Significance
Heart disease is the leading cause of death worldwide, and hypertrophic cardiomyopathy (HCM) is the most common inherited form of heart disease, affecting over 1 in 200 people. Mutations in myosin, the motor protein responsible for contraction of the heart, are a common cause of HCM but have diverse effects on the biomechanics of the myosin protein. We demonstrate that complex biomechanical effects of mutations associated with heart disease can be effectively studied and understood using a multiscale experimental and computational modeling approach. This work confirms an important role for disruption of the super relaxed state for one particular HCM mutation and provides an approach that can be extended to aid in the understanding of disease mechanisms for different mutations.
Keywords: hypertrophic cardiomyopathy, β-cardiac myosin, optical trapping, hiPSC-CMs, super relaxed state
Abstract
Hypertrophic cardiomyopathy (HCM) is the most common inherited form of heart disease, associated with over 1,000 mutations, many in β-cardiac myosin (MYH7). Molecular studies of myosin with different HCM mutations have revealed a diversity of effects on ATPase and load-sensitive rate of detachment from actin. It has been difficult to predict how such diverse molecular effects combine to influence forces at the cellular level and further influence cellular phenotypes. This study focused on the P710R mutation that dramatically decreased in vitro motility velocity and actin-activated ATPase, in contrast to other MYH7 mutations. Optical trap measurements of single myosin molecules revealed that this mutation reduced the step size of the myosin motor and the load sensitivity of the actin detachment rate. Conversely, this mutation destabilized the super relaxed state in longer, two-headed myosin constructs, freeing more heads to generate force. Micropatterned human induced pluripotent derived stem cell (hiPSC)–cardiomyocytes CRISPR-edited with the P710R mutation produced significantly increased force (measured by traction force microscopy) compared with isogenic control cells. The P710R mutation also caused cardiomyocyte hypertrophy and cytoskeletal remodeling as measured by immunostaining and electron microscopy. Cellular hypertrophy was prevented in the P710R cells by inhibition of ERK or Akt. Finally, we used a computational model that integrated the measured molecular changes to predict the measured traction forces. These results confirm a key role for regulation of the super relaxed state in driving hypercontractility in HCM with the P710R mutation and demonstrate the value of a multiscale approach in revealing key mechanisms of disease.
Hypertrophic cardiomyopathy (HCM) is one of the most prevalent genetic diseases of the heart, affecting over 1 in 200 individuals (1, 2), and is a leading cause of sudden cardiac death (3). HCM is characterized by cardiomyocyte hypertrophy, myofibril disarray, hypercontractility, and diastolic dysfunction, although there are substantial heterogeneity and complexity in the presentation of HCM in patients (4, 5). Tissue remodeling, including interstitial fibrosis, can eventually progress to heart failure and death (5–7). Over 1,000 causative mutations have been identified, with the majority in genes encoding sarcomeric proteins responsible for generating and regulating contraction. Roughly a third of mutations are located in β-cardiac myosin, the primary ventricular motor protein in humans (8). Increased ejection fraction has been observed in patients with pathogenic mutations in β-cardiac myosin without left ventricular (LV) hypertrophy, which suggests that hypercontractility may precede hypertrophic remodeling in some patients with HCM mutations in β-cardiac myosin (9).
Because hypercontractility is often observed in HCM patients with mutations in β-cardiac myosin, HCM mutations were hypothesized to increase the activity of myosin at the protein level, resulting in increased force production at the sarcomere and cellular levels that propagates to the whole-organ level (10). Myosin protein activity is characterized by biochemical and biophysical measurements. The activity of actively cycling myosin interacting with actin is characterized by the rate of ATP turnover, the rate of detachment from actin, force production, step size, and actin-sliding velocity. Myosin that is not actively cycling resides in a super relaxed state (SRX) (11), associated with a folded-back conformation not available for actin interaction. Biochemical and biophysical assessments of HCM mutations in purified human β-cardiac myosin have revealed various changes in both actively cycling myosin (12–16) and SRX proportions (17–20).
Myosin is a mechanoenzyme that harnesses the chemical energy from ATP hydrolysis to perform a force-producing power stroke. Its converter domain plays a critical role in facilitating this power stroke by coupling conformational changes originating from its nucleotide pocket to the rotation of its lever arm. This domain is a hot spot for pathogenic mutations (21–24). The P710 residue is located at the proximal edge of the converter domain, and at least three HCM mutations have been identified at this site, including P710R (25–27). The P710R mutation was identified in a cohort of patients with pediatric onset HCM (defined based on LV hypertrophy in patients 13 or younger), and echocardiograms of this cohort found significantly reduced LV end systolic dimension and increased fraction shortening suggesting hypercontractility (27). According to a recent meta-analysis, the average age of disease onset for HCM in patients without a known mutation is 44 y, and the average age of onset with mutations in β-cardiac myosin is 35 y (28). A previous report characterizing both early-onset and late-onset HCM mutations found that actively cycling heads with the P710R mutation have lower ATP turnover activity and duty ratio compared with both wild-type (WT) myosin and other HCM mutations (29). The discrepancy between this reduced function in actively cycling myosin and a potentially hypercontractile, early-onset clinical phenotype invites a comprehensive, multiscale assessment of the effects of the P710R mutation on myosin activity and SRX proportions.
In the past, it has been difficult to determine early mechanisms of HCM disease triggered by mutations in β-MHC because of the inability to culture human cardiomyocytes. Samples from hearts obtained at the time of transplant or myectomy reflect a combination of primary and secondary pathologies. Rodents fail to recapitulate many human heart diseases, and their adult ventricles predominantly express the α-MHC isoform, which has different kinetics from β-MHC (30–32). However, the expansion of CRISPR/Cas-9 protocols for human induced pluripotent derived stem cell (hiPSC) gene editing coupled with efficient differentiation into cardiomyocytes (hiPSC-CMs) provides a valuable model system for studying early mechanisms of disease, especially mechanisms related to contractile alterations, in a controlled context (31). In the past, hiPSC-CM models have been limited by the relative immaturity of the cardiomyocytes and high population heterogeneity (33). In traditional two-dimensional culture, these cells have disorganized myofibrils, impaired calcium handling, and immature cell signaling (34, 35). However, recent advances in microengineered environments can provide external environmental cues that better recapitulate in vivo conditions to promote myofibril alignment and accelerate maturation of both contractile machinery and cell signaling (35, 36). While these engineering solutions can improve the functional maturity of hiPSC-CMs, they are still immature relative to adult cardiomyocytes in the heart (33). We have previously developed a hydrogel platform with rectangles of extracellular matrix (ECM) at a 7:1 aspect ratio (similar to that of cardiomyocytes in the left ventricle) that, combined with traction force microscopy (TFM), allows for single-cell assessment of both cellular organization and biomechanics (35–37). When combined with measurements of myosin function at the molecular level, these cellular measurements can provide validation of the molecular basis of force generation and resultant disease mechanisms of HCM.
Finally, computational models can provide key insights into the interactions between related dynamic parameters and the resultant implications for the total production of force. Computational models incorporating different degrees of detail and different components of sarcomere structure have been used for decades to answer questions relating to fundamental muscle mechanics (38–47) and alterations to thin filament regulation in the context of cardiac disease and HCM (48, 49). A new model of myosin activity that incorporates an OFF state representative of the SRX state has recently been validated against experimental measurements of cardiac muscle (50). This study concluded that force-sensitive regulation of the OFF state significantly improves the fit to experimental data, but this model has not previously been used in the context of MHY7 mutations nor hiPSC-CMs (50).
In this work, we used multiscale experimental techniques to assess the biomechanical effects of the HCM mutation P710R, which demonstrates decreased activity at the level of the motor domain yet increased force generation at the cell level. We furthermore used a computational model to integrate the molecular findings and found that altered regulation of the SRX state is an essential driver of hypercontractility for this mutation. Our multiscale experimental findings combined with computational modeling enabled us to assess the relative contributions of individual molecular parameters to cellular contraction.
Results
The P710R Mutation Reduced Load Sensitivity and Step Size of Single Myosin Molecules.
We first focused on the motor domain of myosin by using the recombinant human β-cardiac myosin sS1 domain (catalytic domain plus the essential light chain binding portion of the lever arm). The properties of individual myosin heads interacting with actin were assessed by optical trapping using harmonic force spectroscopy (HFS). In this technique, the durations of binding events between a single myosin and an actin filament under different load forces are measured in physiological (2 mM) ATP conditions. The sample stage oscillates sinusoidally so that by the randomness of where myosin initially attaches to actin, a range of mean forces are automatically applied over the course of many binding events (51). This technique has been used to quantify changes in the load-dependent detachment rate of β-cardiac myosin due to mutations and myosin inhibitor and activators (14).
