Significance
Immune checkpoint therapy (ICT) has led to durable responses in a subset of cancer patients. Generally, patients who respond to ICT bear tumors with high mutational burden. Radiation is used for treatment of many types of cancers and has been shown to induce new mutations in treated tumor cells and to synergistically facilitate ICT. However, these latter actions have largely been explained by radiation-induced tumor cell death and/or effects on the host. Herein, we show that noncurative irradiation induces mutations in tumor cells lacking neoantigens and that these de novo-generated neoantigens function as targets for CD8+ T cells, resulting in increased immunogenicity of nonimmunogenic tumor cells. This study thus identifies an additional mechanism that explains synergy between immunotherapy and radiation.
Keywords: neoantigens, radiation, immune checkpoint therapy
Abstract
Immunotherapies are a promising advance in cancer treatment. However, because only a subset of cancer patients benefits from these treatments it is important to find mechanisms that will broaden the responding patient population. Generally, tumors with high mutational burdens have the potential to express greater numbers of mutant neoantigens. As neoantigens can be targets of protective adaptive immunity, highly mutated tumors are more responsive to immunotherapy. Given that external beam radiation 1) is a standard-of-care cancer therapy, 2) induces expression of mutant proteins and potentially mutant neoantigens in treated cells, and 3) has been shown to synergize clinically with immune checkpoint therapy (ICT), we hypothesized that at least one mechanism of this synergy was the generation of de novo mutant neoantigen targets in irradiated cells. Herein, we use KrasG12D x p53−/− sarcoma cell lines (KP sarcomas) that we and others have shown to be nearly devoid of mutations, are poorly antigenic, are not controlled by ICT, and do not induce a protective antitumor memory response. However, following one in vitro dose of 4- or 9-Gy irradiation, KP sarcoma cells acquire mutational neoantigens and become sensitive to ICT in vivo in a T cell-dependent manner. We further demonstrate that some of the radiation-induced mutations generate cytotoxic CD8+ T cell responses, are protective in a vaccine model, and are sufficient to make the parental KP sarcoma line susceptible to ICT. These results provide a proof of concept that induction of new antigenic targets in irradiated tumor cells represents an additional mechanism explaining the clinical findings of the synergy between radiation and immunotherapy.
Immune checkpoint therapy (ICT) can lead to durable responses in subsets of cancer patients (1–8). On the basis of computational analyses, the patients who most benefit from ICT are those with cancers that have high mutational burden (9–18). For example, patients bearing tumors with high mutational burden caused by environmental exposure (such as ultraviolet-induced melanoma) or deficiencies in DNA repair (such as microsatellite instability-high colorectal cancers) tend to respond well to immunotherapy (18–26). Presumably the sensitivity of such cancers reflects the increased likelihood of formation of immunogenic, tumor-specific mutant neoantigens (27). We and others previously showed that certain tumor-specific neoantigens are major targets of natural and therapeutically induced antitumor responses in both mice and humans (28–41). Therefore, the presence of immunogenic tumor neoantigens is currently thought to contribute to tumor sensitivity to immunotherapy.
However, many cancer patients do not respond to ICT, suggesting that their neoantigen burden is either of insufficient magnitude or immunogenicity to function as targets for T cell-dependent antitumor mechanisms. Indeed, there are many tumor types, such as acute myeloid leukemia, estrogen receptor-positive breast, and prostate cancers, that have limited mutational burdens and display low response rates to ICT (9, 13, 42, 43). Additionally, tumor cell clones expressing immunogenic neoantigens that develop during tumor evolution may be eliminated from tumors with high mutational burden by the process of cancer immunoediting, resulting in outgrowth of tumor cell clones with reduced immunogenicity that can then grow progressively in the presence of the unmanipulated immune system (33, 44, 45). Therefore, a process by which tumors with low neoantigen burden can acquire immunogenic mutations has the potential to expand the number of patients able to benefit from ICT.
Ionizing radiation has been shown to elicit DNA damage in tumor cells, leading to an increase in overall mutational load (46–52). This damage is thought to occur primarily through generation of reactive oxygen species which induce base pair substitutions by mechanisms involving transitions, transversions, and/or faulty DNA repair (53). Multiple preclinical studies have demonstrated antitumor responses when focal radiation is combined with ICT in tumors that do not respond to ICT alone (54–60) and several clinical studies have demonstrated that human tumor patients have improved responsiveness to ICT following focal radiation (e.g., NCT02303990, NCT02298946, NCT02383212) (61–67). Radiation has been demonstrated to function as an in vivo tumor vaccine by inducing damage-associated molecular patterns (DAMP)-dependent immunogenic cell death (68), inducing DNA damage sensed by pattern recognition receptors (69, 70), enhancing access of immune effector cells to their cognate targets through tumor cell debulking and vasculature changes (71, 72), up-regulating major histocompatibility complex class I (MHC-I) receptors (73), up-regulating cell-surface molecules such as Fas (74), and augmenting tumor antigen cross-presentation by specific subsets of dendritic cells through up-regulation of type I interferon (IFN), which results in increased numbers and action of tumor-specific CD8+ T cells (75–77). However, none of these explanations take into account that following irradiation, tumor cells acquire novel mutations that may function as effective tumor neoantigens. In fact, two groups have demonstrated broadening of the T cell repertoire following radiation treatment of mouse 4T1 mammary tumors and B16F10 melanoma tumors (56, 78). Radiation-induced neoantigens may partially explain the broadening of the T cell repertoire reported during noncurative doses of irradiation.
Given the above observations, we specifically explored whether one dose of in vitro irradiation could increase the immunogenicity of poorly immunogenic tumor cell lines through mechanisms involving the de novo generation of tumor-specific mutant neoantigens. For this purpose, we used a mouse KrasG12D x p53−/− sarcoma cell line as a model system since the R.D.S. and T.J. laboratories have previously shown that these tumor cells express a very limited number of somatic mutations, are essentially devoid of mutational neoantigens, and are nonimmunogenic and grow progressively in syngeneic wild-type (WT) mice either following treatment with control antibody or the combination of anti–PD-1/anti–CTLA-4 (34, 41). We find that treating these cell lines with noncurative doses of irradiation induces expression of somatic mutations, some of which function as neoantigens and render the sarcoma cells susceptible to ICT in vivo. These data support the concept that an additional mechanism underlying the synergy between radiation therapy and immunotherapy is that the former induces immunogenic mutations in tumors that now function as targets for the latter.
Results
KrasG12D x p53−/− Sarcomas Are Nonimmunogenic due to a Lack of Immunogenic Mutations.
