Significance
Depression is a serious disease afflicting an increasing number of people, including adolescents. Animal studies indicate that the neuropeptide galanin and its three receptors, GalR1 to 3, are involved in depression-like behavior, evidenced by increased GalR1 transcript levels in the ventral periaqueductal gray of “depressed” rats. Here, we show coexistence of Scratch2 and GalR1, identifying a possible mechanism underlying control of GalR1 expression in this region: the transcription factor Scratch2 lowers, via binding to an E-box in the GalR1 promoter, expression of the GalR1 receptor. This may represent a further role for Scratch2, earlier associated with embryonic development, cell migration, and neurogenesis. Because GalR1 seems involved in major depression, we discuss whether or not Scratch2 also may play a role in the human disease.
Keywords: galanin, neuropeptides, stress, transcription factor, transmitter coexistence
Abstract
Galanin receptor1 (GalR1) transcript levels are elevated in the rat ventral periaqueductal gray (vPAG) after chronic mild stress (CMS) and are related to depression-like behavior. To explore the mechanisms underlying the elevated GalR1 expression, we carried out molecular biological experiments in vitro and in animal behavioral experiments in vivo. It was found that a restricted upstream region of the GalR1 gene, from −250 to −220, harbors an E-box and plays a negative role in the GalR1 promoter activity. The transcription factor Scratch2 bound to the E-box to down-regulate GalR1 promoter activity and lower expression levels of the GalR1 gene. The expression of Scratch2 was significantly decreased in the vPAG of CMS rats. Importantly, local knockdown of Scratch2 in the vPAG caused elevated expression of GalR1 in the same region, as well as depression-like behaviors. RNAscope analysis revealed that GalR1 mRNA is expressed together with Scratch2 in both GABA and glutamate neurons. Taking these data together, our study further supports the involvement of GalR1 in mood control and suggests a role for Scratch2 as a regulator of depression-like behavior by repressing the GalR1 gene in the vPAG.
Depression is the most prevalent mental disorder that affects over 350 million people worldwide and is associated with a great economic burden (1–3). Underlying mechanisms include environmental stressors (4, 5), a significant heritability (6), and genetic–environmental interactions (7). Increasing evidence shows that conventional antidepressants, such as selective serotonin reuptake inhibitors, can display side effects (8) and unsatisfactory response rates (treatment resistance) (9), which restrict their further application. There is therefore a need to further explore the molecular mechanisms underlying depression and to search for novel therapeutic targets.
Galanin is a 29-amino acid neuropeptide (30 amino acid in humans) (10) that is found in neurons in the peripheral and central nervous system. In the rat brain, galanin is widely distributed and coexpressed with 5-hydroxytryptamine (5-HT) in dorsal raphe nucleus neurons and with noradrenaline/norepinephrine in locus coeruleus neurons (11–13). Animal experiments have shown that stress and exercise robustly increase galanin expression in noradrenergic neurons in the locus coeruleus (14–16). In fact, there is increasing evidence that galanin is involved in the regulation of many physiological and patho-physiological processes, including depression or depression-like behavior and resilience (17–24).
The action of galanin is mediated via three G protein-coupled receptors, GalR1, GalR2, and GalR3 (25). All above-mentioned galanin receptors have unique distribution patterns (26–28) and exert their effects via different signaling pathways (25). GalR1 mainly activates Gi/o proteins to mediate inhibitory actions, such as inhibiting adenylate cyclase (29–32) and opening potassium channels (33). In contrast, GalR2 activates, among others, Gαq/11 protein to relay the stimulatory effects of galanin, and GalR3 has a similar transduction profile to GalR1 (25).
Human genetic studies have shown that GalR1 may play a role in various brain disorders. For example, GalR1 has been associated with smoking and craving (34–36), stress and drug addiction (37), depression (38, 39), and schizophrenia (40). Animal experiments have also suggested involvement of GalR1 in addiction (41) and in depression-like behaviors. For example, GalR1 mRNA levels are significantly increased in the ventral periaqueductal gray (vPAG) of rats exposed to chronic mild stress (CMS) (42). Inhibition of GalR1 in prefrontal cortex (PFC) alleviates the depressive-like behavior in a rat postpartum depression model (43). Thus, up-regulation of GalR1 in selective brain regions is associated with depression-like behaviors.
In contrast to the galanin gene (44), less is known about the promoter region of the GalR1 gene. In the present study, we investigated the upstream sequence of the GalR1 gene and found that Scratch2 (45, 46), a member of the Snail superfamily of zinc-finger (ZF) transcription factors (47, 48), binds to the E-box of the GalR1 promoter. The Scratch family of transcription factors is expressed in the nervous system (49) and is important for embryonic development, cell migration, and neurogenesis (45, 46, 50–52). However, so far there is no report on possible involvement of Scratch2 in the pathophysiology of mood. We carried out in vitro molecular biological experiments and in vivo animal behavioral experiments to explore, if Scratch2 is involved in the depression-like behaviors in the rat by regulating GalR1 expression in the vPAG.
Results
Determination of the Rat GalR1 Gene Transcription Start Site.
To identify the transcription start site of the rat GalR1 gene, we carried out a rapid amplification of cDNA ends (5′-RACE) analysis using total RNA isolated from rat vPAG tissue. Sequence analysis showed that the rat GalR1 gene has one transcription start site, which begins 229 bp upstream of the translation start site (SI Appendix, Fig. S1 A–C). The diagrammatic structure of the rat GalR1 gene is shown in SI Appendix, Fig. S1D, and partial sequences of the rat GalR1 promoter and downstream region are shown in SI Appendix, Fig. S2.
Identification of a Key Negative Region in the GalR1 Promoter.
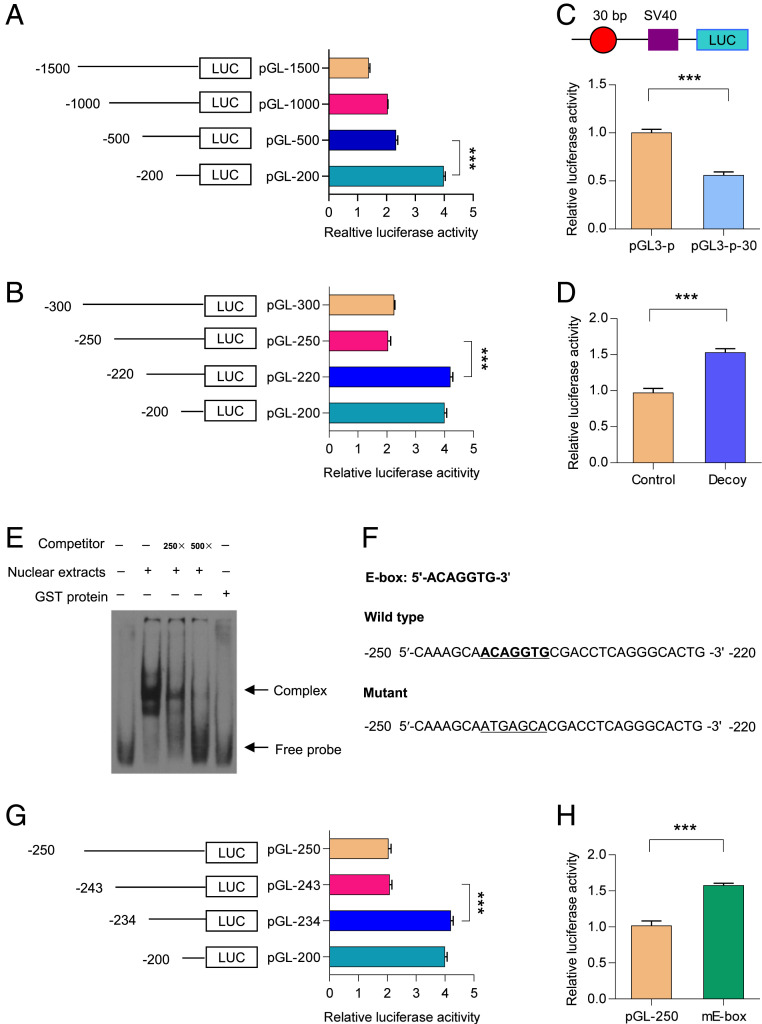
To determine the main regulatory regions in the GalR1 promoter, a 1,500-bp fragment of the GalR1 promoter (−1,500/+1) and its four 5′-deletion segments were cloned into the pGL3-basic vector (SI Appendix, Table S1), and these reporter plasmids were transfected into PC12 cells. As shown in Fig. 1A, deletion of the region from −500 to −200 resulted in a remarkable increase in luciferase activity, indicating the presence of putative negative elements in this region. To determine the key negative elements more accurately, we carried out a detailed 5′-deletion analysis of the −500 to −200 region. As shown in Fig. 1B, deletion of −250 to −220 resulted in a significant increase in luciferase activity, suggesting presence of negative elements in this restricted region.
