Summary
Plants are able to adjust phenotype in response to changes in the environment. This system depends on an internal capacity to sense environmental conditions and to process this information to plant response. Recent studies have pointed to mitochondria and plastids as important environmental sensors, capable of perceiving stressful conditions and triggering gene expression, epigenomic, metabolic and phytohormone changes in the plant. These processes involve integrated gene networks that ultimately modulate the energy balance between growth and plant defense. This review attempts to link several unusual recent findings into a comprehensive hypothesis for the regulation of plant phenotypic plasticity.
Keywords: chloroplast, epigenetics, mitochondria, phenotypic plasticity, retrograde regulation
I. Introduction
Phenotypic plasticity is a concept that encompasses diverse mechanisms to allow plants to adjust their growth behavior in response to environmental change (Schlichting, 1986). In general, plastic growth responses are adaptive and may be heritable, reflecting an accelerated evolution process that facilitates the acclimation of a plant species to new niches. Phenotypic plasticity does not necessarily manifest in morphological changes, and can reflect subtle physiological adjustments in growth rate, day-length response or behavior under stress. The underlying mechanisms that reprogram growth in response to environmental change are only just coming into focus, with a surprising interlinkage of metabolism, stress networks and organellar redox effects on the plant adaptation process (Margalha et al., 2019).
II. Energy-generating organelles as important environmental sensors in plants
Mitochondria and plastids function as signal integrators to link metabolic processes with environmental sensing and can lead to epigenetic changes in the plant. The vast majority of proteins required for mitochondrial and chloroplast function are nuclear-encoded, so nuclear communication with organelles, or anterograde regulation, is essential to adjust organellar properties during development. Retrograde, or organelle-to-nucleus signaling, is mediated by a variety of organelle-generated molecules that direct plant responses (Box 1), many involving environmental sensing (Wang et al., 2020). Not surprisingly, the numerous metabolic intermediates that participate in plastid retrograde signaling are regulated through their organellar export and/or import. For example, plastid signaling involves carotenoid-derived β-cyclocitral and dihydroactiniolide as a consequence of increased singlet oxygen (Ramel et al., 2012), derivatives of the isoprenoid precursor methylerythritol cyclodiphosphate (MEcPP) to influence salicylic acid (SA) and jasmonate pathways (Xiao et al., 2012), or 3′-phosphoadenosine 5′-phosphate, a metabolite affecting 5′–3′ exoribonuclease activity within the nucleus (Estavillo et al., 2011). The spectrum of intermediates identified as components of retrograde signaling point to a vital function of plastid envelope transport systems in the evolution of this intracellular communication (Unal et al., 2020). What remains unclear is how these transport systems partition to specialized plastid types.
Box 1. Signaling metabolites that participate in retrograde regulation.
| Metabolite | References |
|---|---|
| Tetrapyrrole derivatives | |
| Heme | Woodson et al. (2011) |
| GUN4/5 | Mochizuki et al. (2001), Larkin et al. (2003) |
| Mg-ProtoIX | Strand et al. (2003) |
| Carotenoid derivatives | |
| β′ cyclocitral (β-cyc) | Ramel et al. (2012) |
| Dihydroactinidiolide | Shumbe et al. (2014) |
| 3′-Phosphoadenosine 5′-phosphate (PAP) | Estavillo et al. (2011) |
| 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MEcPP) | Xiao et al. (2012) |
| Ca2+ | Guo et al. (2016) |
| Fatty acids (FAs) | |
| Oxylipin | Muñoz & Munné-Bosch (2020) |
| NO2-FAs | Mandal et al. (2012) |
| Free fatty acid (FFAs) | Walley et al. (2013) |
| Dihydroxyacetone phosphate (DHAP) | Alsharafa et al. (2014) |
Retrograde signaling links to nuclear RNA metabolism to broadly impact nuclear gene expression (Zhao et al., 2020). The resulting adaptive responses can be complex, such as repressing photosynthesis-associated nuclear genes while simultaneously inducing photoprotectant anthocyanin accumulation in response to light shifts (Richter et al., 2020) or supporting nuclear microRNA (miRNA) biogenesis under stress conditions to alter development and environmental stress responses (Fang et al., 2019). Similarly expansive nuclear influence on organellar gene expression is accomplished via nuclear-encoded RNA-binding proteins that include organellar ribosome maturation and splicing domain proteins, pentatricopeptide repeat proteins, DEAD-Box RNA helicases and S1-domain containing proteins (Lee & Kang, 2020). The multifarious nature of this coregulation system reflects a post-endosymbiotic evolution process that has equipped the plant system for broad-spectrum responsiveness through coordinated organelle-nuclear networks.
