Abstract
Background
Identifying patients at risk of severe asthma is vitally important given the disproportionate burden of disease imposed by that state. However, biomarkers to support such needs remain elusive.
Methods
In this letter, we assessed whether specific panels of circulating miRNAs (microRNAs) can differentiate between mild and severe asthma patients as well as between healthy subjects and severe asthma patients.
Results
To our knowledge, the miRNAs identified in our work such as miR‐28‐3p, miR‐16‐2‐3p, and miR‐210‐3p have not been previously reported as differentially expressed in the serum of severe asthma patients.
Conclusion
Our findings suggest that miRNA expression profiles may have the capability as potential biomarkers that signal the risk of having severe asthma. As such, these findings have significant novelty and merit wider dissemination to facilitate further work in this field.
To the Editor,
Although one of the commonest chronic diseases, asthma lacks a gold standard diagnostic test. Reliance on a clinical diagnosis alongside the heterogeneous nature of asthma means that asthma diagnosis is often delayed to the detriment of the patient. Therefore, new asthma diagnostic biomarkers are an important research focus. MicroRNAs (miRNAs) might hold promise in that regard. These small noncoding RNAs act through RNA‐induced silencing complexes to post‐transcriptionally regulate mRNA (messenger RNA). A previous study demonstrated differential miRNA expression in blood eosinophils from asthmatics compared with healthy individuals. They clustered asthmatics and healthy subjects showing that miRNA profiles in eosinophils (especially miR‐185‐5p and miR‐1246) are potential serum biomarkers for ranking asthma severity. 1 Another study from our lab demonstrated that miR‐150, miR‐152, and miR‐375 are upregulated in severe asthma. 2
We determined the differential miRNA expression in sera of severe asthmatic patients and compared it to mild asthma and no asthma to assess their potential as biomarkers for the diagnosis of asthma and its discriminative capability for asthma severity.
We investigated the presence of 175 miRNAs in serum samples collected from 12 adult subjects (four healthy, four mild asthma, and four severe asthma) of our cohort studies (summary features and definitions, Table 1). Healthy subjects and mild asthma patients were from the Isle of Wight Birth Cohort, 4 while severe asthma patients were from the Wessex AsThma CoHort of difficult asthma (WATCH) study. 5 We hypothesized that asthma‐specific miRNAs may differentiate asthmatic from healthy subjects, while other miRNAs may differentiate mild from severe asthma.
TABLE 1.
Clinical characteristics for subjects from different groups
| Severe asthma | Mild asthma | Healthy | |
|---|---|---|---|
| N = 4 | N = 4 | N = 4 | |
| Definition of asthma status | Enrolled in WATCH study and taking BTS steps “additional controller therapies/specialist therapies” 3 | Enrolled in IOWBC. Physician diagnosis of asthma at age 18‐years plus either wheeze within the last 12 months or currently taking asthma medication at BTS steps “regular preventer/initial add on therapy” 3 | Enrolled in IOWBC. No documented history of asthma ever at age 18‐years |
| Age (years) | 35.7 (18–52) | 18 (NA) | 18 (NA) |
| Gender (F); % (N) | 75 (3) | 75 (3) | 100 (4) |
| Current smoker; % (N) | 0 (0) | 50 (2) | 25 (1) |
| Never smoked; % (N) | 50 (2) | 25 (1) | 75 (3) |
| ICS dose (BDP equivalent μg/day)* | 2500 (1000–3000) | 400 (0–800) | NA |
| BMI (Kg/m2)* | 31.8 (22.1–39.6) | 23.9 (20.2–26.4) | 21.7 (19.0–24.6) |
| Rescue OCS (in last 12 months); (N)* | 3.7 (1–7) | 0 | 0 |
| On maintenance OCS | No | No | No |
| On asthma biologics; % (N) | 0 (0) | 0 (0) | 0 (0) |
| FEV1 (%)* | 80.84 (68.73–96.76) | 82.90 (77.15–88.92) | 88.91 (90.56–92.43) |
| FEV1/FVC (%)* | 71.67 (65–76) | 84.25 (77–91) | 89 (81–94) |
| Total IgE (KU/L)* | 1463.7 (19.6–2907.9) | 119.4 (5.6–346.0) | 210 (36–384) |
| Atopy; % (N) | 50 (2) | 75 (3) | 50 (2) |
Note: Atopy was defined as positive Skin Prick Test (≥3 mm wheal diameter) to any aeroallergen.
