgR loci (Figure 3C), and breakage fraction of bRSS and fRSS between Igκ and TCRα (Figure S3). t test, has been used to compare 12RSS and 23RSS nonamer scores between broken and not broken fRSS, and to compare RIC scores and deletion scores between different usage groups. Paired t test has been used to compare bRSS and fRSS (Figure 1H) breakage and RAG1 binding (Figure 4D) between WT and ΔJΚ cells. Finally, pearson correlation has been performed between usage and RIC score, and combination of RIC and deletion scores (Figure 5B). All statistical test above were performed using R version 3.6.1 (https://www.r-project.org/).
DATA AND CODE AVAILABILITY
The accession number for the sequencing data reported in this paper is GEO: GSE140677.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE Antibodies | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-RAG1 (mAb 23) | Coster et al., 2012 | N/A |
| Rabbit monoclonal anti-RAG2 (mAb 39) | Coster et al., 2012 | N/A |
| Rabbit polyclonal anti-H3K4me3 | Millipore | Cat# 07-473; RRID: AB_1977252 |
| eFluor 450 Rat monoclonal CD24 (M1/69) | Thermo Fisher Scientific | Cat# 48-0242-82; RRID: AB_1311169 |
| APC Rat monoclonal CD43 (S11) | BioLegend | Cat# 143207; RRID: AB_11149489 |
| APC/Cy7 Rat anti-Mouse/Human B220 (RA3-6B2) | BioLegend | Cat# 103224; RRID: AB_313007 |
| FITC Rat anti-Mouse IgM (II/41) | BD Biosciences | Cat# 553437; RRID: AB_394857 |
| Bacterial and Virus Strains | ||
| Bacteria: TOP10 Chemically Competent E. coli | Thermo Fisher Scientific | Cat# C404006 |
| Lentivirus: pKLV-U6gRNA-EF(BbsI)-PGKpuro2ABFP | Addgene | RRID:Addgene_62348 |
| Mammalian expression: hCas9 | Addgene | RRID:Addgene_41815 |
| Mammalian expression: pcDNA3.1-Hygro-delta-hCD4 | Gift from Eugene Oltz | N/A |
| Mammalian expression: pX330-U6-Chimeric_BB-CBh-hSpCas9 | Addgene | RRID:Addgene_42230 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dynabeads Protein A | Thermo Fisher Scientific | Cat# 10002D |
| CD43 microbeads (Ly-48) | Miltenyi Biotec | Cat# 130-049-801 |
| Imatinib mesylate (STI-571) | Selleckchem | Cat# S1026 |
| DAPI | Thermo Fisher Scientific | Cat# 62248 |
| cOmplete, Mini Protease inhibitor cocktail | Roche Diagnostics | Cat# 11836153001 |
| Puregene Proteinase K enzyme | QIAGEN | Cat# 158920 |
| Puregene RNase A Solution | QIAGEN | Cat# 158924 |
| T4 DNA Polymerase | NEB | Cat# M0203L |
| T4 Polynucleotide Kinase | NEB | Cat# M0201L |
| DNA Polymerase I, Large (Klenow) Fragment | NEB | Cat# M0210L |
| Exonuclease T (ExoT) | NEB | Cat# M0265L |
| Klenow Fragment (3′→5′ exo-) | NEB | Cat# M0212L |
| Quick Ligation Kit | NEB | Cat# M2200L |
| USER enzyme | NEB | Cat# M5505L |
| KAPA HiFi HotStart ReadyMix (2X) | KAPA Biosystems | Cat# KK2600 |
