Significance
The 2015 Korean Middle East respiratory syndrome (MERS) outbreak occurred after an infected traveler returned from the Middle East, allowing virus evolution without further MERS-CoV introduction. We demonstrate that two mutations selected during the outbreak led to attenuation in mice, assessed using isogenic viruses. In Africa, MERS-CoV is endemic among camels without evident human MERS cases. Analysis of isogenic viruses expressing camel Spike proteins revealed insignificant change in murine virulence. These results illustrate differences in coronavirus evolution that occur under conditions of camel endemicity versus a one-time introduction into human populations, providing insight into the evolution of surface glycoproteins of SARS-CoV-2, and other respiratory viruses, in which virus evolves in zoonotic hosts and during human-to-human spread, such as influenza virus.
Keywords: MERS-CoV, Spike, mutation, virulence
Abstract
Human Middle East respiratory syndrome (MERS) cases were detected primarily in the Middle East before a major outbreak occurred in South Korea in 2015. The Korean outbreak was initiated by a single infected individual, allowing studies of virus evolution in the absence of further MERS-CoV introduction into human populations. In contrast, MERS is primarily a camel disease on the Arabian Peninsula and in Africa, with clinical disease in humans only in the former location. Previous work identified two mutations in the South Korean MERS-CoV, D510G and I529T on the Spike (S) protein, that led to impaired binding to the receptor. However, whether these mutations affected virulence is unknown. To address this question, we constructed isogenic viruses expressing mutations found in the S protein from Korean isolates and showed that isogenic viruses carrying the Korean MERS-CoV mutations, D510G or I529T, were attenuated in mice, resulting in greater survival, less induction of inflammatory cytokines, and less severe lung injury. In contrast, isogenic viruses expressing S proteins from African isolates were nearly fully virulent; other studies showed that West African camel isolates carry mutations in MERS-CoV accessory proteins, which may limit human transmission. These data indicate that following a single-point introduction of the virus, MERS-CoV S protein evolved rapidly in South Korea to adapt to human populations, with consequences on virulence. In contrast, the mutations in S proteins of African isolates did not change virulence, indicating that S protein variation likely does not play a major role in the lack of camel-to-human transmission in Africa.
Middle East respiratory syndrome coronavirus (MERS-CoV) is a zoonotic virus first identified in Saudi Arabia in 2012 (1). According to the World Health Organization, as of January 2020, cases of MERS-CoV have been reported in 27 countries, with 2,494 laboratory-confirmed cases and 858 deaths since September 2012 (2). Outbreaks in Middle East countries constitute the vast majority of reported cases (3), while around 10% of the reported cases were documented outside of Middle East countries, initiated by travelers from this location (4). Such an outbreak took place in South Korea, where 186 confirmed cases and 38 deaths occurred from May to July 2015 (5, 6). Intriguingly, the South Korean outbreak is the only documented major outbreak outside of Middle East countries thus far and allows studies of single-point introduction of MERS-CoV into a naïve human population. A possible explanation for the widespread and rapid outbreak in Korea was the emergence of mutant viruses with enhanced infectivity or transmission in humans (5). Two nonsynonymous mutations (D510G and I529T) in the receptor-binding domain (RBD) of the Spike (S) protein, which binds to the dipeptidyl peptidase 4 (DPP4) receptor to enter cells, were identified only in South Korean isolates (5–7) (Fig. 1A).
Fig. 1.
D510G and I529T lead to impaired entry and attenuation in vitro independent of S1/S2 cleavage. (A) Schematic diagram showing MERS-CoV genome. Region of the S protein is magnified. Mutations are shown by upright or inverted triangles. Red and purple inverted triangles represent mutations in Korean isolate and Nigeria camel isolate, respectively; blue and yellow upright triangles represent mutations in Ethiopia (Ethi-126 isolate carries an extra mutation, P515L compared to Ethi-118 isolate) and Morocco camel isolates, respectively; black inverted triangles represent mutations in Burkina Faso camel isolate; white inverted triangle represents V26A mutation that is present in all five Africa camel isolates. (B) Indicated cell lines were transduced with VSV pseudoviruses expressing MERS-CoV Spike variants as shown. At 18 h posttransduction, cells were lysed and luciferase levels were measured. vec, VSV particles with no surface glycoprotein. Data are representative of three independent experiments. Statistical significance was determined by two-tailed Student’s t test, where *P < 0.05, **P < 0.01, ***P < 0.001. Bars represent mean and error bars represent SEM. (C) Vero 81 and Huh-7 cells were infected with WT and mutant MERS-CoV at an MOI of 0.01. Cells and supernatant were frozen at the indicated time points. Virus titers were determined by plaque assay. Data are representative of two independent experiments. Data points represent geometric mean and error bars represent geometric SD. (D) Cell-free WT and mutant MERS-CoV were incubated at 37 °C for the indicated durations. The virus titers were then determined by plaque assay at the end of the incubation period and normalized to initial virus titers (T = 0). Data were pooled from two independent experiments. Data points represent mean and error bars represent SEM. Areas under the curve were calculated. P = 0.0505 represents P value when rMA-WT and rMA-I529T AUC values were compared.
While most isolates contained just one of these mutations, double mutants were occasionally identified, although never as the predominant isolate (6). Paradoxically, structural predictions showed that D510G and I529T mutations are energetically unfavorable for optimal interaction between the S protein and DPP4 (5). In vitro and cell binding assays using mutant S protein showed reduced binding affinity to the human DPP4 (hDPP4) receptor, while pseudotyped viruses carrying mutant S proteins exhibit reduced entry in human cell lines (5, 8). Furthermore, viral isolates encoding T529 formed smaller plaques than WT virus (6). In addition, these mutations were reported to be modestly resistant to anti–MERS-CoV neutralizing antibodies and monoclonal antibodies that target different epitopes of the RBD (5, 8). It was therefore suggested that MERS-CoV tolerated compromised viral fitness in order to evade preexisting antibody responses (5). Characterization of mutations identified in the S protein of the South Korean outbreak in the context of viral infection would directly establish the role of the two mutations in virulence and would contribute to the understanding of virus evolution.