Components of plastid redox regulation, including plastoquinone and tocopherol pools, also influence nuclear gene expression (Havaux, 2020) and reactive oxygen is a vital component of organellar signaling. In plastids, the relationship of plastoquinone to reactive oxygen species (ROS) homeostasis is not yet entirely clear, but PQ-9 can influence photosynthetic acclimation by adjusting ROS signaling with SNT7, a kinase that alters redox homeostasis by phosphorylation of light-harvesting components (Tikkanen et al., 2012). This process of dictating ROS production generates cell-wide signals that alter hormonal networks within the cell (Tikkanen et al., 2012). Likewise, hydrogen peroxide accumulation within the PQ pool can participate directly in retrograde signaling to alter gene expression (Mubarakshina & Ivanov, 2010; Exposito-Rodriguez et al., 2017; Havaux, 2020). This plastid-nuclear ROS signaling can be surprisingly direct through physical interaction of clusters of perinuclear plastids via organellar stromule extensions or plastid-nuclear complexes (Mullineaux et al., 2020).
In mitochondria, potential damage from overactive electron flow is mitigated by alternative oxidase encoded by Alternative oxidase I (AOX1) (Millar, et al., 2011). Investigation of the AOX1 system by forward genetic screens unveils a number of unexpected links between this electron bypass system to cellular growth and metabolism. Regulator of Alternative Oxidase (rao) mutants are identified by their inability to induce AOX1a expression in response to antimycin A, an inhibitor of mitochondrial cytochrome c reductase (Zarkovic, et al., 2005). A diverse set of AOX regulators have been identified through this screen, providing details of mitochondrial association with plant growth functions. RAO1, for example, encodes the Cyclin-Dependent Kinase E1 (CDKE1) (Ng et al., 2013). This gene regulates both AOX1a and Light Harvesting Complex B (LHCB) to integrate mitochondrial and plastid retrograde signals during stress (Blanco et al., 2014). RAO1 also interacts with KIN10 (Ng et al., 2013), a subunit of the Sucrose Nonfermenting-Related Kinase 1 (SnRK1), which balances energy signaling for growth with plant defense response (Baena-González et al., 2007).
Interestingly, RAO mutants also encode components of auxin transport in the plant. These include rao3/big, rao4/pin1, rao5/mdr1/abcb19 and rao6/asymmetric leaves 1 (as1) (Ivanova et al., 2014). Studies of these mutants, together with chemical inhibitors of auxin, reveal an antagonistic relationship between auxin and mitochondrial retrograde signaling, presumably part of the plant’s ability to modulate growth during stress. Auxin can activate the TARGET of RAPAMYCIN (TOR) pathway (Schepetilnikov et al., 2013) and, in plants, the TOR pathway works antagonistically with SnRK1 in adjusting the balance in energy for growth with stress response (Margalha et al., 2019). Discovery of AOX1 in this network serves to interlink these cellular programming controls with mitochondrial status and further elaborates mitochondrial influence at the plant–environment intersection.
Plastid functions appear similarly intrinsic to establishing energy balance and stress perception by the plant. For example, instability of the plastid genome triggers a specific nuclear genome response. Enhanced nuclear endoreplication and altered cell cycle regulation occur in response to chemical and genetic disruption of the plastid genome (Duan et al., 2020). This phenomenon requires the gene SOG1, a putative nuclear transcription factor responsive to DNA damage (Duan, et al., 2020). Plastid genome disruption triggers increased ROS to effect this response and, remarkably, SOG1 interacts with Sucrose Nonfermenting-Related Kinase 1 (SnRK1) (Hamasaki et al., 2019), reiterating the linkage of plastid status with energy homeostasis. Mitochondria maintain their cellular population through fission and fusion, so that functions they perform are relatively uniform in scope across cell types. However, plastids do not fuse and, thus, can undergo spatiotemporal differentiation to acquire specialized properties in various plant tissues (Wise, 2007). These properties include photosynthesis and light perception, cellular metabolism, carbohydrate and lipid storage, and environmental sensing, each requiring specialized plastid proteome components.
