Significance
cGAS (cytosolic DNA sensor cyclic AMP-GMP synthase)-STING (stimulator of interferon genes) signaling is critical for sensing cytosolic DNA to initiate host immune responses against invading pathogens and cancer. However, inappropriate activation of STING signaling causes severe and often fatal autoimmune or autoinflammatory diseases. Hence, STING is an attractive drug target for the treatment of STING-driven autoimmune and inflammatory disorders. Therefore, there is a need to identify lead compounds that effectively inhibit human STING for further drug development. Here, we identified and characterized a STING-specific inhibitor SN-011 with high efficiency, specificity, and safety, paving the way for therapeutically manipulating STING-mediated clinical diseases.
Keywords: STING, type I interferons, antagonist, Aicardi–Goutières syndrome, SAVI
Abstract
Cytosolic DNA activates cGAS (cytosolic DNA sensor cyclic AMP-GMP synthase)-STING (stimulator of interferon genes) signaling, which triggers interferon and inflammatory responses that help defend against microbial infection and cancer. However, aberrant cytosolic self-DNA in Aicardi–Goutière’s syndrome and constituently active gain-of-function mutations in STING in STING-associated vasculopathy with onset in infancy (SAVI) patients lead to excessive type I interferons and proinflammatory cytokines, which cause difficult-to-treat and sometimes fatal autoimmune disease. Here, in silico docking identified a potent STING antagonist SN-011 that binds with higher affinity to the cyclic dinucleotide (CDN)-binding pocket of STING than endogenous 2′3′-cGAMP. SN-011 locks STING in an open inactive conformation, which inhibits interferon and inflammatory cytokine induction activated by 2′3′-cGAMP, herpes simplex virus type 1 infection, Trex1 deficiency, overexpression of cGAS-STING, or SAVI STING mutants. In Trex1−/− mice, SN-011 was well tolerated, strongly inhibited hallmarks of inflammation and autoimmunity disease, and prevented death. Thus, a specific STING inhibitor that binds to the STING CDN-binding pocket is a promising lead compound for STING-driven disease.
Cytoplasmic microbial and host DNAs act as danger signals (1). The cytosolic DNA sensor cyclic AMP-GMP synthase (cGAS, also known as MB21D1 or C6orf150) catalyzes the production of a noncanonical cyclic dinucleotide (CDN) c[G (2′,5′)pA (3′,5′)p] (2′3′-cGAMP) from ATP and GTP (2–5). The 2′3′-cGAMP and bacterial CDNs c-di-GMP and c-di-AMP bind to and activate the endoplasmic reticulum (ER)-associated stimulator of interferon (IFN) genes (STING, also known as MITA, ERIS, or MPYS) (6–9). CDNs bind to the CDN-binding domain (CBD) in a pocket at the interface of the STING dimer (10, 11). CDN binding induces a conformational change that closes the dimer pocket and causes higher-order multimerization and activation of STING (12–15). Activated STING translocates from the ER, through the Golgi apparatus, to perinuclear vesicles (16). During this process, STING recruits and activates the kinase TBK1, which in turn phosphorylates the transcription factor IRF3 (17–20). Dimerization and nuclear translocation of phosphorylated IRF3 potently induces type I IFNs. STING activation also initiates NF-κB signaling through TBK1 and TRAF6, which leads to proinflammatory cytokine expression (21).
The cGAS-STING axis plays an essential role in initiating host immune defense against microbial invasion. STING-deficient mice are more susceptible to some viral infections (22). Genomic DNA extruded into the cytosol because of genomic instability in cancer also stimulates cGAS and STING to enhance antitumor immunity (23, 24). However, inappropriate activation of this signaling pathway by aberrant self-DNAs leads to chronic IFN expression, which has been implicated in the development of some autoimmune disorders (25). Genetic mutations of genes that cause cytosolic accumulation of nucleic acids, including the DNase TREX1, RNASEH2A-C, SAMHD1, ADAR, and MDA5, cause a broad spectrum of inflammatory and autoimmune phenotypes, including Aicardi–Goutières syndrome (AGS), familial chilblain lupus, and systemic lupus erythematosus (SLE) (26–28). Trex1 deficiency in mice causes inflammatory myocarditis, progressive cardiomyopathy, and circulatory failure. Trex1-deficient mice lacking STING are completely protected from otherwise lethal inflammatory diseases, indicating the central role of STING in disease pathogenesis. Moreover, gain-of-function (GOF) mutations in STING cause early-onset vasculopathy and pulmonary inflammation in patients, known as STING-associated vasculopathy with onset in infancy (SAVI) (29, 30). In addition, in some IFN or inflammation-driven disorders, including senescence, lethal sepsis, acute pancreatitis, Parkinson’s disease, nonalcoholic fatty liver disease, and chronic kidney fibrosis, blocking cGAS-STING signaling in mouse models ameliorates disease progression (24, 31–41). Thus, STING plays a key role in the pathogenesis of autoimmune and inflammatory diseases and is an attractive drug target for the treatment of STING-driven autoimmune and inflammatory disorders. Although several STING antagonists—including C-176, C-178, H-151, Astin C, compound 18, and endogenous nitro-fatty acids (NO2-FAs)—have been identified, they all have limited potential for therapeutic applications because of low affinity, inactivity against human STING, and probable lack of specificity (42–45).
In this study, in silico docking was used to identify small molecules that bind to the CDN pocket of STING. A common scaffold structure that specifically inhibited STING signaling was found. One analog (SN-011) is a potent and selective mouse and human STING inhibitor that blocked induction of type I IFNs and proinflammatory cytokines in response to cytosolic DNA. SN-011 competed with CDNs for the binding pocket of the STING dimer, blocking CDN binding and STING activation. SN-011 similarly inhibited activation of WT and SAVI-associated GOF mutants of STING. Consistently, SN-011 ameliorated autoimmune pathology and prevented death in Trex1−/− mice. Taking these data together, this study identified a STING-specific antagonist, SN-011, as an attractive lead compound for developing drugs to treat STING-driven autoimmune and inflammatory disease.
Results
In Silico Docking Screen for STING Inhibitors.
To identify STING inhibitors, we performed in silico docking screening based on the crystal structure of the C-terminal domain of human STING (STINGCTD) (Protein Data Bank [PDB] ID code 4EF5) (46). A library of >500,000 “lead-like” compounds in the ZINC15 database was screened against the STINGCTD structure to identify molecules that bound in the CDN binding pocket of STING (47). A series of compounds containing a benzene-1-sulfonamido-3-amide group scored highly in the docking screen. Eleven high-ranking compounds were chosen to evaluate bioactivity in vitro. Four compounds (SN-001 to SN-004) significantly inhibited STING-triggered Ifnb gene induction in mouse embryonic fibroblasts (MEFs) stimulated with IFN stimulatory DNA (ISD) (SI Appendix, Fig. S1A). None of the compounds significantly affected basal Ifnb gene expression in the absence of ISD (SI Appendix, Fig. S1B) An inactive compound SN-100 was selected as a negative control for subsequent experiments. To confirm bioactivity, the effect of the most active compound SN-001 was assessed in two other models of STING-dependent signaling, triggered by herring testis DNA (HT-DNA) in L929 mouse fibroblasts and herpes simplex virus type 1 (HSV-1) infection of the human monocyte cell line THP-1. SN-001 at concentrations of 5 to 20 μM significantly impaired the induction of Ifnb mRNA, in a dose-dependent manner in both models, whereas SN-100 had no inhibitory effect (SI Appendix, Fig. S1C).