In contrast to South Korea, where the outbreak was caused by introduction of virus through a single patient, MERS-CoV is endemic among camels in Africa (9–12). There have been no reports of clinically apparent human MERS in Africa. Previous reports suggested that camel MERS-CoV in Africa are phylogenetically different from viruses in the Arabian Peninsula but remain antigenically similar in microneutralization tests (11). Clade C1 MERS-CoV in North and West Africa share unique genetic features involving deletions in the ORF4b gene (11). This may account for the inability of African camel viruses to cause disease in humans, as ORF4b is known to suppress innate immune responses (13–16). One study that compared pseudoviruses coated with S proteins from West African isolates to those coated with the prototypic EMC (Erasmus Medical Center) S protein revealed no differences in viral entry (17).
In order to understand virus evolution in humans infected with SARS-CoV-2, or as an example of another type of respiratory virus, influenza A virus (IAV), we must take both interhuman spread and, in the case of IAV, periodic introduction of virus from zoonotic hosts (birds and swine) into consideration. However, these processes cannot easily be separated. The identification of two different patterns of MERS-CoV spread (single-source introduction versus endemic spread in camels with occasional zoonotic spillover) allows studies of virus adaptation not easily performed with SARS-CoV-2 or IAV. Based on SARS-CoV-2 evolution (18–20), one might predict that MERS-CoV in the Korean outbreak would evolve to be more transmissible, with minimal change in virulence. Despite previous reports characterizing the S protein mutations that were selected in South Korean patients and in African camels, their effects on virulence in vivo remain largely unknown.
Here, we investigated the effects of the mutations in the S protein on MERS-CoV virulence. For these experiments, we used mice genetically modified to enable MERS-CoV infection (hDPP4-knockin; hDPP4-KI mice). Using reverse genetics, we generated isogenic recombinant MERS-CoV on a mouse-adapted MERS-CoV (MA-WT) background to pinpoint the role of the Korean and African S protein mutations in pathogenesis in the absence of other mutations (21, 22). Our results indicated that isogenic recombinant mouse-adapted MERS-CoV carrying only the D510G or I529T mutations identified in the Korean isolate S proteins resulted in attenuation of the virus in vivo. In contrast, those carrying the African MERS-CoV mutations did not manifest signs of significant attenuation, suggesting that the mutations on the S protein are not a major cause of the lack of significant clinical infection observed in Africa.
Results
D510G and I529T Mutations Result in Impaired Viral Entry and Compromised Viral Fitness In Vitro.
Initially, we generated and characterized three mutant S proteins on the background of the S protein of MA-WT (MA-S), two of which were singly mutated (D510G or I529T) and a third that was a double mutant carrying both D510G and I529T (DM) (21). MA-WT was developed through repeated lung passage of the parental virus (EMC/2012), which caused only mild disease in susceptible mice, resulting in a virulent virus in mice that encodes two mutations in the S protein (N222D and S749R) that are required for this virulence (21). Upon transfection into HEK293T cells, all proteins were expressed and cleaved as detected by α-S1 antibody (SI Appendix, Fig. S1A), suggesting that the mutations did not affect S1/S2 cleavage. Next, we assayed viral entry using vesicular stomatitis virus (VSV)-pseudotyped viruses carrying the various S mutations. In previous studies, the D510G and I529T mutations were suggested to reduce viral entry by weaker binding to DPP4, but only when DPP4 levels were low (8). However, the role of cell surface protease TTSP family TMPR serine 2 (TMPRSS2) on these mutations was not addressed. We therefore used three different cell lines for our investigations. Huh-7 cells express higher levels of hDPP4 than Vero and Calu-3 cells, but Calu-3 cells express higher levels of TMPRSS2 than Huh-7 cells. Vero cells express very little TMPRSS2 (23, 24). VSV pseudotyped with VSV-G (VSV-G) transduced all three cell lines efficiently, while “bald” VSV particles with no surface glycoprotein transduced cells minimally (mock). D510G and I529T mutations reduced pseudovirus entry in Vero and Calu-3 cells, with the I529T mutation resulting in the greatest reduction. Entry was further impaired in the double mutant in all cell lines compared to the two single mutants (Fig. 1B). Consistent with previous reports, entry into Huh-7 cells was essentially identical for all S protein variants, suggesting that the D510G and I529T mutations had no effects on entry when DPP4 expression was abundant (8). In addition, viral entry was lower in Calu-3 cells than in Huh-7 cells despite higher levels of TMPRSS2, indicating that the abundance of DPP4 largely dictated the entry for both mutants, as TMPRSS2 could not compensate for the impaired entry that was observed in the context of low DPP4 expression.
Next, we characterized the mutations in the context of authentic MERS-CoV. To do this, using reverse genetics, D510G and I529T mutations were individually introduced or introduced in combination into MA-WT resulting in three isogenic recombinant viruses (22). Viruses were propagated in Huh-7 cells and plaqued on Vero 81 cells. S protein cleavage was unaffected by these mutations, since we detected largely equivalent amounts of S1 and S2 subunits when concentrated MERS-CoV carrying the mutant S proteins were analyzed by Western blot (SI Appendix, Fig. S1B). The presence of the D510G and I529T mutations resulted in delayed plaque formation (SI Appendix, Fig. S1C). rMA-WT (MA-WT) infection resulted in clear, round plaques by 3 d postinfection (dpi), with a further increase in size at 4 dpi. However, rMA-D510G infection resulted in plaques only at 4 dpi. rMA-I529T infection showed even more delayed plaque formation wherein plaques were seen only at 5 dpi and these plaques had irregular borders. We detected no visible plaques at any time in cells infected with the double mutant (rMA-DM), indicating that the combination of D510G and I529T significantly compromised viral fitness.