Fig. 1.
Location of the E-box element in the GalR1 promoter. (A) Initial characterization of the GalR1 promoter in PC12 cells using larger truncated fragments from −1,500 to −200. One-way ANOVA, F(3, 8) = 1,219, P < 0.0001. (B) Refined analysis of the GalR1 promoter in PC12 cells using the smaller truncated fragment from −300 to −200. One-way ANOVA, F(3, 8) = 585.3, P < 0.0001. (C) The 30-bp sequence between −250 and −220 of the GalR1 promoter was cloned into pGL3-promoter vector and transfected into PC12 cells for 24 h, followed by monitoring and measuring of luciferase activity. T4 = 14.9, P < 0.0001. pGL3-p: pGL3-promoter vector, pGL3-p-30: the pGL3-promoter vector containing above 30-bp sequence of the GalR1 promoter. (D) The reporter plasmid pGL-250 was transfected into PC12 cells with or without decoy oligodeoxynucleotides for 24 h. Then, cells were collected and luciferase activities were analyzed. T4 = 11.85, P < 0.001. (E) The oligonucleotides from −250 to −220 of the GalR1 promoter were labeled with digoxigenin (dig). Then, dig-labeled oligonucleotides were incubated with PC12 cell nuclear extracts in the absence or presence of 250- or 500-fold excess of unlabled oligonucleotides. DNA–protein complexes were resolved by nondenaturing PAGE. (F) Nucleotide sequences of the region from −250 to −220 of the GalR1 promoter. The E-box element is underlined. The TRANSFAC database was used to identify putative cis-elements in this region. (G) Refined analysis of the GalR1 promoter using the smaller truncated fragments from −250 to −220 in PC12 cells. One-way ANOVA, F(3, 8) = 511.8, P < 0.0001. (H) Effects of mutation of E-box on the activity of pGL-250. T4 = 12.97, P < 0.001. Data are presented as mean ± SD (n = 3 per group). ***P < 0.001.
To further determine the negative role of the −250 to −220 region, this sequence was inserted into the pGL3-promoter vector. Luciferase activity confirmed that the region from −250 to −220 indeed reduced activity of the pGL3-promoter (Fig. 1C). To confirm the above results, we performed a DNA decoy assay, which involves the delivery of double-stranded oligodeoxynucleotides corresponding to a specific promoter region, resulting in changes of target-gene transcription (53), further supporting the findings mentioned above (Fig. 1D). Taking these data together, we find that the region from −250 to −220 may play a critical, negative role in the regulation of the GalR1 promoter activity.
The E-Box Is Involved in the Negative Regulation of the GalR1 Promoter Activity.
Because the region from −250 to −220 plays a negative role in the regulation of GalR1 promoter activity, it may bind some transcription factors. To explore this possibility, we performed a gel-shift assay using nuclear extracts from PC12 cells. As shown in Fig. 1E, a strong DNA complex was observed, which was effectively out-competed by a 500-fold excess of unlabeled probe. Moreover, GST protein did not bind to this probe, which supported the specificity of the DNA–protein complex.
The region from −250 to −220 of the GalR1 promoter was analyzed with the TRANSFAC database (54) to explore which putative cis-element is present in this region, and a putative E-box was found (Fig. 1F). To determine whether the E-box plays a negative role in the regulation of GalR1 promoter activity, a 5′-deletion analysis of the −250 to −220 region was carried out. As shown in Fig. 1G, deletion of the region from −243 to −234 increased the activity of the GalR1 promoter, suggesting a negative regulatory role of the E-box. To further confirm the above results, the E-box was mutated and the resulting increase in luciferase activity suggested that this manipulation increased the activity of pGL-250 in PC12 cells. Taken together, these results demonstrate that the E-box acts as a negative modulator in the regulation of the GalR1 promoter activity (Fig. 1H).
Scratch2 Binds to the E-Box of the GalR1 Promoter.
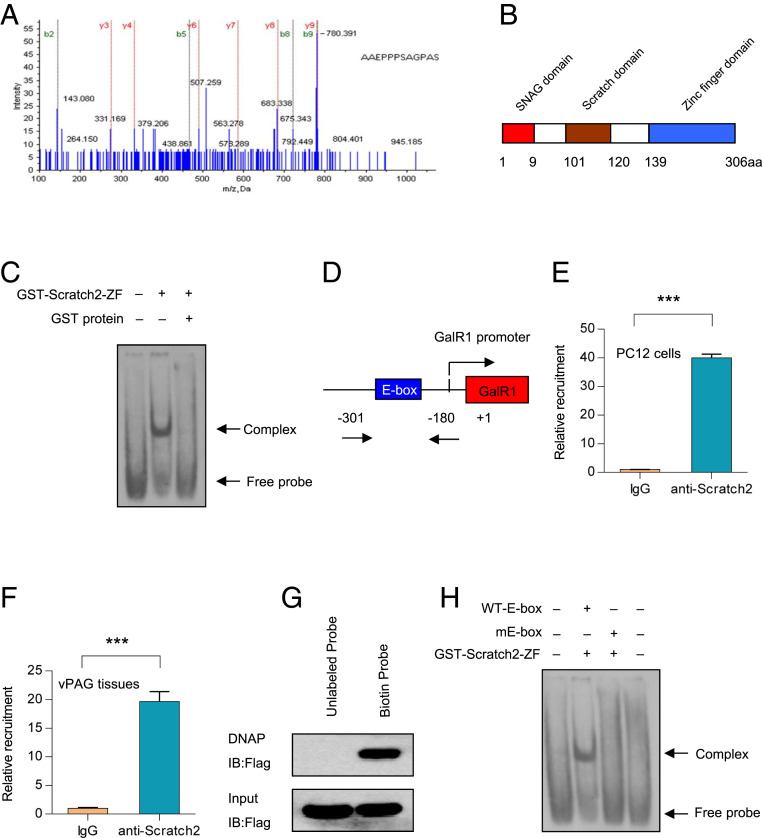
To isolate the putative transcription factors binding to the region from −250 to −220 of the GalR1 promoter, a DNA pull-down assay was applied, and the bound proteins were analyzed by mass spectrometry. The results showed that a transcription factor, Scratch2, is present in the bound proteins (Fig. 2A). The detailed results of the mass spectrometric analysis of Scratch2 can be found in SI Appendix, Table S4. Scratch2 contains an N-terminal SNAG domain, a Scratch domain, and a C-terminal ZF structure domain (Fig. 2B). To determine whether Scratch2 could bind to the −250 to −220 region, a gel-shift assay was performed using recombinant GST–Scratch2–ZF fusion protein and a digoxigenin (dig)-labeled probe spanning the −250 to −220 segment. It was found that the GST–Scratch2–ZF protein binds to the probe, whereas the GST protein does not (Fig. 2C).