The detachment rate is an exponential function of the mean load force (F):
| [1] |
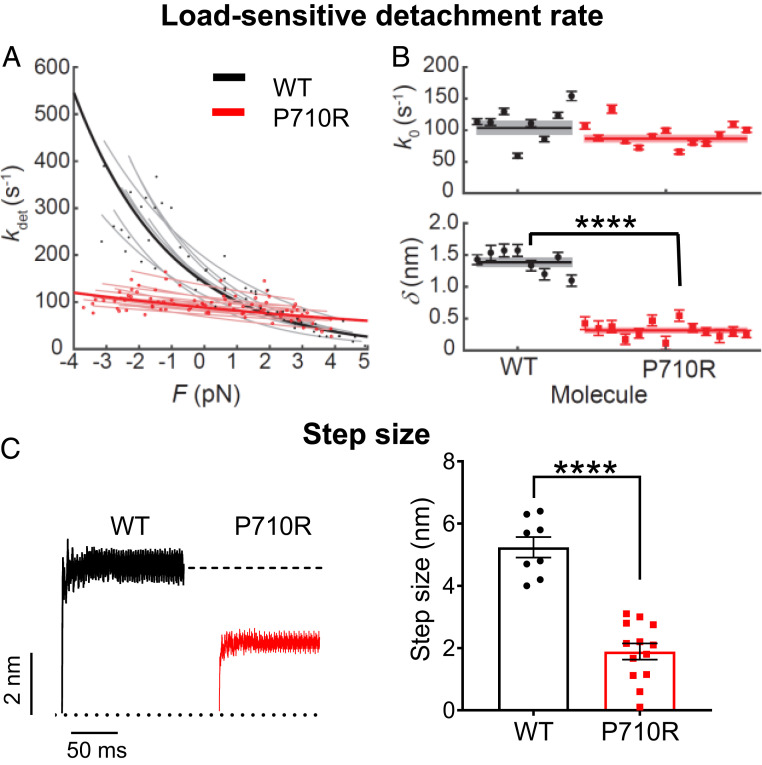
where k0 is the rate at zero load, δ is the distance to the transition state of the rate-limiting step in the bound state (a measure of force sensitivity), kB is the Boltzmann constant, T is temperature, and I0 is the zeroth-order modified Bessel function of the first kind (to correct for the harmonic force with amplitude ΔF). The release of ADP is the rate-limiting step for detachment from actin at high (2 mM) ATP concentrations, and this step is sensitive to load force. Thus, the detachment rate k0 and its load sensitivity δ determined by HFS correspond to the rate of ADP release and its load sensitivity, respectively. WT β-cardiac myosin had detachment rate at zero load k0 = 104 ± 10 s−1 and force sensitivity δ = 1.39 ± 0.06 nm (results from 8 molecules), consistent with previous results (14, 51–53). The unloaded detachment rate of P710R was not significantly changed (87 ± 5 s−1, P = 0.12), while δ was dramatically reduced (0.31 ± 0.03 nm, P < 0.0001) (results from 13 molecules) (Table 1 and Fig. 1 A and B). Consistent with our single-molecule finding of an unchanged unloaded detachment rate, previous ensemble stopped-flow measurements had found that P710R did not alter the rate of ADP release, the rate-limiting step for detachment from actin (29). Our single-molecule trap experiments reveal that this rate of ADP release is rendered much less sensitive to load forces as a result of the mutation.
Table 1.
Parameters measured for β-cardiac myosin sS1
| k0, s−1 | δ, nm | d, nm | kcat, s−1 | Kapp, μM | V, nm⋅s−1 | pCa50 | n | |
| WT | 104 ± 10 | 1.39 ± 0.06 | 5.2 ± 0.3 | 4.1 ± 0.4 | 33 ± 6 | 762 ± 16 | 6.46 ± 0.04 | 2.2 ± 0.2 |
| P710R | 87 ± 5 | 0.31 ± 0.03**** | 1.9 ± 0.3**** | 2.5 ± 0.1* | 14 ± 1* | 239 ± 9**** | 6.48 ± 0.07 | 3.1 ± 0.6 |
The rate of detachment from actin at zero load k0, its force sensitivity δ, and myosin’s step size d were measured from single molecules using the HFS technique (8 WT and 13 P710R molecules). kcat and Kapp were measured using a colorimetric actin-activated ATPase assay and were previously reported (29). Unloaded motility velocities V were measured by the motility assay with actin filaments (nine WT and eight P710R independent experiments). Calcium sensitivity parameters pCa50 and Hill coefficient n were measured by motility assay with regulated thin filaments (five WT and four P710R independent experiments). Data are presented as mean ± SEM. *P < 0.05 and ****P < 0.0001 different between P710R and WT.
Fig. 1.
Single molecules of β-cardiac myosin sS1 with P710R mutation had reduced load sensitivity and step size. (A) Measurements of myosin’s load-sensitive rate of detachment from actin kdet(F) using the HFS technique in a dual-beam optical trap. Positive forces represent load in the opposite direction of the power stroke (resistive), and negative forces represent load in the same direction of the power stroke (assistive). Each light line is a fit of Eq. 1 to data from one molecule, each with a few hundred binding events. (B) The fitted parameters k0 (rate at zero load) and δ (load sensitivity) of each molecule corresponding to light lines in A. Error bars represent the error in the parameter fit for each molecule. Horizontal lines represent weighted means across all molecules, and shaded rectangles represent SEM. (C) Averaged start-aligned position traces of binding events from two example molecules revealing the power stroke of myosin, which occur within milliseconds of actin binding. Step size values of multiple molecules are shown on the Right. Error bars represent SEM. Single-molecule data are from 8 WT molecules and 13 P710R molecules as shown. Values are given in Table 1. See also Materials and Methods and SI Appendix, Fig. S1. ****P < 0.0001.
Further analysis of the average trapped bead displacement during binding events revealed that the mutation decreased myosin’s step size by ∼60% (WT, d = 5.2 ± 0.3 nm; P710R, d = 1.9 ± 0.3 nm; P < 0.0001) (results from 8 WT and 13 P710R molecules) (Table 1 and Fig. 1C and SI Appendix, Fig. S1).
The P710R Mutation Reduced kcat, Duty Ratio, and Velocity but Not Calcium Sensitivity of Myosin in Ensemble.
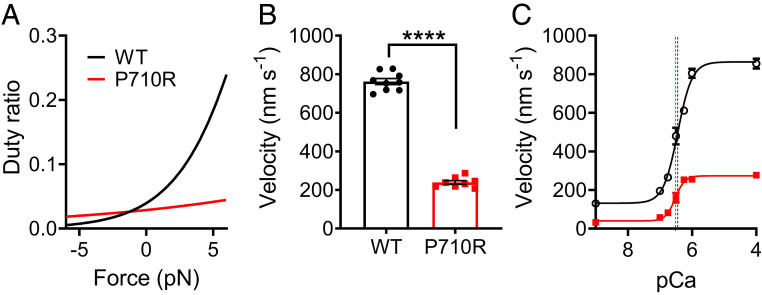
We next assessed the effects of the P710R mutation on properties of myosin sS1 in ensemble. It has been previously reported that myosins with the P710R mutation had significantly reduced actin-activated ATPase activity at saturating actin concentrations (WT, kcat = 4.1 ± 0.4 s−1; P710R, kcat = 2.5 ± 0.1 s−1) but higher apparent actin affinity (WT, Kapp = 33 ± 6 μM; P710R, Kapp = 14 ± 1 μM) (29) (Table 1). Given the single-molecule and ensemble ATPase measurements, the duty ratio can be estimated as a function of load force (F) (14):
| [2] |
where the attachment rate kattach, calculated at saturating actin concentrations and assumed to be independent of force, is given by the following:
| [3] |
Our calculations suggest that P710R has a much lower duty ratio than WT myosin at low or resistive (positive) loads (Fig. 2A), consistent with a previously reported prediction (29). Since duty ratio is the fraction of total cycle time that a myosin head spends in the bound force-producing state, P710R is predicted to have a lower time-averaged force per head than WT at those loads. Our calculations predict that P710R has a slightly higher duty ratio than WT under high assistive (negative) loads, suggesting that P710R heads may not release actin as efficiently in an actively shortening sarcomere.
Fig. 2.
β-Cardiac myosin sS1 ensembles with P710R mutation had altered duty ratio, reduced actin sliding velocity, and unchanged calcium sensitivity. (A) Calculated duty ratio across load forces based on ATPase and actin detachment rate measurements. (B) Actin sliding velocity (mean velocity including stuck filaments [MVIS]; Materials and Methods) in the unloaded motility assay. Each data point represents one independent experiment (n = 9 for WT, n = 8 for P710R). (C) Velocities (MVIS) of regulated thin filaments at various calcium concentrations measured by the motility assay. pCa = −log10[Ca]. The curves are fits to the Hill equation of averaged data from multiple independent experiments (n = 5 for WT, n = 4 for P710R), and the vertical dashed lines represent the fitted pCa50 values. Error bars represent SEM. Values are given in Table 1. See also SI Appendix, Fig. S2. ****P < 0.0001.
Actin sliding velocity in an unloaded motility assay was reduced by ∼60% for P710R compared with WT (WT, V = 762 ± 16 nm/s from 9 experiments; P710R, V = 239 ± 9 nm/s from 8 experiments; P < 0.0001) (Table 1 and Fig. 2B and SI Appendix, Fig. S2 and Movies S1 and S2). The conditions of our motility experiments ensured that myosin heads were present in sufficient numbers on the surface and had short enough tethers such that sliding velocities were limited by filaments’ detachment from myosin rather than by attachment (54, 55) (Materials and Methods). Under these detachment-limiting conditions, velocity can be approximated as the step size divided by the actin-bound time, which is inversely proportional to the actin detachment rate: v = d/tbound = d*kdet. Thus, the 60% reduction in unloaded motility velocity can be explained by the 60% reduction in P710R’s step size and an unchanged unloaded detachment rate k0. A previous study of this mutation failed to predict the measured decrease in velocity likely because the authors did not account for a major reduction in step size (29). We further observed that 5–15% of actin filaments were immobile for P710R compared with 0–5% for WT (SI Appendix, Fig. S2). This suggests that the mutation may induce a less proper or stable protein fold in a small percentage of myosin molecules, which, as a result, have reduced function in moving actin (Materials and Methods and Discussion).