When STING is activated, it is phosphorylated by TBK1, which triggers a kinase cascade that results in phosphorylation of IRF3, IκBα, and p65 and nuclear translocation of p-IRF3 and p-p65 transcription factors. We next evaluated whether SN-001 inhibited each step in STING-activated signaling by immunoblot probed for phosphorylated STING pathway intermediates (SI Appendix, Fig. S1D) and confocal microscopy to assess the nuclear translocation of the phosphorylated transcription factors (SI Appendix, Fig. S1 E and F) in HT-DNA–stimulated L929. SN-001, but not SN-100, decreased cytosolic DNA-induced phosphorylation of STING, TBK1, IRF3, IκBα, and p65, as well as nuclear translocation of IRF3 and p65. Additionally, SN-001 showed no evidence of cytotoxicity in MEFs, L929, or THP-1 cells by MTS viability assay when added at the concentrations that inhibited STING (5 to 20 μM) (SI Appendix, Fig. S1 G–I). Thus, SN-001 inhibits cytosolic DNA-triggered STING signaling in both human and mouse cells without any apparent cytotoxicity.
SN-011 Is a Potent STING Inhibitor.
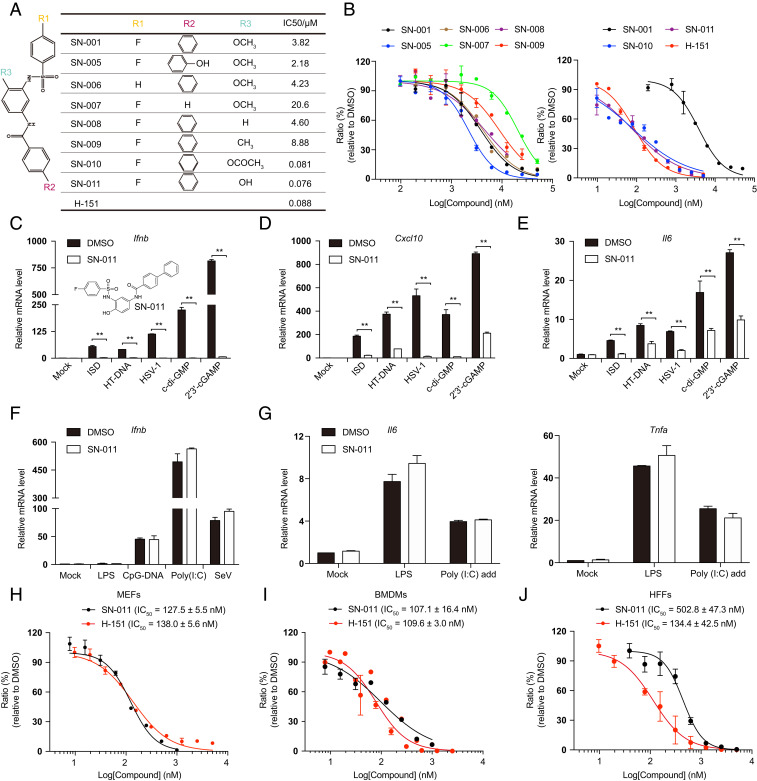
To identify more potent inhibitors, a structure–activity relationship (SAR) study of SN-001 was performed using a set of seven SN-001 analogs, SN-005 to SN-011 (Fig. 1 A and B). Removal of the phenyl group (SN-007) in SN-001 impaired STING inhibition of Ifnb induction in L929 transfected with HT-DNA. When the phenoxy methyl group was substituted by a phenyl acetate (SN-010) or phenol hydroxyl (SN-011), the compounds suppressed STING signaling more efficiently (Fig. 1 A and B) with half maximal inhibitory concentration (IC50) values of 81 nM and 76 nM, respectively, which were comparable to the IC50 of the cysteine-reactive STING covalent inhibitor H-151 (88 nM) (42). None of the series significantly affected basal Ifnb gene expression (SI Appendix, Fig. S1J). SN-011 (1 μM) significantly suppressed the induction of Ifnb, Cxcl10, and Il6 mRNA in MEFs activated by various canonical STING stimulators (ISD, HT-DNA, HSV-1, c-di-GMP, and 2′3′-cGAMP) (Fig. 1 C–E). The effect on STING was specific since IRF3 and NF-κB–responsive gene expression was not inhibited by SN-011 in Tmem173−/− MEFs challenged with LPS (Toll-like receptor 4, TLR4, agonist), CpG-DNA (TLR9 agonist), poly (I:C) (TLR3 agonist), or Sendai virus (SeV, Retinoic acid-inducible gene I, RIG-I, stimulator) (Fig. 1 F and G). Thus, SN-011 is a specific inhibitor of STING-dependent signaling.
Fig. 1.
SN-011 is a potent STING inhibitor. (A and B) Chemical structures of SN-001, SN-005 to SN-011 (A), and dose-dependent inhibitory curves (B). L929 cells, pretreated with different concentrations of the indicated compounds for 6 h, were stimulated by transfection of HT-DNA and Ifnb mRNA was measured by quantitative PCR 6 h later. The dose-dependent inhibitory curve was fit to calculate the IC50s of SN-001, SN-005 to SN-011, and H-151. (C–E) MEFs, pretreated for 6 h with 1 μM SN-011, were stimulated by transfection of ISD or HT-DNA or HSV-1 infection for 6 h, or addition of c-di-GMP or 2′3′-cGAMP for 3 h. Induction of Ifnb (C), Cxcl10 (D), Il6 (E) mRNA was measured by quantitative PCR. (F and G) Tmem173−/− MEFs, pretreated with 1 μM SN-011 for 6 h, were stimulated by adding LPS or poly (I and C) to the medium for 1.5 h or transfection of CpG-DNA or poly (I and C) or infection with SeV for 6 h. Induction of Ifnb (F), Il6, and Tnfa (G) mRNA was measured by quantitative PCR. (H–J) MEFs (H), BMDMs (I), and HFFs (J) were pretreated with the indicated concentrations of SN-011 or H-151 before 2′3′-cGAMP stimulation for 3 h. Induction of Ifnb mRNA was measured by quantitative PCR. The dose-dependent inhibitory curve was fit to calculate the IC50s. Data shown are mean ± SD from three independent experiments. **P < 0.01. Gene expression was normalized to Gapdh.
To further explore the inhibitory property of SN-011, we determined IC50s in different cell lines. SN-011 was active at nanomolar concentrations to inhibit 2′3′-cGAMP-induced Ifnb expression in MEFs, mouse bone marrow-derived macrophages (BMDMs) and human foreskin fibroblasts (HFFs) with IC50 values of 127.5, 107.1, and 502.8 nM, respectively (Fig. 1 H–J). In comparison, the IC50 values of H-151 in MEFs, BMDMs, and HFFs were 138, 109.6, and 134.4 nM (Fig. 1 H–J), indicating SN-011 and H-151 exhibits comparable inhibitory effect on mouse STING-dependent signaling in cell-based assays. Like SN-001, SN-011 was not cytotoxic in BMDMs and HFFs when added at 1.25 to 20 μM (SI Appendix, Fig. S1 K and L). These studies identify SN-011 as a lead compound with nanomolar activity for inhibiting STING in both human and mouse cells.