Fig. 2.
Scratch2 binds to the E-box of the GalR1 promoter. (A) The elution proteins from DNA pull-down assay were analyzed by mass spectrometry, and the resulting peptide of Scratch2 was identified. (B) A schematic structure of Scratch2. (C) Dig-labeled oligonucleotides were incubated with GST–Scratch2–ZF fusion protein, and the DNA–protein complexes were resolved by nondenaturing PAGE. (D) A schematic diagram for the GalR1 promoter harboring the E-box element. (E and F) PC12 cells (E) or vPAG tissues (F) were lysed, and ChIP assays were performed by using anti-Scratch2 antibody. Then, the immunoprecipitated DNA was detected by real-time PCR. In E, T4 = 51.16, P < 0.0001. In F, T4 = 18.39, P < 0.0001. (G) DNA affinity precipitation assays were performed with 293T cells extracts expressing Flag-Scracth2 and a biotin-labeled probe spanning −250 to −220 of the GalR1 promoter. DNA precipitates were detected with anti-Flag antibody. (H) The oligonucleotides described in Fig. 1F were labeled with digoxigenin. Dig-labeled oligonucleotides were incubated with GST–Scratch2–ZF fusion protein, and DNA–protein complexes were detected by nondenaturing PAGE. Data are presented as mean ± SD (n = 3 per group). ***P < 0.001.
To verify the above results in vivo, a chromatin immunoprecipitation (ChIP) assay was carried out in PC12 cells and rat vPAG tissues. As expected, the cross-linked DNA–Scratch2 complexes immunoprecipitated with Scratch2 antibody and were detected by PCR amplification with primers spanning the −250 to −220 region (Fig. 2 E and F). For further confirmation, DNA affinity precipitation was used to identify the DNA–protein interactions (55). In fact, the DNA–Flag–Scratch2 complexes were detected by anti-Flag antibody followed by the precipitation of streptavidin beads (Fig. 2G).
To further determine whether the Scratch2 protein binds to the E-box between −250 and −220, a gel-shift assay was then carried out using recombinant GST–Scratch2–ZF protein and mutant E-box oligonucleotides. As shown in Fig. 2H, mutation of the E-box nearly abolished the formation of the DNA–protein complex. Taken together, all of the above results strongly suggest that Scratch2 binds to the E-box of the GalR1 promoter, both in vitro and in vivo.
Scratch2 Regulates GalR1 Promoter Activity and GalR1 Gene Expression.
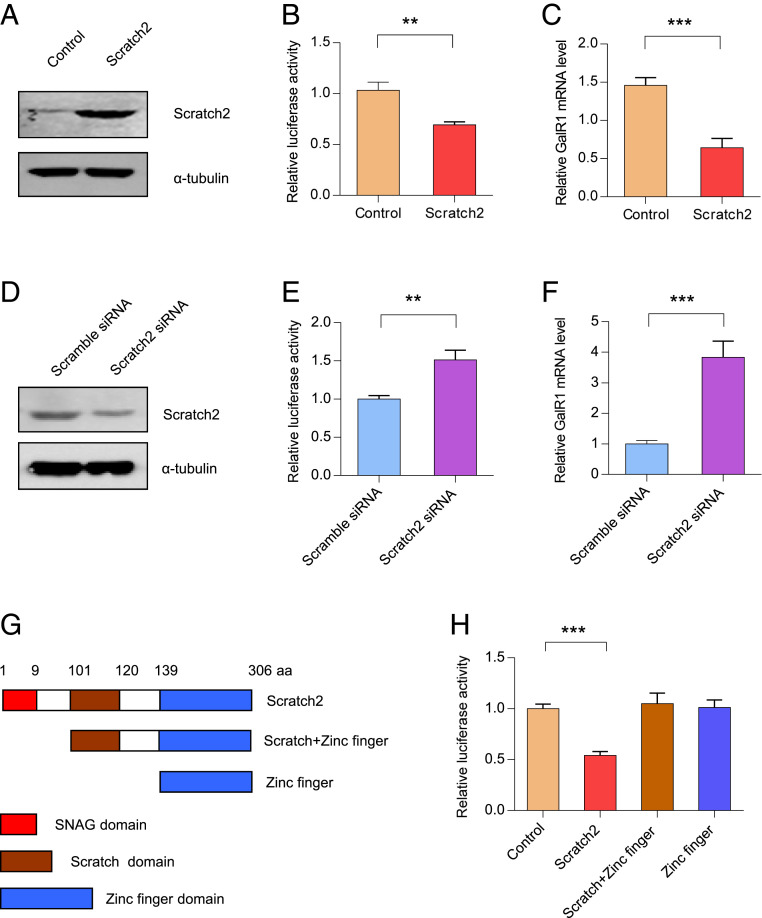
To determine whether Scratch2 is involved in the regulation of GalR1 transcription, PC12 cells were transfected with pcDNA3.1-Scratch2. Overexpression of Scratch2 significantly increased the levels of Scratch2 in PC12 cells (Fig. 3A and SI Appendix, Fig. S3A). Moreover, overexpression of Scratch2 efficiently decreased the activity of pGL-250 and GalR1 mRNA levels (Fig. 3 B and C). In agreement, knockdown of Scratch2 expression in PC12 cells by Scratch2 small-interfering RNA (siRNA) resulted in a lower expression of Scratch2 in the PC12 cells (Fig. 3D and SI Appendix, Fig. S3B). Finally, knockdown of Scratch2 increased the activity of pGL3-250 and the expression of GalR1 at the mRNA level (Fig. 3 E and F). Taken together, these data demonstrate that Scratch2 can decrease expression of GalR1.
Fig. 3.
Scratch2 regulates the expression of the GalR1 gene. (A) PC12 cells were transfected with pcDNA3.1-Scratch2 or control for 24 h. Cells were lysed and cell total proteins were detected by anti-Scratch2 antibody. (B) PC12 cells were cotransfected with pGL-250 and pcDNA3.1-Scratch2 for 24 h, and then luciferase activity was measured. T4 = 6.892, P < 0.01. (C) PC12 cells were transfected with pcDNA3.1-Scratch2 for 48 h, and real-time PCR was performed to detect the GalR1 mRNA level. T4 = 8.933, P < 0.001. (D) PC12 cells were transfected with Scramble-siRNA or Scratch2-siRNA for 48 h. Then, cells were lysed and Western blots were performed with anti-Scratch2 antibody. (E) PC12 cells were cotransfected with pGL-250 and Scratch2-siRNA for 48 h, and luciferase activity was analyzed. T4 = 6.652, P < 0.01. (F) PC12 cells were transfected with Scramble siRNA or Scratch2 siRNA for 48 h, and real-time PCR was performed to detect GalR1 mRNA levels. T4 = 9.013, P < 0.001. (G) Schematic drawings of Scratch2 and its truncation mutants. (H) PC12 cells were cotransfected with pGL-250 and Scratch2 or its truncated mutants as indicated for 24 h, and luciferase activities were measured. One-way ANOVA, F(3, 8) = 35.34, P < 0.0001. Data are presented as mean ± SD (n = 3 per group). **P < 0.01, ***P < 0.001.
