Abstract
Background
COVID-19 has been associated with Interstitial Lung Disease features. The immune transcriptomic overlap between Idiopathic Pulmonary Fibrosis (IPF) and COVID-19 has not been investigated.
Methods
we analyzed blood transcript levels of 50 genes known to predict IPF mortality in three COVID-19 and two IPF cohorts. The Scoring Algorithm of Molecular Subphenotypes (SAMS) was applied to distinguish high versus low-risk profiles in all cohorts. SAMS cutoffs derived from the COVID-19 Discovery cohort were used to predict intensive care unit (ICU) status, need for mechanical ventilation, and in-hospital mortality in the COVID-19 Validation cohort. A COVID-19 Single-cell RNA-sequencing cohort was used to identify the cellular sources of the 50-gene risk profiles. The same COVID-19 SAMS cutoffs were used to predict mortality in the IPF cohorts.
Findings
50-gene risk profiles discriminated severe from mild COVID-19 in the Discovery cohort (P = 0·015) and predicted ICU admission, need for mechanical ventilation, and in-hospital mortality (AUC: 0·77, 0·75, and 0·74, respectively, P < 0·001) in the COVID-19 Validation cohort. In COVID-19, 50-gene expressing cells with a high-risk profile included monocytes, dendritic cells, and neutrophils, while low-risk profile-expressing cells included CD4+, CD8+ T lymphocytes, IgG producing plasmablasts, B cells, NK, and gamma/delta T cells. Same COVID-19 SAMS cutoffs were also predictive of mortality in the University of Chicago (HR:5·26, 95%CI:1·81–15·27, P = 0·0013) and Imperial College of London (HR:4·31, 95%CI:1·81–10·23, P = 0·0016) IPF cohorts.
Interpretation
50-gene risk profiles in peripheral blood predict COVID-19 and IPF outcomes. The cellular sources of these gene expression changes suggest common innate and adaptive immune responses in both diseases.
Keywords: COVID-19, IPF, 50-gene risk profiles, Mortality, Monocytes, Dendritic Cells and Neutrophils
Research in context.
Evidence before this study
We searched the scientific literature using PubMed to identify studies that use gene expression in peripheral blood to identify outcome prediction in COVID-19 and Idiopathic Pulmonary Fibrosis (IPF). We used the search terms “COVID-19″, “gene expression”, “outcome prediction”, and “blood”, and identified 23 studies. When we added the term Idiopathic Pulmonary Fibrosis (IPF) we found no studies investigating this association.
Added value of this study
We have previously identified a transcriptomic signature predictive of IPF mortality in peripheral blood. In this work, we sought to determine whether genomic risk profiles based on 50 genes of this signature could be predictive of COVID-19 outcomes. A 50-gene, high-risk profile predicted ICU admission, need for mechanical ventilation and in-hospital mortality in COVID-19. 50-gene expressing cells with a high-risk profile in COVID-19 mainly included CD14+ monocytes, dendritic cells, and neutrophils while low-risk profile-expressing cells included CD4+, CD8+ T lymphocytes, IgG producing plasmablasts, B cells, NK and gamma/delta T cells
Implications of all the available evidence
The identification of 50-gene risk profiles in COVID-19, in addition to clinical variables, can facilitate healthcare utilization such as triage of patients to the most appropriate location, reduce hospital length-of-stay, and allow for proper allocation of limited resources. It may also allow the identification of patients that are more likely to respond to COVID-19 targeted therapies.
Alt-text: Unlabelled box
1. Introduction
The COVID-19 pandemic has so far caused more than three million deaths worldwide, mainly due to the development of acute respiratory distress syndrome (ARDS). While autopsy data from patients dying early on after ARDS development demonstrate diffuse alveolar damage, endothelial injury, thrombosis, and angiogenesis [1,2]; longer disease courses associate with features of Interstitial Lung Disease (ILD) including tissue remodeling, fibroblast proliferation, airspace obliteration, micro-honeycombing and extracellular matrix deposition [3,4]. Moreover, radiological surrogates of lung fibrosis, including sub-pleural reticulation and fibrotic streaks have also been described in COVID-19 [5]. While an association between COVID-19-induced ARDS and risk for ILD development has been recently suggested [6], no research has focused on immune gene expression profiles shared by COVID-19 and Idiopathic Pulmonary Fibrosis (IPF) patients. That characterization could provide pathophysiological insight to better understand the mechanisms regulating COVID-19-induced pulmonary injury and repair as well as facilitate the identification of molecular predictors of long-term lung damage, mortality, and other relevant outcomes in these patients. In this work, we hypothesized that a peripheral blood transcriptomic signature known to predict mortality in IPF [7,8] could also be associated with COVID-19 outcomes. To address that hypothesis, we analyzed transcriptomic data reported by multiple centers enrolling COVID-19 and IPF patients. Using a previously established bioinformatic pipeline [8] we found a remarkable overlap of an outcome-predicting signature demonstrated by both diseases, and data from single-cell RNA-sequencing (RNA-seq) analyses revealed the cell types accounting for the aforementioned signature in COVID-19.