SN-011 Inhibits STING Activation and STING-Dependent Signal Transduction.
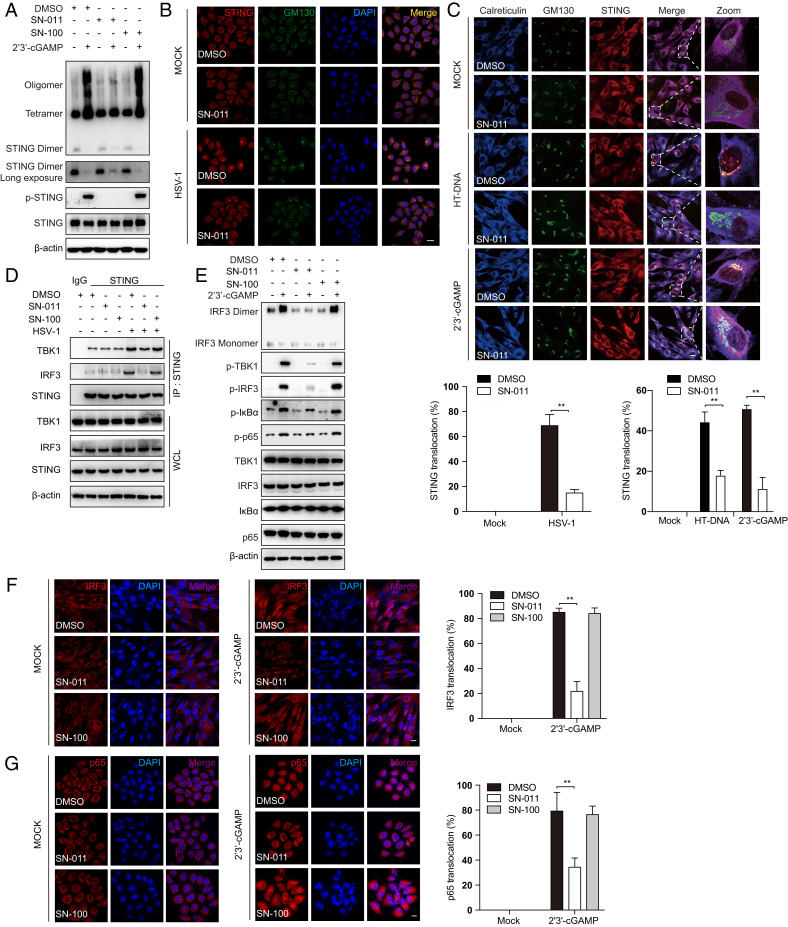
Next, we examined the effects of SN-011 (1 μM) on the key events in STING-dependent signaling in multiple human cell models. Like SN-001, SN-011 substantially suppressed 2′3′-cGAMP-induced STING oligomerization and phosphorylation assessed by immunoblot in HFFs (Fig. 2A). Additionally, STING ER-to-Golgi translocation induced by HSV-1 infection, HT-DNA, or 2′3′-cGAMP stimulation was substantially suppressed (Fig. 2 B and C). Moreover, in HSV-1–infected HFFs and 293T overexpressing tagged STING and IRF3 or TBK-1, SN-011 impaired the recruitment of TBK1 and IRF3 to the STING signalosome in immunoprecipitation experiments (Fig. 2D and SI Appendix, Fig. S2 A–D). Consistently, phosphorylation of TBK1, IRF3, IκBα, and p65, as well as IRF3 dimerization and IRF3 and p65 nuclear translocation (Fig. 2E–G), were all decreased in the presence of SN-011. In contrast, SN-100 had no effect. SN-011 (1 to 10 μM) did not significantly affect cGAS, STING, TBK1, and IRF3 mRNA or protein levels (SI Appendix, Fig. S2 E–G), ruling out the possibility that SN-011 functions by regulating the expression or stability of cGAS-STING signaling proteins. SN-011 also impaired IFNB mRNA induced by ectopic expression of cGAS or STING in HEK293 cells but had no effect on IFNB induction by TBK1 or IRF3-5D overexpression (SI Appendix, Fig. S2 H–K), as expected for a STING inhibitor. Together, these data demonstrate that SN-011 specifically inhibits STING activation and STING-dependent signaling.
Fig. 2.
SN-011 inhibits STING activation and signal transduction. (A) HFFs were pretreated with SN-011 or SN-100 (1 μM) and then stimulated with 2′3′-cGAMP for 1 h. STING phosphorylation and oligomerization were analyzed by immunoblot probed for STING. (B) HeLa, pretreated with SN-011 or SN-100 (1 μM) or DMSO control, were infected with HSV-1 for 4 h before immunostaining for STING and GM130 (Golgi autoantigen) and confocal microscopy imaging. (C) HFFs, pretreated with SN-011, were stimulated with HT-DNA for 1 h or 2′3′-cGAMP for 30 min before immunostaining for STING, calreticulin, and GM130 and subsequent confocal microscopy imaging analysis (Upper). Quantification of cells with STING puncta that colocalize with the Golgi (Lower). (D) HFFs were pretreated with SN-011 or SN-100 (1 μM) before infecting with HSV-1 for 3 h. Cell lysates were immunoprecipitated with STING antibody and immunoblotted with indicated antibodies. (E) HFFs, pretreated or not with SN-011 or SN-100 (1 μM) were stimulated with 2′3′-cGAMP for 2 h and then analyzed for phosphorylated (p)- IRF3, TBK1, IκBα, and p65, and IRF3 dimerization by immunoblot. (F) HFFs, pretreated or not with SN-011 or SN-100 (1 μM), were stimulated with 2′3′-cGAMP for 2 h. Cells were then immunostained with anti-IRF3 and imaged by confocal microscopy. (Left and Center) Representative images; (Right) quantification of cells with nuclear IRF3. (G) HeLa, pretreated or not with SN-011 or SN-100 (1 μM), were stimulated with 2′3′-cGAMP for 2 h and immunostained with anti-p65 and imaged by confocal microscopy. (Left and Center) Representative images; (Right) quantification of cells with nuclear p65. Data in B, C, F, and G are mean ± SD from three independent experiments. The images are 63× magnification with a digital scan zoom of 3.0 that was used to enhance magnification. **P < 0.01. (Scale bars, 25 μm.)
SN-011 Binds to the CDN Binding Domain of STING.