To identify which domain of Scratch2 is responsible for the negative regulatory effect on the activity of the GalR1 promoter, two eukaryotic truncated mutants, Scratch-ZF (amino acids 101 to 306) and ZF (amino acids 139 to 306), were constructed (Fig. 3G). Subsequently PC12 cells were transfected with either of the two constructs, in both cases together with pGL3-250. Luciferase activities revealed that deletion of the Scratch2 region from 1 to 100 abolished the repressive effect on GalR1 promoter activity, suggesting that it is the SNAG domain that is involved in the negative regulation of Scratch2 (Fig. 3H).
Scratch2 Is Expressed in GABAergic and Glutamatergic Neurons in the vPAG.
It has been reported that Scratch2 is expressed in the mouse nervous system (51). We used a commercial Scratch2 antibody to explore the distribution of this transcription factor in the rat brain. The specificity of the antibody was determined using 1) Western blotting together with overexpression or knockdown of Scratch2 (Fig. 3 A and D) and 2) immunofluorescence staining carried out after preincubation with synthetic peptide derived from the region from 1 to 130 of rat Scratch2 protein (SI Appendix, Fig. S4). Our results show that Scratch2 is indeed expressed in the rat brain (SI Appendix, Fig. S5). Meanwhile, our results also showed the presence and expression in most neurons of Scratch2 in the rat vPAG (SI Appendix, Fig. S6).
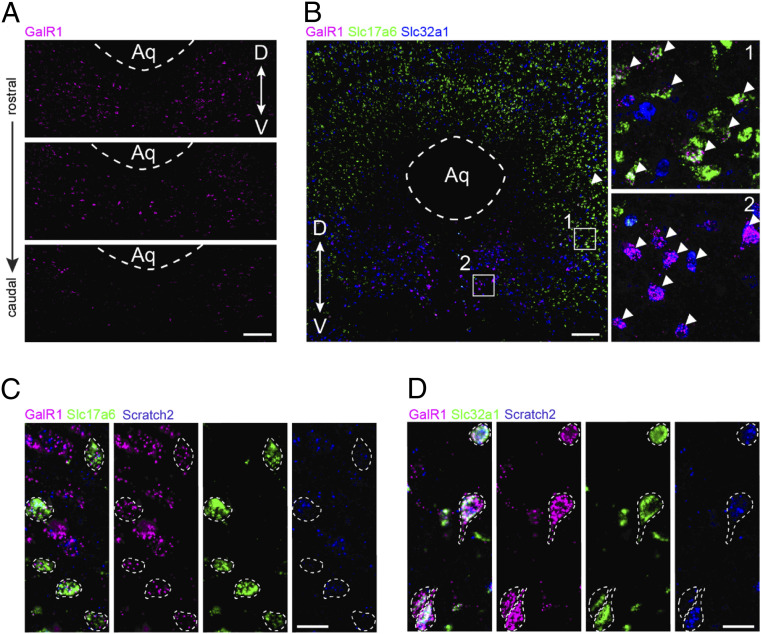
To further phenotype the cellular expression of GalR1 and Scratch2 in the vPAG, we carried out a series of multiplexed in situ hybridization (RNAscope) assays. GalR1 transcripts were observed throughout the vPAG, while more abundantly expressed rostrally (Fig. 4A). We colabeled the GalR1+ neurons with major neurotransmitter markers and identified two subgroups, expressing Slc17a6 (coding vGlut2, glutamatergic neurons) and Slc32a1 (coding VGAT, GABAergic neurons), respectively (Fig. 4B, shows a rostral section). Notably, the GalR1+/Slc17a6+ subgroup often expressed lower levels of GalR1 transcript and located more laterally (Fig. 4B, box 1), while the GalR1+/Slc32a1+ subgroup often expressed higher GalR1 transcript levels and occupied a medial localization (Fig. 4B, box 2). In both GalR1+ subgroups, the Scratch2 transcript was detected in the majority of the neurons (Fig. 4 C and D).
Fig. 4.
The expression of GalR1 and Scratch2 in the vPAG. GalR1 transcript expression in the vPAG detected by RNAscope assay at three different levels of adult rat brain sections. (A) The density of GalR1+ cells is highest in the rostral PAG. (Scale bar, 200 µm.) (B) A rostral section of adult rat PAG triple-labeled for GalR1, Slc17a6 (VGLUT2), and Slc32a1 (VGAT) transcripts using multiplexed RNAscope assay. Arrowheads point to GalR1+ neurons expressing Slc17a6 (selected region 1: lateral vPAG) and Slc32a1 (selected region 2: medial vPAG), respectively. Note very high density of VGLU2 cells in the dorsal PAG. (Scale bar, 200 µm in main figure, 30 μm in selected regions.) (C) RNAscope image showing expression of Scratch2 in the GalR1+/Slc17a6+ subgroup in the vPAG. (Scale bar, 20 µm.) (D) RNAscope image showing expression of Scratch2 in the GalR1+/Slc32a1+ subgroup in the vPAG. (Scale bar, 20 µm.)
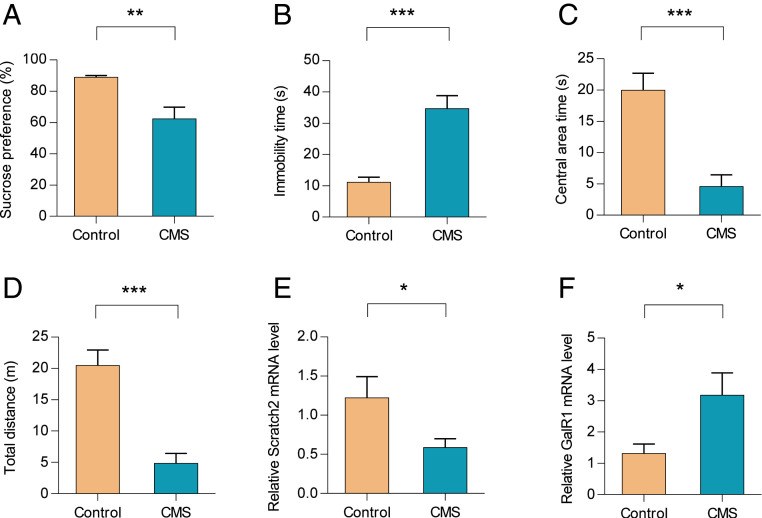
CMS Decreases the Expression of Scratch2 in the vPAG.
We then determined the effect of CMS on the expression of Scratch2. This treatment resulted in a decreased sucrose preference in the sucrose preference test (SPT) (Fig. 5A), an increased immobility time in the forced swim test (FST) (Fig. 5B), a shorter time spent in the central area of an open field test (OFT) (Fig. 5C), and a reduced total traveled distance in the OFT (Fig. 5D). We next examined the expression of Scratch2 and GalR1 in the vPAG using laser-capture microdissection from CMS and control groups (SI Appendix, Fig. S7). Our real-time PCR results showed that CMS treatment reduced the expression of Scratch2 (Fig. 5E), but increased the expression of GalR1 in the rat vPAG (Fig. 5F). However, CMS did not change the expression of Scratch2 significantly in the CA3 of the dorsal hippocampus (SI Appendix, Fig. S8).
Fig. 5.