In addition to determining motility velocity, myosin’s actin-detachment kinetics may affect the calcium sensitivity of sarcomeres through the activation of the thin filament by bound myosin heads (47). In the thin filament, binding sites on actin are blocked by the regulatory proteins troponin and tropomyosin when calcium levels are low (in diastole) and are activated and accessible to myosin heads when calcium levels rise (in systole). However, heads that stay bound to actin longer may cooperatively activate the thin filament at lower calcium concentrations (56, 57), thereby altering the calcium sensitivity. While P710R had an unchanged unloaded actin-detachment rate k0, the effect of its reduced force sensitivity δ on this mechanism of calcium sensitivity is unclear. To this end, we measured the sliding velocities of regulated thin filaments (RTFs) (actin decorated with troponin and tropomyosin) at various calcium concentrations in the motility assay. RTF velocities increased as calcium concentration increased through the physiological range (pCa7-6, or 100 nM to 1 μM), and velocities saturated at high calcium levels (pCa4) (Fig. 2C). We found that the P710R mutation did not significantly affect the calcium sensitivity; neither the calcium concentration for half-maximum velocity (P710R, pCa50 = 6.48 ± 0.07; WT, pCa50 = 6.46 ± 0.04; P = 0.78) nor the Hill coefficient (P710R, n = 3.1 ± 0.6; WT, n = 2.2 ± 0.2; P = 0.23) were significantly different from WT (results from five WT and four P710R experiments) (Table 1 and Fig. 2C).
Taken together, the above measurements of the myosin motor domain at the single-molecule and ensemble levels do not suggest a clear mechanism of hypercontractility by the HCM mutation P710R; in contrast, they would appear to suggest hypocontractility.
The P710R Mutation Disrupted the SRX State of Myosin In Vitro.
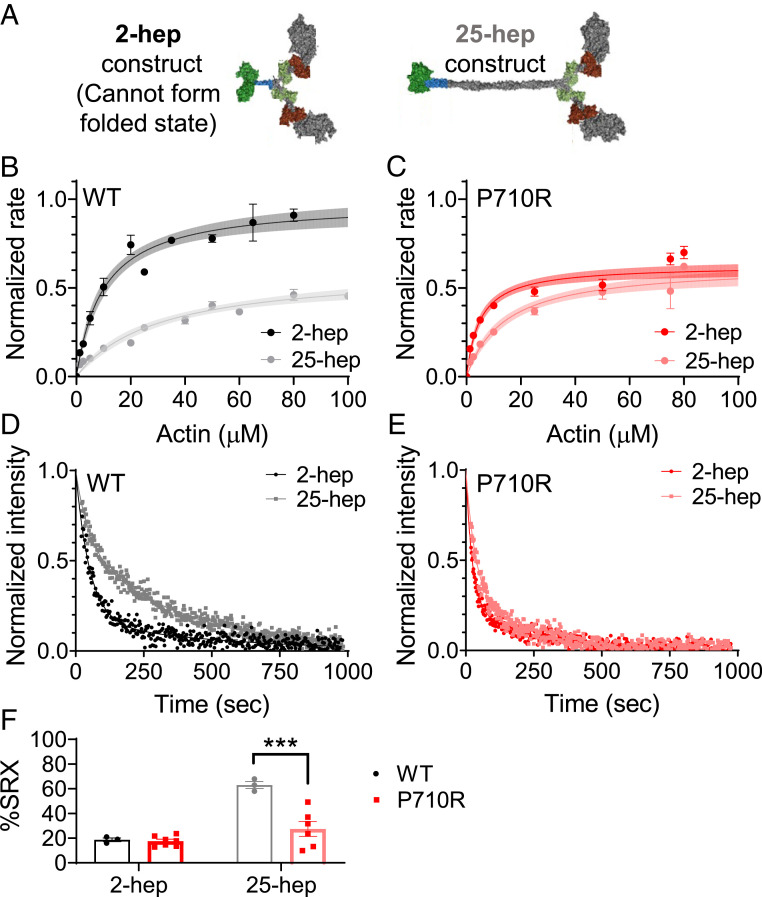
We and others have previously shown that the ability of myosin to form the folded-back interacting-heads motif structure is critical for regulating myosin activity, and many HCM-causing mutations appear to disrupt the ability of myosin to enter this state (17–20, 58). To assess the ability of myosin to form this folded-back “off” state in vitro, we used two-headed myosin constructs as previously described (20). We measured a significant 37% reduction in the actin-activated ATPase rate (kcat) of a long-tailed (25-hep) WT myosin construct compared with a short-tailed (2-hep) WT construct (2-hep, kcat = 5.6 ± 0.3 s−1⋅head−1; 25-hep, kcat = 3.5 ± 0.2 s−1⋅head−1; P < 0.001) (results fit to data from four and two separate experiments, respectively, with three replicates per experiment) (Fig. 3 A and B), consistent with our previous observations (17, 19, 20). This difference can be attributed to the sequestration of a large fraction of the myosin in the folded-back state in the long-tailed population, thus preventing the heads from binding to actin and entering the ATPase cycle. In contrast, we found no significant difference between the ATPase rate of the P710R short- and long-tailed myosins (P710R 2 hep, kcat = 3.7 ± 0.3 s−1⋅head−1; P710R 25 hep, kcat = 3.6 ± 0.3 s−1⋅head−1; P = 0.76) (results fit to data from two experiments with three replicates each) (Fig. 3C). This suggests that P710R has a greatly reduced ability to access the sequestered folded-back state.
Fig. 3.
β-Cardiac myosin ensembles with P710R mutation had reduced SRX state. (A) Schematic of protein constructs show 2 head (S1) domains (with light chains) and a 2- or 25-heptad tail domain. (B and C) Ensemble measurement of actin-activated ATPase rate normalized to the kcat for WT 2-hep from preparations performed on the same day. Each plot shows summary data from two independent experiments with triplicate measurements, for a total of n = 6. See also Materials and Methods and SI Appendix, Fig. S3. Error bars represent SEM of measurements, and shading represents error of the fit. (D and E) Representative traces of mant-ATP single turnover in the short (2-hep)- and long (25-hep)-tailed protein constructs. (F) Quantification of the %SRX in WT and P710R myosins (n = 3 for WT, n = 7 for P710R 2-hep, and n = 6 for P710R 25-hep). Error bars represent SEM. WT data (D and F) was previously reported (17). ***P < 0.001. A, D–E: Adapted from ref. 17, which is licensed under CC BY 4.0.
We have previously correlated the folded-back structural state with the proportion of basal ATP turnover at the SRX rate (∼0.003 s−1), as opposed to the disordered relaxed state (DRX) rate (∼0.01–0.03 s−1) (18, 58). These rates are measured using a single mant-ATP turnover assay in the absence of actin, in which the release of fluorescent mant-ADP from myosin is measured over time. The signal shows a biexponential decay characterized by two rates: The faster rate corresponds to the DRX rate, and the slower rate corresponds to the SRX rate (Fig. 3D). Lower fractions of SRX correlate with a reduced ability to form the folded-back state [via EM imaging and ATPase (18)]. We have previously shown that the percentage of WT 25-hep myosin hydrolyzing ATP at the slower SRX rate was 55–65% (at a salt concentration of 5 mM KOAc), in contrast to WT 2-hep myosin, which shows 15–25% SRX (regardless of salt concentration) due to its inability to form the folded-back state (results from three experiments) (Fig. 3F) (17). Here, we found no significant difference in the percentage of molecules in the SRX state between WT and P710R 2-hep myosins (P = 0.12; Fig. 3F). In contrast to WT 25-hep, only 27 ± 6% of the P710R 25-hep myosin hydrolyzed ATP at the slow rate (results from seven experiments) (P = 0.0001 vs. WT 25-hep; Fig. 3F). The fast rate of P710R 2-hep myosin was higher than that of WT 2-hep myosin (P = 0.004; SI Appendix, Table S1), while the fast rate of P710R 25-hep myosin was not significantly different from that of WT 25-hep myosin. Consistent with our 25-hep ATPase findings, these single-turnover results confirm that the P710R mutation reduced myosin’s ability to form the folded-back SRX state.
To summarize our molecular studies, the P710R mutation significantly reduced the load sensitivity of the detachment rate from actin, step size, and unloaded motility, suggesting hypocontractility. However, the mutation also reduced the ability of myosin to form the SRX state, suggesting hypercontractility. Thus, to understand these competing effects, we next investigated the effects of this mutation in human cardiomyocytes.
The P710R Mutation Increased Traction Force Generation and Cell Size in Micropatterned hiPSC-CMs.