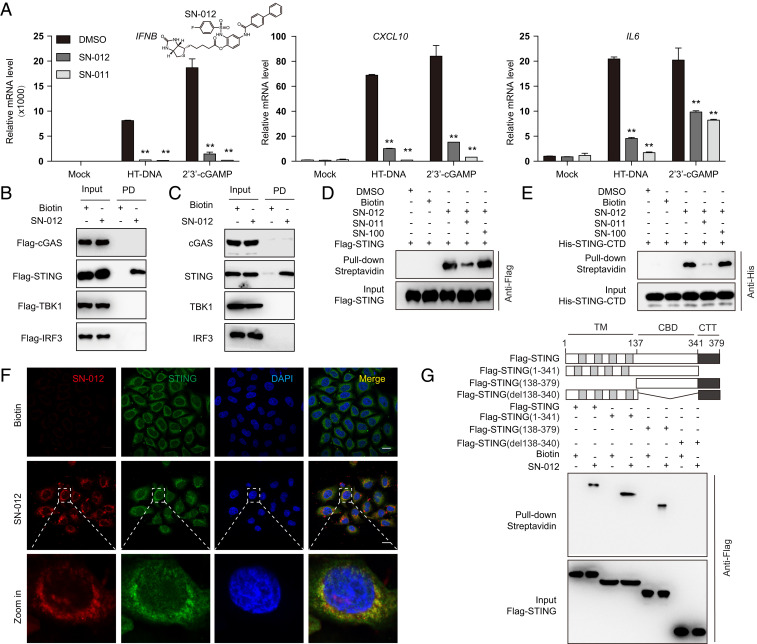
To confirm that SN-011 functions by targeting the STING CDN binding domain, a biotinylated SN-011 derivative (SN-012) was synthesized by chemically attaching biotin to the hydroxyl group of SN-011. Biotinylation had little effect on STING inhibition (Fig. 3A). A biotin pull-down confirmed that SN-012 bound to STING, but not other components of the STING pathway (Fig. 3 B and C). Moreover, an excess of SN-011, but not SN-100, competed with SN-012 for binding to tagged STING or tagged soluble STING lacking the transmembrane domain (TM) region (amino acids 149 to 379) (Fig. 3 D and E). Confocal microscopy revealed cytoplasmic perinuclear colocalization of STING and SN-012 (Fig. 3F). To confirm the STING binding site of SN-012, full-length STING and STING truncation mutants lacking the TM domain (amino acids 1 to 137), CBD (amino acids 138 to 340), or the C-terminal tail (CTT, amino acids 341 to 379) were expressed in HEK293T cells. A streptavidin pull-down of cell lysates was performed after adding SN-012. Loss of the TM domain or CTT did not affect STING binding to SN-012, but deletion of the CBD abolished the interaction (Fig. 3G), confirming that SN-012 binds to the STING CBD.
Fig. 3.
Biotinylated SN-011 specifically binds to the STING CDN binding domain. (A) HFFs, pretreated with SN-011 or biotinylated SN-011 (SN-012; 1 μM), were transfected with HT-DNA for 6 h or treated with 2′3′-cGAMP for 3 h. Induction of IFNB, CXCL10, and IL6 mRNA was measured by quantitative PCR assay. (B) Cell lysates of HEK293T cells, transfected for 24 h with expression plasmids for the indicated Flag-tagged proteins, were incubated with biotin (5 μM) or SN-012 (5 μM) for 1 h, followed by pull-down with streptavidin-conjugated beads and immunoblot with anti-Flag. (C) HFF lysates were incubated with biotin (5 μM) or SN-012 (5 μM) for 1 h, followed by pull-down with streptavidin-conjugated beads, and immunoblot with the indicated antibodies. (D) Cell lysates of HEK293T cells overexpressing Flag-hSTING were incubated with SN-012 (5 μM) in the absence or presence of a 10-fold excess (50 μM) of SN-011 or SN-100, before pull-down with streptavidin-conjugated beads and immunoblot with anti-Flag. (E) Recombinant His-tagged hSTING-CTD (149-379) protein was incubated with SN-012 (5 μM) in the absence or presence of a 10-fold excess (50 μM) of SN-011 or SN-100, before pull-down with streptavidin-conjugated beads and immunoblot with anti-His. (F) HeLa cells were incubated with Biotin or SN-012 (1 μM) and then immunostained with anti-STING and Alexa Fluor 594-conjugated streptavidin. Nuclei were stained with DAPI. (Scale bars, 25 μm.) (G) Schematic of STING and its truncation mutants (Upper). HEK293T cells were transfected to express Flag-tagged hSTING or hSTING-CTD (138–379), hSTING-∆CTT (1–341) or hSTING-∆CBD (∆138–340) truncations and 24 h later, cell lysates were collected and incubated with biotin or SN-012 (5 μM), followed by pull-down with streptavidin-conjugated beads, and immunoblot with anti-Flag (Lower). Data in A are mean ± SD from three independent experiments. The images in F are 63× magnification with a digital scan zoom of 3.0 that was used to enhance magnification. **P < 0.01. Gene expression was normalized to Gapdh.
Mutation at the STING–SN-011 Interface Decreases the Inhibitory Effect of SN-011.
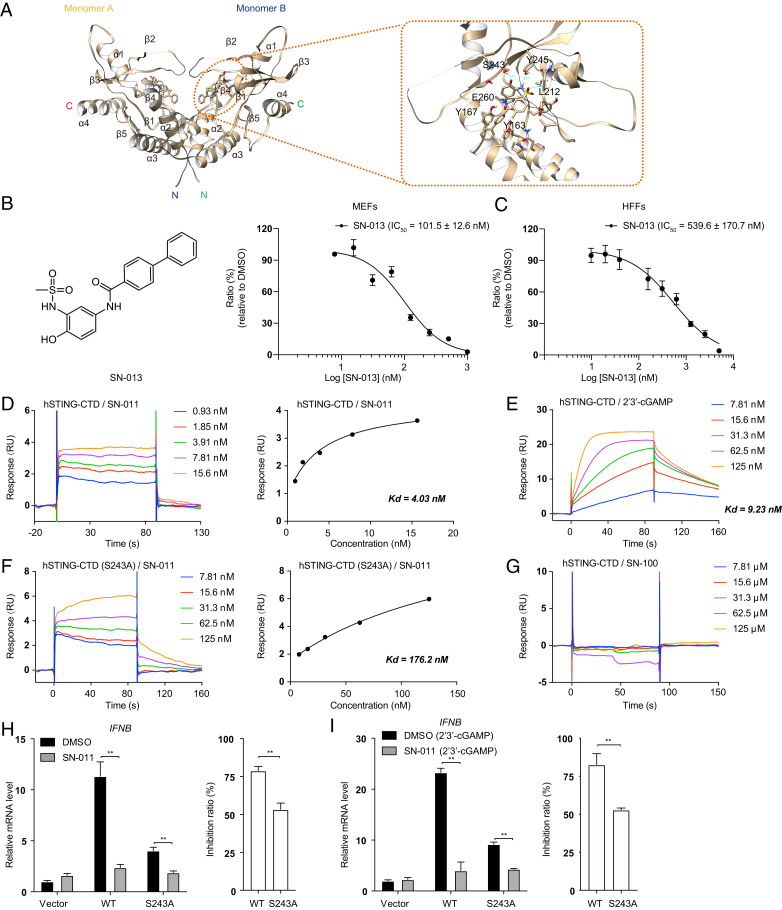
In silico docking was used to determine the interaction between human STING CTD (PDB ID code 4EF5) and SN-011. The docking structure suggested that two molecules of SN-011 were in the STING CDN binding pockets (Fig. 4A). Each SN-011 aligned along the STING dimer interface with the biphenyl group at the base (Fig. 4A). In the docking structure, SN-011 was anchored to the STING dimer interface by a combination of hydrogen bonds and a stacking interaction (Fig. 4A). Notably, the biphenyl base group of SN-011 stacked against the aromatic ring of Tyr167 and was further stabilized by Glu-260. The phenolic hydroxyl and sulfonamide groups of SN-011 formed several hydrogen bonds with Leu212, Ser243, and Tyr245. Consistently, substitution of the hydroxyl group, which interacted with Ser243 via hydrogen bonding, with hydrogen (SN-007) or a methyl group (SN-008), significantly decreased inhibitory activity (Fig. 1 A and B). As the fluorobenzene group in SN-011 did not interact with soluble human STING in the structure, we synthesized an analog of SN-011, named SN-013, without the fluorobenzene group. SN-013 inhibited 2′3′-cGAMP-induced Ifnb gene expression with IC50 values of 101.5 nM, and 539.6 nM in MEF and HFF cells, respectively, which were comparable to those of SN-011 (Fig. 4 B and C).