To determine replication kinetics of these viruses, Vero and Huh-7 cells were infected at a multiplicity of infection (MOI) of 0.01 with the indicated viruses and samples were obtained up to 72 h postinfection (hpi). The double mutant was not further evaluated as no plaques could be observed even after 5 d of incubation. rMA-WT replicated to the highest titers at all time points. rMA-WT replicated 100- to 1,000-fold better than either rMA-D510G or rMA-I529T at all time points in Huh-7 cells. In Vero cells, rMA-D510G replicated to similar levels as rMA-WT except at 48 hpi, while the titers of rMA-I529T were around 10- to 100-fold lower at all time points. The apparent discrepancy between levels of S protein (SI Appendix, Fig. S1B) and infectious virus titers (Fig. 1C) suggested that there was a discrepancy between numbers of particles and PFUs, consistent with an effect of the mutations on virus stability in addition to impacting DPP4 binding.
MA-I529T Is More Resistant to Temperature-Driven Impairment.
Since the mutant viruses showed compromised viral fitness in cell culture and an increased particle/PFU ratio, we next assessed whether factors other than reduced binding affinity for hDPP4 contributed to attenuation. We tested the thermostability of the mutant viruses by incubating viruses at 37 °C for up to 6 h and measuring virus titers. We observed a greater reduction in infectivity of rMA-WT and rMA-D510G compared to rMA-I529T, suggesting that rMA-I529T was more thermostable (Fig. 1D). Together, these results suggest that I529T results in increased rMA-I529T stability, possibly enhancing survival in the human airway but at the cost of diminishing entry into host cells (25).
MA-D510G and MA-I529T Are Attenuated In Vivo.
To assess the virulence of the mutant viruses in vivo, we infected hDPP4-KI mice with a nonlethal dose of 200 PFU of virus. rMA-WT– and rMA-D510G–infected mice lost around 15% of their weight by day 5 postinfection. rMA-D510G–infected mice started to recover by 6 dpi and reached close to 100% of initial weight by 9 dpi, while rMA-WT–infected mice showed delayed recovery at 8 dpi and reached only 90% of initial weight by 12 dpi. rMA-I529T-infected mice did not show any weight loss during the course of the experiment (Fig. 2A). We next challenged hDPP4-KI mice with 1,000 PFU of viruses per mouse. rMA-WT–infected mice showed significant weight loss and all succumbed to infection by 7 dpi. rMA-D510G–infected mice lost around 20% of weight and fewer than 50% of these mice survived the infection. In stark contrast, rMA-I529T–infected mice did not exhibit any weight loss or death after challenge with an equivalent dose of virus (Fig. 2B).
Fig. 2.
D510G and I529T result in attenuation in vivo. hDPP4-KI mice were infected i.n. with WT and mutant MERS-CoV and daily weight changes and survival were as shown at a nonlethal dose of 200 PFU (n = 4 to 6 mice per group) (A) and a lethal dose of 1,000 PFU (n = 6 to 8 mice per group) (B). Lungs of infected mice were harvested and homogenized at the specified time point for RNA isolation and qRT-PCR for (C) viral genomic RNA as measured by ORF1a and N transcripts relative to HPRT (n = 5 to 7 mice per group per time point) or (D) determination of viral titers by plaque assay (n = 5 mice per group). Statistical significance for survival analysis was determined by log-rank (Mantel–Cox) test, where ***P < 0.001 (B). Statistical significance was determined by two-tailed Student’s t test, where *P < 0.05 (C and D). Data points represent mean and error bars represent SEM in weight curves. Bars represent mean and geometric mean while error bars represent SEM and geometric SD in C and D, respectively. (E and F) hDPP4-KI mice were infected i.n. with 1,000 PFU of WT or mutant virus. Lungs were harvested at 4 dpi for H&E staining. (E) Summary scores for edema and inflammation (n = 6 to 8 mice per group). Bars represent mean and error bar represent SEM. Statistical significance was determined by two-tailed Student’s t test, where *P < 0.05. (F) Representative H&E staining of lungs demonstrating lung edema and inflammation (low magnification, 4×, Upper; high magnification, 20×; Lower). Asterisks indicate edema and arrows point to regions with hypercellularity. (Scale bars, 525 μm in 4× and 105 μm in 20×.)
To further characterize the infection, we harvested lungs at 2 and 5 dpi for virus titers and pathological analysis after infection with 1,000 PFU. The levels of viral genomic RNA (gRNA) measured by ORF1a abundance and of subgenomic mRNA assessed by N mRNA abundance were largely the same on day 2 postinfection in mice infected with the three variants. At 5 dpi, the levels of gRNA in rMA-D510G–infected mice were only 2-fold lower than rMA-WT, while those of gRNA and N mRNA in rMA-I529T–infected lungs were 10-fold lower than rMA-WT (Fig. 2 C, Left). Similar results were also obtained when the abundance of nucleocapsid (N) transcripts was measured (Fig. 2 C, Right). Levels of infectious virus were determined by plaque assay at 2 and 5 dpi. rMA-D510G– and rMA-I529T–infected lungs had 103- and 106-fold less virus than rMA-WT–infected lungs at 2 dpi, respectively. By 5 dpi, virus was detected at high levels in the lungs of mice infected with rMA-WT, while rMA-I529T virus was mostly cleared from the lungs, as low levels of virus were detected only rarely. rMA-D510G virus-infected mice had virus levels intermediate to those of rMA-WT and rMA-I529T (Fig. 2D). H&E staining revealed significantly more edema (exudate in alveolar spaces) as a sign of diffuse alveolar damage and hypercellularity, in rMA-WT–infected lungs when compared to lungs infected with either rMA-D510G or rMA-I529T (Fig. 2 E and F). In summary, these results indicate that the S protein mutations that were selected during the Korean outbreak resulted in substantial virus attenuation, at least in mice.
rMA-I529T Induces Minimal Inflammatory Responses and Protects Mice from Lethal rMA-WT Challenge.