III. The potential role of sensory plastids in stress signaling
Sensory plastids reside within epidermal and vascular tissues and differ in size, thylakoid structure and proteome composition from the mesophyll chloroplast of neighboring cells (Beltrán et al., 2018). The sensory plastid, while presumably photosynthetic (Barton et al., 2016), contains a proteome enriched in components of shikimate pathway-associated metabolism and stress response. Estimating the extent to which sensory plastids participate in retrograde signaling and environmental sensing is confounded by the fact that past and present studies of chloroplast functions are generally carried out in experiments that pool mesophyll and sensory plastids.
Perhaps the most well-detailed plastid retrograde signaling pathway, mediated by SAL1 and 3′-phosphoadenosine 5′-phosphate (PAP), appears to be sensory plastid-associated. The redox-regulated phosphatase SAL1 localizes to both mitochondria and plastids, with predominant expression in vascular tissues, and regulates the concentration of PAP by dephosphorylation to adenosine monophosphate (Estavillo et al., 2011). A by-product of sulfur metabolism, PAP is transferred to the nucleus where it influences gene expression by inhibiting XRN type exoribonucleases (Estavillo et al., 2011; Litthauer & Jones, 2018). These exoribonucleases target miRNAs that can broadly influence plant response. Tocopherols, derived from tyrosine in the sensory plastid, serve to upregulate PAP and, consequently, nuclear miRNA biogenesis (Fang et al., 2019). What links this process to the sensory plastid is its dependence on CUE1 (Fang et al., 2019), a phosphoenolpyruvate import protein that resides on the inner membrane of the sensory plastid (Lundquist, et al., 2014; Beltrán et al., 2018). Disruption of CUE1 (also named PPT1) results in a reticulata green venation phenotype because of its association with vascular plastids, while its counterpart, PPT2, serves to import PEP within mesophyll chloroplasts (Hilgers et al., 2018). Genetic evidence of CUE1 dependence for the tocopherol-influenced XRN activity locates SAL1-PAP signaling to the sensory plastid and to vascular tissues of the plant, but parallel association of this pathway to mesophyll chloroplasts remains unclear.
One means of rescuing the cue1 mutant, and other sensory plastid-associated reticulata mutants, is by supplementing with aromatic amino acids (Streatfield et al., 1999). This observation is consistent with proteome studies showing sensory plastid enrichment for components of the shikimate pathway (Beltrán et al., 2018). Aromatic amino acid metabolism plays a significant role in plant defense, and effector-triggered immunity is characterized by significant increases in phenylalanine pools, and phenylpropanoid metabolism more generally (Yoo et al., 2020). Thus, specialized plastids may also play a previously undetailed role in biotic stress response.
ICS2, a gene encoding isochorismate synthase, converts chorismate to isochorismate during the biosynthesis of phylloquinone, a component of electron transport. This enzyme is also encoded by ICS1, and both participate in SA biosynthesis, an important component of plant defense (Garcion et al., 2008). The genes are differentially regulated by environmental factors (Macaulay et al., 2017), but ICS2 predominantly localizes to vascular tissue. Consistent with this localization, sensory plastid perturbation induces upregulation of the SA pathway (Shao et al., 2017; Yang et al., 2020).
Fluctuating light is an important environmental factor that elicits plastid retrograde signaling in plants. In response, epidermal plastids display dynamic stress-response morphological behaviors that are distinct from neighboring mesophyll chloroplasts. These visible changes involve abundant stromule production, perinuclear association and H2O2 plastid-nuclear transfer (Brunkard et al., 2015; Exposito-Rodriguez et al., 2017). Studies of this signal, using an elegant fluorescent sensor, show H2O2 within plastids of epidermal cells that align in physical association with the nucleus (Exposito-Rodriguez et al., 2017). While aspects of this light response, as well as plastid stromule production, can also occur in mesophyll chloroplasts, epidermal and mesophyll plastids are distinct systems as evidenced in their response to sucrose and photosynthesis inhibitors (Brunkard et al., 2015; Virdi et al., 2016; Exposito-Rodriguez et al., 2017).