Abbreviations: BDP, beclometasone dipropionate; BMI, body mass index; BTS, British Thoracic Society; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; ICS, inhaled corticosteroids; IOWBC, Isle of Wight Birth Cohort; NA, not applicable; OCS, oral corticosteroids; WATCH, Wessex AsThma CoHort.
*Mean (range).
We extracted miRNAs from stored serum using miRNeasy Serum/Plasma Kit (QIAGEN). After extraction, RNA was reverse‐transcribed to cDNA. We then evaluated miRNA expression by quantitative PCR using Human Serum/Plasma Focus, miRCURY LNA miRNA Focus PCR Panel kit (QIAGEN) which is designed for profiling 175 human miRNAs commonly found in serum and plasma. Results were analyzed using QIAGEN web portal GeneGlobe. At least twofold change in expression and p ≥ 0.05 were required to consider the miRNA of diagnostic value.
Figure 1 shows volcano plots for comparisons of the studied miRNAs in severe asthmatic, mild asthmatic, and healthy groups. We found that, miR‐193a‐5p was significantly increased while miR‐181a‐5p, miR‐146a‐5p, and miR‐16‐2‐3p were significantly decreased in serum from severe asthmatics compared to healthy subjects. Furthermore, miR‐197‐3p, miR‐223‐3p, miR‐151a‐3p, miR‐191‐5p, and miR‐28‐3p were significantly increased and miR‐451a, miR‐16‐2‐3p, miR‐210‐3p, miR‐133a‐3p, miR‐660‐5p, and miR‐144‐3p significantly decreased in serum from severe asthmatics compared to mild asthmatics. There were no significant differences in expression of the studied miRNAs in serum between healthy subjects and mild asthma patients.
FIGURE 1.
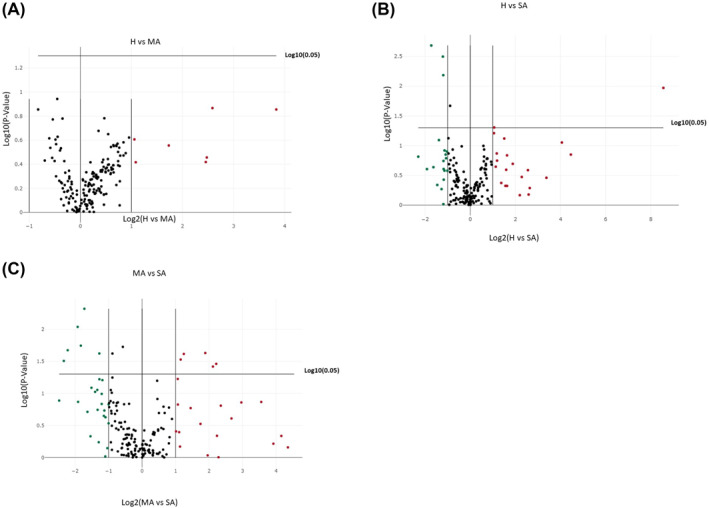
Volcano plots for the 175 miRNAs tested in the sera of different groups of patients. (A) Healthy (H) against mild asthma (MA) patients. (B) Healthy (H) against severe asthma (SA) patients. (C) mild asthma (MA) against severe asthma (SA) patients
Our preliminary study demonstrated differential sera miRNA expression in severe asthma compared to both mild asthma and healthy subjects. We also found that different miRNAs respectively distinguished severe asthma from mild asthma than from healthy controls. Such findings emphasize the distinctive nature of severe asthma and may aid understanding of mechanistic differences between mild and severe asthma. To our knowledge, miR‐28‐3p, miR‐16‐2‐3p, and miR‐210‐3p have not been previously reported as differentially expressed miRNAs in asthma while miR‐151a‐3p has been inversely correlated with Bronchoalveolar lavage eosinophil percentage in asthma patients. 6 We tested 175 miRNAs within a standard panel but other miRNAs previously reported in the literature may differentiate asthmatic patients from healthy subjects. 1 , 2 Furthermore, asthma is a heterogeneous condition so different miRNAs may be relevant to different phenotypes and endotypes. Differential miRNA expression may be seen in different tissues too. Our pilot study shows that miRNA expression may help to distinguish severe asthma status. However, the numbers studied were small. Furthermore, there were factors other than severity that differed between the groups in our study including age, body mass index and current smoking status which might also have plausibly impacted on differential miRNA expression. It is worth noting though that more difficult‐to‐treat asthma in our WATCH cohort is typically a multimorbid state associated with older age, obesity and absence of current smoking 7 as seen in the severe asthma subgroup analyzed in this study. Larger scale work studying miRNA expression in different asthma subgroups, different tissue compartments and using a wider array of miRNAs is therefore warranted as we seek to build on these initial findings.