CMS decreases the expression of Scratch2 in the rat vPAG. (A–D) CMS treatment resulted in depression-like behaviors in CMS group compared with control group. In A, T14 = 3.518, P < 0.01. In B, T14 = 5.203, P < 0.0001. In C, T14 = 4.652, P < 0.001. In D, T14 = 5.285, P < 0.0001. (E and F). The expression of Scratch2 (E) and GalR1 (F) was detected with real-time PCR in rat vPAG tissues after CMS treatment. In E, T14 = 2.159, P < 0.05. In F, T14 = 2.406, P < 0.05. Data are presented as mean ± SE (n = 8 per group). *P < 0.05, **P < 0.01, ***P < 0.001.
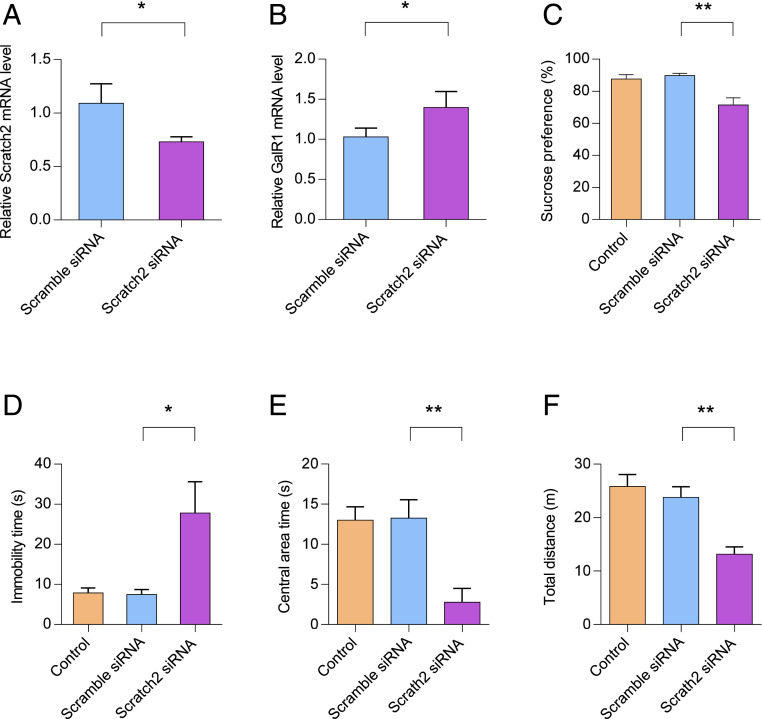
Knockdown of Scratch2 in the vPAG Induces Depression-like Behaviors.
Our previous study has shown that knockdown of GalR1 in the vPAG attenuates depression-like behaviors in CMS rats (42). Given the fact that Scratch2 decreases the expression of GalR1, we speculated that knockdown of Scratch2 in the vPAG might induce depression-like behavior in rats. To this end, rats were divided into three groups: controls, scramble-siRNA–treated rats, and Scratch2-siRNA–treated rats. Control rats were left undisturbed in their home cages except for general handling once daily. Of the other two groups, one was injected with scramble-siRNA lentivirus and the second with Scratch2-siRNA lentivirus. The injection site of the vPAG was described in our previous study (42). To further determine the effect of Scratch2-siRNA on the expression of Scratch2 in vivo, the injected vPAG tissues were collected by laser-capture microdissection for real-time PCR analyses. As expected, the Scratch2-siRNA significantly reduced the expression of Scratch2 in the vPAG (Fig. 6A), whereas knockdown of Scratch2 efficiently increased the expression of GalR1 (Fig. 6B). However, Scratch2-siRNA did not significantly change the expression of GalR2 and GalR3 in vPAG (SI Appendix, Fig. S9).
Fig. 6.
Knockdown of Scratch2 in rat vPAG results in depression-like behaviors. (A and B) The mRNA level of Scratch2 (A) or GalR1 (B) was determined by real-time PCR in the vPAG after the injection of Lenti-Scratch2-siRNA virus. Data are presented as mean ± SD (n = 3 per group). In A, T14 = 3.171, P < 0.05. In B, T14 = 2.807, P < 0.05. (C) Decreased sucrose intake in the sucrose preference test was observed after injection of Lenti-Scratch2-siRNA virus. One-way ANOVA, F(2, 21) = 10.01, P < 0.001. (D) Increased immobility time in the FST was induced after injection of Lenti-Scratch2-siRNA virus. One-way ANOVA, F(2, 21) = 6.387, P < 0.01. (E and F) Decreased central area time (E) and total distance (F) in the OFT was observed after injection of Lenti-Scratch2-siRNA virus. In E, One-way ANOVA, F(2, 21) = 9.776, P < 0.001. In F, One-way ANOVA, F(2, 21) = 12.81, P < 0.001. Data are presented as mean ± SE (n = 8 per group). *P < 0.05, **P < 0.01.
To explore a possible role of Scratch2 in mood control, we evaluated the behavior after lentivirus injection. As shown in Fig. 6C, the Scratch2-siRNA group showed a decrease in the SPT compared with controls, whereas such a decrease was not observed after injection of scramble-siRNA (Fig. 6C). In the FST, the Scratch2-siRNA group showed a significant increase of immobility time compared with the scramble-siRNA group as well as the control group (Fig. 6D). In the OFT, the Scratch2-siRNA group showed a significant decrease both of total traveled distance and of central area time as compared with the scramble-siRNA and control groups (Fig. 6 E and F).
Discussion
In the present study, we have identified a restricted region in the GalR1 promoter that exerts a negative effect on its activity. We demonstrate that the transcription factor Scratch2 binds to the E-box of the GalR1 promoter and decreases the expression of GalR1. Moreover, CMS exposure decreases the expression of Scratch2 in the rat vPAG, and knockdown of Scratch2 selectively in this region induces depression-like behaviors.
GalR1, one of three galanin receptor subtypes, has been shown to be involved in depression-like behaviors, both in the rat PFC (43) and vPAG (42). Because specific anti-GalR1 antibodies for rodents are lacking (56, 57), the expression of GalR1 in the brain has only been shown at the mRNA level. In the rat brain, GalR1 mRNA has a wide distribution, including the hypothalamus, amygdala, locus coeruleus, ventral hippocampus, and dorsal raphe nucleus (26–28). Our previous study has shown that the GalR1 is expressed in the vPAG, and that CMS treatment results in a significant increase of GalR1 mRNA levels in vPAG and of depression-like behavior (42). However, little is known about the mechanisms underlying these changes in GalR1 expression.
Using a 5′-RACE assay, we identified the transcription start site of the GalR1 gene and found a longer 5′-UTR in this gene compared with the one previously reported in the National Center for Biotechnology Information database. In fact, a similar phenomenon has also been observed in the 5′-UTR of the GalR2 gene (58). We speculate that the previously reported 5′-UTR sequence might not be intact because of RNA degradation. Moreover, we carried out a series of deletion analyses and found that the region from −250 to −220 of the GalR1 promoter plays an important negative role in the regulation of the GalR1 promoter activity. This inhibitory effect was confirmed by the DNA decoy assay, a powerful tool for gene-regulation analysis (53). Although deletion of the region from −1,500 to −1,000 also increases the activity of the GalR1 promoter, this effect is less pronounced compared with the deletion of the −250 to −220 region, suggesting that the latter is the key negative region.