Given the variability in clinical disease and kinetics of virus clearance after infection with the three viruses, we next examined the host immune responses in mice infected with rMA-WT and the two variants. The expression in the lung of several proinflammatory cytokines and chemokines at 2 and 5 dpi in mice infected with 1,000 PFU of rMA-WT or its variants revealed an initial immune response that was greatest in mice infected with rMA-WT. We observed significant reduction in the expression of interferon (IFN)-I (IFN-α and IFN-β), CCL5, ISG15, TNF, and CCL2, but not IFN-γ, when rMA-D510G– or rMA-I529T–infected mice were compared to those infected with rMA-WT (Fig. 3A). Upon analysis of immune cell infiltration at 4 dpi (gating strategy shown in SI Appendix, Fig. S2), we found significantly fewer infiltrating neutrophils (CD45+CD11b+Ly6G+) and inflammatory monocytes-macrophages (IMM; CD45+CD11b+Ly6G−Ly6Chi) in the lungs of rMA-I529T– compared to rMA-WT–infected mice (Fig. 3 B–E). The frequency and number of plasmacytoid dendritic cells (pDC; CD45+CD11b−CD11c+Siglec H+) in the lungs were similar in rMA-WT– and mutant-infected mice while there were more CD11b− DCs (CD45+CD11b−CD11c+MHC class II+) and alveolar macrophages (AM; CD45+CD11b−CD11c+Siglec F+) in the lungs of rMA-I529T–infected mice when compared to rMA-WT– and rMA-D510G–infected mice (SI Appendix, Fig. S3).
Fig. 3.
D510G and I520T induce less cytokines expression and lung injury. (A) hDPP4-KI mice were infected i.n. with 1,000 PFU of WT and mutant MERS-CoV. Lungs were homogenized in TRIzol for RNA isolation and qRT-PCR at the indicated time point. Levels of cytokines and chemokines induced are shown relative to HPRT by the ΔCt method (n = 5 to 6 mice per group per time point). Lungs from infected mice were processed for flow cytometric analysis at 4 dpi. Frequency and number of neutrophils (B) and IMMs (C) in the lungs are shown (n = 7 to 10 mice per group). Statistical significance was determined by two-tailed Student’s t test where *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Bars represent mean and error bars represent SEM. Representative flow plots for neutrophils (D) and IMMs (E).
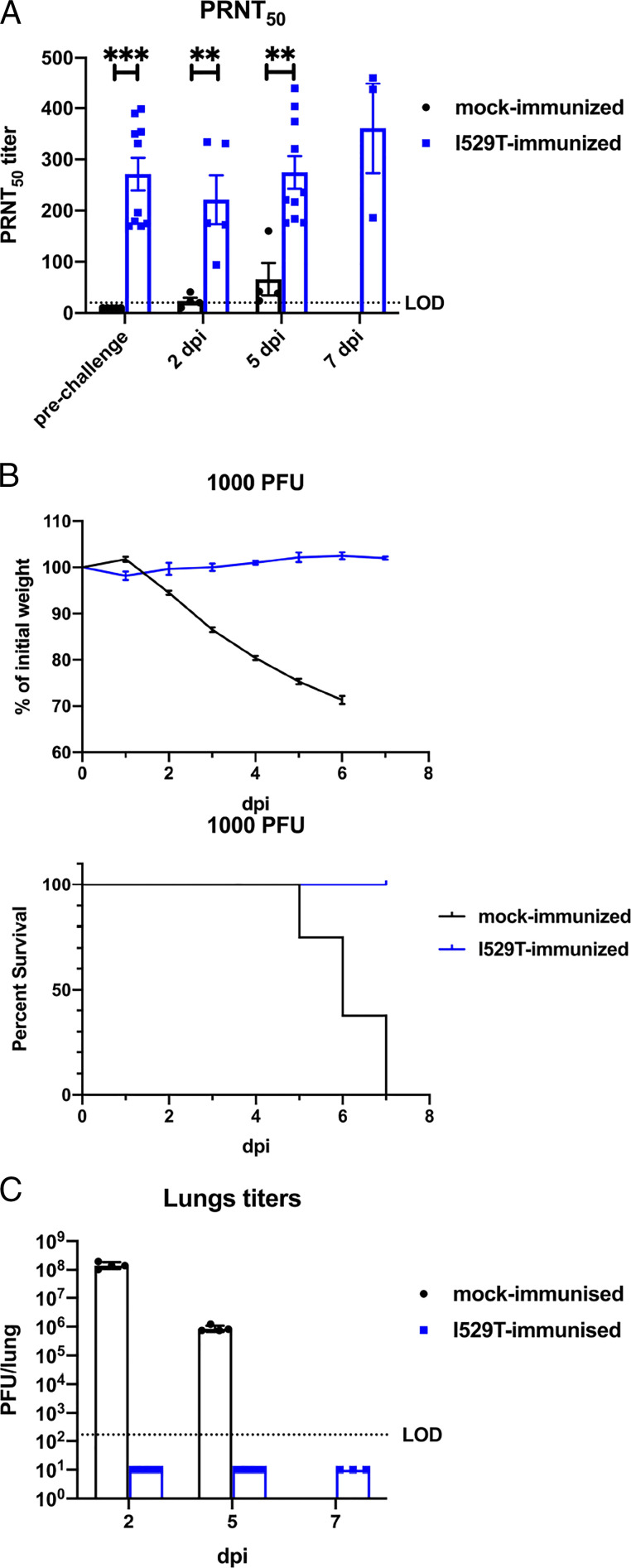
Since the rMA-I529T variant infected mice without causing clinical disease and with only minimal inflammatory cytokine induction, immune cell infiltration, and histopathological changes (Figs. 2 and 3), we tested whether rMA-I529T was a potential live-attenuated vaccine candidate. For this purpose, hDPP4-KI mice were intranasally (i.n.) immunized with 1,000 PFU of rMA-I529T. This elicited a virus-specific antibody response (Fig. 4A, prechallenge). We then challenged these and mock-immunized mice with a lethal dose of rMA-WT (1,000 PFU) at 28 d postimmunization. Mock-immunized mice succumbed to the infection; however, rMA-I529T-immunized mice showed no weight loss and all survived lethal challenge (Fig. 4B). Infectious virus was not detected in the lungs of rMA-I529T-immunized mice at 2, 5, and 7 dpi (Fig. 4C). We also measured levels of neutralizing antibodies by plaque reduction neutralization test (PRNT50) assay at 2, 5, and 7 d postlethal rMA-WT challenge. Neutralizing antibody levels did not change appreciably after challenge when compared to the prechallenge samples but were significantly higher in rMA-I529T–immunized mice than in mock-immunized mice (Fig. 4A). Together, these results suggest that the diminished host immune response following attenuated rMA-I529T infection was sufficiently robust to protect mice from subsequent lethal rMA-WT challenge.