2. Methods
2.1. Study design and subjects
In this retrospective, multicentre cohort study, we analyzed gene expression and clinical data from five independent cohorts: (1) COVID-19 Discovery cohort (N = 8 subjects). Peripheral Blood Mononuclear Cells (PBMC) were obtained twice from three of these subjects at two different time points during hospitalization. PBMC specimens from patients with COVID-19 were assigned to severe (N = 6) or mild (N = 5) disease groups according to the National Early Warning Score [9] (NEWS; mild < 5, severe ≥ 5) evaluated on the day of blood sampling [10] (PBMC, Single-cell RNA-seq data, GEO Accession: GSE14968910); (2) COVID-19 Validation cohort (N = 100 subjects, bulk leukocyte RNA-seq data, GEO Accession: GSE15710311). This study was designed to enroll all hospitalized patients older than 18 years of age with COVID-19 diagnosis who were not anticipated to die imminently (3) COVID-19 Single-cell cohort (N = 7 subjects, N = 155 single cells , single-cell RNA-seq data, GEO accession: GSE15072812); (4) IPF-University of Chicago cohort (N = 45, Bulk PBMC, Affymetrix Human Exon 1.0 ST RNA Array data, GEO Accession: GSE282217); 5) IPF-Imperial College London cohort (N = 55, Bulk whole blood, Affymetrix Human Gene 1·1 ST RNA Array data, GEO Accession: GSE9360613). Transcriptomics data collection from all cohorts have been previously described [7,10-13].
2.2. Ethics
As these are publicly available and de-identified datasets, no institutional review board's approvals were warranted.
2.3. Data extraction, pre-processing and statistical analysis
All analyses were performed in R software (version 4·0·2) [14]. For the COVID-19 Discovery cohort, we used the R package “Seurat” to pre-process the feature-barcode matrices of the single-cell RNA-seq data. Cells expressing less than 200 genes or more than 15% of mitochondrial genes of their total gene expression were excluded. Genes expressed in less than 10 cells were also excluded from the analysis. NormalizeData® function was used to normalize gene expression levels. The subject-level expression profile was estimated using the average expression level across all cells. For bulk RNA-seq data in the COVID-19 validation cohort, Transcripts Per Million (TPM) matrix was analyzed using log(1+TPM) to normalize gene expression levels. For the COVID-19 Single-cell cohort dataset, pre-processed and normalized data were provided directly according to the published report [10].
The Scoring Algorithm of Molecular Subphenotypes (SAMS) was used to identify genomic risk profiles as previously described [8]. SAMS, Up and Down scores were calculated in each cohort using the product of two variables: the proportion of genes expected to be increased or decreased per subject (or single-cells) and their median normalized expression levels. In this study, we calculated Up and Down scores based on the expression levels of seven increased genes (PLBD1, TPST1, MCEMP1, IL1R2, HP, FLT3, S100A12) and 43 decreased genes (LCK, CAMK2D, NUP43, SLAMF7, LRRC39, ICOS, CD47, LBH, SH2D1A, CNOT6L, METTL8, ETS1, P2RY10, TRAT1, BTN3A1, LARP4, TC2N, GPR183, MORC4, STAT4, LPAR6, CPED1, DOCK10, ARHGAP5, HLA-DPA1, BIRC3, GPR174, CD28, UTRN, CD2, HLA-DPB1, ARL4C, BTN3A3, CXCR6, DYNC2LI1, BTN3A2, ITK, CD96, GBP4, S1PR1, NAP1L2, KLF12, IL7R) from a gene signature previously found to be predictive of IPF mortality [7,8]. Two non-coding transcripts (SNHG1, C2orf27A) of the original gene signature were excluded because they were not consistently present across COVID-19 datasets. The Scoring Algorithm for Molecular Subphenotypes (SAMS) was applied as follows:
-
(1)
Gene normalization: The expression of each gene was normalized to the median of all the samples in each independent cohort. This step is performed to determine whether the expression of a gene is either increased or decreased in a subject or single-cell when compared to other subjects or single-cells in the same cohort.
-
(2)
Calculation of the proportion of up and down-regulated genes: Given that 50-gene risk profiles are based on seven increased and 43 decreased genes, the proportion of genes expected to be either increased or decreased can be estimated per subject or single-cell to calculate up and down scores. That is if a subject or single-cell X has five increased genes out of the seven genes expected to be increased, then the proportion of increased genes for this subject or single-cell is 0·714.