The E-box element is a canonical sequence to which Snail superfamily members bind (47, 48). In the present study, we found that a putative E-box (5′-ACAGGTG-3′) (59) is present in the region from −250 to −220 of the GalR1 promoter using the TRANSFAC database. Moreover, mutation of the E-box in the GalR1 promoter increases the reporter activity in PC2 cells. These findings strongly suggest that the E-box is involved in the negative regulation of the GalR1 promoter. A previous study has shown that human Scratch2 binds to the E-box in vitro (45). Here, we confirmed that rat Scratch2 binds to the E-box of the GalR1 promoter both in vitro and in vivo and functions as a transcriptional repressor to regulate the expression of GalR1. This is similar to the function of the E-box of the Puma promoter (51). In addition to acting as transcriptional repressors, Snail superfamily members also work as a gene activator. It has been shown that Snail, another member of the Snail superfamily, is involved in the up-regulation of MMP-9 in the prototypic epithelial MDCK cell lines (60). We also found that rat Snail increased the activity of GalR1 promoter in PC12 cells (SI Appendix, Fig. S10). Therefore, Snail superfamily members exert different transcriptional effects in different tissue or cell lines, which may be due to the manner of transcriptional complex formation, including the recruitment of corepressors or coactivators.
Scratch proteins belong to Snail superfamily, whose N terminus has a basic amino acid-rich domain (SNAG domain) that is necessary for the transcriptional repression (61). Here, we found that deleting the SNAG domain of Scratch2 abolished the repressive effect of Scratch2 on the activity of GalR1 promoter, further confirming a key role of the SNAG domain. Therefore, like other Snail family members (62), the SNAG domain of Scratch2 is crucial for the ability to decrease expression of GalR1.
The PAG is a midbrain region with a columnar organization coordinating a multitude of functions, including defensive reactions, analgesia, and autonomic processes (63, 64). Here, the focus is on its ventral part, the vPAG, which in rodents contains subpopulations of neurons expressing classic neurotransmitters, like serotonin (65), GABA (66), and glutamate (67), as well as diverse neuropeptides (68–70) (see ref. 71 for mouse). Our previous study showed that the level of GalR1 transcript is selectively increased in the vPAG after exposure to CMS, and that knockdown of GalR1 in the vPAG attenuates the depression-like behaviors of CMS rats, further confirming the involvement of the vPAG in response for aversive stimuli (42). Therefore, we focus here on the vPAG to investigate localization and possible functions of Scratch2 in CMS rats.
Both Western blotting and immunofluorescence showed a wide distribution of Scratch2 in the vPAG, as did the in situ hybridization analysis. Our and others’ in situ hybridization analyses showed that the GalR1 mRNA signal is also present in the entire PAG (33, 34, 42). Through examining adjacent sections of the vPAG, we previously found that the GalR1 mRNA signal partly overlaps with Tph2 mRNA and more extensively with Gad-67 and Vglut2 mRNAs (42). In the present study, using multiplex RNAscope analysis, we now can definitely say that in the rostral vPGA GalR1 and Scratch2 are colocalized both in GABA (high GalR1 levels, located medially) and glutamate (low GalR1 levels, located laterally) neurons. GalR1 mRNA could not be detected in serotonin neurons.
Symptoms of depression have, for a long time, been associated with a deficient serotonin signaling (72) that can often be alleviated by drug-induced elevation of extracellular serotonin levels (73). We therefore hypothesize that the depression-like behavior in our CMS mouse is due to decreased 5-HT levels in the forebrain and that this is associated with activation of GalR1 expressed either on GABA or glutamate neurons in the vPAG. Regarding GABA neurons, there is a population that directly innervates (74, 75) and inhibits (76, 77) the 5-HT neurons. Since GalR1 has mainly been associated with (postsynaptic) inhibition (30, 78, 79), activation of GalR1 on GABA interneurons synapsing on serotonin neurons would result in disinhibition and increased 5-HT release in the forebrain: that is, elevated mood. This agrees with a study by Sharkey et al. (80). If, on the other hand, GalR1 levels/activation are increased in glutamatergic interneurons (75, 77), this would reduce glutamate-induced stimulation of 5-HT neurons, and possibly lead to depression-like behavior. Key questions are therefore whether 1) increased GalR1 levels equals increased signaling, and 2) the depression-induced GalR1 increase is associated with GalR1 in the GABA or in the glutamate population, or both?
It has been shown that GalR1 single nucleotide polymorphisms are associated with self-reported heavy smoking, tobacco craving during a previous quit attempt, and smoking cessation (37–39). In the human brain, GalR1 mRNA levels are significantly increased in the PFC of patients with major depression (42). Interestingly, a candidate gene study has reported that variants in all four genes of the galanin system (i.e., also GalR1) confer increased risk of depression and anxiety in people who experienced childhood adversity or negative life events. Two GalR1 single nucleotide polymorphisms interacted with recent life events and two with childhood adversity to influence phenotypes, and three GALR1 haplotypes interacted with childhood adversity and one with recent life events (38). It has been suggested that GALR1 might modulate neurodevelopmental processes relevant to the effects of childhood adversity. How, and if, these human data may relate to a possible involvement of GalR1 and Scratch2 in depression-like behaviors in rat, remains to be analyzed.
Taking these data together, we find that Scratch2 is widely expressed in rat brain, as previously shown for the Snail superfamily of transcription factors in other species, ranging from Drosophila and Caenorhabditis elegans to vertebrates (50, 51). Moreover, knockdown of Scratch2 in the vPAG resulted in up-regulated expression of GalR1 along with increased depression- and anxiety-like behaviors. Therefore, in addition to regulation of neurogenesis, neuronal migration, and neuronal survival already reported (51, 52, 81), we demonstrate a role for this transcription factor: Scratch2 is in rats involved in the regulation of depression-like behaviors by repressing GalR1. However, we do not exclude the possibility that Scratch2 might also regulate other depression-related genes containing E-box, and further investigations are therefore required.
Materials and Methods
All experimental procedures involving animals were approved by the Animal Care Committee at Capital Medical University. The complete description of materials and methods is available in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81671345, 31271154 and 31171032); the Beijing Municipal Science & Technology Commission (Z181100001518001); the Beijing Natural Science Foundation (7162016); the Special Project on Natural Chronic Noninfectious Diseases (2016YFC1307202); the Swedish Foundation for International Cooperation in Research and Higher Education (IG2013-5166); and the Swedish Research Council (04X-2887).
Footnotes
Competing interest statement: T.G.M.H. owns stock in Lundbeck and Bioarctic.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922586118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.Kessler R. C.et al.; National Comorbidity Survey Replication , The epidemiology of major depressive disorder: Results from the National Comorbidity Survey Replication (NCS-R). JAMA 289, 3095–3105 (2003). [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization , “Mental health and older adults” (WHO, Geneva, 2017). https://www.who.int/news-room/fact-sheets/detail/mental-health-of-older-adults. Accessed 12 December 2017.