Fig. 4.
rMA-I529T immunization protects mice from subsequent lethal rMA-WT challenge. (A) Serum was collected at the indicated time point after rMA-WT lethal challenge. PRNT50 titers were determined by plaque reduction neutralization assay. Bars represent mean and error bars represent SEM. (B) hDPP4-KI mice were infected i.n. with 1,000 PFU of I529T virus or mock-infected and rested for 28 d before lethal i.n. challenge with 1,000 PFU of WT virus. Daily weights and survival are shown (n = 6 to 8). Data points represent mean and error bars represent SEM. (C) Mock- or rMA-I529T–immunized mice were challenged with 1,000 PFU of rMA-WT. Virus titers in the lungs were measured at the indicated times postchallenge. Data points represent geometric mean and error bars represent geometric SD. LOD: limit of detection. Statistical significance was determined by two-tailed Student’s t test, where **P < 0.01; ***P < 0.001.
African Mutations on Spike Have Minimal Effects in Virulence.
MERS-CoV widely infects camels in Africa but no human clinical disease has been recorded. Recombinant MERS-CoV expressing the East African S protein on an EMC/2012 background are modestly attenuated in vitro (26). We introduced mutations from Burkina Faso (BF), Morocco (Mor), and Nigeria (Nga) (clade C1) and Ethiopia (Ethi-118S, Ethi-126S) (clade C2) isolates into the MA S protein (Table 1), as described for the South Korean mutations. Upon transfection into cells, all mutated S proteins were expressed and cleaved (SI Appendix, Fig. S4A). The effects of these mutant S proteins on viral entry were determined utilizing VSV-pseudotyped viruses in three cell lines. The relative ability of pseudoviruses coated with the different S proteins to enter cells was consistent among all three cell lines. Clade C2 mutants showed the greatest reduction in entry, followed by Nga- and Mor-S, compared to WT-S, while BF-S entered cells as efficiently as WT-S (Fig. 5A). Next, we examined the effects of the mutations present in the East African and West African isolates on disease by engineering isogenic rMA viruses expressing clade C1 and clade C2 S proteins. After challenge with 200 PFU or 1,000 PFU, all mutant viruses induced comparable weight loss and mortality to rMA-WT (Fig. 5 B and C). We further measured virus titers in the lungs of rMA-Ethi-118S– (clade C2) and rMA-BF– (clade C1) infected mice since infection with these two viruses resulted in the greatest differences in weight (Fig. 5B). There were no significant differences in virus titers in mouse lungs at 2 and 5 dpi (Fig. 5D). However, rMA-Ethi-118S and rMA-Ethi-126S formed smaller plaques than did rMA-WT and rMA-BF viruses (SI Appendix, Fig. S4B), consistent with the modest reduction in viral entry (Fig. 5A).
Table 1.
Mutations in African camel S proteins compared to EMC/2012
| Residue no. | Domain within S | EMC | BF | Ethi-118S | Ethi-126S | Mor | Nga |
| 26 | S1-NTD | V | A | A | A | A | A |
| 89 | S1-NTD | A | S | . | . | S | . |
| 139 | S1-NTD | T | . | I | I | . | . |
| 158 | S1-NTD | D | . | Y | Y | . | . |
| 167 | S1-NTD | H | . | . | . | . | Y |
| 194 | S1-NTD | H | Y | Y | Y | . | Y |
| 390 | S1-RBD | S | . | F | F | . | . |
| 424 | S1-RBD | T | I | . | . | I | . |
| 431 | S1-RBD | A | . | . | . | . | . |
| 457 | S1-RBD | S | . | . | . | . | . |
| 495 | S1-RBD | L | . | . | . | . | F |
| 515 | S1-RBD | P | . | . | L | . | . |
| 588 | S1-RBD | L | . | . | . | . | F |
| 597 | S1-RBD | A | . | V | V | . | . |
| 626 | S1-SD1 | R | . | P | P | . | . |
| 677 | S1-SD2 | V | . | . | . | . | . |
| 773 | S2-L | L | . | . | . | . | . |
| 810 | S2-L | V | . | . | . | . | . |
| 851 | S2-UH | A | . | E | E | . | . |
| 856 | S2-L | S | Y | . | . | Y | Y |
| 884 | S2-L | R | . | . | . | L | . |
| 1020 | S2-HR1 | Q | . | . | . | . | . |
| 1158 | S2-CH | A | S | S | S | S | L |
| 1193 | S2-SD3 | A | . | . | . | . | . |
| 1200 | S2-SD3 | L | . | . | . | . | F |
| 1209 | S2 | V | . | . | . | L | . |
| 1302 | S2-TM | G | . | S | S | . | . |
. Represents no changes in amino acid compared to EMC/2012 strain. Amino acids are identified using single-letter codes. CH, central helix; HR, heptad repeat; L, linker region; NTD, N-terminal domain; SD, subdomain; TM, transmembrane region/domain; UH, upstream helix (45).
Fig. 5.