Unique to the sensory plastid proteome are components that contribute to phenotypic plasticity by triggering epigenetic changes in the plant (Mackenzie & Kundariya, 2020). The plant-specific gene MSH1 encodes a dual-targeted mitochondrial and plastid protein that resides within the sensory plastid but not the mesophyll chloroplast (Xu et al., 2011). Downregulation or disruption of MSH1 causes sensory plastid perturbation, a reduced plastoquinone pool, and enhanced expression of abiotic and biotic stress response pathways (Xu et al., 2011, 2012; Virdi et al., 2016; Shao et al., 2017). Progeny from MSH1-suppressed plants, when restored for MSH1 expression, can display heritable msh1 stress memory (Xu et al., 2012; Virdi et al., 2015) that depends on RNA-directed DNA methylation (RdDM) pathway components (Yang et al., 2020), The msh1 memory phenomenon alters gene expression and DNA methylation in auxin response, phytohormone signaling, circadian rhythm and alternative RNA splicing pathways (Yang et al., 2020). Recapitulation of these memory effects in other plant species (Xu et al., 2012; Yang et al., 2015; Raju et al., 2018) suggests this to be a conserved process in plants.
Incorporation of msh1 mutants as rootstocks in graft experiments to wild-type scions produces progeny that are also altered heritably. Whereas the parental rootstock displays stress effects, the graft progeny plants are enhanced in growth vigor and resilience relative to the wild-type (Virdi et al., 2015; Kundariya et al., 2020). This unexpected acquired vigor is similarly small interfering RNA- and RdDM-dependent (Kundariya et al., 2020).
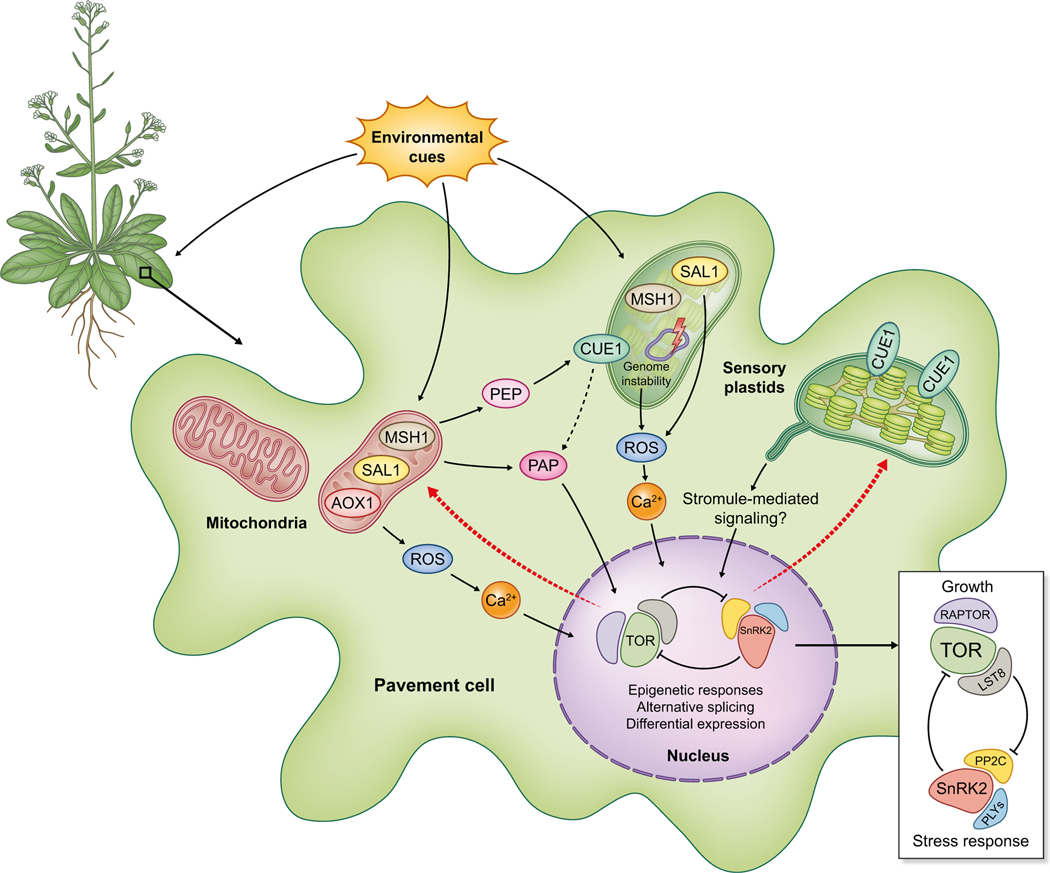
DNA methylation and gene expression in msh1 graft progeny reveal a pronounced auxin response signal that is phenotypically evident in vigorous lateral root growth (Kundariya et al., 2020). Network-based analysis of DNA methylation repatterning for the msh1-derived vigor phenotype identifies TOR, SnRK1 and PP2C as putative network hubs integrating growth with stress response (Kundariya et al., 2020). Extending upon previous reports of mitochondrial AOX1-directed stress behaviors, these observations support mito-plastid coordination in environmental sensing and reveal a capacity to influence key integrators of growth and stress response towards reprogramming of plant phenotype (Fig. 1).