-
(3)
Sum of the median normalized expression values of increased and decreased genes: the sum of the median normalized expression values is calculated per subject or single-cell for the entire set of increased and decreased genes.
-
(4)
Calculation of the product between the sum of normalized expression values and the proportion of increased or decreased genes: for this step, the sum of increased genes calculated in step three is multiplied by the proportion of increased genes calculated in step two.
To determine 50-gene risk profiles in the COVID-19 Discovery cohort, up scores above the median and Down scores below the median value within this cohort were classified as high-risk. Subjects without this pattern of expression were classified as low-risk. In the 50-gene, high-risk group of the COVID-19 Discovery cohort, the lowest Up score (0·41) and the highest Down score (−0·41) were used as cutoffs to identify a 50-gene, high-risk profile (subjects or single-cells with Up score >0·41 and Down score <−0·41) in the COVID-19 Validation, COVID-19 Single-cell cohort, IPF-University of Chicago and IPF-Imperial College London cohorts.
Two-sided Fisher's exact test was used to identify differences in disease severity between risk profiles in the COVID-19 Discovery cohort. Categorical variables and continuous clinical variables were analyzed using Two-sided Fisher's exact and two-sample t-test, respectively. The Area Under the Curve (AUC) was used to assess the prediction accuracy of 50-gene risk profiles to determine ICU admission, use of mechanical ventilation and in-hospital mortality in the COVID-19 Validation cohort. These patients were followed for 45 days after hospitalization. We used logistic regression to determine the relationship between 50-gene risk profiles and studied outcomes after adjusting for Age, Charlson comorbidity index, absolute lymphocyte count, corticosteroid therapy and convalescent plasma use at the time of enrollment. To determine whether adding the Charlson comorbidity index [14] to 50-gene risk profiles could improve outcome prediction in COVID-19, we compared three AUC models (50-gene risk profiles alone, Charlson index alone and 50-gene risk profiles combined with Charlson index) using logistic regression with 10-fold cross-validation. Kaplan-Meier curves were used to evaluate the association between 50-gene risk profiles and Mortality in IPF cohorts. Significance was defined as P < 0.05 for all tests.
2.4. 50-gene, single-cell type analysis
To determine cell types expressing either 50-gene high or low-risk profiles in COVID-19, we conducted a cell-type-specific analysis using eight single-cell data measurements from seven subjects with COVID-19 (COVID-19 Single-cell cohort). We estimated the average expression levels of each gene, for each cell type, producing 155 cell-type-specific expression profiles. An Up score >0·41 and a Down score <−0·41 were used to classify 50-gene risk profiles into High and Low-risk groups. The estimated proportion of specific cell types was compared between risk profiles (High versus Low). The cell type definition and classification has been previously described [12]. We tested the overall difference in cell proportions between high and low-risk subgroups using a chi-square test.
2.5. Role of funding source
The Funders had no role in study design, data collection, data analyses, interpretation of results, or manuscript writing.
3. Results
3.1. 50-gene risk profiles in peripheral blood distinguish COVID-19 severity subgroups in a discovery cohort
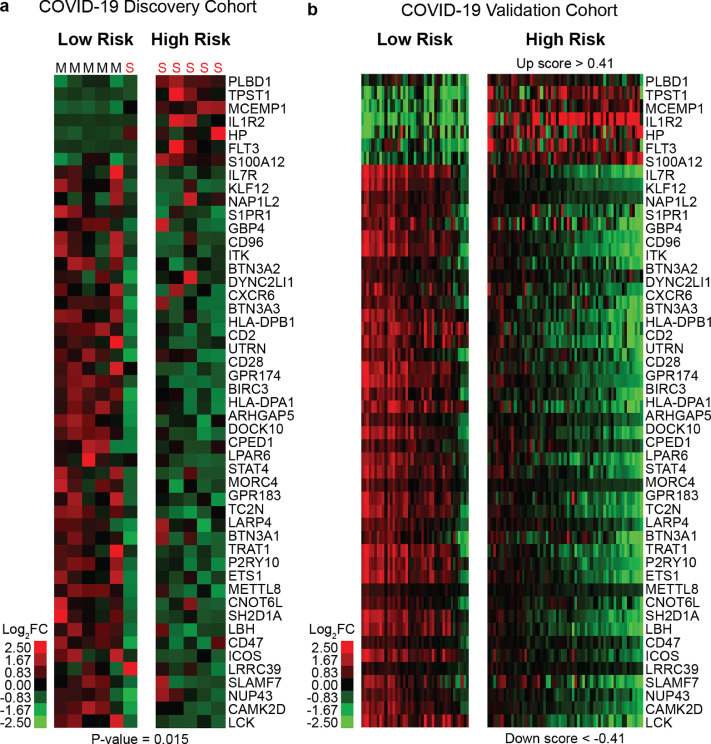