A key requirement for this study was the identification of a mouse tumor model that was insensitive to ICT as a result of low mutational burden and absence of inherent immunogenicity. For this purpose, we chose cell lines derived from genetically engineered KrasLSL-G12D x p53fl/fl mice. In a past study, injection of a lentiviral vector encoding Cre recombinase into the muscle of these mice led to deletion of the p53 gene and expression of an active mutant Kras oncogene (KrasG12D), giving rise to a set of sarcomas (KP sarcomas) that developed at the site of injection (79). Using this model, the R.D.S. and T.J. laboratories have reported that the resulting sarcoma cells, for example KP9025, expressed a very limited number of somatic mutations and were nonimmunogenic (34, 41). To expand upon and confirm the generality of our previous observations, we selected two additional independent KP sarcoma cell lines (KP9093 and KP9032) and assessed their mutational burdens and immunogenicities. Like our previously reported KP9025 sarcoma cells that express only 4 nonimmunogenic somatic mutations (41), KP9093 and KP9032 expressed only 14 and 12 somatic mutations, respectively. Moreover, using our validated neoepitope prediction algorithms, none of the mutations were predicted to form MHC-I neoantigens and neither sarcoma line displayed any immunogenicity. Specifically, both lines grew progressively in WT syngeneic mice treated with either control monoclonal antibody (mAb) or the combination of anti–CTLA-4/anti–PD-1 (SI Appendix, Fig. S1A) and neither sarcoma line induced immunologic memory in challenge–resection–rechallenge experiments (SI Appendix, Fig. S1B). To verify that the lack of immunogenicity of KP9093 tumor cells was due to the absence of tumor antigens rather than expression of a novel immunosuppressive moiety, we ectopically expressed full-length ovalbumin (OVA) (that contains both MHC-I– and MHC-II–restricted epitopes) into KP9093 cells and tested the immunogenicity of the transfected cell lines. Whereas KP9093 cells transduced with empty vector grew progressively in syngeneic mice, KP9093.OVA tumor cells were spontaneously rejected in a T cell-dependent manner (SI Appendix, Fig. S1C). Thus, KP sarcoma cells are poorly immunogenic because they do not express tumor antigens of either sufficient quality or quantity to mediate response to ICT.
Generation of KP9093 Sarcoma Cells Expressing the Pan MHC-II Antigen PADRE.
Recent work from our laboratory and others has shown that in vivo tumor rejection requires both MHC-I– and MHC-II–restricted tumor antigens (41, 80, 81). We therefore engineered KP9093 cells to be specifically sensitive to the presence of MHC-I antigens by enforcing expression of the pan MHC-II–restricted epitope PADRE (82). KP9093.PADRE cells grew progressively either in WT mice treated with anti–CTLA-4 or anti–PD-1, either alone or in combination, or after challenge–resection–rechallenge (Fig. 1A and SI Appendix, Fig. S1D). These data reveal that the KP9093.PADRE sarcoma line is a good model for poorly immunogenic tumors that express very limited numbers of immunogenic neoepitopes and thus could be useful in assessing the effects of radiation on inducing immunogenic MHC-I tumor neoepitopes.
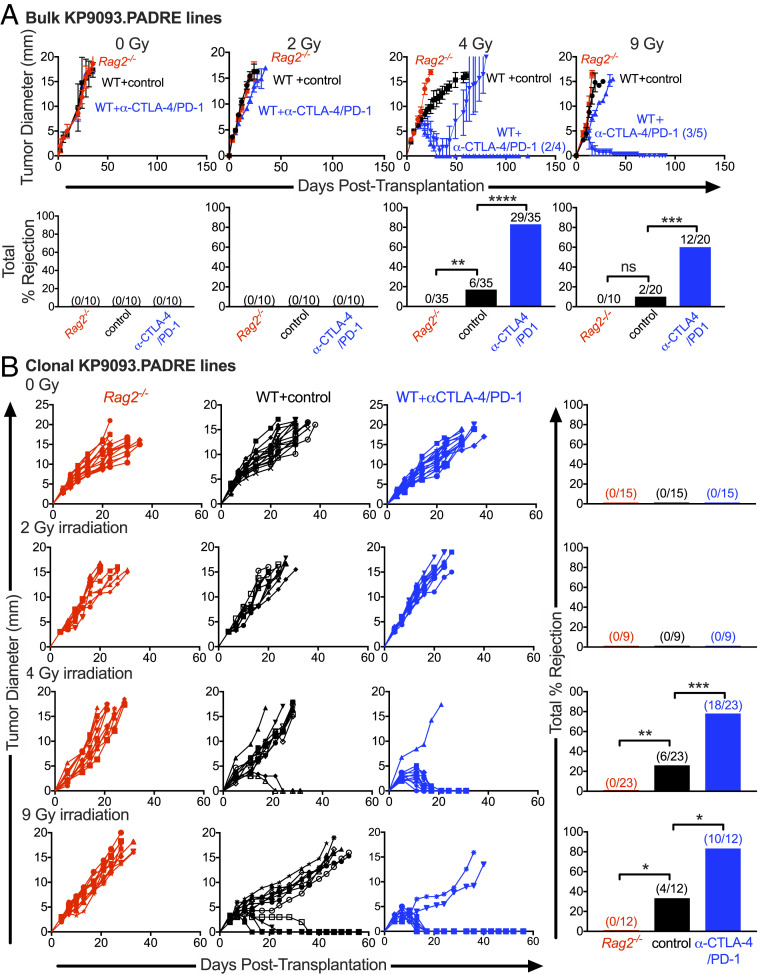
Fig. 1.
Radiation sensitizes nonimmunogenic tumors to immune checkpoint therapy. (A) Growth kinetics of KP9093.PADRE cells irradiated in vitro with 0, 2, 4, or 9 Gy. Numbers on the growth curves indicate the numbers of tumor clone lines that were rejected (Top). Cells were injected into mice at >95% viability. Representative data are shown as average tumor diameter ± SEM; n ≥ 4 per group. Bar graphs summarize results from multiple biological repeats (Bottom). (B) Bulk tumor lines receiving the indicated dose of radiation were single-cell-cloned by limiting dilution, and multiple clones from each bulk line were inoculated into Rag2−/− mice (red), syngeneic WT mice (black), or WT mice treated with anti–CTLA-4/anti–PD-1 (blue). Representative data are shown as the tumor diameter of individual clones. Bar graphs show cumulative percent rejection from multiple biological replicates. ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Radiation Sensitizes KP9093.PADRE Cells to ICT in a Dose-Dependent Manner.