Fig. 1.
Highly simplified diagram of nuclear-organellar communication in an epidermal pavement cell in plants. An example of retrograde regulation involving epidermal sensory plastids and mitochondria is shown in the SAL1–3′-phosphoadenisine-5′-phosphate (SAL1-PAP) pathway. Anterograde signaling is depicted by red dashed arrows, emanating from components linked to the growth-stress network represented by TARGET of RAPAMYCIN (TOR), SnRK1/2 and associated components. MSH1, shown within the sensory plastid, is suppressed by environmental stress and its depletion can induce organellar genome changes that are postulated to trigger plastid-nuclear signaling. ROS, reactive oxygen species; PEP, phosphoenolpyruvate.
IV. Alternative RNA splicing as a nuclear response to organellar signaling
One means of broad and rapid influence on nuclear gene expression, following organellar perturbation, occurs through alternative RNA splicing (Staiger, 2015). The light environment of a plant appears to be a strong determinant of alternative splicing behavior as a likely means of enhancing growth plasticity (Tognacca et al., 2020). Changing light conditions leads to reduction of the plastoquinone pool in plastids, which alters particular splicing factors for light-responsive nuclear genes (Petrillo et al., 2014). Auxin response factor regulation by alternative splicing is likewise an important component of phenotypic resilience during environmental change (Lanctot & Nemhauser, 2020). DNA methylation repatterning following MSH1 suppression shows changes in numerous components of the RNA spliceosome pathway (Yang et al., 2020).
During abiotic stress, alternative splicing occurs predominantly in regulatory loci, with outcomes that range from nonsense mediated decay to activation of otherwise nonfunctional transcripts (Mastrangelo et al., 2012). This association of environment-induced alternative splicing activity with regulators offers an elegant means of rapidly deploying phenotypic plasticity to a system under stress. It is perhaps not surprising, then, that gene-associated DNA methylation repatterning occurs within gene networks that align with stress response pathways and phenotype changes (Yang et al., 2020; Kundariya et al., 2020); subtle methylation changes may be sufficient to adjust or respond to local splicing changes that relax phenotypic constraints (Zhang et al., 2020).
V. Conclusions
Mitochondria and plastids, as energy-generating, light-sensing, phytohormone-producing and metabolic regulators of the cell, function as central integrators of environmental information for the plant (Fig. 1). Thus, organellar signaling is multifaceted and broadly targeted. Yet, cell- and tissue-level resolution of plant processes make it feasible to localize organellar sensing and signaling functions, with epidermal and vascular tissues appearing particularly important.
Implementation of forward genetic mutant, transcriptomic, epigenomic and physiological studies by numerous groups reveal striking intersection of organellar signaling with central gene networks to regulate bioenergetics for growth and plant defense. This emerging research provides the most comprehensive view yet of the components underpinning phenotypic plasticity in plants and offers a preliminary road map for understanding the genotype-to-phenotype relationship under dynamic change.
Acknowledgements
Owing to formatting constraints, we were not able to cite all of the highly relevant work in this review. Research cited from our own group was supported by awards from NSF (1853519) and NIH (R01 GM134056-01) to SAM.