The COVID-19 Discovery cohort included PBMC samples from eight subjects, three of them (Subjects C3, C6, and C7) with two repeated measurements during hospitalization. These samples were classified as mild (N = 5) and severe (N = 6) COVID-19, based on the NEWS score as previously published [10]. To identify 50-gene risk profiles in this cohort, SAMS Up and Down scores were calculated for each sample. All of the samples with a 50-gene, high-risk profile were classified as severe COVID-19 while 83·3% of samples with a low-risk profile were classified as mild COVID-19 (P = 0·015) (Fig. 1A). Table 1 describes the clinical characteristics of the COVID-19 Discovery cohort. Subjects in the low-risk profile had radiological evidence of pneumonia while subjects from the high-risk profile had evidence of multifocal pneumonia with ground glass opacities. 50-gene, high-risk samples had significantly higher NEWS score (mean of 9·2 versus 1·8, P < 0·001), C-reactive protein (mean of 16·9 mg/dl versus 3·3 mg/dl, P = 0·047) and lower absolute lymphocyte counts (mean of 802 cells/μL versus 1430 cells/μL, P = 0·033) when compared to low-risk samples. Regarding the three subjects with baseline samples and repeated measurements, the high-risk profile remained the same in subject C3 (Table 1) after four days of follow up which associated with an increase in NEWS score from eight to ten. Subjects C6 and C7 (Table 1) changed their 50-gene risk profile from High to Low-risk during follow up (mean: 5·5 days) which associated with a mean decline in NEWS score from seven to four.
Fig. 1.
50-gene risk profiles are predictive of COVID-19 outcomes. Clustering of COVID-19 subjects based on 50-gene risk profiles (High versus Low) determined by SAMS in Discovery (a) and Validation cohorts (b). Every column represents a subject and every row represents a gene. Log-based two-color scale is shown next to the heatmaps. Red denotes increase expression over the median of the sample and green denotes decrease expression. M: Mild, S; Severe cases. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Table 1.
Clinical Variables of the COVID-19 Discovery Cohort.
| Patient ID | C2 | C4 | C5 | C8 | C1 | C3 | C6 | C7 | C3 | C6 | C7 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Time point | Baseline | Baseline | Baseline | Baseline | Baseline | Baseline | Baseline | Baseline | Follow up | Follow up | Follow up |
| Hospital day | 10 | 13 | 3 | 5 | 16 | 3 | 5 | 3 | 7 | 9 | 10 |
| Disease group | mild COVID-19 | mild COVID-19 | mild COVID-19 | mild COVID-19 | severe COVID-19 | severe COVID-19 | severe COVID-19 | severe COVID-19 | severe COVID-19 | severe COVID-19 | mild COVID-19 |
| NEWS score | 0 | 0 | 2 | 1 | 14 | 8 | 8 | 6 | 10 | 5 | 3 |
| 50-gene risk profile | low | low | low | low | high | high | high | high | high | low | low |
| Comorbidity | hypertension | hypertension | diabetes mellitus, dyslipidemia | hypothyroidism, dyslipidemia | none | hypertension | hypertension, asthma, atrial fibrillation | history of tuberculous pleuritis | |||
| Absolute Lymphocyte count/μL | 1547 | 2057 | 1834 | 984 | 1055 | 763 | 600 | 647 | 945 | 644 | 1519 |
| C-reactive protein (mg/dL) | 0•05 | 0•19 | 0•59 | 0•83 | 7•58 | 31•41 | 30•52 | 7•07 | 8•2 | 16•6 | 1•53 |
| Chest X-ray | pneumonia | pneumonia | pneumonia | no lesion | pneumonia | multifocal consolidations in both lungs | multifocal patchy opacities in both lungs | no gross change of consolidation and GGO in both lungs | multifocal consolidations in both lungs | diffuse increased lung opacity and multifocal consolidation | unchanged extent of consolidation and GGO in both lungs |