- 3.Smith K., Mental health: A world of depression. Nature 515, 181 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Kendler K. S., What psychiatric genetics has taught us about the nature of psychiatric illness and what is left to learn. Mol. Psychiatry 18, 1058–1066 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Lutz P. E., et al., Association of a history of child abuse with impaired myelination in the anterior cingulate cortex: Convergent epigenetic, transcriptional, and morphological evidence. Am. J. Psychiatry 174, 1185–1194 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Sullivan P. F., Neale M. C., Kendler K. S., Genetic epidemiology of major depression: Review and meta-analysis. Am. J. Psychiatry 157, 1552–1562 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Nestler E. J., et al., Neurobiology of depression. Neuron 34, 13–25 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Trivedi M. H.et al.; STAR*D Study Team , Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: Implications for clinical practice. Am. J. Psychiatry 163, 28–40 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Akil H., et al., Medicine. The future of psychiatric research: Genomes and neural circuits. Science 327, 1580–1581 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tatemoto K., Rökaeus A., Jörnvall H., McDonald T. J., Mutt V., Galanin—A novel biologically active peptide from porcine intestine. FEBS Lett. 164, 124–128 (1983). [DOI] [PubMed] [Google Scholar]
- 11.Merchenthaler I., López F. J., Negro-Vilar A., Anatomy and physiology of central galanin-containing pathways. Prog. Neurobiol. 40, 711–769 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Xu Z. Q., Hökfelt T., Expression of galanin and nitric oxide synthase in subpopulations of serotonin neurons of the rat dorsal raphe nucleus. J. Chem. Neuroanat. 13, 169–187 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Xu Z. Q., Shi T. J., Hökfelt T., Galanin/GMAP- and NPY-like immunoreactivities in locus coeruleus and noradrenergic nerve terminals in the hippocampal formation and cortex with notes on the galanin-R1 and -R2 receptors. J. Comp. Neurol. 392, 227–251 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Sweerts B. W., Jarrott B., Lawrence A. J., Expression of preprogalanin mRNA following acute and chronic restraint stress in brains of normotensive and hypertensive rats. Brain Res. Mol. Brain Res. 69, 113–123 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Holmes P. V., Blanchard D. C., Blanchard R. J., Brady L. S., Crawley J. N., Chronic social stress increases levels of preprogalanin mRNA in the rat locus coeruleus. Pharmacol. Biochem. Behav. 50, 655–660 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Sciolino N. R., Dishman R. K., Holmes P. V., Voluntary exercise offers anxiolytic potential and amplifies galanin gene expression in the locus coeruleus of the rat. Behav. Brain Res. 233, 191–200 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuxe K., et al., “Galanin/5-HT interactions in the rat central nervous system. relevance for depression” in Galanin: A New Multifunctional Peptide in the Neuro-Endocrine System, Hökfelt T., Bartfai T., Jacobowitz D., Ottoson D., Eds. (Macmillan Education, London, 1991), pp. 221–235. [Google Scholar]
- 18.Weiss J. M., Bonsall R. W., Demetrikopoulos M. K., Emery M. S., West C. H., Galanin: A significant role in depression? Ann. N. Y. Acad. Sci. 863, 364–382 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Lu X., Sharkey L., Bartfai T., The brain galanin receptors: Targets for novel antidepressant drugs. CNS Neurol. Disord. Drug Targets 6, 183–192 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Kuteeva E., Hökfelt T., Wardi T., Ogren S. O., Galanin, galanin receptor subtypes and depression-like behaviour. Exp Suppl 102, 163–181 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Picciotto M. R., Brabant C., Einstein E. B., Kamens H. M., Neugebauer N. M., Effects of galanin on monoaminergic systems and HPA axis: Potential mechanisms underlying the effects of galanin on addiction- and stress-related behaviors. Brain Res. 1314, 206–218 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sciolino N. R., et al., Galanin mediates features of neural and behavioral stress resilience afforded by exercise. Neuropharmacology 89, 255–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinshenker D., Holmes P. V., Regulation of neurological and neuropsychiatric phenotypes by locus coeruleus-derived galanin. Brain Res. 1641, 320–337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hökfelt T., et al., Neuropeptide and small transmitter coexistence: Fundamental studies and relevance to mental illness. Front. Neural Circuits 12, 106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang R., et al., Physiology, signaling, and pharmacology of galanin peptides and receptors: Three decades of emerging diversity. Pharmacol. Rev. 67, 118–175 (2015). [DOI] [PubMed] [Google Scholar]
- 26.O’Donnell D., Ahmad S., Wahlestedt C., Walker P., Expression of the novel galanin receptor subtype GALR2 in the adult rat CNS: Distinct distribution from GALR1. J. Comp. Neurol. 409, 469–481 (1999). [PubMed] [Google Scholar]
- 27.Burazin T. C., Larm J. A., Ryan M. C., Gundlach A. L., Galanin-R1 and -R2 receptor mRNA expression during the development of rat brain suggests differential subtype involvement in synaptic transmission and plasticity. Eur. J. Neurosci. 12, 2901–2917 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Waters S. M., Krause J. E., Distribution of galanin-1, -2 and -3 receptor messenger RNAs in central and peripheral rat tissues. Neuroscience 95, 265–271 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Habert-Ortoli E., Amiranoff B., Loquet I., Laburthe M., Mayaux J. F., Molecular cloning of a functional human galanin receptor. Proc. Natl. Acad. Sci. U.S.A. 91, 9780–9783 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker E. M., et al., Cloning and characterization of the rat GALR1 galanin receptor from Rin14B insulinoma cells. Brain Res. Mol. Brain Res. 34, 179–189 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Fitzgerald L. W., Patterson J. P., Conklin D. S., Horlick R., Largent B. L., Pharmacological and biochemical characterization of a recombinant human galanin GALR1 receptor: Agonist character of chimeric galanin peptides. J. Pharmacol. Exp. Ther. 287, 448–456 (1998). [PubMed] [Google Scholar]
- 32.Wang S., Hashemi T., Fried S., Clemmons A. L., Hawes B. E., Differential intracellular signaling of the GalR1 and GalR2 galanin receptor subtypes. Biochemistry 37, 6711–6717 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Smith K. E., et al., Cloned human and rat galanin GALR3 receptors. Pharmacology and activation of G-protein inwardly rectifying K+ channels. J. Biol. Chem. 273, 23321–23326 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Jackson K. J., Chen X., Miles M. F., Harenza J., Damaj M. I., The neuropeptide galanin and variants in the GalR1 gene are associated with nicotine dependence. Neuropsychopharmacology 36, 2339–2348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lori A., et al., The galanin receptor 1 gene associates with tobacco craving in smokers seeking cessation treatment. Neuropsychopharmacology 36, 1412–1420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gold A. B., et al., Pharmacogenetic association of the galanin receptor (GALR1) SNP rs2717162 with smoking cessation. Neuropsychopharmacology 37, 1683–1688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levran O., et al., Drug addiction and stress-response genetic variability: Association study in African Americans. Ann. Hum. Genet. 78, 290–298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juhasz G., et al., Brain galanin system genes interact with life stresses in depression-related phenotypes. Proc. Natl. Acad. Sci. U.S.A. 111, E1666–E1673 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barde S., et al., Alterations in the neuropeptide galanin system in major depressive disorder involve levels of transcripts, methylation, and peptide. Proc. Natl. Acad. Sci. U.S.A. 113, E8472–E8481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., et al., Association between polymorphisms in the 5′ region of the GALR1 gene and schizophrenia in the Northern Chinese Han population: A case-control study. Neuropsychiatr. Dis. Treat. 