Fig. 4.
Mutations of the STING-SN-011 interface reduce inhibition by SN-011. (A) Intermolecular contacts of SN-011 bound to hSTING-CTD (amino acids 149 to 379) in virtual docking structure (Left). SN-011 and contacted STING amino acids are shown as a stick model. Hydrogen-bounds with distance are labeled in green (Right). (B and C) MEFs (B) and HFFs (C) were pretreated with the indicated concentrations of SN-013 before 2′3′-cGAMP stimulation for 3 h. Induction of Ifnb mRNA was measured by quantitative PCR. The dose-dependent inhibitory curve was fit to calculate the IC50s. (D) Biacore analysis of hSTING-CTD and SN-011 binding (Left). The binding affinity (Kd) was determined by fitting the binding data to a steady-state 1:1 binding model (Right). RU, response units. (E) Biacore analysis of hSTING-CTD and 2′3′-cGAMP binding. The binding affinity (Kd) was determined by fitting the binding data to a kinetics 1:1 binding model. (F) Biacore analysis of hSTING-CTD (S243A) and SN-011 binding (Left). Kd was determined by fitting the binding data to a steady-state 1:1 binding model (Right). (G) Biacore analysis of hSTING-CTD and SN-100 binding. (H) HEK293T cells, transfected with Flag-hSTING or its mutants, were treated 12 h later with DMSO or SN-011 (10 μM) for 12 h. mRNA was measured by quantitative PCR assay (Left). The inhibition ratio of SN-011 relative to DMSO control was calculated based on Ifnb mRNA levels (Right). (I) HEK293T cells, transfected with Flag-hSTING or its mutant, were treated 12 h later with DMSO or SN-011 (10 μM) for 9 h and then stimulated with 2′3′-cGAMP for 3 h. mRNA was measured by quantitative PCR assay (Left). The inhibition ratio of SN-011 relative to DMSO was calculated based on Ifnb mRNA levels (Right). Data from B, C, H, and I are mean ± SD from three independent experiments. **P < 0.01. Gene expression was normalized to Gapdh.
To confirm that the key STING residues identified by structural analysis mediate SN-011 binding, surface plasmon resonance (SPR) was used to measure the binding affinity and kinetics between SN-011 and soluble human STING. Soluble human STING bound SN-011 with an affinity (Kd) of 4.03 nM (Fig. 4D), which is lower than that of the natural 2′3′-cGAMP ligand (Kd = 9.23 nM) (Fig. 4E). Notably, the binding kinetics of SN-011 (fast association and dissociation) and 2′3′-cGAMP (slow association and dissociation) were markedly different. In contrast to WT STING, the S243A mutant had lower binding affinities to SN-011 (Kd = 176 nM) (Fig. 4F) and the control compound SN-100 did not bind (Fig. 4G). Next, we examined the effect of the mutants on STING signaling and SN-011 inhibition. Overexpression of S243A mutant induced less IFNB than WT STING both basally and in the presence of 2′3′-cGAMP and SN-011 was less active at inhibiting what IFNB was produced (Fig. 4 H and I). Taken together, these data confirm that the identified residues in the STING dimer interface mediate binding of SN-011 and its inhibitory activity.
SN-011 Abrogates SAVI-Associated Mutant STING Signal Activation.
To evaluate the therapeutic potential of SN-011, we tested its in vitro ability to inhibit the autoactivation of human STING carrying GOF mutations N154S, V155M, G166E, C206Y, R281Q, and R284G linked to SAVI, an autoinflammatory disease caused by GOF mutations in TMEM173, the gene encoding STING (29, 30, 48, 49). WT and SAVI-linked mutated human STING were expressed in HEK293T. Cells expressing the GOF human STING mutants exhibited high levels of IFNB, CXCL10, and TNFA mRNA, which was strongly inhibited by incubation with SN-011 (SI Appendix, Fig. S3A). SN-011 incubation also markedly decreased the recruitment of TBK1 and IRF3 to the STING signalosome, assessed by STING pull-down (SI Appendix, Fig. S3B), and also reduced TBK1 and IRF3 phosphorylation induced by STING SAVI mutants (SI Appendix, Fig. S3C). These GOF mutants spontaneously oligomerize more than WT STING when expressed in HEK293T cells (13, 14). SN-011 disrupted oligomerization by WT as well as mutant STING (SI Appendix, Fig. S3D). Notably, SN-011 significantly inhibited the spontaneous accumulation of STING GOF mutants with the Golgi apparatus, suggesting that SN-011 locks STING GOF mutants into a conformation that is incompatible with ER-to-Golgi translocation (SI Appendix, Fig. S4 A and B). Together, these data suggest that SN-011 could be used to inhibit STING activation caused by either WT STING or mutant STING in SAVI patients.
SN-011 Suppresses Systemic Inflammation in Trex1−/− Mice.