| Treatment | lopinavir/ritonavir, ceftriaxone | ciclesonide inhalor | lopinavir/ritonavir, hydroxychloroquine | none | lopinavir/ritonavir, hydroxychloroquine, nafamostat | lopinavir/ritonavir, levofloxacin | lopinavir/ritonavir, linezolid, cefepime, vancomycin, meropenem, colistin, tigecycline, anidulafungin, hydrocortisone | lopinavir/ritonavir | lopinavir/ritonavir, levofloxacin | lopinavir/ritonavir, linezolid, cefepime, vancomycin, meropenem, colistin, tigecycline, anidulafungin, hydrocortisone | lopinavir/ritonavir |
3.2. 50-gene risk profiles in peripheral blood predict poor COVID-19 outcomes in a validation cohort
To assess the reproducibility of our findings, we analyzed 50-gene risk profiles in the COVID-19 Validation cohort. SAMS cutoffs derived from the COVID-19 Discovery cohort (Up score >0·41 and Down score <−0·41) distinguished High versus Low-risk subjects in the COVID-19 Validation cohort (Fig. 1B). High-risk subjects in the validation cohort were significantly older (64·8 versus 55 years, P = 0·002), had higher APACHE-II severity score (22·5 versus 14·1, P = 0·006), Charlson Comorbidity Index (4 versus 2·3, P < 0·001), C-reactive protein (165·7 mg/l versus 101·3 mg/l, P = 0·003), and Ferritin levels (1215·6 ng/ml versus 497 ng/ml, 0·002) when compared to low-risk subjects. They also had lower absolute lymphocyte counts (838·2 cells/ versus 1550, P < 0·001) and albumin levels (2·8 mg/L versus 3·2 mg/L, P < 0·001) (Table 2). High-risk subjects were more likely to have a prior history of myocardial infarction (16·9% versus 2.4%, P = 0·02) and were more likely to receive convalescent plasma (32·2% versus 12·2%, P = 0·02), and corticosteroid therapy (64·4% versus 14·6%, P < 0·001). There was no significant difference in the incidence of venous thromboembolism between risk subgroups. A 50-gene, high-risk profile predicted ICU admission (AUC:0·77, 95%CI:0·686–0·844, P < 0·001), mechanical ventilation (AUC:0·75, 95%CI:0·67–0·827, P < 0·001) and in-hospital mortality (AUC:0·74, 95%CI:0·678–0·815, P < 0·001) in the COVID-19 Validation cohort (Table 2). Prediction based on 50-gene risk profiles remained statistically significant (P < 0·05) for each outcome measure after adjusting for age, Charlson index, absolute lymphocyte count, corticosteroid therapy and convalescent plasma use. The addition of the Charlson index to 50-gene risk profiles modestly improved the in-hospital mortality prediction accuracy of the genomic classifier by 3% (AUC went from 0·74 to 0·77) (Table 3).
Table 2.
Clinical Variables of the COVID-19 Validation cohort.
| Demographics and Baseline Characteristics of COVID-19 Validation Cohort | ||||
|---|---|---|---|---|
| Total (n = 100) | Low Risk (n = 41) | High Risk (n = 59) | P-Value | |
| Outcome measures | ||||
| ICU Admission | 50 (50%) | 7 (17·1%) | 43 (72·9%) | < 0·001 |
| Mechanical Ventilation - n (%) | 42 (42·0%) | 5 (11·9%) | 37 (62·7%) | < 0·001 |
| Death - n (%) | 24 (24·0%) | 1 (2·4%) | 23 (39·0%) | < 0·001 |
| Ventilator-Free Days - mean (SD) | 19·8 (11·5) | 21·9 (6·6) | 15·5 (12·3) | < 0·001 |
| Hospital Length of Stay - mean (SD) | 16·2 (12·7) | 9 (12·7) | 21·1 (8·9) | < 0·001 |
| Sex - n (%) | ||||
| Male | 62 (62·0%) | 26 (63·4%) | 36 (61·0%) | 0·81 |
| Female | 38 (38·0%) | 15 (36·6%) | 23 (39·0%) | 0·81 |
| Age and BMI - mean (SD) | ||||
| Age | 60·7 (16·1) | 54·8 (16·6) | 64·9 (14·6) | 0·002 |
| BMI | 30·4 (10·3) | 30·5 (7·6) | 30·3 (11·8) | 0·95 |
| Ethnicity - n (%) | ||||
| White | 45 (45·0%) | 18 (43·9%) | 27 (45·8%) | 0·86 |