Using KP9093.PADRE sarcoma cells, we asked whether noncurative irradiation in vitro could sensitize these cells to spontaneous or ICT-induced immune rejection in vivo. KP9093.PADRE tumor cells were irradiated one time in vitro with either 2, 4, or 9 Gy, cultured for 2 wk to reestablish >95% viable cell cultures (SI Appendix, Fig. S2), and then monitored for induction of tumor cell immunogenicity in vivo. This approach allowed us to separate the effects of radiation on tumor cell antigenicity versus generalized effects on the host immune system and tumor microenvironment. All irradiated tumor lines grew progressively when transplanted into either immunodeficient Rag2−/− mice or immunocompetent WT mice treated with control mAb, validating that the radiation doses used did not generate cell lines inherently incapable of in vivo growth (Fig. 1A). Moreover, bulk KP9093.PADRE tumor cells treated with 0- or 2-Gy irradiation were also not rejected in WT mice treated with anti–CTLA-4/anti–PD-1. In contrast, bulk tumor lines that received 4- or 9-Gy doses of radiation acquired heterogeneous sensitivity to therapeutic anti–CTLA-4/anti–PD-1 treatment in vivo as 29/35 and 12/20 mice rejected their tumors, respectively (Fig. 1A). This finding shows that there is a threshold irradiation dose required for the induction of immunogenicity in the bulk irradiated KP9093.PADRE cell line.
Exploring the Immunogenic Heterogeneity of KP9093.PADRE Cells following Noncurative Doses of Radiation.
To determine whether the heterogeneous responses of the bulk cell lines derived from noncurative doses (4 or 9 Gy) of radiation were due to varying, distinct immunogenicities of the component tumor cell clones, each bulk line was cloned (SI Appendix, Fig. S2) and single-cell clones were expanded into cell lines that were then tested for tumorigenicity and immunogenicity by transplantation into either immunodeficient Rag2−/− mice or into WT mice that were subsequently treated with or without ICT (Fig. 1B). All clonal lines, regardless of irradiation dose used, formed progressively growing tumors when transplanted into immunodeficient Rag2−/− mice. All clonal lines derived from either untreated or 2-Gy-treated KP9093.PADRE cells also grew progressively in WT mice with or without ICT. In contrast, 6/23 (26%) and 18/23 (78%) of clonal lines generated from 4-Gy-irradiated KP9093.PADRE cells were rejected in WT mice either spontaneously or following ICT, respectively. Similarly, 4/12 (33%) and 10/12 (83%) of clonal cell lines generated following 9-Gy irradiation were rejected by WT mice either spontaneously or following ICT, respectively. Thus, single doses of radiation that do not induce complete killing of KP sarcoma cells are capable of rendering nonimmunogenic tumor cells susceptible to ICT in a manner that is independent of irradiation-induced changes in either the tumor microenvironment or the host immune system.
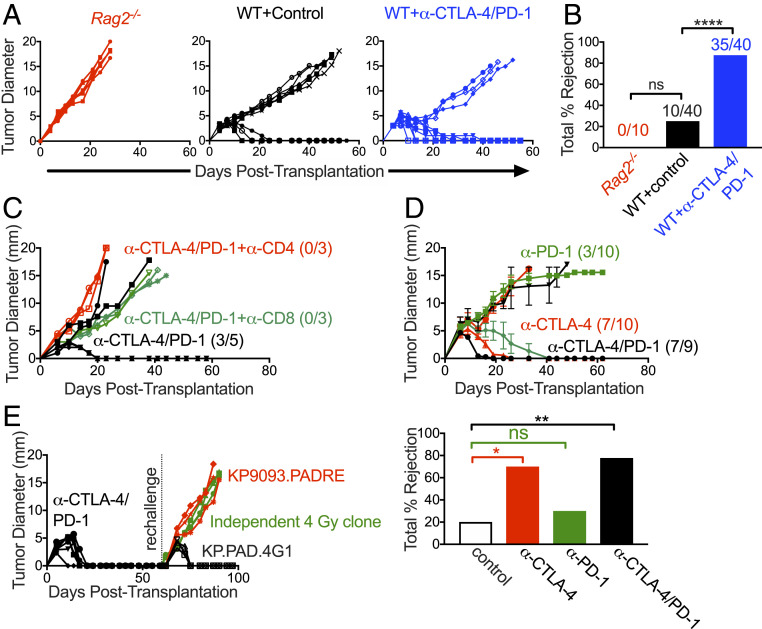
To more deeply assess the immune response against the irradiated KP9093.PADRE tumor lines, we selected a representative clonal line derived from the 4-Gy-treated KP9093.PADRE bulk line (KP.PAD.4G1) and extensively characterized its in vivo growth and immunogenicity. This clonal tumor line grew progressively in immunodeficient Rag2−/− mice and immunocompetent WT mice treated with control mAb but was rejected when tumor-bearing immunocompetent mice were treated with ICT (Fig. 2 A and B). Sensitivity to ICT required both CD4+ and CD8+ T cells, as antibody-mediated depletion of either T cell subset led to tumor outgrowth during ICT (Fig. 2C). KP.PAD.4G1 tumors were sensitive to anti–CTLA-4 treatment alone but were not sensitive to anti–PD-1 monotherapy (Fig. 2D). Given that anti–CTLA-4 therapy functions predominantly by lowering the threshold for T cell activation and thus enhancing T cell priming (83), these findings may suggest that the enhanced immunogenicity observed following radiation is dependent on the enhanced induction of new T cell responses rather than alleviation of dysfunction. Mice that rejected KP.PAD.4G1 cells following anti–CTLA-4/anti–PD-1 ICT resisted rechallenge with the same cell line but did not reject the parental KP9093.PADRE line or an independently derived, antigenically irrelevant clonal cell line from a different 4-Gy-treated bulk population (Fig. 2E). Taken together, these data show that the 4-Gy-irradiated KP9093.PADRE bulk line consists of a heterogeneous set of clones with distinct immunogenicities. Furthermore, given that 1) the enhanced immunogenicity of the KP.PAD.4G1 clonal line was cell line-specific and not due to a generalized characteristic of irradiated cells and 2) ICT-mediated rejection of KP.PAD.4G1 required both CD4+ and CD8+ T cells, these findings strongly suggest that the enhanced immunogenicity observed after radiation is driven by the acquisition of tumor-specific antigens.
Fig. 2.