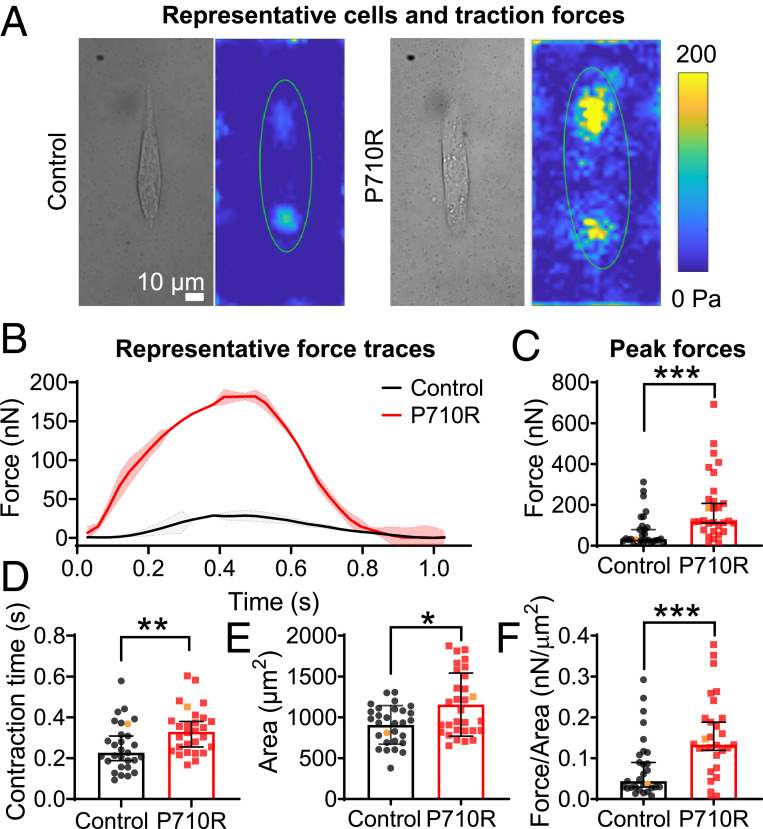
We used CRISPR/Cas9 gene editing to insert a point mutation of proline to arginine in one allele of the MYH7 gene (SI Appendix, Fig. S4). We first measured the effects of the P710R mutation on cellular force generation using TFM. Isolated hiPSC-CMs at 35–40 d after cardiac differentiation were cultured for 4–5 d on hydrogels of physiologic stiffness (10 kPa) with matrigel bioprinted in a mature cardiomyocyte aspect ratio of 7:1 (35, 37). We have previously shown that this platform optimizes force generation and dramatically enhances hiPSC-CM maturation (35). Patterned hiPSC-CMs with the P710R mutation produced significantly higher total peak force than WT controls (Fig. 4 A–C). Our single-cell platform allowed assessment of 29 WT and 30 P710R cells from three differentiation batches (Fig. 4 C–F) and provides spatially and temporally resolved measurements of traction force for each cell. Contraction duration—measured as the time between peak velocity of contraction and peak velocity of relaxation as previously described (37)—was significantly lengthened in the P710R cells (Fig. 4D). P710R cells were significantly larger than the WT (Fig. 4E), and the significant increase in force persisted after normalizing peak force to cell spread area (Fig. 4F).
Fig. 4.
P710R mutation in micropatterned single hiPSC-cardiomyocytes significantly increased contractile function. (A) Representative single cells shown in bright field and as peak traction force plots. (B) Multiple (two to four) beats were captured per cell and the averaged traces for representative cells are shown, with shading representing the SD of force between beats. (C) Peak total force and (D) contraction time of single control (n = 29) and P710R (n = 30) cells collected from three differentiation batches. (E) Cell spread areas were measured and used to calculate (F) cell force normalized to cell area. Representative cells are identified with orange markers in plots of population. Data are presented as median ± 95% confidence interval. *P < 0.05, **P < 0.01, and ***P < 0.001.
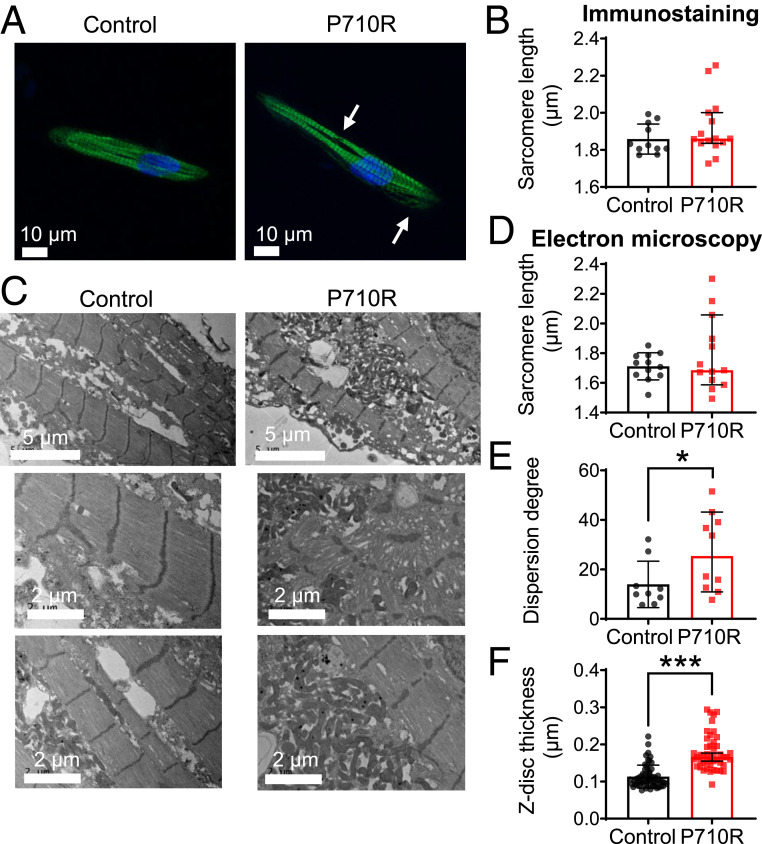
The P710R Mutation Caused Myofibril Disruption and z-Disk Thickening in Patterned hiPSC-CMs.
The ability of myosin to generate contractile forces in vivo depends on its incorporation into sarcomeres and myofibrils. Staining for β-MHC revealed organization into sarcomeres in cells grown on patterned hydrogels (Fig. 5A). Sarcomere lengths were not significantly different between control and mutant cells when quantified from immunostained cells (n = 11 control and n = 15 P710R) (Fig. 5B). We confirmed this result using transmission electron microscopy (TEM) (Fig. 5 C and D). With both imaging techniques, while the median values were not significantly different, there was a significant increase in the range and variability (determined by F test) of sarcomere lengths in P710R cells compared with controls, consistent with our finding of myofiber disarray. TEM of patterned cells showed aligned myofibrils in the WT controls and varying degrees of myofibril and sarcomeric disruption in many P710R cells. The degree of myofiber disarray was quantified using the directionality tool in ImageJ, and the degree of dispersion of directionality across the image was significantly increased in P710R cells (n = 12 control and n = 13 P710R regions of interest) (Fig. 5E). We also observed significant thickening (measured along fiber direction) of the z disks in P710R cells (Fig. 5F). This cytoskeletal disruption may be an indication of cellular remodeling that ultimately leads to hypertrophy.
Fig. 5.
P710R mutation in micropatterned hiPSC-cardiomyocytes significantly disrupted myofibril organization. (A) Immunostaining of β-cardiac myosin in micropatterned cells (sarcomere disruption is marked with arrows) enabled (B) quantification of sarcomere length in micropatterned cells (n = 11 control and n = 15 P710R cells). (C) Representative TEM images of patterned cells (n = 12 control and n = 13 P710R images) enabled (D) quantification of sarcomere length, (E) dispersion degree, a measure of myofibril alignment, and (F) z-disk thickness from TEM images (57 control and 46 P710R z disks). Data are presented as median ± 95% confidence interval. *P < 0.05.
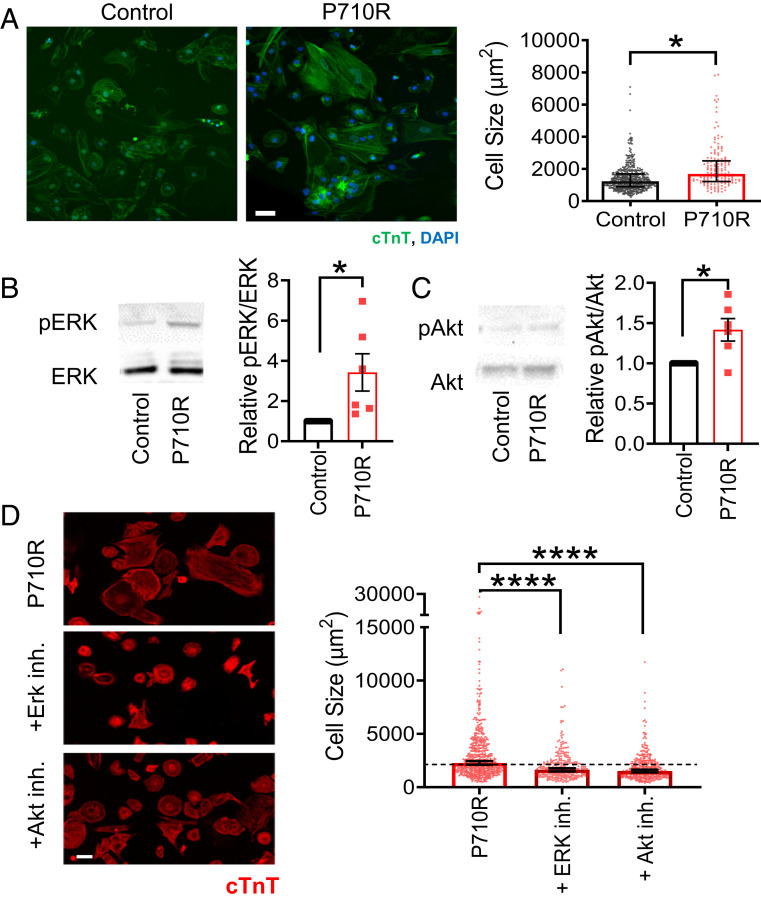
The P710R Mutation Caused Hypertrophic Growth of hiPSC-CMs via Akt and ERK.