Spike of African isolates mediate impaired entry but not virulence in vivo. (A) Indicated cell lines were transduced with VSV pseudoviruses expressing MERS-CoV Spike variants as shown. At 18 h posttransduction, cells were lysed and luciferase level were measured. Bars represent mean and error bars represent SEM (Clade C2 and C1 colored with blue and red respectively). Data are representative of three independent experiments. Statistical significance was determined by two-tailed Student’s t test, where *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. hDPP4-KI mice were infected i.n. with WT or mutant MERS-CoV and daily weight changes and survival are shown after infection with 200 PFU (n = 7 to 10 mice per group) (B) or with 1,000 PFU (n = 7 to 10 mice per group) (C). Bars represent mean and error bars represent SEM in weight curves. Statistical significance for survival analysis was determined by log-rank (Mantel–Cox) test. (D) hDPP4-KI mice were infected i.n. with WT and mutant MERS-CoV at 200 PFU. Lungs were harvested and homogenized at 2 and 5 dpi. Virus titers were determined by plaque assay. Bars represent geometric mean and error bars represent geometric SD.
Discussion
Our results demonstrate the stark differences in evolution of the MERS-CoV S protein that occurred in Africa, where the virus is endemic in camels, and in Korea, where the 2015 outbreak was initiated by a single infected patient. Throughout much of the world, dromedary camels are infected with MERS-CoV and spillover to human populations occurs rarely (27). MERS-CoV that cross species to infect humans on the Arabian Peninsula are the dominant strains prevalent in camels at the time (28). The patient who was the source for the Korean outbreak was infected with a MERS-CoV strain circulating in Saudi Arabia at the time. The numbers and origins of the different camel strains and the nature of MERS-CoV evolution in camels are not well understood. When MERS-CoV crosses species to infect humans in Africa, it only causes subclinical disease (29), as far as is known. Our results show that while there are subtle differences in virulence in isogenic viruses expressing African camel S proteins, these differences do not appear to account for this lack of clinical MERS in Africa. They support the notion that other factors, such as mutations and deletions in accessory proteins such as ORF4b (11), are major factors explaining the absence of symptomatic human MERS cases in West Africa; the basis for the lack of human transmission in East Africa will require further investigation. Our results are also in agreement with previous reports demonstrating no (clade C1) or modest (clade C2) effects of the African S protein mutations when compared to the reference EMC/2012 S protein in the context of pseudoviruses or isogenic viruses, respectively (8, 17).
In contrast, MERS-CoV in Korea evolved solely in human populations, since the outbreak was initiated by a single infected patient and there was no periodic reintroduction from an infected zoonotic (e.g., camel) source (5). Under these conditions, viruses are predicted to evolve to better infect humans, with selection of mutations that enhance transmission. This evolutionary process was well illustrated when SARS-CoVs isolated at different times during the 2002 to 2003 SARS epidemic were analyzed. Mutations that increased binding to the human ACE2 receptor were identified over time, consistent with human adaptation (30). Our results with isogenic mouse-adapted viruses expressing the Korean MERS-CoV S proteins indicate that the S protein did not evolve to enhance virulence. Consistent with this, modest attenuation was observed in human patients: the average fever duration in patients infected with WT virus was generally longer than in I529T-infected patients (6), although changes in morbidity or mortality were not observed. These results raise the possibility that infection of a larger number of patients might have revealed a more profoundly attenuated phenotype. However, one caveat is that only 48 patients were recruited in the study in which both clinical disease and virus sequence analysis were described previously (5, 6). The lack of diminution in clinical severity may have reflected infection by mixed populations of viruses encoding both WT and mutant S sequences (31).
It is possible that the enhanced thermostability exhibited by rMA-I529T makes viruses more durable in external or human airway environments. Thermostability is frequently correlated with the integrity of S protein heterodimers. MERS-CoV S proteins are proteolytically cleaved into noncovalently associated S1 and S2 fragments, making the RBD-containing S1 fragments prone to heat-induced shedding. S1 shedding before receptor interaction could diminish entry; however, S1 shedding after receptor binding is required for subsequent refolding of fusion machinery (25, 32). It is therefore important for the virus to achieve an optimal balance in S1 shedding before and after receptor binding for efficient entry and transmission. Whether the D510G or I529T variants could be transmitted more stably and effectively than the parental strain is not known, since only a small number of patients were infected with the different variants during the 2015 Korean outbreak. Transmission might be examined in experimentally infected animals when a useful model for studying MERS-CoV transmission is established.
Kim et al. (6) also found that the two mutations appeared sequentially. The more attenuated I529T mutation emerged before the less attenuated D510G mutation (6). The I529T mutation emerged in the index patient (P001) and was spread to P014. P014 harbored mixed populations of viruses including WT, I529T, D510G, and the double mutant. The D510G variant was transferred from P014 to additional contacts. Our results show that the I529T/D510G virus was viable but unable to form plaques. The double mutant could represent an intermediary in virus evolution in infected patients, which could be a consequence of recombination between singly mutated viruses or could be a strain that happened to be present in the groups of viruses (quasispecies) that infected the patient in question. In any case, there is little evidence that the I529T/D510G virus was transmitted efficiently during the 2015 outbreak.
A remarkable feature of the Korean outbreak is that the attenuation of the S protein occurred in only a few virus generations. This rapid selection has been postulated to reflect pressure from the human antibody response since the variant Korean viruses are slightly less susceptible to neutralization by MERS-CoV polyclonal plasma (5). Arguing against a role for human immune pressure is that no Korean should have been immune to MERS-CoV since it was newly introduced into the country. While there is some evidence for cross-reactivity between common cold coronaviruses (CCCoV) and SARS-CoV-2 (33–35), this has not been demonstrated for MERS-CoV. Even if present, based on the SARS-CoV-2 studies, it is unlikely that the response to CCCoV would be sufficient to exert appreciable immune pressure. Thus, we speculate that MERS-CoV in Korea was actually adapting to enhance transmission between humans, with attenuation as a by-product. This was only observable in the absence of periodic introduction of zoonotic MERS-CoV. A limitation of this study is that conclusions about attenuation were based on studies that used mouse-adapted MERS-CoV. The effects of apparent virus attenuation on transmission or clinical disease cannot be validated in patient populations because MERS-CoV was eradicated from the Korean peninsula soon after its introduction.