References
- Alsharafa K, Vogel MO, Oelze ML, Moore M, Stingl N, König K, Friedman H, Mueller MJ, Dietz KJ. 2014. Kinetics of retrograde signalling initiation in the high light response of Arabidopsis thaliana. Philosophical Transactions of the Royal Society B: Biological Sciences 369: 201304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. 2007. A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942. [DOI] [PubMed] [Google Scholar]
- Barton KA, Schattat MH, Jakob T, Hause G, Wilhelm C, Mckenna JF, Máthé C, Runions J, Van Damme D, Mathur J. 2016. Epidermal pavement cells of Arabidopsis have chloroplasts. Plant Physiology 171: 723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán J, Wamboldt Y, Sanchez R, LaBrant EW, Kundariya H, Virdi KS, Elowsky C, Mackenzie SA. 2018. Specialized plastids trigger tissue-specific signaling for systemic stress response in plants. Plant Physiology 178: 672–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco NE, Guinea-Diaz M, Whelan J, Strand A. 2014. Interaction between plastid and mitochondrial retrograde signaling pathways during changes to plastid redox status. Philosophical Transactions of the Royal Society B: Biological Sciences 369: 20130231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkard JO, Runkel AM, Zambryski PC. 2015. Chloroplasts extend stromules independently and in response to internal redox signals. Proceedings of the National Academy of Sciences, USA 112: 10044–10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, Hu L, Dong B, Jin H-L, Wang H-B. 2020. Signaling from plastid genome stability modulates endoreplication and cell cycle during plant development. Cell Reports 32: 108019. [DOI] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P et al. 2011. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. The Plant Cell 23, 3992 LP–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Rodriguez M, Laissue PP, Yvon-Durocher G, Smirnoff N, Mullineaux PM. 2017. Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nature Communications 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Zhao G, Zhang S, Li Y, Gu H, Li Y, Zhao Q, Qi Y. 2019. Chloroplast-to-nucleus signaling regulates MicroRNA biogenesis in Arabidopsis. Developmental Cell 48: 371–382.e4. [DOI] [PubMed] [Google Scholar]
- Garcion C, Lohmann A, Lamodière E, Catinot J, Buchala A, Doermann P, Métraux J-P. 2008. Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiology 147: 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Feng P, Chi W, Sun X, Xu X, Li Y, Ren D, Lu C, David Rochaix J, Leister D et al. 2016. Plastid-nucleus communication involves calcium-modulated MAPK signalling. Nature Communications 7: 12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki H, Kurihara Y, Kuromori T, Kusano H, Nagata N, Yamamoto YY, Shimada H, Matsui M. 2019. SnRK1 kinase and the NAC transcription factor SOG1 are components of a novel signaling pathway mediating the low energy response triggered by ATP depletion. Frontiers in Plant Science 10: 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M. 2020. Plastoquinone in and beyond photosynthesis. Trends in Plant Science. 25: 1252–1265. [DOI] [PubMed] [Google Scholar]
- Hilgers EJA, Staehr P, Flügge U-I, Häusler RE. 2018. The xylulose 5-phosphate/phosphate translocator supports triose phosphate, but not phosphoenolpyruvate transport across the inner envelope membrane of plastids in Arabidopsis thaliana mutant plants. Frontiers in Plant Science 9: 1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A, Law SR, Narsai R, Duncan O, Lee J-H, Zhang B, Aken OV, Radomiljac JD, van der Merwe M, Yi K et al. 2014. A functional antagonistic relationship between auxin and mitochondrial retrograde signaling regulates alternative oxidase1; expression in Arabidopsis. Plant Physiology 165: 1233 LP–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundariya H, Yang Y, Morton K, Sanchez R, Axtell MJ, Hutton SF, Fromm M, Mackenzie SA. 2020. Heritable enhanced growth vigor through grafting is associated with the RdDM pathway in plants. Nature Communications 11: 5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot A, Nemhauser JL. 2020. It’s Morphin’ time: how multiple signals converge on ARF transcription factors to direct development. Current Opinion in Plant Biology 57: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J. 2003. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906. [DOI] [PubMed] [Google Scholar]
- Lee K, Kang H. 2020. Roles of organellar RNA-binding proteins in plant growth, development, and abiotic stress responses. International Journal of Molecular Sciences 21: 4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litthauer S, Jones MA. 2018. SAL1-PAP retrograde signalling extends circadian period by reproducing the loss of exoribonuclease (XRN) activity. Plant Signaling & Behavior 13: e1500066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist PK, Rosar C, Bräutigam A, Weber APM. 2014. Plastid signals and the bundle sheath: mesophyll development in reticulate mutants. Molecular Plant 7: 14–29. [DOI] [PubMed] [Google Scholar]