We next tested whether the P710R mutation increases cell size across the broader population of cells (and not only in paceable, patterned cells). After 45 d of culture in confluent monolayers, hiPSC-CMs were replated sparsely onto Matrigel-coated tissue culture plastic, allowed to attach and spread for 2 d, and then fixed and stained to quantify their spread area (n = 474 control cells and n = 151 P710R cells). P710R cells were significantly larger than control cells (Fig. 6A). Western blot analysis of two known hypertrophic growth signaling pathways showed increased ERK and Akt basal activation (Fig. 6 B and C) in P710R cells (n = 6 differentiation batches). Inhibition of these signaling proteins with previously characterized specific inhibitors (SCH772984 and Akti 1/2) (59, 60) between days 26 and 46 resulted in significantly reduced cell area in P710R cells relative to untreated P710R cells (n = 538, 294, and 370 P710R cells, no treatment, ERK inhibitor, and Akt inhibitor, respectively) (Fig. 6D). However, treatment of P710R cells with either inhibitor did not fully reduce their area to the size of the corresponding inhibitor-treated unedited control cells (SI Appendix, Fig. S5).
Fig. 6.
P710R mutation in hiPSC-cardiomyocytes significantly increases cell size and activation of proteins involved in hypertrophic signaling. (A) Size of unpatterned cells was quantified from immunostaining for cardiac troponin T (cTnT) (n = 474 control cells and n = 151 P710R cells). (B and C) Western blots and densitometry analysis of phosphorylation of the hypertrophic signaling pathway proteins ERK (B) and Akt (C) in unpatterned cells with each P710R sample normalized to matched isogenic control. Data are presented as mean ± SD (n = 6 differentiation batches). (D) Quantification of cell size after treatment of P710R cells with specific inhibitors of ERK and Akt (n = 538, 294, and 370 P710R cells). (Scale bar: 50 μm.) Graphs of cell area are presented as median ± 95% confidence interval. *P < 0.05, and ***P < 0.001.
We also quantified the relative expression of β-MHC and α-MHC in P710R cells compared with isogenic control cells with both Western blot and qPCR (SI Appendix, Fig. S6 B–D). We found no significant difference in β-MHC protein or transcript levels between P710R and control cells, while α-MHC protein and transcript levels were significantly reduced in P710R cells compared with control. However, the relative transcript levels of α-MHC to β-MHC was less than 0.1, suggesting that the majority of myosin heavy chain in both isogenic control and P710R cells was β-MHC.
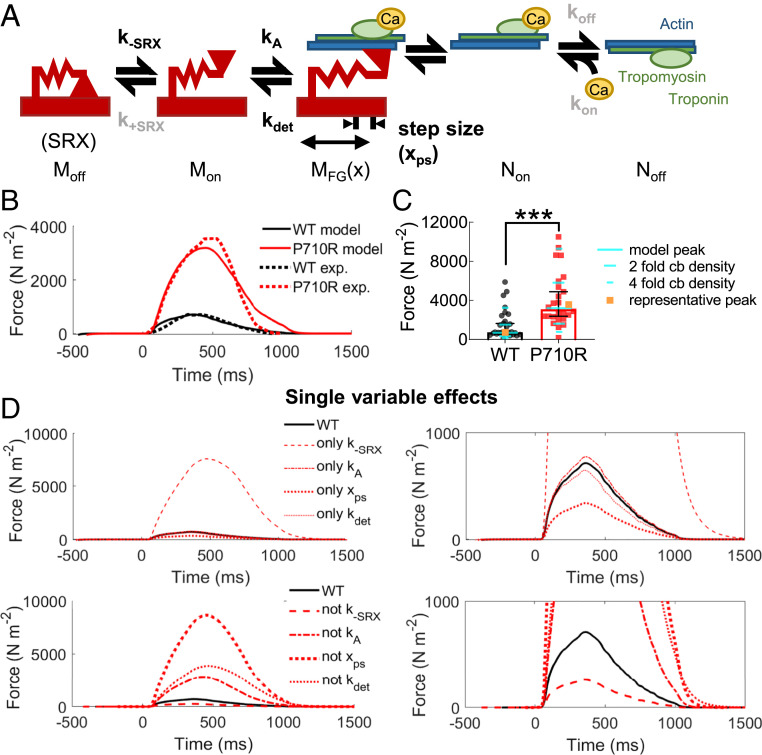
Computational Modeling of P710R Myosin Compared with WT Predicted an Increase in Force When SRX Disruption Was Included.
To determine the predicted effects of individual and combined changes in myosin kinetics on total force production, we used a previously validated computational model of cardiac muscle contraction (50), modified to contain an exponential load-dependent detachment rate (Fig. 7A and SI Appendix, Fig. S7). This model includes an OFF state to represent the SRX state of myosin, an ON state of unbound, available myosin, and a single bound, force generating state. Actin availability is regulated by calcium availability and the fraction bound to myosin and does not include specific states for blocked and free binding sites. This kinetic scheme was chosen for the sake of model parsimony and correspondence with both previously published work and our experimental measurements, but it limits the insights this analysis can provide into details of bound myosin transitions and thin filament regulation. We are also assuming a pure population of either WT or P710R myosin, whereas the genotype of our P710R cells is heterozygous, and the cells likely express a mixture of WT and P710R myosin, limiting the assessment of cooperative effects of heterozygous myosins. We measured calcium transients in monolayers of isogenic control and P710R cells when paced at 1 Hz (SI Appendix, Fig. S8), scaled the transients to the range of calcium concentration previously published for this model, and used the scaled transients as inputs for the calculations (61) (SI Appendix, Fig. S9A). We incorporated in this model the measured unloaded detachment rate (k0), its load sensitivity (δ), and step size (xcb). We defined the rate of myosin attachment (kA) from the previous modeling analysis that used our steady-state actin-activated ATPase measurements for sS1 (29) (see Materials and Methods for more details). We assumed that all other parameters would be unchanged between WT and P710R cells.
Fig. 7.
Modeling predicted that P710R-induced modulation of SRX/DRX transitions is essential for hypercontractility. (A) Model schematic with adaptation of detachment rate to incorporate an exponential dependence on load. Moff, Mon, and MFG(x) represent the different states of myosin. Non and Noff represent available and deactivated thin filaments, respectively. Parameters estimated or fit from in vitro experiments are shown in black, while rates held constant between groups are shown in gray. The model predicted (B) contractile force (solid line), which was fit to force traces from representative cells (dashed lines) normalized to cell cross-sectional area. (C) The predicted peak force (teal) fell close to the median measured force in our cell population when normalized to cross-sectional area (n = 29 WT and n = 30 P710R), and twofold to fourfold changes to cross-bridge (cb) density relatively match the measured distribution of data. Representative cells highlighted in orange. (D) Simulations of independent effects of the four parameters (by inclusion with WT parameters and replacing from the best-fit mutant parameters). Zoomed-in plots are included to improve visualization of low-force traces.
We did not directly measure the rates of transitioning into and out of the SRX state, but we did measure significant differences in the percentage of myosins in the SRX state between WT and P710R in the presence or absence of actin (Fig. 3). We assumed that the % SRX of the control cells at baseline would be no more than 90% based on some experimental measurements of SRX regulation in cells and filaments (18, 62, 63), and we defined the rate of opening from the OFF state (k−SRX) for control cells to be 20 s−1. After identifying a set of parameters that successfully predicted the measured force output of a representative control cell (normalized to cross-sectional area), we found a best fit for the relative increase in k−SRX to fit the force output of a representative P710R cell (Materials and Methods and SI Appendix, Fig. S9B). This process determined that a 12.9-fold (confidence limits 10.8- to 14.6-fold) increase in k−SRX provided the optimal fit to the representative data. An increase in k−SRX is consistent with the decrease in functional SRX state we measured experimentally, and the model predicted SRX% to be lower for the P710R cell at both contracted and relaxed periods of the cardiac cycle (SI Appendix, Fig. S9F). The rate of entering the SRX state (k+SRX) was set to 200 s−1 for both groups based on published data and previous modeling (50, 64). All other parameters were held constant between control and P710R, including the rates of thin filament activation (kon), inactivation (koff), and the cooperativity coefficient (kcoop). The changes to myosin kinetics (k−SRX, k0, δ, and step size) were sufficient to increase the percentage of thin filament activation and binding over the time course of contraction (SI Appendix, Fig. S9C).
The force predicted by the model after fitting parameters (SI Appendix, Table S2) agreed well with the representative force traces (Fig. 7B) and with the median force normalized to cross-sectional area of the population of cells (Fig. 7C). Sensitivity analysis (SI Appendix, Fig. S10) suggested that fourfold changes in cross-bridge density up and down spans the range of 90% of the cell forces we measured and matches the distribution of cell data. This parameter incorporates both the number of myosin heads per myofibril (which may vary with cell maturity and myofibril disruption) and the density of myofibrils in a cell (which we observed to be variable in these cells; SI Appendix, Fig. S11). Further simulations using the best fit parameter set and independently adding or removing each of the four measured parameters revealed that modulation of the rate of exiting the SRX state accounts for the majority of the measured increase in force (Fig. 7D). An important limitation of this analysis is that it does not distinguish between the possibility that the basal rate of opening from the SRX state is increased, as opposed to an equivalent reduction in the rate of returning to the SRX state (SI Appendix, Fig. S9). Both our sensitivity analysis (SI Appendix, Fig. S10) and additional fitting suggest (SI Appendix, Fig. S9B) that the effects of changing k−SRX and k+SRX are nearly mirrored. Our results support the hypothesis that a change in the equilibrium between the SRX state and actively cycling states is essential for the P710R mutation to produce significant hypercontractility.