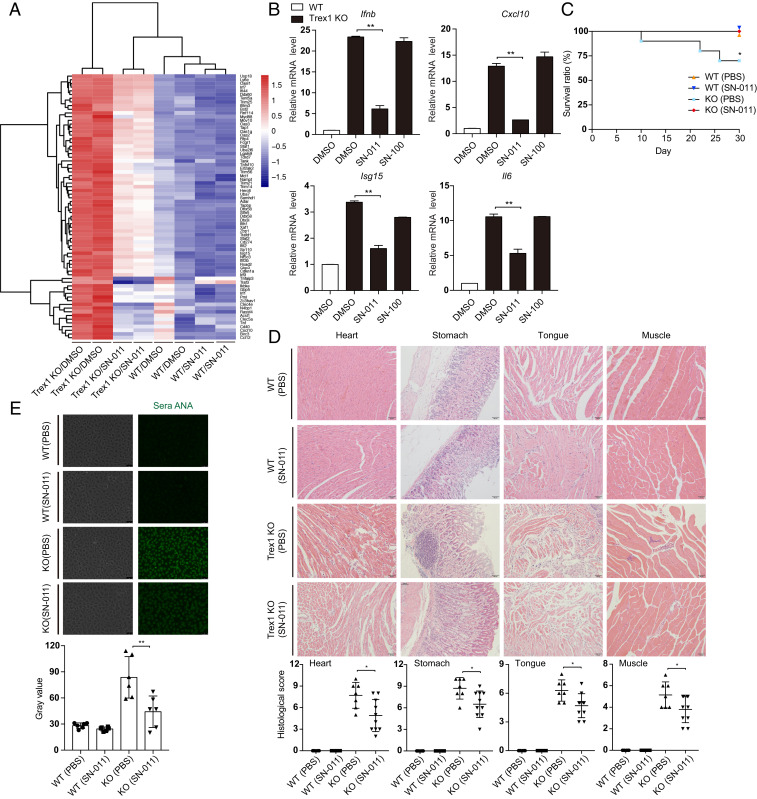
Accumulation of cytosolic self-DNA causes severe and fatal STING-dependent IFN-mediated autoinflammatory disease especially in the heart and other muscles in Trex1−/− mice (26, 50, 51). To begin to evaluate whether SN-011 could be used therapeutically to inhibit STING signaling, BMDMs, harvested from WT and Trex1−/− mice, were treated for 12 h with 500 nM SN-011 or DMSO and then analyzed by RNA-sequencing (RNA-seq) (Fig. 5A). SN-011 treatment significantly reduced the IFN signature of Trex1−/− BMDMs, which was confirmed by measuring expression of Ifnb and representative IFN-stimulated genes (ISGs) by quantitative PCR (Cxcl10, Isg15, and Il6) (Fig. 5B). To test the ability of SN-011 to protect Trex1−/− mice, SN-011 (5 mg/kg) or medium was injected intraperitoneally three times per week for a month into 4-wk-old WT and Trex1−/− mice. During the course of treatment, 3 of 10 untreated Trex1−/− mice died, while none of the 10 mice that received SN-011 died (P = 0.018) (Fig. 5C). At the end of the month, surviving mice were killed and heart, stomach, tongue, and muscle were analyzed by hematoxylin and eosin (H&E) staining, which showed severe multiorgan inflammation in untreated Trex1−/− mice, which was reduced by SN-011 treatment (Fig. 5D). Moreover, Ifnb and representative ISG mRNA levels assessed by quantitative PCR of RNA isolated from whole tissues at the time of killing were also significantly reduced (SI Appendix, Fig. S5A). Moreover, serum antinuclear antibody was markedly reduced by SN-011 treatment (Fig. 5E). SN-011 also significantly reduced the number of activated CD69+ CD8 T cells and memory CD44highCD62Llow CD4 and CD8 T cells to near normal levels in the spleens of treated Trex1−/− mice (SI Appendix, Fig. S5 B and C). SN-011 had no significant effect on the number of splenic activated CD4 T cells. Thus, SN-011 strongly reduced inflammation and protected Trex1−/− mice from death.
Fig. 5.
SN-011 suppresses systemic inflammation and death in Trex1−/− mice. (A) Heat map of RNA-seq of BMDMs from WT or Trex1−/− mice that were treated or not with 0.5 μM SN-011. The top 72 differentially expressed ISGs are shown. (B) Trex1−/− BMDMs were treated with SN-011 (0.5 μM), SN-100 (0.5 μM) or DMSO. Induction of Ifnb, Cxcl10, Isg15, and Il6 mRNA was measured by quantitative PCR. Fold-changes are relative to WT BMDM controls. (C) Survival curves of WT and Trex1−/− mice treated or not with SN-011 (5 mg/kg) three times per week for a month. *P < 0.05, statistical analysis was performed using the log-rank test (10 mice per group). (D) Representative H&E-stained tissue sections from WT or Trex1−/− mice treated or not with SN-011 (5 mg/kg) three times per week for a month (Upper). Results of blinded histological scoring of individual animal samples is shown (Lower). (E) Antinuclear antibodies (ANA) in the serum of WT or Trex1−/− mice treated or not with SN-011 (5 mg/kg) detected by antinuclear antibody antigen substrate slide kit (Upper). Fluorescence intensity was analysis by calculating mean gray value of individual animal sera (Lower). Data in B are mean ± SD from three independent experiments. Data from D and E are presented as mean ± SD from at least six mice in each group. Images in D and E are 20× magnification. *P < 0.05; **P < 0.01. Gene expression was normalized to Gapdh.
SN-011 Exhibits Low Cytotoxicity and High Specificity in Comparison with H-151.
To compare potential cytotoxicity of SN-011 and H-151, cell viability and cell death were examined after adding different concentrations of each compound (1 to 10 μM) for 12 to 36 h to MEFs, Tmem173−/− MEFs, and 3T3 cells (SI Appendix, Fig. S6 A–F). SN-011 had no significant effect on cell viability, whereas H-151 significantly impaired cell viability and caused cell death (SI Appendix, Fig. S6 A–F). Moreover, H-151 was less specific for STING signaling since it more potently than SN-011 suppressed Ifnb and inflammatory cytokine (Tnfa and Il6) mRNA induction triggered by STING-independent stimuli, LPS, poly (I:C) and hp-RNA, in L929, HFFs, and Tmem173−/− MEFs (SI Appendix, Fig. S6 G–I). These data indicate that SN-011 is a more specific inhibitor of STING-dependent signaling than H-151 in vitro.
Next, we compared the inhibitory effects of SN-011 and H-151 on self-DNA–triggered inflammatory responses in Trex1−/− BMDMs by RNA-seq analysis. Both SN-011 and H-151 suppressed differentially expressed (DE) ISG genes to a comparable extent independently of the false-discovery rate (FDR) cutoff used for the analysis (0.05, 0.01, 0.001) (SI Appendix, Fig. S7A). The number, overlap and magnitude of significantly down-regulated and up-regulated DE ISG genes after SN-011 or H-151 treatment were not significantly different (SI Appendix, Fig. S7 B–D). To further assess and compare the efficacy of the two compounds in vivo, 10 mg/kg SN-011 or H-151 was injected intraperitoneally into 6-wk-old Trex1−/− mice daily for 2 wk. Ifnb and ISGs expression in the affected tissues of Trex1−/− mice were comparably suppressed by SN-011 and H-151 (SI Appendix, Fig. S7E). H&E staining also showed comparable amelioration of heart inflammation by both compounds (SI Appendix, Fig. S7 F and G). Taken together, these data show that SN-011 and H-151 comparably inhibit STING-mediated inflammation in both cell-based and Trex1−/− mouse models.
Discussion
In this study, we performed a large-scale virtual screen against the STING CDN-binding pocket to identify STING antagonists. Structural optimization identified SN-011 as a potent STING inhibitor that impedes STING oligomerization, trafficking and activation in response to cytosolic DNA and markedly decreases STING-driven expression of type I IFNs and proinflammatory cytokines. SN-011 binds to the CDN-binding pocket with higher affinity than the endogenous cGAS product 2′3′-cGAMP and locks the STING dimer in an open, inactive conformation. SN-011 inhibits STING-dependent signaling, with IC50 values of ∼100 nM and ∼500 nM for mouse and human cells, respectively. SN-011 inhibits induction of interferons and inflammatory cytokines in multiple in vitro mouse and human models of STING activation, including stimulation with 2′3′-cGAMP, HSV-1 infection, Trex1 deficiency, or overexpression of cGAS-STING proteins. Importantly SN-011 also inhibits the spontaneous activation of GOF STING mutants that cause SAVI. In Trex1−/− mice, SN-011 was well tolerated, strongly inhibited hallmarks of inflammation and autoimmunity, and prevented death. Collectively, these studies suggest that SN-011 is an attractive lead compound for developing drugs to inhibit STING-dependent signaling.