| MyOne Streptavidin C1 Beads | ThermoFisher | Cat# 650-01 |
| Agencourt AMPure XP beads | Beckman Coulter | Cat# A63881 |
| Hexaamminecobalt(III) chloride | Sigma-Aldrich | Cat# 481521 |
| T4 DNA Ligase | Promega | Cat# M1804 |
| Phusion High-Fidelity PCR Master Mix with HF buffer | NEB | Cat# M0531L |
| Critical Commercial Assays | ||
| KAPA Library Quantification Kit | Kapa Biosciences | Cat# KK4824 |
| CHEF Mammalian Genomic DNA plug kit | Bio-Rad | Cat# 1703591 |
| EasySep Human CD4 Positive Selection Kit II | STEMCELL technologies | Cat# 18052 |
| EasySep Human B Cell Isolation Kit | STEMCELL technologies | Cat# 17954 |
| Human B cell Nucleofector Kit | Lonza | Cat# VPA-1001 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GSE140677 |
| ChIP-seq for RAG1 | Teng et al., 2015 | GSE69478 |
| Experimental Models: Cell Lines | ||
| Pre-B cell lines | Bredemeyer et al., 2006 | N/A |
| Pre-B cell lines: ΔJk | This paper | N/A |
| Pre-B cell lines: ΔJk-RS | This paper | N/A |
| Pre-B cell lines: ΔRag1−/− | Bredemeyer et al., 2006 | N/A |
| Pre-B cell lines: Lig4−/− | Bredemeyer et al., 2008 | N/A |
| Pre-B cell lines: Artemis−/− clone1 | Bredemeyer et al., 2008 | N/A |
| Pre-B cell lines: Artemis−/− clone2 | Bredemeyer et al., 2008 | N/A |
| Experimental Models: Organisms/Strains | ||
| C57BL/6NCr mice | Charles River | Strain code# 027 |
| Mouse: Vavp-Bcl2 Tg | Ortega-Molina et al., 2015 | N/A |
| Oligonucleotides | ||
| sgRNA to generate ΔJk v-Abl cells: Sense 5′-AAGCATGCGTGGAAGCGCTT-3′; Antisense 5′-GGGCTCATTATCAGTTGACG-3′ | This paper | N/A |
| sgRNA to generate ΔJk-RS v-Abl cells: Sense 5′-ATCACACGTATAGAGTAAGC-3′; Antisense 5′-CCTGCCCACACGACTCCTTC-3′ | This paper | N/A |
| Primers for genotyping ΔJk allele: ΔJk-Fw, 5′-ACTAACTGCTGAGCCACCTC-3′; ΔJk-Rv, 5′-GCAGTCAGACCCAGATCTCAA-3′; ΔJk-Intact-Rv, 5′-AGCCACAGACATAGACAACGG-3′ | This paper | N/A |
| Primers for genotyping ΔJk-RS allele: ΔJk-RS-Fw, 5′-ACCTGGGGAACAAAACTGGA-3′; ΔJk-RS-Rv, 5′-AATCTGCCTGTCTGAAGCCC-3′; ΔJk-RS-Intact-Rv, 5′-GGAAGACAAAGGAGGCCACG-3′ | This paper | N/A |
| sgRNA to downstream of Vk1–117: Vk1–117 CRISPR-Cas9 5′-TTGCTACATATCTGGCACCG-3′ | This paper | N/A |
| END-seq adaptor 1, 5′-phosphate -GATCGGAAGAGCGTCGTGTAGGGAAAGAGTGUU[Biotin-dT]U[Biotin-dT]UUACACTCTTTCCCTACACGACGCTCTTCCGATC*T-3′ | Canela et al., 2016 | N/A |
| END-seq adaptor 2, 5′-phosphate -GATCGGAAGAGCACACGTCUUUUUUUUAGACGTGTGCTCTTCCGATC*T-3′ | Canela et al., 2016 | N/A |
| TruSeq barcoded primer p5, 5′-AATGATACGGCGACCACCGAGATCTACACNNNNNNNNACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ (N = barcode) | Canela et al., 2019 | N/A |