In summary, our cellular experiments confirm a hypercontractile and hypertrophic phenotype in hiPSC-CMs harboring the P710R mutation. Furthermore, our modeling results suggest that the measured molecular effects of P710R are consistent with hypercontractility if and only if SRX regulation is included.
Discussion
The diversity of molecular alterations associated with HCM have made predictions of disease penetrance, severity, and response to therapeutics challenging. Our study demonstrates the power of an integrated, multiscale approach to study the biomechanical effects of mutations in myosin known to cause HCM. The P710R mutation was previously reported to have reduced ATPase activity and motility velocity (29), suggesting in isolation that it should reduce duty ratio and cause hypocontractility at the cellular level. In the present work, integrating molecular measurements of single molecules and ensembles of single and two-head constructs provides a more complete picture of the molecular mechanisms that contribute to the hypercontractility observed in single hiPSC-CMs with this mutation. By combining both a cellular model system and computational modeling, we were able to reveal the hypercontractile effect of this mutation, demonstrate additional HCM-associated phenotypes, and provide key insights into the driving factors by which HCM mutations can result in hypercontractility.
Single molecules of β-cardiac myosin with the P710R mutation have reduced step size d, preserved actin detachment rate under zero load k0, and reduced load sensitivity δ of the actin detachment rate (Fig. 1). This combination of findings suggests a possible molecular mechanism described as follows. In WT myosin, the converter domain, where P710 resides, couples the swing and forces at the lever arm to the conformational changes and forces at the nucleotide pocket. Through this coupling, load on the lever arm affects the rate of release of ADP from the pocket, which determines the actin detachment rate kdet. In P710R myosin, the proline-to-arginine substitution may disrupt the local structure of the converter domain, which would result in a reduction in the described coupling. Consequently, the rate of ADP release (the actin detachment rate, kdet) would be less dependent on load force (smaller δ) because load placed on the lever arm would no longer be fully conveyed to the nucleotide pocket. Similarly, the step size d would be reduced because the conformational change at the pocket would no longer be fully conveyed to the lever arm. The unloaded ADP release rate (unloaded actin detachment rate k0) would not be affected by the mutation presumably because the local structure of the nucleotide pocket would remain intact. Due to inherent variability among individual molecules, as evident in our single-molecule data (Fig. 1), the extent of disruption by P710R may vary. Thus, P710R may cause a small percentage (on the order of 1%) of myosin molecules to be fully dysfunctional in moving actin (“deadheads”), resulting in the observed higher numbers of immobile actin filaments in the motility assay (SI Appendix, Fig. S2; Materials and Methods).
We have previously observed reductions in the load sensitivity of myosin’s actin detachment rate by cardiac myosin effectors but not by cardiomyopathy causing mutations (14). For example, the investigational heart failure drug omecamtiv mecarbil (OM) dramatically reduces the detachment rate, its load sensitivity (14), and myosin’s step size (53). As the effects of OM and P710R share some similarities, it is important to note that residue P710 is part of the OM binding pocket in the converter domain (65). A recent preprint reported a reduction in myosin’s step size due to a different HCM mutation in the same region (P712L) (66). Taken together, these results emphasize the converter domain’s critical facilitation of myosin’s power stroke and load sensitivity and reveal various consequences of perturbing this domain.
A major finding of this study is that the disruption of the folded, SRX state is a critical driver of hypercontractility with the P710R mutation. The results of the actin-activated ATPase and single-turnover experiments using the longer tailed 25-hep construct (Fig. 3) demonstrate that a significantly larger fraction of myosins with this mutation are in an open, more active state than WT control. This increases the number of heads available for binding to actin, which we hypothesized would increase force production. We have measured similar changes in SRX across a range of HCM-causing mutations (17, 19), and others have confirmed these effects in cellular and animal models (18, 62), suggesting that SRX regulation may be a major cause of hypercontractility in HCM. This is further corroborated by the finding that mavacamten, a small-molecule inhibitor of myosin that is known to increase the SRX state (18), is effective in increasing exercise performance, reducing ejection fraction and LV mass index, and reducing symptoms in adult patients with obstructive HCM (67). Treatment with mavacamten has also been shown to reduce sarcomere shortening percentage, prevent cellular hypertrophy, and increase SRX% in hiPSC-CMs with three other HCM mutations in β-myosin (62). Our computational modeling analysis also supports the importance of destabilization of the SRX for increasing net force even when combined with a hypocontractile change like decreased step size.
Interestingly, in contrast with previous mutations implicated in the folded SRX state, the P710R mutation is buried inside the converter domain and is thus unlikely to be directly involved in interactions required to form the folded state. This suggests that, instead, it may result in allosteric changes that indirectly reduce myosin’s ability to form the folded SRX state. While it is impossible to parse exactly how such allostery might operate without a high-resolution structure of human cardiac myosin in the folded SRX state, we speculate based on models (20, 68, 69) that perhaps the converter domain requires a uniquely bent conformation to access the folded state, particularly in the “blocked” head, which might be precluded when the P710R mutation is present. Alternatively, it has previously been suggested that the converter domain of the “free” head is a docking site for the “blocked” head (16); this surface interaction could also be disrupted allosterically by the P710R mutation in the internal region of the converter domain. Ultimately, these highly speculative hypotheses can only be resolved through a high-resolution structure of human β-cardiac myosin in the folded SRX state. Regardless, this finding importantly demonstrates that even mutations distal from predicted head–tail and head–head interaction sites in the folded state may result in reduced SRX.
We confirmed that the P710R mutation causes hypercontractility and other key features of HCM in cardiomyocytes using a cellular experimental model in parallel with molecular level experiments. Using a micropatterned culture system also allowed for measurement of cellular-scale forces that are enhanced by the alignment of myofibrils and balanced by a physiologic-range substrate stiffness (10 kPa). This system has also been used to quantify hypercontractility in hiPSC-CMs with HCM-causing troponin mutations (70). The force per cross-sectional area we measured in the cells was significantly lower than what has been reported for mature cardiac tissue, but this is a known sign of the relative immaturity of hiPSC-derived CMs (71). Changes in myofibril force production between stem cell-derived and mature tissue has been shown to correspond with expression of different light chain and troponin and tropomyosin isoforms (71, 72). The presence of α-myosin in a fraction of these cells also likely reflects relative immaturity and contributes to the heterogeneity of force production observed in these cells. Despite this limitation, cells with the P710R mutation produce significantly greater force even after normalizing for the increase in cell area (Fig. 4). There was significant variability in force production within the cell populations, which may be due in part to differences in myofibril density between cells, as suggested by our modeling analysis. The P710R mutation caused significant perturbation in alignment of myofibrils in the mutant cells, a well-described phenotype in HCM (73), as well as an increase in the thickness of the z disk (Fig. 5). Increased z-disk thickness has been described in many cardiomyopathies (74–76), which suggests that z-disk thickening is a conserved phenotype associated with hypercontractility and hypertrophic remodeling. Finally, we observed cellular hypertrophy, which was mediated in part by Akt and ERK. ERK activation has been specifically linked to concentric hypertrophy in response to an increase in force index (force production integrated over time of contraction) (77). By comparing cellular responses with an isogenic control, we have more confidence that the altered biomechanical consequences of the P710R mutation are driving both hypercontractility and additional cellular dysfunction.
The P710R mutation induces a number of profound changes to myosin activity whose effects become easier to interpret when integrated into a computational model. Even with similar driving calcium transients and identical rates of thin filament activation by calcium, the model simulations predict an increase in thin filament activation and a lengthening of contraction time resulting solely from the changes in myosin kinetics (specifically the change in regulation of SRX). In simulations where only step size, attachment and detachment rates were changed (as is true for the sS1 construct), there were minimal effects on actin availability (±15% change), which matches the observed similarity in pCa50 in sS1 motility with or without the mutation. When the disruption of SRX caused by the mutation is included, the predicted percentage of actin kept in the active state by myosin increases by 8- to 10-fold. This large magnitude of increase is likely due in part to the low percentage of active actin found when fitting the control cell forces. We used a simplified model framework with only one attached state for this initial analysis of mechanisms of force generation, but extension of this work could include additional myosin and thin filament states that may more accurately capture thin filament dynamics. The model used in this study, while by no means comprehensive, provides valuable insights into the expected individual and combined effects of changes to myosin kinetics and biomechanical function on force production, actin activation, and potential sources of variability.
Several important limitations of this project should be addressed. We measured the SRX state using shortened constructs that do not contain the full thick filament backbone and thus likely underestimate the additional stabilization of the SRX in thick filaments in vivo. Hypertrophy measurements and assessment of protein activation were performed on unpatterned cells grown on tissue culture plastic or glass because it would be difficult to quantify phosphorylation in small populations of cells recovered from patterned gel platforms. This modeling work shows the singular effects of a mutation, but does not fully capture potential synergistic or emergent consequences of a mixture of control and mutant myosins acting together. Furthermore, this modeling approach assumes that muscle force is felt uniformly throughout the muscle, while in reality, the spatial distribution of myosins within a sarcomere can have profound effects on the force experienced by individual myosin heads. Future modeling could incorporate spatially explicit information that could provide even more detail on the effects of local loads and heterotypic myosin effects.
In summary, the P710R mutation reduces several functional parameters of the isolated myosin molecule; however, dysregulation of the SRX state appears to be the major driver of hypercontractility. When expressed as a heterozygous mutation in human stem cell-derived cardiomyocytes, this mutation causes hypercontractility, myofibril disruption, and hypertrophy mediated in part through ERK and Akt. This project also demonstrates the value of computational modeling integrated with multiscale measurements to give insights into the interactions of load-sensitive, functional parameters and provides a platform to study other mutations.