KP.PAD.4G1 induces a tumor cell-specific immune response. (A) KP.PAD.4G1 was injected into Rag2−/− (red), syngeneic WT mice (black), and WT mice treated with anti–CTLA-4/anti–PD-1 (blue) at 95% viability. Representative data of individual tumor growth curves are shown; 5 ≤ n ≤ 10 for all groups. (B) The bar graph summarizes cumulative percent tumor rejection of four biological repeats shown in A, with numbers above each bar indicating the number of tumors that were rejected in each group. (C) WT mice were implanted with KP.PAD.4G1 tumor cells and treated with anti–CTLA-4/anti–PD-1 (black), anti–CTLA-4/anti–PD-1 and anti-CD8–depleting antibody (green), or anti–CTLA-4/anti–PD-1 and anti-CD4–depleting antibody (red). Representative data are shown as individual tumor growth curves; n = 3 to 5 per group. Numbers in the graph represent the number of mice that rejected their tumors. (D) KP.PAD.4G1 tumor growth in WT mice treated with anti–CTLA-4 (red), anti–PD-1 (green), or dual anti–PD-1/anti–CTLA-4 ICT (black). Representative data are shown as the average tumor diameter ± SEM; n ≥ 9 per group. Numbers in the graph represent the number of mice that rejected their tumors following treatment. The bar graph represents the cumulative percent tumor rejection within each treatment group. (E) Mice that rejected KP.PAD.4G1 following treatment with dual anti–CTLA-4/anti–PD-1 ICT were challenged ≥30 d later with either the same cell line (black), parental KP9093.PADRE cells (red), or a KP9093.PADRE clone derived from an independent 4-Gy irradiation (green). Representative data are shown as individual tumor growth curves; n = 3 per group. ns, not significant; *P < 0.05, **P < 0.01, ****P < 0.0001.
Noncurative Doses of Irradiation Induce Tumor-Specific Mutant MHC-I Neoantigens Capable of Eliciting Functional CD8+ T Cell Responses.
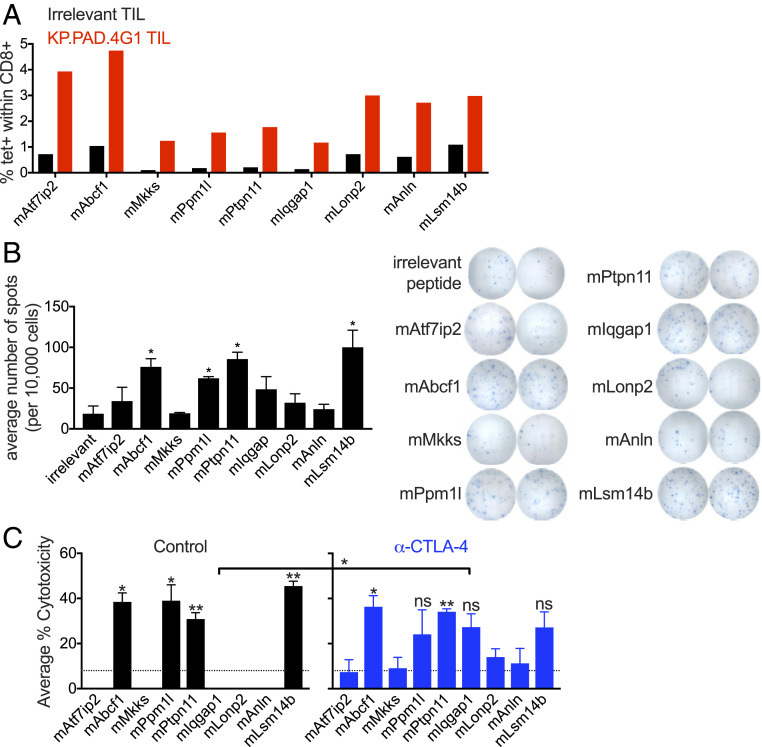
To formally address the possibility that the irradiation process had induced immunogenic de novo mutations in the tumor line, KP.PAD.4G1 cells were subjected to whole-exome and complementary DNA (cDNA) capture sequencing and variant calling. By comparing sequences of the KP.PAD.4G1 clone with parental cells, 19 irradiation-induced missense mutations were identified. Due to the limited number of radiation-induced missense mutations, we screened all of them for the ability to elicit mutation-specific CD8+ T cell populations via H-2-Kb p-MHC-I tetramer staining of tumor-infiltrating lymphocytes (TILs) from untreated KP.PAD.4G1 tumors 11 d post tumor transplantation. Tetramer staining revealed endogenously occurring CD8+ T cell populations specific for nine of the irradiation-induced mutations, while only background levels of CD8+ T cell staining were observed using the same KP.PAD.4G1-specific tetramers on TIL derived from irrelevant methylcholanthrene (MCA)-induced T3 sarcomas (Fig. 3A and SI Appendix, Fig. S3). Peptides encompassing the irradiation-induced mutations expressed in KP.PAD.4G1 were incapable of folding H-2-Db tetramers.
Fig. 3.
Tumor cell clone KP.PAD.4G1 contains unique radiation-induced mutations that lead to de novo antitumor CD8+ T cell responses. (A) TIL was isolated from KP.PAD.4G1 and irrelevant MCA-induced T3 tumors 11 d post transplantation and representative quantification of H-2-Kb p-MHC-I tetramer staining is shown as the percentage of CD8+ T cells of viable CD45+ cells within the tetramer-positive gate for the indicated KP.PAD.4G1 irradiation-induced mutations. (B) CD8+ T cells were enriched from KP.PAD.4G1 TIL and stimulated with splenocytes pulsed with the indicated peptides. IFNγ secretion was measured using ELISpot. Representative data are shown as the average number of spots per 10,000 CD8+ T cells ± SEM. (C) LDH cytotoxicity ELISA from splenocytes at day 12 post KP.PAD.4G1 tumor inoculation in control mAb-treated (black) and anti–CTLA-4–treated (blue) mice. Data represent the average percent cytotoxicity from two independent biological repeats ± SEM. Asterisks above bars indicate significance compared with the level of cytotoxicity observed against an irrelevant antigen (denoted by dashed lines). ns, not significant; *P < 0.05, **P < 0.01.
To determine if the irradiation-induced, mutation-specific CD8+ T cell populations were functional cytotoxic T lymphocytes (CTLs), we stimulated CD8+ TIL isolated 11 d post tumor transplantation with splenocytes pulsed with each tetramer-positive peptide, and measured IFNγ secretion by enzyme-linked immunosorbent spot assay (ELISPOT). Four peptides (mAbcf1, mPpm1l, mPtpn11, and mLsm14b) elicited IFNγ production significantly above background levels (Fig. 3B). Lactate dehydrogenase (LDH) enzyme-linked immunosorbent assay (ELISA) of splenocytes from KP.PAD.4G1 tumor-bearing mice revealed cytotoxic T cell populations against these same four antigens. Treatment of KP.PAD.4G1 tumor-bearing mice with anti–CTLA-4 monotherapy resulted in significantly more cytotoxicity compared with the level seen in control-treated mice for one additional antigen (mIqgap1) (Fig. 3C). Together, these results indicate that KP.PAD.4G1 cells express four irradiation-induced mutations that elicit endogenous neoantigen-specific CD8+ CTL populations.