| Black | 10 (10·0%) | 4 (9·8%) | 6 (10·2%) | 0·94 |
| Asian | 2 (2·0%) | 0 (0%) | 2 (3·4%) | 0·23 |
| Hispanic | 21 (21·0%) | 9 (21·9%) | 12 (20·3%) | 0·85 |
| Other | 22 (22·0%) | 10 (24·4%) | 12 (20·3%) | 0·63 |
| Comorbidities - n (%) | ||||
| Smoking history | 17 (17·0%) | 8 (19·5%) | 9 (15·2%) | 0·58 |
| Myocardial infarction | 11 (11·0%) | 1 (2·4%) | 10 (16·9%) | 0·02 |
| Congestive heart failure | 4 (4·0%) | 1 (2·4%) | 3 (5·1%) | 0·51 |
| Peripheral vascular disease | 1 (1·0%) | 0 (0·0%) | 1 (1·7%) | 0·4 |
| Cerebrovascular accident | 2 (2·0%) | 1 (2·4%) | 1 (1·7%) | 0·79 |
| Dementia | 6 (6·0%) | 1 (2·4%) | 5 (8·5%) | 0·21 |
| Pulmonary disease | 21 (21·0%) | 6 (14·6%) | 15 (25·4%) | 0·19 |
| Rheumatic disease | 3 (3·0%) | 1 (2·4%) | 2 (3·4%) | 0·79 |
| Peptic ulcer disease | 1 (1·0%) | 1 (2·4%) | 0 (0·0%) | 0·23 |
| Diabetes mellitus | 35 (35·0%) | 11 (26·8%) | 24 (40·7%) | 0·15 |
| Renal disease | 10 (10·0%) | 2 (4·9%) | 8 (13·6%) | 0·16 |
| Cancer (solid) | 4 (4·0%) | 1 (2·4%) | 3 (5·1%) | 0·51 |
| HIV/AIDS | 2 (2·0%) | 1 (2·4%) | 1 (1·7%) | 0·79 |
| Severity Indexes - mean (SD) | ||||
| APACHEII | 21·4 (8·2) | 14·1 (3·5) | 22·5 (8·1) | 0·006 |
| SOFA | 8·1 (4·0) | 5·6 (2·3) | 8·5 (4·1) | 0·06 |
| Charlson comorbidity index | 3·3 (2·5) | 2·3 (1·9) | 4·0 (2·6) | <0·001 |
| Biomarkers - mean (SD) | ||||
| Ferritin (ng/mL) | 932·8 (1094·0) | 497·0 (403·8) | 1215·6 (1294·6) | 0·002 |
| C-reactive protein (mg/L) | 140·5 (103·6) | 101·3 (83·7) | 165·7 (108·0) | 0·003 |
| D-dimer (mg/L FEU) | 11·7 (22·5) | 6·7 (19·5) | 14·3 (23·7) | 0·14 |
| Procalcitonin (ng/mL) | 3·2 (10·4) | 2·4 (7·0) | 3·7 (12·0) | 0·56 |
| Lactate (nmol/L) | 1·2 (0·5) | 1·1 (0·4) | 1·3 (0·5) | 0·09 |
| Fibrinogen (mg/dL) | 543·8 (196·9) | 545·5 (188·4) | 542·9 (203·3) | 0·96 |
| Albumin (mg/L) | 2·9 (0·5) | 3·2 (0·5) | 2·8 (0·4) | <0·001 |
| Absolute Lymphocyte count/μL | 1130 (794) | 1550 (893) | 838·2 (561) | <0·001 |
| Treatment - n (%) | ||||
| Hydroxychloroquine | 86 (86·0%) | 36 (87·8%) | 50 (84·7%) | 0·67 |
| Antibiotics | 97 (97·0%) | 39 (95·1%) | 58 (98·3%) | 0·36 |
| Antivirals | 1 (1·0%) | 0 (0·0%) | 1 (1·7%) | 0·4 |
| IL6 antagonist | 4 (4·0%) | 1 (2·4%) | 3 (5·1%) | 0·51 |
| Convalescent plasma | 24 (24·0%) | 5 (12·2%) | 19 (32·2%) | 0·02 |
| Steroids | 44 (44·0%) | 6 (14·6%) | 38 (64·4%) | <0·001 |
| Anticoagulation | 98 (98·0%) | 41 (100·0%) | 57 (96·6%) | 0·23 |
Table 3.
Prediction accuracy of 50-gene risk profiles to predict outcomes in COVID-19.
| Prediction models | ICU admission (AUC, 95% CI) | Mechanical Ventilation (AUC, 95% CI) | In-Hospital Mortality (AUC, 95%) |
|---|---|---|---|
| 50-Gene Risk Profiles (High versus Low) | 0·77, 95% CI (0·686–0·844) | 0·75, 95% CI (0·67–0·827) | 0·74, 95% CI (0·678–0·815) |
| Charlson Index | 0·54, 95% CI (0·432–0·648) | 0·48, 95% CI (0·37–0·576) | 0·69, 95% CI (0·553–0·797) |
| 50-Gene Risk Profiles and Charlson index | 0·78, 95% CI (0·597–0·847) | 0·79, 95% CI (0·634–0·864) | 0·77, 95% CI (0·531–0·866) |
High-risk patients spent more days on mechanical ventilation (21·9 versus 15·5 days, P < 0·001) and had longer hospitalizations (21·1 versus 9 days, P < 0·001) compared to low-risk patients. Only one patient in the 50-gene, low-risk profile group died while 23 patients in the 50-gene, high-risk profile group died during hospitalization (P = < 0·001) (Table 2). All deceased patients in the validation cohort were in severe ARDS and on mechanical ventilation. Refractory respiratory failure was the cause of death in all the patients who died from COVID-19 in the validation cohort.