Materials and Methods
Molecular Measurements of Myosin Function in Transgenic Human Proteins with P710R Mutation.
Recombinant human β-cardiac myosin protein constructs (short subfragment 1 [sS1], short-tailed [2-hep], and long-tailed [25-hep]) were expressed in C2C12 mouse myoblast cells and purified as previously described (13, 20). The load-dependent detachment rates of WT and P710R sS1-eGFP molecules were measured in a dual-beam optical trap using the HFS method previously described (14, 51, 78) with slight modifications. The step sizes of myosins were determined from the same HFS data by adapting the ensemble averaging method (79) to HFS’s oscillatory data. For each molecule, position traces of all events were aligned at the start of binding, extended to the length of the longest event, and averaged. Then the oscillations were removed by subtracting a fitted sine function. The total step size for each molecule was taken as the difference between the initial position and the end position of the extended, averaged, sine-subtracted traces. Motility measurements of WT and P710R sS1-AC were performed using our previously described motility assay (32, 80) with some modifications. Motility measurements with regulated thin filaments were performed using recombinant human troponin complex and bovine cardiac tropomyosin (81). NADH-coupled ATPases were used to compare the activity of the 2-hep and 25-hep constructs of WT and P710R β-cardiac myosin as previously described (82). Single-turnover experiments were performed as previously described (17, 18), using WT and mutant versions of human β-cardiac 2-hep and 25-hep. Additional details are included in the SI Appendix.
Measurements of Functional Consequences of P710R Mutation in Gene Edited hiPSC-CMs.
The control human iPSC line (Stanford Cardiovascular Institute [SCVI-113]) was obtained from the Stanford CVI iPSC Biobank. The P710R mutation was edited into these cells using a method that has been described previously (83, 84). Cells were differentiated and their contractile function was assessed on micropatterned hydrogels gels as previously described (35, 37). Cells on micropatterened gels were fixed and stained for β-cardiac myosin and their sarcomere length quantifed. Cells were also plated onto micropatterned film and imaged with TEM. Unpatterned cells were stained for cardiac troponin T to measure cell area. To clarify the effect of ERK and Akt, the cells were treated with either Akt inhibitor (Akti 1/2; Tocris; 5 µM) or and ERK inhibitor (SCH772984; SelleckChem; 1 µM) every 3 d between day 26 and day 46 (a time frame over which hiPSC-CMs have been shown to increase in area). Additional details are included in the SI Appendix.
Implementation of Computational Model.
We used a previously validated continuum model that incorporates discretized flux equations to describe transitions of myosin heads and thin filaments from inactive to active/bound states and provides prediction of active and passive force (50, 85). The rate of myosin detachment from actin was adapted to match the measured exponential load dependence, and the relative change in k-SRX was fit using experimental force measurements from representative hiPSC-CMs. Additional details about model implementation are included in the SI Appendix.
Supplementary Material
Acknowledgments
This work was funded by NIH Grants RM1GM131981 to J.A.S., D.B., and B.L.P.; 1R21HL13099301 to D.B. and B.L.P.; HL117138 and GM033289 to J.A.S.; HL123655 to D.B.; and HL133359 and HL149164 to K.S.C. The microscope was funded by NIH Grant S10RR026775 to J.A.S. This work was funded by the Stanford Translational and Clinical Innovation Award (to D.B. and J.A.S.). A.S.V.R. and K.B.K. were supported by NIH Grant T32 HL094274. The study was also funded by American Heart Association Grant 17CSA33590101 to B.L.P. and D.B. and a American Heart Association Postdoctoral Fellowship (20POST35211011) to A.S.V.R. M.M.M. was supported by Stanford Cellular and Molecular Biology Training Grant T32GM007276. G.P. was supported by the Swiss National Science Foundation, the Early Postdoc Mobility Fellowship (P2SKP2_164954), the Postdoc Mobility Fellowship (P400PM_180825), and the American Heart Association Postdoc Fellowship (American Heart Association Award 18POST34080160). The hiPSC line(s) were obtained from Joseph C. Wu, MD, PhD, at the Stanford Cardiovascular Institute funded by NIH Grant 75N92020D00019. We acknowledge John Perrino and the electron microscopy imaging core for their help with fixing, sectioning, and imaging of the cell samples; Alexandre Ribeiro for his initial help with micropatterning experiments; Arjun Adhikari for his preliminary work on the molecular data; a generous gift of an α-myosin antibody from Theresia Kraft; and Oscar Abilez for his help with imaging calcium transients.
Footnotes
Competing interest statement: J.A.S. is cofounder and J.A.S. and D.B. are on the Scientific Advisory Board of Cytokinetics, Inc., a company developing small molecule therapeutics for treatment of hypertrophic cardiomyopathy.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025030118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. Previously published data were used for this work (17).
References
- 1.Virani S. S.et al.; American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee , Heart disease and stroke statistics-2020 update: A report from the American Heart Association. Circulation 141, e139–e596 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Semsarian C., Ingles J., Maron M. S., Maron B. J., New perspectives on the prevalence of hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 65, 1249–1254 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Jordà P., García-Álvarez A., Hypertrophic cardiomyopathy: Sudden cardiac death risk stratification in adults. Glob. Cardiol. Sci. Pract. 2018, 25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marian A. J., Braunwald E., Hypertrophic cardiomyopathy: Genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circ. Res. 121, 749–770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arad M., Seidman J. G., Seidman C. E., Phenotypic diversity in hypertrophic cardiomyopathy. Hum. Mol. Genet. 11, 2499–2506 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Ho J. E., et al., Biomarkers of cardiovascular stress and fibrosis in preclinical hypertrophic cardiomyopathy. Open Heart 4, e000615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho C. Y., et al., Myocardial fibrosis as an early manifestation of hypertrophic cardiomyopathy. N. Engl. J. Med. 363, 552–563 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho C. Y., et al., Genotype and lifetime burden of disease in hypertrophic cardiomyopathy: Insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation 138, 1387–1398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho C. Y., et al., Assessment of diastolic function with Doppler tissue imaging to predict genotype in preclinical hypertrophic cardiomyopathy. Circulation 105, 2992–2997 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Spudich J. A., Hypertrophic and dilated cardiomyopathy: Four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys. J. 106, 1236–1249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooijman P., Stewart M. A., Cooke R., A new state of cardiac myosin with very slow ATP turnover: A potential cardioprotective mechanism in the heart. Biophys. J. 100, 1969–1976 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nag S., et al., Contractility parameters of human β-cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of motor function. Sci. Adv. 1, e1500511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sommese R. F., et al., Molecular consequences of the R453C hypertrophic cardiomyopathy mutation on human β-cardiac myosin motor function. Proc. Natl. Acad. Sci. U.S.A. 110, 12607–12612 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C., Kawana M., Song D., Ruppel K. M., Spudich J. A., Controlling load-dependent kinetics of β-cardiac myosin at the single-molecule level. Nat. Struct. Mol. Biol. 25, 505–514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adhikari A. S., et al., Early-onset hypertrophic cardiomyopathy mutations significantly increase the velocity, force, and actin-activated ATPase activity of human β-cardiac myosin. Cell Rep. 17, 2857–2864 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawana M., Sarkar S. S., Sutton S., Ruppel K. M., Spudich J. A., Biophysical properties of human β-cardiac myosin with converter mutations that cause hypertrophic cardiomyopathy. Sci. Adv. 3, e1601959 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adhikari A. S., et al., β-cardiac myosin hypertrophic cardiomyopathy mutations release sequestered heads and increase enzymatic activity. Nat. Commun. 10, 2685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson R. L., et al., Deciphering the super relaxed state of human β-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers. Proc. Natl. Acad. Sci. U.S.A. 115, E8143–E8152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarkar S. S., et al., The hypertrophic cardiomyopathy mutations R403Q and R663H increase the number of myosin heads available to interact with actin. Sci. Adv. 6, eaax0069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nag S., et al., The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat. Struct. Mol. Biol. 24, 525–533 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Homburger J. R., et al., Multidimensional structure-function relationships in human β-cardiac myosin from population-scale genetic variation. Proc. Natl. Acad. Sci. U.S.A. 113, 6701–6706 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore J. R., Leinwand L., Warshaw D. M., Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ. Res. 111, 375–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buvoli M., Hamady M., Leinwand L. A., Knight R., Bioinformatics assessment of beta-myosin mutations reveals myosin’s high sensitivity to mutations. Trends Cardiovasc. Med. 18, 141–149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rayment I., Holden H. M., Sellers J. R., Fananapazir L., Epstein N. D., Structural interpretation of the mutations in the beta-cardiac myosin that have been implicated in familial hypertrophic cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 92, 3864–3868 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landrum M. J., et al., ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kindel S. J., et al., Pediatric cardiomyopathy: Importance of genetic and metabolic evaluation. J. Card. Fail. 18, 396–403 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaski J. P., et al., Prevalence of sarcomere protein gene mutations in preadolescent children with hypertrophic cardiomyopathy. Circ. Cardiovasc. Genet. 2, 436–441 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Sedaghat-Hamedani F., et al., Clinical outcomes associated with sarcomere mutations in hypertrophic cardiomyopathy: A meta-analysis on 7675 individuals. Clin. Res. Cardiol. 