CONFLICTS OF INTEREST
The authors have no conflicts of interest related to this article to disclose.
AUTHOR CONTRIBUTION
Mohammed A. Kyyaly contributed to study design, undertook laboratory experiments, data analysis, and wrote the first draft of the manuscript. Tilman Sanchez‐Elsner supervised the experiments, guided data interpretation, and revised the manuscript. S. Hasan Arshad helped supervise the work and revised the manuscript. Ramesh J. Kurukulaaratchy supervised the work, helped with patient selection, manuscript writing, and acts as Guarantor for the manuscript. Peijun He and Collin L. Sones critically reviewed and revised the manuscript.
ACKNOWLEDGMENTS
We would like to extend our gratitude to participants in both the WATCH and Isle of Wight Birth Cohort studies as well as the wider WATCH and Isle of Wight Birth Cohort study teams. We wish to acknowledge the support of the Southampton NIHR Biomedical Research Centre and the Southampton NIHR Clinical Research Facility. The BRC and CRF are funded by the NIHR and are a partnership between the University of Southampton and University Hospital Southampton NHS Foundation Trust. We also wish to acknowledge funding contributions from the Asthma, Allergy & Inflammation Research (AAIR) Charity and Asthma UK in supporting this work. This work was funded by Asthma, Allergy & Inflammation Research (AAIR) Charity and Asthma UK (Grant: AUK‐PG‐2019‐419).
Kyyaly MA, Sanchez‐Elsner T, He P, Sones CL, Arshad SH, Kurukulaaratchy RJ. Circulating miRNAs—A potential tool to identify severe asthma risk?. Clin Transl Allergy. 2021; 1–XXX. 10.1002/clt2.12040
REFERENCES
- 1. Rodrigo‐Muñoz JM, Cañas JA, Sastre B, et al. Asthma diagnosis using integrated analysis of eosinophil microRNAs. Allergy. 2019;74(3):507‐517. 10.1111/all.13570 [DOI] [PubMed] [Google Scholar]
- 2. Rupani H, Martinez‐Nunez RT, Dennison P, et al. Toll‐like receptor 7 is reduced in severe asthma and linked to an altered MicroRNA profile. Am J Respir Crit Care Med. 2016;194(1):26‐37. 10.1164/rccm.201502-0280OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. British Thoracic Society/NHS Scotland. British Guideline for the Management of Asthma. A National Clinical Guideline. British Thoracic Society/NHS Scotland; 2019. [Google Scholar]
- 4. Arshad SH, Holloway JW, Karmaus W, et al. Cohort profile: the Isle of Wight whole population birth cohort (IOWBC). Int J Epidemiol. 2018;47(4):1043‐1044i. 10.1093/ije/dyy023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Azim A, Mistry H, Freeman A, et al. Protocol for the Wessex AsThma CoHort of difficult asthma (WATCH): a pragmatic real‐life longitudinal study of difficult asthma in the clinic. BMC Pulm Med. 2019;19(1):99. 10.1186/s12890-019-0862-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Francisco‐Garcia AS, Garrido‐Martín EM, Rupani H, et al. Small RNA species and microRNA profiles are altered in severe asthma nanovesicles from broncho alveolar lavage and associate with impaired lung function and inflammation. Noncoding RNA, 2019;5(4):51. 10.3390/ncrna5040051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Azim A, Freeman A, Lavenu A, et al. New perspectives on difficult asthma; sex and age of asthma‐onset based phenotypes. J Allergy Clin Immunol Pract. 2020;8:3396‐3406. 10.1016/j.jaip.2020.05.053 [DOI] [PubMed] [Google Scholar]