Responses against Irradiation-Induced Neoantigens Can Promote Rejection of Nonimmunogenic Tumor Cells.
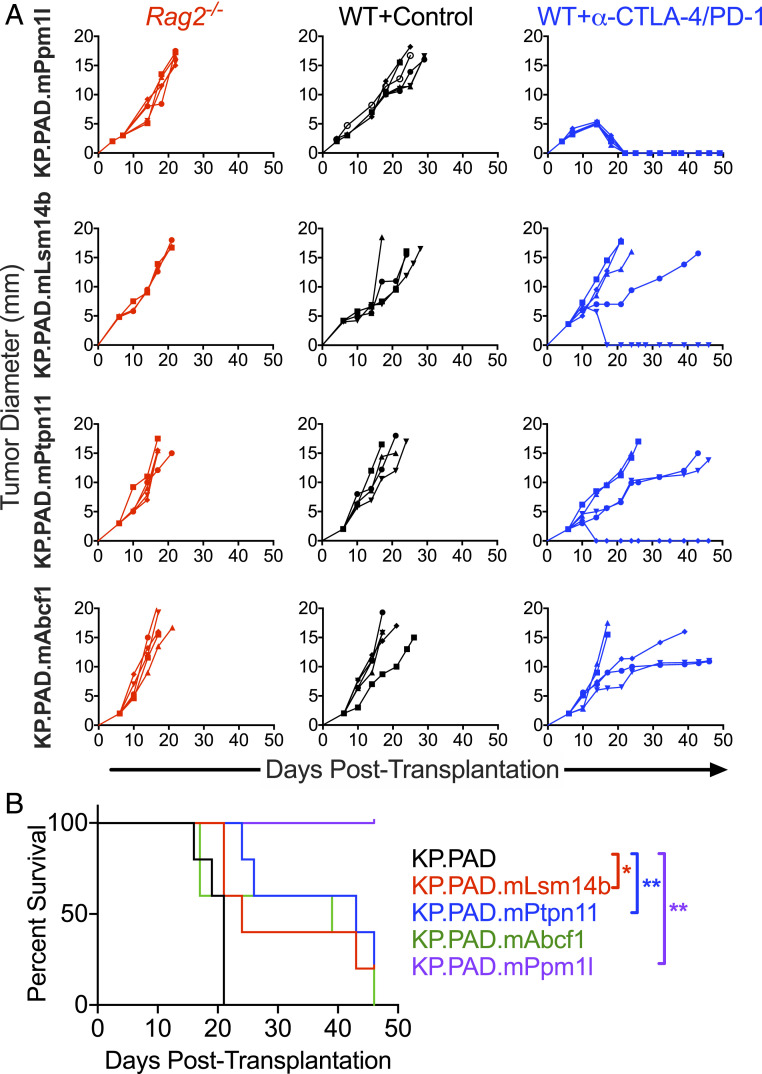
Radiation induces a variety of changes within tumor cells. To determine whether neoantigens induced after irradiation can promote tumor rejection independent from other cellular changes, we enforced expression of the four validated neoantigens in parental KP9093.PADRE cells and tested each resulting cell line for susceptibility to ICT in vivo (Fig. 4A). Of the four neoantigens screened, mPpm1l was sufficient to drive checkpoint sensitivity of the transduced KP9093.PADRE line. Additionally, checkpoint-treated mice bearing tumors expressing mLsm14b or mPtpn11 experienced significantly prolonged survival compared with mice bearing control KP.PAD tumors (Fig. 4B).
Fig. 4.
Radiation-induced neoantigens are sufficient to sensitize KP9093.PADRE tumors to ICT. (A) Parental KP9093.PADRE lines ectopically expressing the indicated single, radiation-induced neoantigen were implanted subcutaneously into Rag2−/− mice (red), immunocompetent WT mice (black), or anti–CTLA-4/anti–PD-1–treated WT mice (blue). Representative data are shown as individual tumor growth curves from one of two independent experiments; n = 5 per group. (B) Survival plots of the experiment described in A. *P < 0.05, **P < 0.005.
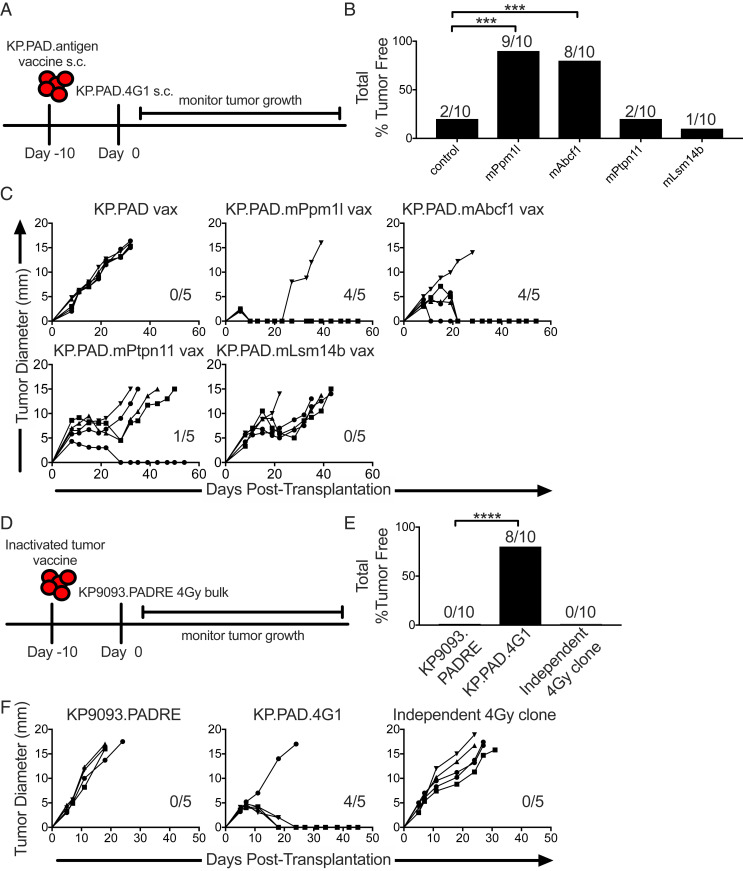
We next sought to validate the ability of irradiation-induced mutations expressed by KP.PAD.4G1 to drive tumor rejection using a vaccine approach. To this end, KP9093.PADRE tumor lines expressing the four individual neoantigens were lethally irradiated for use as inactivated tumor vaccines and injected into mice. Ten days later, mice were challenged with KP.PAD.4G1 (Fig. 5A). Vaccination against mPpm1l and mAbcf1 resulted in 80 and 60% of mice being protected against subsequent challenge with KP.PAD.4G1, respectively. While not sufficient to drive tumor rejection, vaccination against mPtpn11 or mLsm14b led to delayed KP.PAD.4G1 tumor outgrowth (Fig. 5 B and C).