| TruSeq barcoded primer p7, 5′-CAAGCAGAAGACGGCATACGAGANNNNNNNGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′ (N = barcode) | Canela et al., 2019 | N/A |
| Illumina truncated adaptor 1, 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATC*T-3′ | Canela et al., 2017 | N/A |
| Illumina truncated adaptor 2, 5′-phosphate-GATCGGAAGAGCACACGTCT-3′ | Canela et al., 2017 | N/A |
| Adaptor-upper 5′-GGTACACGACGCTCTTCCGATCTNNNNNN/3AmMO/-3′ | Canela et al., 2019 | N/A |
| Adaptor-lower 5′-phosphate-AGATCGGAAGAGCGTCGTGTACC/3AmMO/-3′ | Canela et al., 2019 | N/A |
| I5-bridge 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ | Canela et al., 2019 | N/A |
| P5-I5c 5′-AATGATACGGCGACCACCGAGATCTACACTCTTT*C-3′ | Canela et al., 2019 | N/A |
| P7-I7c 5′-CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTT*C-3′ | Canela et al., 2019 | N/A |
| Jk1-bio 5′-/Biosg/TTCCCAGCTTTGCTTACGGAG-3′ | Lin et al., 2016 | N/A |
| I7-Jk1 -nested-barcode 5′-CTCGGCATTCCTGCTGAACCGCTCTTCCGATCTNNNNNNNNAGTGCCAGAATCTGGTTTCAGAG-3′ (N = barcode) | This paper | N/A |
| Vk1–117-bio 5′-/5Biosg/CAGAAGCCTTCAGTATGCACCA-3′ | This paper | N/A |
| I7-Vk1–117-nested-barcode 5′-CTCGGCATTCCTGCTGAACCGCTCTTCCGATCTNNNNNNNNCAGGGACAGATTTCACACTCAAG-3′ (N = barcode) | This paper | N/A |
| Vk8–34-bio 5′-/5Biosg/CAGAAACCAGGACGATCTCCT-3′ | This paper | N/A |
| I7-Vk8–34-nested-barcode5′-CTCGGCATTCCTGCTGAACCGCTCTTCCGATCTNNNNNNNNACTAGGGTATCTGGAGTCCCTG-3′ | This paper | N/A |
| bio-human-IGJK1 5′-/5Biosg/TCCCCAGGACATTTCTGAAG-3′ | This paper | N/A |
| I7-human-IGJK1-barcode 5′-CTCGGCATTCCTGCTGAACCGCTCTTCCGATCNNNNNNNNGGGCTGATTGCAGAGTCACCT-3′ | This paper | N/A |
| bio-human-IGVK2–28 5′-/5Biosg/TAACTTTGCAATTCATTATTTCAGGA-3′ | This paper | N/A |
| I7-human-IGVK2–28-barcode5′-CTCGGCATTCCTGCTGAACCGCTCTTCCGATCNNNNNNNNCTGGATACAACTATTTGGATTGG-3′ | This paper | N/A |
| bio-human-IGVK3–20 5′-/5Biosg/GCACCCTGTCTTTGTCTCCA-3′ | This paper | N/A |
| I7-human-IGVK3–20-barcode5′-CTCGGCATTCCTGCTGAACCGCTCTTCCGATCNNNNNNNNTATCAGCAGACTGGAGCCTGAA-3′ | This paper | N/A |
| Software and Algorithms | ||
| Prism 8 | GraphPad | https://www.graphpad.com/ |
| RStudio | RStudio Team | https://rstudio.com/ |
| Bowtie 1.1.2 | Langmead et al., 2009 | https://sourceforge.net/projects/bowtie-bio/files/bowtie/1.1.2/ |
| MACS 1.4.3 | Zhang et al., 2008 | https://pypi.org/pypi/MACS/1.4.3 |
| UCSC database | Karolchik et al., 2004 | https://genome.ucsc.edu |
| UCSC genome browser | Kent et al., 2002 | https://genome.ucsc.edu |
| Bedtools | Quinlan and Hall, 2010 | https://github.com/arq5x/bedtools2 |
| R 3.6.1 | R Core Team | https://www.r-project.org/ |
| FlowJo (10.1) | FlowJo LLC | https://www.flowjo.com/ |
| Other | ||
| Amaxa Nucleofector II | Lonza | N/A |
| FACSAria II cell sorter | BD Biosciences | N/A |
Highlights.