In comparison, other recently identified STING antagonists are either inactive (C-176, C-178) or have low bioactivity against human STING (NO2-FAs, Astin C, compound 18) (42–45). Compound 18 binds deep in the cleft of the human STING dimer, but with an IC50 of ∼11 μM, about 20-fold higher than SN-011, even after optimization by analyzing SARs (44). The most promising reported STING antagonist, H-151, is a covalent inhibitor that binds to a reactive Cys91 in the transmembrane domain of STING to inhibit activation-induced palmitoylation (42). SN-011 and H-151 equivalently inhibit STING in mouse cell lines, with an IC50 value of ∼100 nM. Elevated ISG expression in Trex1−/− BMDMs were similarly ameliorated by in vitro exposure to SN-011 or H-151. When SN-011 and H-151 were administered at the same dose (10 mg/kg), both compounds comparably suppressed inflammation in the affected tissues (heart, stomach, tongue, and muscle) in Trex1−/− mice. Future preclinical experiments to compare pharmacokinetics and dynamics, efficacy, and safety of both drugs administered back-to-back at different doses in mouse AGS and SAVI models will be needed to further explore the relative advantages of each compound. Like other Cys-reactive drugs, which are notoriously nonspecific and covalently bind to unpaired Cys in many proteins, H-151 likely has more off-target activities than SN-011 (52, 53). H-151 significantly impaired cell viability and induced cell death while SN-011 was not cytotoxic. In addition, H-151 showed potent inhibitory effects on TLR- or RIG-I-mediated signaling in cell-based assays compared to SN-011. All of these data indicate that SN-011 shows better specificity and safety than H-151. The ability to modify these lead compounds to optimize STING inhibition and their pharmacological properties will ultimately determine which compounds can lead to effective new treatments of STING-mediated diseases.
Insights into the structural basis for SN-011 activity was provided by docking analysis. In the docking structure, SN-011 bound to the STING dimer interface in the CDN-binding pocket to lock STING in an inactive open conformation. One molecule of SN-011 bound to each STING monomer with an extended binding site along the pocket. SPR confirmed the stoichiometry of STING monomer to SN-011 as 1:1 with a binding dissociation constant of 4.03 nM. By comparison, compound 18 lies in the bottom of the cleft of the STING dimer (PDB ID code 6MXE) in a 1:2 binding stoichiometry with an affinity of ∼50 μM for the binding of the first molecule and an affinity of ∼2 nM for the binding of the second molecule (44). This spatial and affinity difference may explain the different inhibitory activities between SN-011 and compound 18. Moreover, the interaction between STING and SN-011 over an extended binding site is mediated by residues Tyr167, Leu212, Ser243, Tyr245, and Glu260 in STING, all of which are evolutionarily conserved between species. Tyr167 and Glu260 are essential for CDN binding and Ser243 and Tyr245 are in the lid region, which involves lid formation upon 2′3′-cGAMP binding (11, 54, 55). Thus, association of SN-011 with STING leads to the occupation of the binding pocket and steric hindrance that inhibits the approach of CDNs to STING.
Our SAR analysis of SN-001 to SN-011 revealed that the phenolic hydroxyl group is essential for bioactivity and substitution with a hydrogen, methyl, or oxymethyl group impairs inhibitory activity, but the fluorobenzene group is dispensable. The virtual docking structure of SN-011 with the STING dimer showed biphenyl group stacking at the bottom of the STING dimer pocket, which may allow us to improve SN-011 by introducing a linker to connect benzene rings of two molecules of SN-011 (to yield a single dimeric SN-011, di-SN-011). di-SN-011 may enhance binding affinity and bioactivity as was shown for the STING agonists diABZI and MSA-2 (56, 57). Future work will seek to develop more potent SN-011 analogs and define their pharmacokinetics and safety as a prerequisite for clinical development.
Aberrant activation of cGAS-STING signaling by self-DNA causes severe autoimmune diseases, such as AGS, SLE, and other lupus-like diseases, called interferonopathies because they are largely caused by chronic overexpression of type I IFNs (58, 59). Trex1−/− mice are an experimental AGS and SLE model. SN-011 treatment inhibited the interferon signature in Trex1−/− BMDMs and alleviated the systemic autoinflammatory phenotype, extending mouse survival. Future studies will examine the inhibitory effect of SN-011 on peripheral blood mononuclear cells from AGS and SLE patients to evaluate the therapeutic potential of SN-011 and its derivatives on human diseases that require bone marrow transplantation or are inadequately treated with current therapies. SN-011 also blocked the type I IFN and proinflammatory gene expression induced by SAVI-associated STING mutants by preventing spontaneous STING auto-oligomerization and activation. Future structural studies will further probe how SN-011 inhibits GOF mutants of STING. It will also be worth testing the in vivo and ex vivo efficacy of SN-011 in SAVI-related mouse models (V154M or N153S) and primary cells from SAVI patients. As of now, no standard immunosuppressive and curative approach has been identified to treat SAVI. Inhibiting type I IFN signaling with Janus kinase (JAK) inhibitors (e.g., tofacitinib, ruxolitinib, and baricitinib) controls disease progression in a subset of SAVI patients (29, 49, 60). However, lung inflammation, myeloid cell expansion, and T cell cytopenia developed in STING N153S or V154M knockin mouse models of SAVI do not depend on IRF3 or IFN signaling, suggesting that NF-κB–regulated induction of inflammatory genes is also important in disease pathogenesis (61–63). A STING inhibitor, such as SN-011, that blocks the first step of STING activation is an especially attractive drug candidate for treating SAVI, in comparison to JAK inhibitors. Previous studies showed that STING inhibitor tool compounds control tissue inflammation by blocking both IFN and NF-κB signaling, inhibit STING-triggered apoptosis of T cells, monocytes and endothelial cells of SAVI patients and reduce the risk of opportunistic infection by not inhibiting other pattern recognition receptor pathways (59, 64). SN-011 specifically inhibited both IFN and inflammatory downstream consequences of STING activation without interfering with other innate immune pathways. Our findings support drug development of STING inhibitors to treat STING-associated autoimmune diseases.
Materials and Methods
Details of the materials and methods, including immunoblot analysis, immunofluorescence, real-time RT-PCR, animal experiments, flow cytometry, biotin pull-down assay, molecular docking and virtual screening, SPR, RNA-seq experiments, and chemical synthesis are presented in SI Appendix, SI Materials and Methods. All animal experiments were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals (65), with the approval of the Center for New Drug Safety Evaluation and Research, China Pharmaceutical University.
Supplementary Material
Acknowledgments
We thank Dr. Tomas Lindahl and Dr. Deborah Barnes (Cancer Research UK, London) and Dr. Nan Yan (University of Texas Southwestern Medical Center) for kindly sharing Trex1+/− mice; Dr. Min-Hua Luo (Wuhan Institute of Virology) for kindly sharing the human foreskin fibroblast cell line; Dr. Wentao Qiao (Nankai University), Dr. Chunfu Zheng (Suzhou University), Dr. Youcun Qian (Institute of Health Sciences), and Dr. Yong Yang (China Pharmaceutical University) for providing reagents; Xiaonan Ma from the Cellular and Molecular Biology Center of China Pharmaceutical University for providing technical assistance for using the Carl Zeiss LSM700; and Wei Jiang from the State Key Laboratory of Natural Medicines of China Pharmaceutical University for providing technical assistance for surface plasmon resonance data collection. This study was supported by the National Key R&D Program of China (2016YFA0501800), the National Natural Science Foundation of China (31730018, 81672029, 3173000227, 31470428), Project Program of State Key Laboratory of Natural Medicines, China Pharmaceutical University No. SKLNMZZ202002), the Jiangsu Innovative and Entrepreneurial Talents Program, the National New Drug Innovation Major Project of China (2017ZX09309027), and the Fund of Chinese Academy of Sciences (XDA09030301-4, Hundred Talents Program). Z.H. is sponsored by China Postdoctoral Research Program (2019TQ0356). C.L. is sponsored by National Natural Science Foundation of China for the Youth (31800724).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105465118/-/DCSupplemental.