Fig. 5.
Targeting radiation-induced mutations through vaccination is sufficient to promote tumor rejection. (A) WT mice were injected with inactivated tumor cell vaccines consisting of KP9093.PADRE cells ectopically expressing single, radiation-induced neoantigens identified from KP.PAD.4G1 10 d prior to KP.PAD.4G1 tumor challenge. s.c., subcutaneous. (B) Cumulative percentage of mice that rejected KP.PAD.4G1 following treatment as in A. (C) Individual KP.PAD.4G1 tumor growth curves from mice treated as in A. Numbers indicate the number of mice that rejected KP.PAD.4G1 tumors. Representative data from three independent experiments are shown; n = 5 per group. (D) WT mice were treated with parental KP9093.PADRE, KP.PAD.4G1, or an antigenically irrelevant clone derived from an independent 4-Gy-treated bulk cell line as inactivated tumor vaccines followed by challenge with the bulk KP9093.PADRE cell line from which KP.PAD.4G1 was derived 10 d later. (E) Cumulative percentage of mice that rejected the 4-Gy-irradiated KP9093.PADRE bulk line following treatment as in D. (F) Representative data are shown as individual bulk 4-Gy-irradiated KP9093.PADRE tumor growth curves as in D; n = 5 per group. Numbers in the graphs indicate the number of mice that rejected their tumors. ***P < 0.001, ****P < 0.0001.
These results indicate that some of the neoantigens expressed in the clonal KP.PAD.4G1 cell line are capable of promoting sensitivity to ICT. We then asked if neoantigens expressed by a clone could promote rejection of the bulk irradiated tumor cell line from which it was derived. To answer this question, we performed vaccine experiments where mice were challenged with the bulk 4-Gy-irradiated KP9093.PADRE cell line from which KP.PAD.4G1 was obtained (described in SI Appendix, Fig. S2) following prophylactic inoculation with an inactivated tumor cell vaccine consisting of parental KP9093.PADRE cells, KP.PAD.4G1, or an antigenically irrelevant clonal cell line generated from an independent 4-Gy-treated bulk cell population (Fig. 5D). Similar to results shown in Fig. 2E, mice vaccinated with the parental KP9093.PADRE cell line or with a clonal cell line derived from an independently generated 4-Gy-irradiated bulk population were not able to control outgrowth of the 4-Gy-irradiated bulk KP9093.PADRE tumor cell line (Fig. 5 E and F). In contrast, mice were capable of completely rejecting the 4-Gy-irradiated KP9093.PADRE bulk tumor cell line following vaccination with the KP.PAD.4G1 clone derived from it. Therefore, irradiation-induced neoantigens expressed in a clonal cell line are able to promote rejection of the corresponding bulk irradiated tumor cell line.
Discussion
Together, this work demonstrates that irradiation of the parental KP9093.PADRE cell line produced a tumor clone that expressed 19 de novo mutations, 4 of which generated neoantigens capable of eliciting spontaneous CD8+ CTL responses. Additionally, one neoantigen (mPpm1l) was sufficient to drive ICT-mediated rejection when expressed in parental KP9093.PADRE cells. While this work specifically explores the use of radiation to induce MHC-I neoantigens, we expect that radiation similarly induces MHC-II neoantigens especially because CD4+ T cell responses have been shown to be more abundant against neoepitopes in vaccine settings (81, 84). ICT-mediated rejection of irradiated tumor cells was dependent on the activity of both CD4+ and CD8+ T cells and induced immunologic memory that was tumor cell-specific and not dependent on other radiation-induced cellular changes. These findings demonstrate a proof of concept that noncurative irradiation is capable of sensitizing poorly immunogenic tumors to ICT through, in part, induction of de novo neoantigen-specific T cell responses.
While synergy between radiation therapy and ICT has been previously demonstrated, herein we show that one unappreciated factor contributing to this synergy is enhanced tumor cell immunogenicity. Our data suggest that, in addition to neoantigen generation as a result of genomic instability in developing cancers, neoantigens may also be formed by the effects of radiation on the tumor cells themselves. Specifically, we have demonstrated that noncurative doses of radiation can induce newly expressed mutations in the parent tumor that can function as MHC-I neoepitopes. These radiation-induced neoantigens elicit cytolytic tumor-specific CD8+ T cell populations and sensitize nonimmunogenic tumors to ICT in a T cell-dependent manner. In the future it will be important to assess the contribution of these newly derived neoantigens in providing synergy between noncurative radiation and ICT in the context of radiation-induced changes to the tumor microenvironment. It is also interesting to note that the parental KP.9093.PADRE cell line is p53−/−, which may enhance the likelihood of mutation accumulation following radiation. It will be important to assess how tumor genome instability mediated by, for example, p53 deficiency contributes to the accumulation of irradiation-induced mutations and patient p53 status may be an important prognostic indicator for successful combination of radio- and immunotherapies.
Recently published work has demonstrated the requirement for clonal rather than subclonal neoantigens in driving antitumor immune responses in patients treated with ICT (85–88). McGranahan et al. demonstrated that cytotoxic chemotherapy-induced mutations were detectable in patients that were nonresponsive to ICT, and thus concluded that treatment-induced subclonal neoantigens were irrelevant to effective antitumor immune responses (85). However, our results indicate that at least under some conditions subclonal radiation-induced mutations have the ability to drive antitumor immunity. We suggest that contingent on the dose of irradiation being sufficient to form new antigens throughout the tumor population, the initial immune response against subclonal neoantigens may drive epitope spreading and result in rejection of mutationally heterogeneous tumors.
Radiation treatment duration, timing, and dose will need to be carefully considered when targeting treatment-induced mutations in combination with ICT. Neoantigen generation following radiation-induced DNA damage requires cell proliferation; therefore, radiation may need to be used prior to ICT to maximize synergy. Our results raise the exciting possibility that immunogenic neoantigens that are produced as a result of radiation during cancer therapy may create new immunotherapeutic opportunities that did not exist before treatment at the time of patient diagnosis.