Vκ gene segments rearrange within the Vκ cluster independent of Jκ gene segments
Intra-Vκ recombination is mediated by flipped RSS embedded in bona fide RSS
An independent RAG recombination center assembles in the Vκ cluster
Intra-Vκ cluster recombination influences ultimate Vκ gene segment usage
ACKNOWLEDGMENTS
We thank Barry P. Sleckman for discussions and the pre-B cells lines; and Frederick W. Alt for suggestions and HTGTS protocols. D.G.S. was supported by NIH grant R01 AI32524 and K.S. was supported by a fellowship from the Uehara Memorial Foundation. Sequencing was performed by NCI CCR Genomics Core and flow cytometry analyses and sorting were performed at the NCI CCR LGI Flow Cytometry Core. The A.N. laboratory is supported by the Intramural Research Program of the NIH; an Ellison Medical Foundation Senior Scholar in Aging Award (AG-SS-2633-11); the Department of Defense Idea Expansion (W81XWH-15-2-006) and Breakthrough (W81XWH-16-1-599) awards; the Alex Lemonade Stand Foundation Award; and an NIH Intramural FLEX Award.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.11.088.
REFERENCES
- Aoki-Ota M, Torkamani A, Ota T, Schork N, and Nemazee D. (2012). Skewed primary Igκ repertoire and V-J joining in C57BL/6 mice: implications for recombination accessibility and receptor editing. J. Immunol 188, 2305–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas-Mora EM, Kleiman E, Xu J, Carrico NC, Lu H, Oltz EM, Murre C, and Feeney AJ (2019). A B-Cell-Specific Enhancer Orchestrates Nuclear Architecture to Generate a Diverse Antigen Receptor Repertoire. Mol. Cell 73, 48–60.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer AL, Helmink BA, Innes CL, Calderon B, McGinnis LM, Mahowald GK, Gapud EJ, Walker LM, Collins JB, Weaver BK, et al. (2008). DNA double strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature 456, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, and SleCκman BP (2006). ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature 442, 466–470. [DOI] [PubMed] [Google Scholar]
- Canela A, Sridharan S, Sciascia N, Tubbs A, Meltzer P, SleCκman BP, and Nussenzweig A. (2016). DNA Breaks and End Resection Measured Genome-wide by End Sequencing. Mol. Cell 63, 898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Huang SN, Wutz G, Tang W, Zagnoli-Vieira G, Callen E, Wong N, Day A, Peters JM, et al. (2019). Topoisomerase IIInduced Chromosome Breakage and Translocation Is Determined by Chromosome Architecture and Transcriptional Activity. Mol. Cell 75, 252–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Jung S, Wong N, Callen E, Day A, Kieffer-Kwon KR, Pekowska A, Zhang H, Rao SSP, et al. (2017). Genome Organization Drives Chromosome Fragility. Cell 170, 507–521.e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carico ZM, Roy Choudhury K, Zhang B, Zhuang Y, and Krangel MS (2017). TCRδ Rearrangement Redirects a Processive TCRα Recombination Program to Expand the TCRα Repertoire. Cell Rep. 19, 2157–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coster G, Gold A, Chen D, Schatz DG, and Goldberg M. (2012). A dual interaction between the DNA damage response protein MDC1 and the RAG1 subunit of the V(D)J recombinase. J. Biol. Chem 287, 36488–36498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell LG, Davila M, Kepler TB, and Kelsoe G. (2002). Identification and utilization of arbitrary correlations in models of recombination signal sequences. Genome Biol 3, research0072.1–research0072.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiderio S. (2010). Temporal and spatial regulatory functions of the V(D)J recombinase. Semin. Immunol 22, 362–369. [DOI] [PubMed] [Google Scholar]
- Durdik J, Moore MW, and Selsing E. (1984). Novel kappa light-chain gene rearrangements in mouse lambda light chain-producing B lymphocytes. Nature 307, 749–752. [DOI] [PubMed] [Google Scholar]