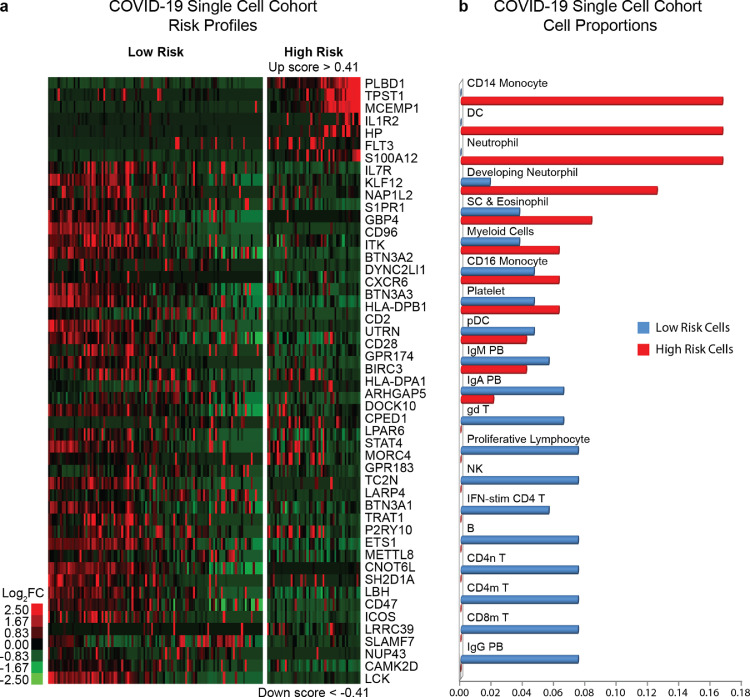
3.3. Single-cell analysis in COVID-19 reveals the cellular sources of the 50-gene risk profiles
A COVID-19 Single-cell cohort [11] was used to identify the cellular origin of 50-gene risk profiles. SAMS cutoffs derived from the COVID-19 Discovery cohort (Up score>0·41 and Down<−0·41) classified 47 cells with a high-risk profile and 108 cells with a low-risk profile (Fig. 2A). 50-gene expressing cells with a high-risk profile mainly included CD14+ monocytes (16·7%), dendritic cells (16·7%) and neutrophils (16·7%), while 50-gene expressing cells with a low-risk profile mainly included IgG producing plasmablasts (7·48%), mature (7·48%) and naïve (7·48%) CD4 T cells, CD8 mature T cells (7·48%), B cells (7·48%), NK cells (7·48%), proliferative lymphocytes (7·48%), gamma/delta T cells (6·54%) and Interferon stimulated CD4-T cells (5·41%) (Fig. 2B). Cells with overlapping 50-gene risk profiles included: developing neutrophils, stem cells, eosinophils, myeloid cells, CD16 monocytes, platelets, plasmacytoid dendritic cells, IgA and IgM producing plasmablasts. The overall difference of cell proportions between 50-gene risk profiles (High versus Low) was statistically significant (P < 0·001). The full list of 50-gene expressing cells can be seen in Table 4. These findings provide evidence of the cellular source of 50-gene expression changes in peripheral blood and point at specific cell types potentially associated with increased risk of mortality, and other poor outcomes in COVID-19.
Fig. 2.
Gene expression analysis of 50-genes in single cells from the COVID-19-Single cell cohort demonstrating differences in risk profiles between cell types. (a) Heatmap shows cell types with 50-gene, low versus high-risk expression profiles. Every column represents a single cell and every row represents a gene. Log-based two-color scale is shown next to heatmap; red denotes increase expression over the median of samples and green, decrease. (b) Proportion of 50-gene expressing cells in low versus high-risk profiles. Y-axis represents cell types and X-axis represents cell proportions. B: B Cell, CD4m T: Memory CD4 T Cell, CD4n T: Naive CD4 T Cell, CD8m T: Memory CD8 T Cell, DC: Conventional Dendritic Cell, gd T: Gamma Delta T cells, IFN-stim CD4 T: Interferon-stimulated CD4 T cell, IgA PB: IgA (Immunoglobulin-A) Plasmablast, IgG PB: IgG (Immunoglobulin-G) Plasmablast, IgM PB: IgM (Immunoglobulin-M) Plasmablast, NK: Natural Killer Cell, pDC: Plasmacytoid Dendritic Cell, Myeloid cells, SC & Eosinophil: Stem Cells and Eosinophil. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 4.