Materials and Methods
Detailed information regarding the mice, tumor cell lines, and experimental procedures and materials (resection–rechallenge, antibodies, tetramer staining, ELISpot, retroviral expression of radiation-induced neoantigens, LDH ELISA, sequencing, vaccination, and statistical analysis) used throughout this report is available in SI Appendix.
Female WT and Rag2−/− mice on a 129S4 background were bred in an animal barrier facility. Mice used in the study were between 8 and 12 wk of age and maintained in accordance with procedures approved by the Association for Assessment and Accreditation of Laboratory Animal Care-accredited Animal Studies Committee of Washington University in St. Louis.
Supplementary Material
Acknowledgments
Use of the XStrahl Small Animal Radiation Research Platform was supported by Grant S10 OD020136. We thank all members of the R.D.S. laboratory for helpful discussions and technical support. This work was supported by grants to R.D.S. from the National Cancer Institute of the NIH (R01CA190700), Parker Institute for Cancer Immunotherapy, Cancer Research Institute, Janssen Pharmaceutical Company of Johnson & Johnson, and Prostate Cancer Foundation, and by a Stand Up to Cancer–Lustgarten Foundation Pancreatic Cancer Foundation Convergence Dream Team Translational Research Grant. Stand Up to Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research. D.M.L. and E.A. were supported by a postdoctoral training grant (T32 CA00954729) from the National Cancer Institute. D.M.L. was supported by an Irvington Postdoctoral Fellowship from the Cancer Research Institute. J.P.W. is supported by the National Cancer Institute of the NIH Paul Calabresi Career Development Award in Clinical Oncology (K12CA167540).
Footnotes
Competing interest statement: R.D.S. is a cofounder, scientific advisory board member, stockholder, and royalty recipient of Jounce Therapeutics, Neon Therapeutics, and Asher Biotherapeutics and is a scientific advisory board member for A2 Biotherapeutics, BioLegend, Codiak Biosciences, Constellation Pharmaceuticals, Lytix Biopharma, NGM Biopharmaceuticals, and Sensei Biotherapeutics. T.J. is a member of the Board of Directors of Amgen and Thermo Fisher Scientific. He is also a cofounder of Dragonfly Therapeutics and T2 Biosystems. T.J. serves on the Scientific Advisory Board of Dragonfly Therapeutics, SQZ Biotech, and Skyhawk Therapeutics. None of these affiliations represent a conflict of interest with respect to the design or execution of this study or interpretation of data presented in this manuscript. T.J.’s laboratory currently also receives funding from the Johnson & Johnson Lung Cancer Initiative and Calico, but this funding did not support the research described in this manuscript.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102611118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Larkin J., et al., Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373, 23–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Postow M. A., et al., Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N. Engl. J. Med. 372, 2006–2017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robert C., et al., Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 372, 320–330 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Weber J. S., et al., Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): A randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 16, 375–384 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Borghaei H., et al., Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 373, 1627–1639 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motzer R. J.et al.; CheckMate 025 Investigators , Nivolumab versus everolimus in advanced renal-cell carcinoma. N. Engl. J. Med. 373, 1803–1813 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herbst R. S., et al., Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 387, 1540–1550 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Ribas A., et al., Pembrolizumab versus investigator-choice chemotherapy for ipilimumab-refractory melanoma (KEYNOTE-002): A randomised, controlled, phase 2 trial. Lancet Oncol. 16, 908–918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizvi N. A., et al., Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snyder A., et al., Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugo W., et al., Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Allen E. M., et al., Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellmann M. D., et al., Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 33, 843–852.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rooney M. S., Shukla S. A., Wu C. J., Getz G., Hacohen N., Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizvi H., et al., Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J. Clin. Oncol. 36, 633–641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown S. D., et al., Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 24, 743–750 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cristescu R., et al., Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, eaar3593 (2018). Correction in: Science 363, eaax1384 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le D. T., et al., PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Network , Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network , Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). Correction in: Nature 514, 262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinbrot E., et al., Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 24, 1740–1750 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govindan R., et al., Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 150, 1121–1134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Network , Genomic classification of cutaneous melanoma. Cell 161, 1681–1696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Germano G., et al., Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 552, 116–120 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Le D. T., et al., Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim T.-M., Laird P. W., Park P. J., The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell 155, 858–868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gubin M. M., Schreiber R. D., CANCER. The odds of immunotherapy success. Science 350, 158–159 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Robbins P. F., et al., A mutated beta-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J. Exp. Med. 183, 1185–1192 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coulie P. G., et al., A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc. Natl. Acad. Sci. U.S.A. 92, 7976–7980 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kripke M. L., Antigenicity of murine skin tumors induced by ultraviolet light. J. Natl. Cancer Inst. 53, 1333–1336 (1974). [DOI] [PubMed] [Google Scholar]
- 31.Wölfel T., et al., A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 269, 1281–1284 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Gubin M. M., et al., Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsushita H., et al., Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 482, 400–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DuPage M., Mazumdar C., Schmidt L. M., Cheung A. F., Jacks T., Expression of tumour-specific antigens underlies cancer immunoediting. Nature 482, 405–409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prehn R. T., Main J. M., Immunity to methylcholanthrene-induced sarcomas. J. Natl. Cancer Inst. 18, 769–778 (1957). [PubMed] [Google Scholar]