107, 30–41 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Vera C. D., et al., Myosin motor domains carrying mutations implicated in early or late onset hypertrophic cardiomyopathy have similar properties. J. Biol. Chem. 294, 17451–17462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milani-Nejad N., Janssen P. M., Small and large animal models in cardiac contraction research: Advantages and disadvantages. Pharmacol. Ther. 141, 235–249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroer A., Pardon G., Castillo E., Blair C., Pruitt B., Engineering hiPSC cardiomyocyte in vitro model systems for functional and structural assessment. Prog. Biophys. Mol. Biol. 144, 3–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aksel T., Choe Yu E., Sutton S., Ruppel K. M., Spudich J. A., Ensemble force changes that result from human cardiac myosin mutations and a small-molecule effector. Cell Rep. 11, 910–920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X., Pabon L., Murry C. E., Engineering adolescence: Maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 114, 511–523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karakikes I., Termglinchan V., Wu J. C., Human-induced pluripotent stem cell models of inherited cardiomyopathies. Curr. Opin. Cardiol. 29, 214–219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ribeiro A. J., et al., Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proc. Natl. Acad. Sci. U.S.A. 112, 12705–12710 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung G., et al., Time-dependent evolution of functional vs. remodeling signaling in induced pluripotent stem cell-derived cardiomyocytes and induced maturation with biomechanical stimulation. FASEB J. 30, 1464–1479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ribeiro A. J. S., et al., Multi-imaging method to assay the contractile mechanical output of micropatterned human iPSC-derived cardiac myocytes. Circ. Res. 120, 1572–1583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell K. S., Compliance accelerates relaxation in muscle by allowing myosin heads to move relative to actin. Biophys. J. 110, 661–668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell S. G., Hatfield P. C., Campbell K. S., A mathematical model of muscle containing heterogeneous half-sarcomeres exhibits residual force enhancement. PLoS Comput. Biol. 7, e1002156 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powers J. D., Williams C. D., Regnier M., Daniel T. L., A spatially explicit model shows how titin stiffness modulates muscle mechanics and energetics. Integr. Comp. Biol. 58, 186–193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanner B. C., Daniel T. L., Regnier M., Filament compliance influences cooperative activation of thin filaments and the dynamics of force production in skeletal muscle. PLoS Comput. Biol. 8, e1002506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanner B. C., Daniel T. L., Regnier M., Sarcomere lattice geometry influences cooperative myosin binding in muscle. PLoS Comput. Biol. 3, e115 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams C. D., Regnier M., Daniel T. L., Elastic energy storage and radial forces in the myofilament lattice depend on sarcomere length. PLoS Comput. Biol. 8, e1002770 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams C. D., Regnier M., Daniel T. L., Axial and radial forces of cross-bridges depend on lattice spacing. PLoS Comput. Biol. 6, e1001018 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams C. D., Salcedo M. K., Irving T. C., Regnier M., Daniel T. L., The length-tension curve in muscle depends on lattice spacing. Proc. Biol. Sci. 280, 20130697 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mijailovich S. M., et al., Modeling the actin.myosin ATPase cross-bridge cycle for skeletal and cardiac muscle myosin isoforms. Biophys. J. 112, 984–996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKillop D. F., Geeves M. A., Regulation of the interaction between actin and myosin subfragment 1: Evidence for three states of the thin filament. Biophys. J. 65, 693–701 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aboelkassem Y., Bonilla J. A., McCabe K. J., Campbell S. G., Contributions of Ca2+-independent thin filament activation to cardiac muscle function. Biophys. J. 109, 2101–2112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barrick S. K., Clippinger S. R., Greenberg L., Greenberg M. J., Computational tool to study perturbations in muscle regulation and its application to heart disease. Biophys. J. 116, 2246–2252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campbell K. S., Janssen P. M. L., Campbell S. G., Force-dependent recruitment from the myosin off state contributes to length-dependent activation. Biophys. J. 115, 543–553 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sung J., et al., Harmonic force spectroscopy measures load-dependent kinetics of individual human β-cardiac myosin molecules. Nat. Commun. 6, 7931 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greenberg M. J., Shuman H., Ostap E. M., Inherent force-dependent properties of β-cardiac myosin contribute to the force-velocity relationship of cardiac muscle. Biophys. J. 107, L41–L44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woody M. S., et al., Positive cardiac inotrope omecamtiv mecarbil activates muscle despite suppressing the myosin working stroke. Nat. Commun. 9, 3838 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uyeda T. Q., Kron S. J., Spudich J. A., Myosin step size. Estimation from slow sliding movement of actin over low densities of heavy meromyosin. J. Mol. Biol. 214, 699–710 (1990). [DOI] [PubMed] [Google Scholar]
- 55.Brizendine R. K., et al., A mixed-kinetic model describes unloaded velocities of smooth, skeletal, and cardiac muscle myosin filaments in vitro. Sci. Adv. 3, eaao2267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bremel R. D., Weber A., Cooperation within actin filament in vertebrate skeletal muscle. Nat. New Biol. 238, 97–101 (1972). [DOI] [PubMed] [Google Scholar]
- 57.Kad N. M., Kim S., Warshaw D. M., VanBuren P., Baker J. E., Single-myosin crossbridge interactions with actin filaments regulated by troponin-tropomyosin. Proc. Natl. Acad. Sci. U.S.A. 102, 16990–16995 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rohde J. A., Roopnarine O., Thomas D. D., Muretta J. M., Mavacamten stabilizes an autoinhibited state of two-headed cardiac myosin. Proc. Natl. Acad. Sci. U.S.A. 115, E7486–E7494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindsley C. W., et al., Allosteric Akt (PKB) inhibitors: Discovery and SAR of isozyme selective inhibitors. Bioorg. Med. Chem. Lett. 15, 761–764 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Morris E. J., et al., Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 3, 742–750 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Monasky M. M., Varian K. D., Davis J. P., Janssen P. M., Dissociation of force decline from calcium decline by preload in isolated rabbit myocardium. Pflugers Arch. 456, 267–276 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Toepfer C. N., et al., Myosin sequestration regulates sarcomere function, cardiomyocyte energetics, and metabolism, informing the pathogenesis of hypertrophic cardiomyopathy. Circulation 141, 828–842 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brunello E., et al., Myosin filament-based regulation of the dynamics of contraction in heart muscle. Proc. Natl. Acad. Sci. U.S.A. 117, 8177–8186 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fusi L., et al., Minimum number of myosin motors accounting for shortening velocity under zero load in skeletal muscle. J. Physiol. 595, 1127–1142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Planelles-Herrero V. J., Hartman J. J., Robert-Paganin J., Malik F. I., Houdusse A., Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil. Nat. Commun. 8, 190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Snoberger A.et al., Myosin with hypertrophic cardiac mutation R712L has a decreased working stroke which is rescued by omecamtiv mecarbil. eLife 10, e63691 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olivotto I.et al.; EXPLORER-HCM Study Investigators , Mavacamten for treatment of symptomatic obstructive hypertrophic cardiomyopathy (EXPLORER-HCM): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 396, 759–769 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Alamo L., et al., Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. eLife 6, e24634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robert-Paganin J., Auguin D., Houdusse A., Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition. Nat. Commun. 9, 4019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clippinger S. R., et al., Disrupted mechanobiology links the molecular and cellular phenotypes in familial dilated cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 116, 17831–17840 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pioner J. M., et al., Isolation and mechanical measurements of myofibrils from human induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Reports 6, 885–896 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iorga B., et al., Differences in contractile function of myofibrils within human embryonic stem cell-derived cardiomyocytes vs. adult ventricular myofibrils are related to distinct sarcomeric protein isoforms. Front. Physiol. 8, 1111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Machackova J., Barta J., Dhalla N. S., Myofibrillar remodeling in cardiac hypertrophy, heart failure and cardiomyopathies. Can. J. Cardiol. 22, 953–968 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Eldik W., et al., Z-disc protein CHAPb induces cardiomyopathy and contractile dysfunction in the postnatal heart. PLoS One 12, e0189139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luther P. K., Padrón R., Ritter S., Craig R., Squire J. M., Heterogeneity of Z-band structure within a single muscle sarcomere: Implications for sarcomere assembly. J. Mol. Biol. 332, 161–169 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Manalo A., et al., Loss of CENP-F results in dilated cardiomyopathy with severe disruption of cardiac myocyte architecture. Sci. Rep. 8, 7546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Davis J., et al., A tension-based model distinguishes hypertrophic versus dilated cardiomyopathy. Cell 165, 1147–1159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sung J., Mortensen K. I., Spudich J. A., Flyvbjerg H., How to measure load-dependent kinetics of individual motor molecules without a force-clamp. Methods Enzymol. 582, 1–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Veigel C., et al., The motor protein myosin-I produces its working stroke in two steps. Nature 398, 530–533 (1999). [DOI] [PubMed] [Google Scholar]
- 80.Kron S. J., Spudich J. A., Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. U.S.A. 83, 6272–6276 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sommese R. F., et al., Effects of troponin T cardiomyopathy mutations on the calcium sensitivity of the regulated thin filament and the actomyosin cross-bridge kinetics of human β-cardiac myosin. PLoS One 8, e83403 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De La Cruz E. M., Ostap E. M., Kinetic and equilibrium analysis of the myosin ATPase. Methods Enzymol. 455, 157–192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kodo K., et al., iPSC-derived cardiomyocytes reveal abnormal TGF-β signalling in left ventricular non-compaction cardiomyopathy. Nat. Cell Biol. 18, 1031–1042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang G., et al., Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 20, 616–623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mann C. K., Lee L. C., Campbell K. S., Wenk J. F., Force-dependent recruitment from myosin OFF-state increases end-systolic pressure-volume relationship in left ventricle. Biomech. Model. Mechanobiol. 19, 2683–2692 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. Previously published data were used for this work (17).