- Macaulay KM, Heath GA, Ciulli A, Murphy AM, Abell C, Carr JP, Smith AG. 2017. The biochemical properties of the two Arabidopsis thaliana isochorismate synthases. Biochemical Journal 474: 1579–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie SA, Kundariya H. 2020. Organellar protein multi-functionality and phenotypic plasticity in plants. Philosophical Transactions of the Royal Society B: Biological Sciences 375: 20190182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal MK, Chandra-Shekara AC, Jeong RD, Yu K, Zhu S, Chanda B, Navarre D, Kachroo A, Kachroo P. 2012. Oleic acid-dependent modulation of NITRIC OXIDE ASSOCIATED1 protein levels regulates nitric oxide-mediated defense signaling in arabidopsis. The Plant Cell 24: 1654–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalha L, Confraria A, Baena-González E. 2019. SnRK1 and TOR: modulating growth–defense trade-offs in plant stress responses. Journal of Experimental Botany 70: 2261–2274. [DOI] [PubMed] [Google Scholar]
- Mastrangelo AM, Marone D, Laidò G, De Leonardis AM, De Vita P. 2012. Alternative splicing: enhancing ability to cope with stress via transcriptome plasticity. Plant Science 185–186: 40–49. [DOI] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Soole KL, Day DA. 2011. Organization and regulation of mitochondrial respiration in plants. Annual Review of Plant Biology 62: 79–104. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. 2001. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of MG-chelatase H subunit in plastid-to-nucleus signal transduction. Proceedings of the National Academy of Sciences, USA 98: 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mubarakshina MM, Ivanov BN. 2010. The production and scavenging of reactive oxygen species in the plastoquinone pool of chloroplast thylakoid membranes. Physiologia Plantarum 140: 103–110. [DOI] [PubMed] [Google Scholar]
- Mullineaux PM, Exposito-Rodriguez M, Laissue PP, Smirnoff N, Park E. 2020. Spatial chloroplast-to-nucleus signalling involving plastid-nuclear complexes and stromules. Philosophical Transactions of the Royal Society of London. Series B, Biological sciences 375: 20190405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz P, Munné-Bosch S. 2020. Oxylipins in plastidial retrograde signaling. Redox Biology 37: 101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Giraud E, Duncan O, Law SR, Wang Y, Xu L, Narsai R, Carrie C, Walker H, Day DA et al. 2013. Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. Journal of Biological Chemistry 288: 3449–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo E, Godoy Herz MA, Fuchs A, Reifer D, Fuller J, Yanovsky MJ, Simpson C, Brown JWS, Barta A, Kalyna M et al. 2014. A chloroplast retrograde signal regulates nuclear alternative splicing. Science (New York, N.Y.) 344: 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju SKK, Shao M-R, Sanchez R, Xu Y-Z, Sandhu A, Graef G, Mackenzie S. 2018. An epigenetic breeding system in soybean for increased yield and stability. Plant Biotechnology Journal 16: 1836–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triataphylides C, Havaux M. 2012. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proceedings of the National Academy of Sciences, USA 109: 5535–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter AS, Tohge T, Fernie AR, Grimm B. 2020. The genomes uncoupled-dependent signalling pathway coordinates plastid biogenesis with the synthesis of anthocyanins. Philosophical Transactions of the Royal Society B: Biological Sciences 375: 20190403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA. 2013. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO Journal 32: 1087–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting CD. 1986. The evolution of phenotypic plasticity in plants. Annual Review of Ecology and Systematics 17: 667–693. [Google Scholar]
- Shao M-R, Kumar Kenchanmane Raju S, Laurie JD, Sanchez R, Mackenzie SA. 2017. Stress-responsive pathways and small RNA changes distinguish variable developmental phenotypes caused by MSH1 loss. BMC Plant Biology 17: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumbe L, Bott R, Havaux M. 2014. Dihydroactinidiolide, a high light-induced β-carotene derivative that can regulate gene expression and photoacclimation in Arabidopsis. Molecular Plant 7: 1248–1251. [DOI] [PubMed] [Google Scholar]