- 36.Robbins P. F., et al., Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med. 19, 747–752 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lennerz V., et al., The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc. Natl. Acad. Sci. U.S.A. 102, 16013–16018 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Rooij N., et al., Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol. 31, e439–e442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gubin M. M., Artyomov M. N., Mardis E. R., Schreiber R. D., Tumor neoantigens: Building a framework for personalized cancer immunotherapy. J. Clin. Invest. 125, 3413–3421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sensi M., Anichini A., Unique tumor antigens: Evidence for immune control of genome integrity and immunogenic targets for T cell-mediated patient-specific immunotherapy. Clin. Cancer Res. 12, 5023–5032 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Alspach E., et al., MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 574, 696–701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrence M. S., et al., Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexandrov L. B.et al.; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain , Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). Correction in: Nature 502, 258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunn G. P., Old L. J., Schreiber R. D., The immunobiology of cancer immunosurveillance and immunoediting. Immunity 21, 137–148 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Dunn G. P., Old L. J., Schreiber R. D., The three Es of cancer immunoediting. Annu. Rev. Immunol. 22, 329–360 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Grosovsky A. J., de Boer J. G., de Jong P. J., Drobetsky E. A., Glickman B. W., Base substitutions, frameshifts, and small deletions constitute ionizing radiation-induced point mutations in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 85, 185–188 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallace S. S., Detection and repair of DNA base damages produced by ionizing radiation. Environ. Mutagen. 5, 769–788 (1983). [DOI] [PubMed] [Google Scholar]
- 48.Ward J. F., “DNA damage produced by ionizing radiation in mammalian cells: Identities, mechanisms of formation, and reparability” in Progress in Nucleic Acid Research and Molecular Biology, Cohn W. E., Moldave K., Eds. (Academic Press, 1988), pp. 95–125. [DOI] [PubMed] [Google Scholar]
- 49.Breimer L. H., Ionizing radiation-induced mutagenesis. Br. J. Cancer 57, 6–18 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans H. H., “Mutagenic effects of ultraviolet and ionizing radiation” in Photobiology, Riklis E., Ed. (Springer, 1991), pp. 83–95. [Google Scholar]
- 51.Cadet J., Berger M., Radiation-induced decomposition of the purine bases within DNA and related model compounds. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 47, 127–143 (1985). [DOI] [PubMed] [Google Scholar]
- 52.Breimer L. H., Nalbantoglu J., Meuth M., Structure and sequence of mutations induced by ionizing radiation at selectable loci in Chinese hamster ovary cells. J. Mol. Biol. 192, 669–674 (1986). [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y., et al., Ionizing radiation-induced bystander mutagenesis and adaptation: Quantitative and temporal aspects. Mutat. Res. 671, 20–25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deng L., et al., Irradiation and anti-PD-L1 treatment synergistically promote antitumor immunity in mice. J. Clin. Invest. 124, 687–695 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharabi A. B., Lim M., DeWeese T. L., Drake C. G., Radiation and checkpoint blockade immunotherapy: Radiosensitisation and potential mechanisms of synergy. Lancet Oncol. 16, e498–e509 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Twyman-Saint Victor C., et al., Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 520, 373–377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Demaria S., Golden E. B., Formenti S. C., Role of local radiation therapy in cancer immunotherapy. JAMA Oncol. 1, 1325–1332 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Stone H. B., Peters L. J., Milas L., Effect of host immune capability on radiocurability and subsequent transplantability of a murine fibrosarcoma. J. Natl. Cancer Inst. 63, 1229–1235 (1979). [PubMed] [Google Scholar]
- 59.Gandhi S. J., et al., Awakening the immune system with radiation: Optimal dose and fractionation. Cancer Lett. 368, 185–190 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Luo L. Y., et al., Combining radiation with immunotherapy: The University of Pennsylvania experience. Semin. Radiat. Oncol. 30, 173–180 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Barker C. A., Postow M. A., Combinations of radiation therapy and immunotherapy for melanoma: A review of clinical outcomes. Int. J. Radiat. Oncol. Biol. Phys. 88, 986–997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Postow M. A., et al., Immunologic correlates of the abscopal effect in a patient with melanoma. N. Engl. J. Med. 366, 925–931 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Escorcia F. E., Postow M. A., Barker C. A., Radiotherapy and immune checkpoint blockade for melanoma: A promising combinatorial strategy in need of further investigation. Cancer J. 23, 32–39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Formenti S. C., et al., Radiotherapy induces responses of lung cancer to CTLA-4 blockade. Nat. Med. 24, 1845–1851 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tazi K., Hathaway A., Chiuzan C., Shirai K., Survival of melanoma patients with brain metastases treated with ipilimumab and stereotactic radiosurgery. Cancer Med. 4, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reynders K., Illidge T., Siva S., Chang J. Y., De Ruysscher D., The abscopal effect of local radiotherapy: Using immunotherapy to make a rare event clinically relevant. Cancer Treat. Rev. 41, 503–510 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Theelen W. S. M. E., et al., Pembrolizumab with or without radiotherapy for metastatic non-small-cell lung cancer: A pooled analysis of two randomised trials. Lancet Respir. Med. 9, 467–475 (2021). [DOI] [PubMed] [Google Scholar]
- 68.Zitvogel L., Galluzzi L., Smyth M. J., Kroemer G., Mechanism of action of conventional and targeted anticancer therapies: Reinstating immunosurveillance. Immunity 39, 74–88 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Harding S. M., et al., Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mackenzie K. J., et al., cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ganss R., Ryschich E., Klar E., Arnold B., Hämmerling G. J., Combination of T-cell therapy and trigger of inflammation induces remodeling of the vasculature and tumor eradication. Cancer Res. 62, 1462–1470 (2002). [PubMed] [Google Scholar]
- 72.Klug F., et al., Low-dose irradiation programs macrophage differentiation to an iNOS+/M1 phenotype that orchestrates effective T cell immunotherapy. Cancer Cell 24, 589–602 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Reits E. A., et al., Radiation modulates the peptide repertoire, enhances MHC class I expression, and induces successful antitumor immunotherapy. J. Exp. Med. 203, 1259–1271 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chakraborty M., et al., Irradiation of tumor cells up-regulates Fas and enhances CTL lytic activity and CTL adoptive immunotherapy. J. Immunol. 170, 6338–6347 (2003). [DOI] [PubMed] [Google Scholar]
- 75.Vanpouille-Box C., et al., DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat. Commun. 8, 15618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Diamond J. M., et al., Exosomes shuttle TREX1-sensitive IFN-stimulatory dsDNA from irradiated cancer cells to DCs. Cancer Immunol. Res. 6, 910–920 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodríguez-Ruiz M. E., Vanpouille-Box C., Melero I., Formenti S. C., Demaria S., Immunological mechanisms responsible for radiation-induced abscopal effect. Trends Immunol. 39, 644–655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rudqvist N.-P., et al., Radiotherapy and CTLA-4 blockade shape the TCR repertoire of tumor-infiltrating T cells. Cancer Immunol. Res. 6, 139–150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kirsch D. G., et al., A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat. Med. 13, 992–997 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Knocke S., et al., Tailored tumor immunogenicity reveals regulation of CD4 and CD8 T cell responses against cancer. Cell Rep. 17, 2234–2246 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Kreiter S., et al., Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520, 692–696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alexander J., et al., Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity 1, 751–761 (1994). [DOI] [PubMed] [Google Scholar]
- 83.Wei S. C., et al., Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 170, 1120–1133.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ott P. A., et al., An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McGranahan N., et al., Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McGranahan N., Swanton C., Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27, 15–26 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Morris L. G. T., et al., Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 7, 10051–10063 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andor N., et al., Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 22, 105–113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.