16, 1519–1532 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zachariou V., et al., The neuropeptide galanin modulates behavioral and neurochemical signs of opiate withdrawal. Proc. Natl. Acad. Sci. U.S.A. 100, 9028–9033 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang P., et al., Depression-like behavior in rat: Involvement of galanin receptor subtype 1 in the ventral periaqueductal gray. Proc. Natl. Acad. Sci. U.S.A. 113, E4726–E4735 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., et al., Inhibition of GALR1 in PFC alleviates depressive-like behaviors in postpartum depression rat model by upregulating CREB-BNDF and 5-HT Levels. Front. Psychiatry 9, 588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bacon A., Kerr N. C., Holmes F. E., Gaston K., Wynick D., Characterization of an enhancer region of the galanin gene that directs expression to the dorsal root ganglion and confers responsiveness to axotomy. J. Neurosci. 27, 6573–6580 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakakura E. K., et al., Mammalian scratch: A neural-specific snail family transcriptional repressor. Proc. Natl. Acad. Sci. U.S.A. 98, 4010–4015 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manzanares M., Locascio A., Nieto M. A., The increasing complexity of the Snail gene superfamily in metazoan evolution. Trends Genet. 17, 178–181 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Boulay J. L., Dennefeld C., Alberga A., The Drosophila developmental gene Snail encodes a protein with nucleic acid binding fingers. Nature 330, 395–398 (1987). [DOI] [PubMed] [Google Scholar]
- 48.Nieto M. A., Bennett M. F., Sargent M. G., Wilkinson D. G., Cloning and developmental expression of Sna, a murine homologue of the Drosophila Snail gene. Development 116, 227–237 (1992). [DOI] [PubMed] [Google Scholar]
- 49.Roark M., et al., Scratch, a pan-neural gene encoding a zinc finger protein related to Snail, promotes neuronal development. Genes Dev. 9, 2384–2398 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Marín F., Nieto M. A., The expression of Scratch genes in the developing and adult brain. Dev. Dyn. 235, 2586–2591 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Rodríguez-Aznar E., Nieto M. A., Repression of Puma by Scratch2 is required for neuronal survival during embryonic development. Cell Death Differ. 18, 1196–1207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paul V., et al., Scratch2 modulates neurogenesis and cell migration through antagonism of bHLH proteins in the developing neocortex. Cereb. Cortex 24, 754–772 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Mann M. J., Dzau V. J., Therapeutic applications of transcription factor decoy oligonucleotides. J. Clin. Invest. 106, 1071–1075 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matys V., et al., TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–D110 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki T., Hirai H., Fujisawa J., Fujita T., Yoshida M., A trans-activator Tax of human T-cell leukemia virus type 1 binds to NF-kappa B p50 and serum response factor (SRF) and associates with enhancer DNAs of the NF-kappa B site and CArG box. Oncogene 8, 2391–2397 (1993). [PubMed] [Google Scholar]
- 56.Lu X., Bartfai T., Analyzing the validity of GalR1 and GalR2 antibodies using knockout mice. Naunyn Schmiedebergs Arch. Pharmacol. 379, 417–420 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brunner S. M., et al., Validation of antibody-based tools for galanin research. Peptides 120, 170009 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Yang Y., et al., Characterization of the rat GAL2R promoter: Positive role of ETS-1 in regulation of the rat GAL2R gene in PC12 cells. Mol. Neurobiol. 54, 4421–4431 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Metzstein M. M., Horvitz H. R., The C. elegans cell death specification gene ces-1 encodes a Snail family zinc finger protein. Mol. Cell 4, 309–319 (1999). [DOI] [PubMed] [Google Scholar]
- 60.Jordà M., et al., Upregulation of MMP-9 in MDCK epithelial cell line in response to expression of the Snail transcription factor. J. Cell Sci. 118, 3371–3385 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Grimes H. L., Chan T. O., Zweidler-McKay P. A., Tong B., Tsichlis P. N., The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. 16, 6263–6272 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barrallo-Gimeno A., Nieto M. A., Evolutionary history of the Snail/Scratch superfamily. Trends Genet. 25, 248–252 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Bandler R., Shipley M. T., Columnar organization in the midbrain periaqueductal gray: Modules for emotional expression? Trends Neurosci. 17, 379–389 (1994). [DOI] [PubMed] [Google Scholar]
- 64.Behbehani M. M., Functional characteristics of the midbrain periaqueductal gray. Prog. Neurobiol. 46, 575–605 (1995). [DOI] [PubMed] [Google Scholar]
- 65.Steinbusch H. W., Distribution of serotonin-immunoreactivity in the central nervous system of the rat-cell bodies and terminals. Neuroscience 6, 557–618 (1981). [DOI] [PubMed] [Google Scholar]
- 66.Mugnaini E., Oertel W. H., “An atlas of the distribution of GABAergic neurons and terminals in the rat CNS as revealed with GAD immunohistochemistry” in Handbook of Chemical Neuroanatomy, Bjorklund A., Hökfelt T., Eds. (Elsevier, 1985), pp. 436–608. [Google Scholar]
- 67.Hioki H., et al., Vesicular glutamate transporter 3-expressing nonserotonergic projection neurons constitute a subregion in the rat midbrain raphe nuclei. J. Comp. Neurol. 518, 668–686 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Van den Bergh P., Wu P., Jackson I. M., Lechan R. M., Neurons containing a N-terminal sequence of the TRH-prohormone (preproTRH53-74) are present in a unique location of the midbrain periaqueductal gray of the rat. Brain Res. 461, 53–63 (1988). [DOI] [PubMed] [Google Scholar]
- 69.Smith G. S., et al., Distribution of messenger RNAs encoding enkephalin, substance P, somatostatin, galanin, vasoactive intestinal polypeptide, neuropeptide Y, and calcitonin gene-related peptide in the midbrain periaqueductal grey in the rat. J. Comp. Neurol. 350, 23–40 (1994). [DOI] [PubMed] [Google Scholar]
- 70.Fu W., et al., Chemical neuroanatomy of the dorsal raphe nucleus and adjacent structures of the mouse brain. J. Comp. Neurol. 518, 3464–3494 (2010). [DOI] [PubMed] [Google Scholar]
- 71.Lein E. S., et al., Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Maes M., Meltzer H. Y., “The serotonin hypothesis of major depression” in Psychopharmacology: The Fourth Generation of Progress, Bloom F., Kupfer D., Eds. (Raven, 1995), pp. 933–944. [Google Scholar]
- 73.Millan M. J., Multi-target strategies for the improved treatment of depressive states: Conceptual foundations and neuronal substrates, drug discovery and therapeutic application. Pharmacol. Ther. 110, 135–370 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Wang Q. P., Ochiai H., Nakai Y., GABAergic innervation of serotonergic neurons in the dorsal raphe nucleus of the rat studied by electron microscopy double immunostaining. Brain Res. Bull. 29, 943–948 (1992). [DOI] [PubMed] [Google Scholar]
- 75.Weissbourd B., et al., Presynaptic partners of dorsal raphe serotonergic and GABAergic neurons. Neuron 83, 645–662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gallager D. W., Aghajanian G. K., Effect of antipsychotic drugs on the firing of dorsal raphe cells. II. Reversal by picrotoxin. Eur. J. Pharmacol. 39, 357–364 (1976). [DOI] [PubMed] [Google Scholar]
- 77.Jolas T., Aghajanian G. K., Opioids suppress spontaneous and NMDA-induced inhibitory postsynaptic currents in the dorsal raphe nucleus of the rat in vitro. Brain Res. 755, 229–245 (1997). [DOI] [PubMed] [Google Scholar]
- 78.Bai Y. F., et al., Activation of galanin receptor 1 inhibits locus coeruleus neurons via GIRK channels. Biochem. Biophys. Res. Commun. 503, 79–85 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Xu Z. Q., Zheng K., Hökfelt T., Electrophysiological studies on galanin effects in brain—progress during the last six years. Neuropeptides 39, 269–275 (2005). [DOI] [PubMed] [Google Scholar]
- 80.Sharkey L. M., Madamba S. G., Siggins G. R., Bartfai T., Galanin alters GABAergic neurotransmission in the dorsal raphe nucleus. Neurochem. Res. 33, 285–291 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Itoh Y., et al., Scratch regulates neuronal migration onset via an epithelial-mesenchymal transition-like mechanism. Nat. Neurosci. 16, 416–425 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.