- Feddersen RM, Martin DJ, and Van Ness BG (1990). Novel recombinations of the IG kappa-locus that result in allelic exclusion. J. Immunol 145, 745–750. [PubMed] [Google Scholar]
- Helmink BA, and SleCκman BP (2012). The response to and repair of RAG-mediated DNA double-strand breaks. Annu. Rev. Immunol 30, 175–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zhang Y, Zhao L, FroCκ RL, Du Z, Meyers RM, Meng FL, Schatz DG, and Alt FW (2015). Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell 163, 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Meyers RM, Dong J, Panchakshari RA, Alt FW, and FroCκ RL (2016). Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat. Protoc 11, 853–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SE, McLaren W, Gil L, Thormann A, Schuilenburg H, Sheppard D, Parton A, Armean IM, Trevanion SJ, Flicek P, and Cunningham F. (2018). Ensembl variation resources. Database (Oxford) 2018, bay119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Ba Z, Zhang Y, Dai HQ, and Alt FW (2018). CTCF-Binding Elements Mediate Accessibility of RAG Substrates During Chromatin Scanning. Cell 174, 102–116.e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Resch W, Corbett E, Yamane A, Casellas R, and Schatz DG (2010). The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell 141, 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S, Kennedy DE, Mclean K, Grzybowski AT, Maienschein-Cline M, Banerjee S, Xu H, Davis E, Mandal M, Labno C, et al. (2018). Regulated Capture of Vk Gene Topologically Associating Domains by Transcription Factories. Cell Rep. 24, 2443–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, and Kent WJ (2004). The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D. (2002). The human genome browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, and Karolchik D. (2010). BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu W, Li Y, Zhao S, Liu C, Hu M, Yue W, Liu Y, Wang Y, Yang R, et al. (2016). Contribution of secondary Igκappa rearrangement to primary immunoglobulin repertoire diversification. Mol. Immunol 78, 193–206. [DOI] [PubMed] [Google Scholar]
- Lieberman AE, Kuraoka M, Davila M, Kelsoe G, and Cowell LG (2009). Conserved cryptic recombination signals in Vκappa gene segments are cleaved in small pre-B cells. BMC Immunol. 10, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, and Murre C. (2012). Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat. Immunol 13, 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SG, Ba Z, Du Z, Zhang Y, Hu J, and Alt FW (2016). Highly sensitive and unbiased approach for elucidating antibody repertoires. Proc. Natl. Acad. Sci. USA 113, 7846–7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SG, Ba Z, Alt FW, and Zhang Y. (2018). RAG Chromatin Scanning During V(D)J Recombination and Chromatin Loop Extrusion are Related Processes. Adv. Immunol 139, 93–135. [DOI] [PubMed] [Google Scholar]
- Livak F, and Schatz DG (1996). T-cell receptor alpha locus V(D)J recombination by-products are abundant in thymocytes and mature T cells. Mol. Cell. Biol 16, 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak F, Burtrum DB, Rowen L, Schatz DG, and Petrie HT (2000). Genetic modulation of T cell receptor gene segment usage during somatic recombination. J. Exp. Med 192, 1191–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, and Oettinger MA (2009). RAG: a recombinase diversified. Nat. Immunol 10, 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merelli I, Guffanti A, Fabbri M, Cocito A, Furia L, Grazini U, Bonnal RJ, Milanesi L, and McBlane F. (2010). RSSsite: a reference database and prediction tool for the identification of cryptic Recombination Signal Sequences in human and murine genomes. Nucleic Acids Res. 38, W262–W267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo SA, and Schlissel MS (2003). A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat. Immunol 4, 31–37. [DOI] [PubMed] [Google Scholar]