These results may aid in understanding coronavirus evolution in general. In the ongoing COVID-19 pandemic, several mutations on the SARS-CoV-2 S protein have been identified. Some of these mutations appear to enhance transmission without affecting virulence. One mutation, D614G, is associated with higher infectious titers in the upper respiratory, but not the lower respiratory tract of golden Syrian hamsters and with enhanced transmissibility (19, 36). In addition, the D614G mutation also reduced S1 shedding and higher levels of S protein incorporated into virions (19, 36, 37). In the recently emerged B1.1.7 SARS-CoV-2 variant, 17 mutations were identified in the genome with 8 mutations found on the S protein (38). This variant exhibited enhanced transmissibility without clear effects on virulence. N501, one of the six residues in the receptor-binding interface, key for binding to ACE2 (39), is mutated in B1.1.7. N501Y may enhance human ACE2 binding (40). Of note, the N501Y mutation has been detected in virus populations since the beginning of the pandemic (20, 38) but is only now becoming fixed, in context with other S protein changes that are presumably required for the increased transmissibility shown by the B1.1.7 variant.
IAV is another virus in which isolates have been extensively sequenced and shown to exhibit variability. In general, sequence changes are believed to commonly occur, since these viruses, like CoV, do not have high stringency proofreading capabilities. In response to a variety of pressures including human immune pressure, IAV isolates that exhibit antigen drift but maintain virulence are selected (41). Because of the large size of the human pool and the potential for zoonotic introduction of recombinant and mutated viruses, it is difficult to sort out the details of this process, unlike the situation in Korea, in which MERS-CoV was introduced only once and in Africa, in which only intercamel spread occurs.
Together, these results indicate the complex relationship of S binding to its host cell receptor and transmissibility. SARS-CoV-2 S protein variants appear to exhibit increased affinity for hACE2 while the Korean S protein of MERS-CoV evolved to have lower affinity for hDPP4 (5). Overall, the sequential emergence of the two mutations during the Korean MERS outbreak provides insight into how mutations are selected that affect virulence and potentially transmission during a human coronavirus epidemic.
Materials and Methods
Mouse Infection.
Human DPP4-KI mice were generated and propagated as described previously (21). Both male and female mice of 6 to 10 wk of age were used in this study. Mice were infected as described previously (42). In brief, mice were anesthetized with ketamine-xylazine and infected i.n. with 200 PFU (nonlethal) and 1,000 PFU (lethal) of virus in 50 μL of DMEM (Gibco). All work with MERS-CoV was performed in the University of Iowa’s Biosafety level (BSL3) laboratories. All animal studies were approved by the University of Iowa Animal Care and Use Committee and meet stipulations of the Guide for the Care and Use of Laboratory Animals (43).
Lung Harvest for Virus Titers.
Mice were anesthetized by intraperitoneal injection of ketamine-xylazine. Lungs were transcardially perfused with 10 mL PBS. Lungs were removed and homogenized in 1 mL PBS. Lung homogenates were clarified by centrifugation and stored at −80 °C for titering by plaque assay on Vero 81 cells (CCL-81; ATCC) as described previously (42).
Collection of Serum.
Mice were anesthetized by intraperitoneal injection of ketamine-xylazine. Blood was collected through retro-orbital bleed with a capillary tube (Fisher Scientific). Blood was allowed to clot at room temperature for 30 min. Serum was clarified by centrifugation and transferred to a new tube for storage at −80 °C. Plaque reduction neutralization assays were performed as previously described (42).
Plaque Reduction Neutralization Test.
Serum or antibody samples were serially diluted in DMEM and mixed 1:1 with 40 PFU MERS-CoV for 1 h at 37 °C. The mixtures were then added to Vero 81 cells for an additional 1 h at 37 °C. After removing the culture medium, cells were overlaid with 1.2% agarose, and cultured for 3 d. Plaques were visualized by 0.1% crystal violet staining. PRNT50 was determined as the concentration of serum required to reduce the number of plaques by 50% compared to control serum-exposed virus.
Lung Histology.
Mice were anesthetized by intraperitoneal injection of ketamine-xylazine. Lungs were transcardially perfused with 10 mL PBS. Lungs were removed, fixed in zinc formalin and paraffin-embedded. Sections were prepared, stained with H&E, and scored in a blinded fashion (44) for edema and inflammation with scores of 0, 1, 2, 3, and 4 representing lung areas with 0%, less than 3%, 6 to 33%, 33 to 66%, and more than 66% detectable edema and inflammation, respectively.
Lung Cell Preparation and Antibodies for Flow Cytometric Analysis.
Animals were anesthetized with ketamine-xylazine and perfused transcardially with 10 mL PBS. Lungs were removed, minced, and digested in HBSS buffer consisting of 2% fetal calf serum, 25 mM Hepes, 1 mg/mL collagenase D (Roche) and 0.1 mg/mL DNase (Roche) at 37 °C for 30 min. Single-cell suspensions were prepared by passage through a 70-μm cell strainer. Cells were enumerated with a Scepter 2.0 cell counter (MilliporeSigma). Cells were then washed and blocked with 1 μg α-CD16/α-CD32 (2.4G2) antibody at 4 °C for 20 min and surface stained with the following antibodies at 4 °C for 30 min: PE-Cy7 α-CD45 (30-F11; BioLegend); PE α-Siglec F (S17007L; BioLegend); APC α-Siglec H (551; BioLegend); PerCP/Cyanine 5.5 α-mouse I-A/I-E (M5/114.15.2; BioLegend); eFluor 450 α-CD11b (M1/70; eBioscience); FITC α-Ly6G (1A8; BioLegend); APC/Cyanine 7 α-Ly6C (HK1.4; BioLegend); LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Invitrogen). Cells were washed, fixed, and permeabilized with Cytofix/Cytoperm (BD Biosciences). Flow cytometric data were acquired using a BD FACSVerse (BD Biosciences) analyzer and interpreted with FlowJo (Tree Star).