- Staiger D. 2015. Shaping the Arabidopsis transcriptome through alternative splicing. Advances in Botany 2015: 419428. [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J. 2003. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrin. Nature 421: 79–83. [DOI] [PubMed] [Google Scholar]
- Streatfield SJ, Weber A, Kinsman EA, Häusler RE, Li J, Post-Beittenmiller D, Kaiser WM, Pyke KA, Flugge UI, Chory J. 1999. The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. The Plant Cell 11: 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Gollan PJ, Suorsa M, Kangasjärvi S, Aro E-M. 2012. STN7 operates in retrograde signaling through controlling redox balance in the electron transfer chain. Frontiers in Plant Science 3: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognacca RS, Kubaczka MG, Servi L, Rodríguez FS, Godoy Herz MA, Petrillo E. 2020. Light in the transcription landscape: chromatin, RNA polymerase II and splicing throughout Arabidopsis thaliana’s life cycle. Transcription 11: 117–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal D, García-Caparrós P, Kumar V, Dietz KJ. 2020. Chloroplast-associated molecular patterns as concept for fine-tuned operational retrograde signalling. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 375: 20190443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virdi KS, Laurie JD, Xu Y-Z, Yu J, Shao M-R, Sanchez R, Kundariya H, Wang D, Riethoven JJM, Wamboldt Y et al. 2015. Arabidopsis MSH1 mutation alters the epigenome and produces heritable changes in plant growth. Nature Communications 6: 6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virdi KS, Wamboldt Y, Kundariya H, Laurie JD, Keren I, Kumar KRS, Block A, Basset G, Luebker S, Elowsky C et al. 2016. MSH1 is a plant organellar DNA binding and thylakoid protein under precise spatial regulation to alter development. Molecular Plant 9: 245–260. [DOI] [PubMed] [Google Scholar]
- Walley JW, Kliebenstein DJ, Bostock RM, Dehesh K. 2013. Fatty acids and early detection of pathogens. Current Opinion in Plant Biology 16: 520–526. [DOI] [PubMed] [Google Scholar]
- Wang Y, Selinski J, Mao C, Zhu Y, Berkowitz O, Whelan J. 2020. Linking mitochondrial and chloroplast retrograde signalling in plants. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 375: 20190410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RR. 2007. The diversity of plastid form and function. In: Wise RR, Hoober JK, eds. The structure and function of plastids. Advances in photosynthesis and respiration, vol. 23. Dordrecht, the Netherlands: Springer, 3–26. [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J. 2011. Heme synthesis by plastid ferrochelatase i regulates nuclear gene expression in plants. Current Biology 21: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EE, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K. 2012. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149: 1525–1535. [DOI] [PubMed] [Google Scholar]
- Xu Y-Z, Arrieta-Montiel MP, Virdi KS, de Paula WBM, Widhalm JR, Basset GJ, Davila JI, Elthon TE, Elowsky CG, Sato SJ et al. 2011. MutSHOMOLOG1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. The Plant Cell 23: 3428–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, De R, Santamaria R, Virdi KS, Arrieta-montiel MP, Razvi F, Li S, Ren G, Yu B, Alexander D et al. 2012. The chloroplast triggers developmental reprogramming when MUTS HOMOLOG1 is suppressed in plants 1. Plant Physiology 159: 710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kundariya H, Xu Y-Z, Sandhu A, Yu J, Hutton SF, Zhang M, Mackenzie SA. 2015. Muts HOMOLOG1-derived epigenetic breeding potential in tomato. Plant Physiology 168: 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Sanchez R, Kundariya H, Maher T, Dopp I, Schwegel R, Virdi K, Axtell M, Mackenzie SA. 2020. Segregation of an MSH1 RNAi transgene produces heritable non-genetic memory in association with methylome reprogramming. Nature Communications 11: 2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, Greene GH, Yuan M, Xu G, Burton D, Liu L, Marqués J, Dong X. 2020. Translational regulation of metabolic dynamics during effector-triggered immunity. Molecular Plant 13: 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkovic J, Anderson SL, Rhoads DM. 2005. A reporter gene system used to study developmental expression of alternative oxidase and isolate mitochondrial retrograde regulationmutants in Arabidopsis. Plant Molecular Biology 57: 871–888. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang Y-Z, Jiang J, Duan C-G. 2020. The crosstalk between epigenetic mechanisms and alternative RNA processing regulation. Frontiers in Genetics 11: 998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Huang J, Chory J. 2020. Unraveling the linkage between retrograde signaling and RNA metabolism in plants. Trends in Plant Science 25: 141–147. [DOI] [PubMed] [Google Scholar]