- Nemazee D. (2017). Mechanisms of central tolerance for B cells. Nat. Rev. Immunol 17, 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Molina A, Boss IW, Canela A, Pan H, Jiang Y, Zhao C, Jiang M, Hu D, Agirre X, Niesvizky I, et al. (2015). The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat. Med 21, 1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudhon C, Hao B, Raviram R, Chaumeil J, and Skok JA (2015). LongRange Regulation of V(D)J Recombination. Adv. Immunol 128, 123–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubelt F, Bolen CR, McGuire HM, Vander Heiden JA, Gadala-Maria D, Levin M, Euskirchen GM, Mamedov MR, Swan GE, Dekker CL, et al. (2016). Individual heritable differences result in unique cell lymphocyte receptor repertoires of naïve and antigen-experienced cells. Nat. Commun 7, 111–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H, Huppi K, Heinrich G, and Tonegawa S. (1979). Sequences a€ t the somatic recombination sites of immunoglobulin light-chain genes. Nature 280, 288–294. [DOI] [PubMed] [Google Scholar]
- Schatz DG, and Ji Y. (2011). Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol 11, 251–263. [DOI] [PubMed] [Google Scholar]
- Shimizu T, and Yamagishi H. (1992). Biased reading frames of pre-existing DH–JH coding joints and preferential nucleotide insertions at VH–DJH signal joints of excision products of immunoglobulin heavy chain gene rearrangements. EMBO J. 11, 4869–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siminovitch KA, Moore MW, Durdik J, and Selsing E. (1987). The human kappa deleting element and the mouse recombining segment share DNA sequence homology. Nucleic Acids Res. 15, 2699–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng G, and Schatz DG (2015). Regulation and Evolution of the RAG Recombinase. Adv. Immunol 128, 1–39. [DOI] [PubMed] [Google Scholar]
- Teng G, Maman Y, Resch W, Kim M, Yamane A, Qian J, Kieffer-Kwon KR, Mandal M, Ji Y, Meffre E, et al. (2015). RAG Represents a Widespread Threat to the Lymphocyte Genome. Cell 162, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. (1983). Somatic generation of antibody diversity. Nature 302, 575–581. [DOI] [PubMed] [Google Scholar]
- van Gent DC, Ramsden DA, and Gellert M. (1996). The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell 85, 107–113. [DOI] [PubMed] [Google Scholar]
- Victor KD, Vu K, and Feeney AJ (1994). Limited junctional diversity in kappa light chains. Junctional sequences from CD43+B220+ early B cell progenitors resemble those from peripheral B cells. J. Immunol 152, 3467–3475. [PubMed] [Google Scholar]
- Vinocur JM, Fesnak AD, Liu Y, Charan D, and Prak ET (2009). Violations of the 12/23 rule at the mouse immunoglobulin kappa locus, including V kappa-V kappa rearrangement. Mol. Immunol 46, 2183–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Bassing CH, Jung D, Woodman BB, Foy D, and Alt FW (2003). Dramatically increased rearrangement and peripheral representation of Vbeta14 driven by the 3′Dbeta1 recombination signal sequence. Immunity 18, 75–85. [DOI] [PubMed] [Google Scholar]
- Yu K, Taghva A, and Lieber MR (2002). The cleavage efficiency of the human immunoglobulin heavy chain VH elements by the RAG complex: implications for the immune repertoire. J. Biol. Chem 277, 5040–5046. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cheng TC, Huang G, Lu Q, Surleac MD, Mandell JD, Pontarotti P, Petrescu AJ, Xu A, Xiong Y, and Schatz DG (2019). Transposon molecular domestication and the evolution of the RAG recombinase. Nature 569, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, EeCκhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, FroCκ RL, Du Z, Hu J, Chen L, Krangel MS, and Alt FW (2016). Orientation-specific RAG activity in chromosomal loop domains contributes to TCRδ V(D)J recombination during T cell development. J. Exp. Med 213, 1921–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the sequencing data reported in this paper is GEO: GSE140677.