Data Availability
All original RNA-seq data are deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE143830). All other study data are included in the article and SI Appendix.
References
- 1.Soni C., Reizis B., DNA as a self-antigen: Nature and regulation. Curr. Opin. Immunol. 55, 31–37 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun L., Wu J., Du F., Chen X., Chen Z. J., Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ablasser A., et al., cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao P., et al., Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J., et al., Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishikawa H., Barber G. N., STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin L., et al., MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 28, 5014–5026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong B., et al., The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Sun W., et al., ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U.S.A. 106, 8653–8658 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burdette D. L., et al., STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X., et al., Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao P., et al., Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 154, 748–762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ergun S. L., Fernandez D., Weiss T. M., Li L., STING polymer structure reveals mechanisms for activation, hyperactivation, and inhibition. Cell 178, 290–301.e10 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Shang G., Zhang C., Chen Z. J., Bai X. C., Zhang X., Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature 567, 389–393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y. H., Liu X. Y., Du X. X., Jiang Z. F., Su X. D., The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat. Struct. Mol. Biol. 19, 728–730 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Mukai K., et al., Activation of STING requires palmitoylation at the Golgi. Nat. Commun. 7, 11932 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C., et al., Structural basis of STING binding with and phosphorylation by TBK1. Nature 567, 394–398 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao B., et al., A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1. Nature 569, 718–722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu S., et al., Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Zhao B., et al., Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins. Proc. Natl. Acad. Sci. U.S.A. 113, E3403–E3412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abe T., Barber G. N., Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J. Virol. 88, 5328–5341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Z., Damania B., The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe 19, 150–158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackenzie K. J., et al., cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bakhoum S. F., et al., Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Theofilopoulos A. N., Kono D. H., Baccala R., The multiple pathways to autoimmunity. Nat. Immunol. 18, 716–724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gall A., et al., Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity 36, 120–131 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao D., et al., Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. U.S.A. 112, E5699–E5705 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crow Y. J., Manel N., Aicardi-Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 15, 429–440 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Liu Y., et al., Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 371, 507–518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeremiah N., et al., Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Invest. 124, 5516–5520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dou Z., et al., Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glück S., et al., Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19, 1061–1070 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heipertz E. L., Harper J., Walker W. E., STING and TRIF contribute to mouse sepsis, depending on severity of the disease model. Shock 47, 621–631 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Zeng L., et al., ALK is a therapeutic target for lethal sepsis. Sci. Transl. Med. 9, eaan5689 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sliter D. A., et al., Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Zhao Q., Wei Y., Pandol S. J., Li L., Habtezion A., STING signaling promotes inflammation in experimental acute pancreatitis. Gastroenterology 154, 1822–1835.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai J., et al., DsbA-L prevents obesity-induced inflammation and insulin resistance by suppressing the mtDNA release-activated cGAS-cGAMP-STING pathway. Proc. Natl. Acad. Sci. U.S.A. 114, 12196–12201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo X., et al., Expression of STING is increased in liver tissues from patients with NAFLD and promotes macrophage-mediated hepatic inflammation and fibrosis in mice. Gastroenterology 155, 1971–1984.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benmerzoug S., et al., STING-dependent sensing of self-DNA drives silica-induced lung inflammation. Nat. Commun. 9, 5226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Q., et al., Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 533, 493–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung K. W., et al., Mitochondrial damage and activation of the STING pathway lead to renal inflammation and fibrosis. Cell Metab. 30, 784–799.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haag S. M., et al., Targeting STING with covalent small-molecule inhibitors. Nature 559, 269–273 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Li S., et al., The cyclopeptide Astin C specifically inhibits the innate immune CDN sensor STING. Cell Rep. 25, 3405–3421.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Siu T., et al., Discovery of a novel cGAMP competitive ligand of the inactive form of STING. ACS Med. Chem. Lett. 10, 92–97 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen A. L., et al., Nitro-fatty acids are formed in response to virus infection and are potent inhibitors of STING palmitoylation and signaling. Proc. Natl. Acad. Sci. U.S.A. 115, E7768–E7775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ouyang S., et al., Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 36, 1073–1086 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sterling T., Irwin J. J., ZINC 15—Ligand discovery for everyone. J. Chem. Inf. Model. 55, 2324–2337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melki I., et al., Disease-associated mutations identify a novel region in human STING necessary for the control of type I interferon signaling. J. Allergy Clin. Immunol. 140, 543–552.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 49.König N., et al., Familial chilblain lupus due to a gain-of-function mutation in STING. Ann. Rheum. Dis. 76, 468–472 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Lee-Kirsch M. A., et al., Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet. 39, 1065–1067 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Stetson D. B., Ko J. S., Heidmann T., Medzhitov R., Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weerapana E., et al., Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh J., Petter R. C., Baillie T. A., Whitty A., The resurgence of covalent drugs. Nat. Rev. Drug Discov. 10, 307–317 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Shang G., et al., Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat. Struct. Mol. Biol. 19, 725–727 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Shu C., Yi G., Watts T., Kao C. C., Li P., Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat. Struct. Mol. Biol. 19, 722–724 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramanjulu J. M., et al., Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature 564, 439–443 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Pan B. S., et al., An orally available non-nucleotide STING agonist with antitumor activity. Science 369, eaba6098 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Crowl J. T., Gray E. E., Pestal K., Volkman H. E., Stetson D. B., Intracellular nucleic acid detection in autoimmunity. Annu. Rev. Immunol. 35, 313–336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ablasser A., Chen Z. J., cGAS in action: Expanding roles in immunity and inflammation. Science 363, eaat8657 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Frémond M. L., et al., Efficacy of the Janus kinase 1/2 inhibitor ruxolitinib in the treatment of vasculopathy associated with TMEM173-activating mutations in 3 children. J. Allergy Clin. Immunol. 138, 1752–1755 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Warner J. D., et al., STING-associated vasculopathy develops independently of IRF3 in mice. J. Exp. Med. 214, 3279–3292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bouis D., et al., Severe combined immunodeficiency in stimulator of interferon genes (STING) V154M/wild-type mice. J. Allergy Clin. Immunol. 143, 712–725.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Motwani M., et al., Hierarchy of clinical manifestations in SAVI N153S and V154M mouse models. Proc. Natl. Acad. Sci. U.S.A. 116, 7941–7950 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gulen M. F., et al., Signalling strength determines proapoptotic functions of STING. Nat. Commun. 8, 427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original RNA-seq data are deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE143830). All other study data are included in the article and SI Appendix.