Statistical Analysis.
A Student’s t test was used to analyze differences in mean values between groups. All results are expressed as mean ± SEM P < 0.05 were considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001. Differences in mortality were analyzed using Kaplan–Meier log-rank survival tests.
Supplementary Material
Acknowledgments
This work was supported in part by NIH Grants R01 AI129269 and P01 AI060699 (to S.P.). L.-Y.R.W. was supported by a Training Grant (T32 AI007511).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102983118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.Zaki A. M., van Boheemen S., Bestebroer T. M., Osterhaus A. D. M. E., Fouchier R. A. M., Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012). [DOI] [PubMed] [Google Scholar]
- 2.WHO , Middle East respiratory syndrome coronavirus (MERS-CoV). https://www.who.int/health-topics/middle-east-respiratory-syndrome-coronavirus-mers#tab=tab_1. Accessed 8 February 2021.
- 3.Oboho I. K., et al., 2014 MERS-CoV outbreak in Jeddah—A link to health care facilities. N. Engl. J. Med. 372, 846–854 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hui D. S., et al., Middle East respiratory syndrome coronavirus: Risk factors and determinants of primary, household, and nosocomial transmission. Lancet Infect. Dis. 18, e217–e227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim Y., et al., Spread of mutant Middle East respiratory syndrome coronavirus with reduced affinity to human CD26 during the South Korean outbreak. mBio 7, e00019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim Y. S., et al., Sequential emergence and wide spread of neutralization escape Middle East respiratory syndrome coronavirus mutants, South Korea, 2015. Emerg. Infect. Dis. 25, 1161–1168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D. W., et al., Variations in Spike glycoprotein gene of MERS-CoV, South Korea, 2015. Emerg. Infect. Dis. 22, 100–104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleine-Weber H., et al., Mutations in the Spike protein of Middle East respiratory syndrome coronavirus transmitted in Korea increase resistance to antibody-mediated neutralization. J. Virol. 93, e01381-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu D. K. W., et al., MERS coronaviruses in dromedary camels, Egypt. Emerg. Infect. Dis. 20, 1049–1053 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu D. K., et al., Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Nigeria, 2015. Euro Surveill. 20, 30086 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Chu D. K. W., et al., MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc. Natl. Acad. Sci. U.S.A. 115, 3144–3149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kandeil A., et al., Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Africa and Middle East. Viruses 11, 717 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canton J., et al., MERS-CoV 4b protein interferes with the NF-κB-dependent innate immune response during infection. PLoS Pathog. 14, e1006838 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thornbrough J. M., et al., Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio 7, e00258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y., et al., Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 5, 17554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthews K. L., Coleman C. M., van der Meer Y., Snijder E. J., Frieman M. B., The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J. Gen. Virol. 95, 874–882 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleine-Weber H., Pöhlmann S., Hoffmann M., Spike proteins of novel MERS-coronavirus isolates from North- and West-African dromedary camels mediate robust viral entry into human target cells. Virology 535, 261–265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korber B.et al.; Sheffield COVID-19 Genomics Group , Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 182, 812–827.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou Y. J., et al., SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Toole Á., et al., Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2. Virological (2021). https://virological.org/t/tracking-the-international-spread-of-sars-cov-2-lineages-b-1-1-7-and-b-1-351-501y-v2/592. Accessed 8 February 2021.
- 21.Li K., et al., Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc. Natl. Acad. Sci. U.S.A. 114, E3119–E3128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almazán F., et al., Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio 4, e00650-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirato K., Kawase M., Matsuyama S., Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 87, 12552–12561 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J. E., et al., Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. U.S.A. 113, 12262–12267 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walls A. C., et al., Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 114, 11157–11162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirato K., et al., Middle East respiratory syndrome coronavirus in dromedaries in Ethiopia is antigenically different from the Middle East isolate EMC. Front. Microbiol. 10, 1326 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dudas G., Carvalho L. M., Rambaut A., Bedford T., MERS-CoV spillover at the camel-human interface. eLife 7, e31257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabir J. S. M., et al., Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 351, 81–84 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Mok C. K. P., et al., T-cell responses to MERS coronavirus infection in people with occupational exposure to dromedary camels in Nigeria: An observational cohort study. Lancet Infect. Dis. 21, 385–395 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chinese SARS Molecular Epidemiology Consortium , Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303, 1666–1669 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Park D., et al., Analysis of intrapatient heterogeneity uncovers the microevolution of Middle East respiratory syndrome coronavirus. Cold Spring Harb. Mol. Case Stud. 2, a001214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai Y., et al., Distinct conformational states of SARS-CoV-2 spike protein. Science 369, 1586–1592 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braun J., et al., SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 587, 270–274 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Lipsitch M., Grad Y. H., Sette A., Crotty S., Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 20, 709–713 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mateus J., et al., Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 370, 89–94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plante J. A., et al., Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592, 116–121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L., et al., SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 11, 6013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rambaut A., et al., COVID-19 Genomics Consortium UK, Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of Spike mutations. Virological (2020). https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563. Accessed 7 January 2021.
- 39.Yi C., et al., Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 17, 621–630 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Starr T. N., et al., Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hensley S. E., et al., Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 326, 734–736 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong L.-Y. R., Sensitization of non-permissive laboratory mice to SARS-CoV-2 with a replication-deficient adenovirus expressing human ACE2. STAR Protocol. 1, 100169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 44.Meyerholz D. K., Beck A. P., Principles and approaches for reproducible scoring of tissue stains in research. Lab. Invest. 98, 844–855 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Yuan Y., et al., Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 8, 15092 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.