Estimated percentage of 50-gene expressing cells with High versus Low-risk profiles.
| Cell Type | Low-risk (%) | High-risk (%) |
|---|---|---|
| IgG PB | 7.48 | 0 |
| CD8m T | 7.48 | 0 |
| CD4m T | 7.48 | 0 |
| CD4n T | 7.48 | 0 |
| B | 7.48 | 0 |
| NK | 7.48 | 0 |
| Proliferative Lymphocytes | 7.48 | 0 |
| gd T | 6.54 | 0 |
| IFN-stim CD4 T | 5.61 | 0 |
| IgA PB | 6.54 | 2.08 |
| IgM PB | 5.61 | 4.17 |
| pDC | 4.67 | 4.17 |
| Platelet | 4.67 | 6.25 |
| CD16 Monocyte | 4.67 | 6.25 |
| Myeloid cells | 3.74 | 6.25 |
| SC & Eosinophil | 3.74 | 8.33 |
| Developing Neutrophil | 1.87 | 12.5 |
| Neutrophil | 0 | 16.67 |
| DC | 0 | 16.67 |
| CD14 Monocyte | 0 | 16.67 |
B: B Cell, CD4m T: Memory CD4 T Cell, CD4n T: Naive CD4 T Cell, CD8m T: Memory CD8 T Cell, DC: Conventional Dendritic Cell, gd T: Gamma Delta T cells, IFN-stim CD4 T: Interferon-stimulated CD4 T cell, IgA PB: IgA (Immunoglobulin-A) Plasmablast, IgG PB: IgG (Immunoglobulin-G) Plasmablast, IgM PB: IgM (Immunoglobulin-M) Plasmablast, NK: Natural Killer Cell, pDC: Plasmacytoid Dendritic Cell, Myeloid cells, SC & Eosinophil: Stem Cells and Eosinophil.
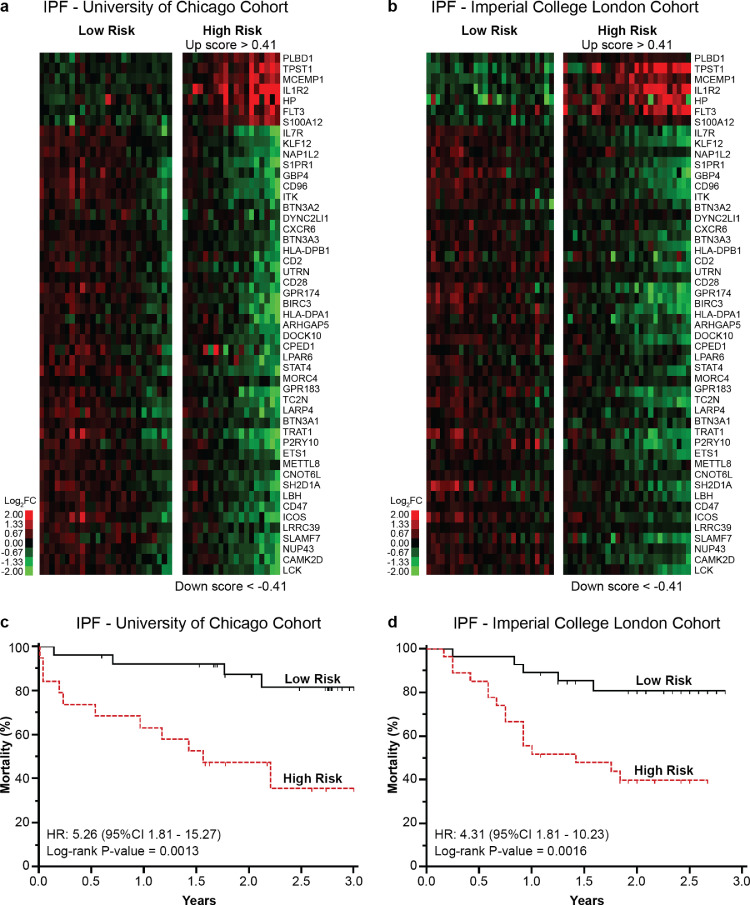
3.4. 50-gene risk profiles in COVID-19 are predictive of mortality in IPF
To determine whether the same SAMS cutoffs used to distinguish a 50-gene, high-risk profile in COVID-19 could also be applied to predict IPF mortality, we reanalyzed peripheral blood 50-gene expression data from two independent IPF cohorts (IPF-University of Chicago and IPF-Imperial College London). An Up Score >0·41 and a Down Score <−0·41 distinguished 50-gene high versus low-risk profiles in both IPF cohorts (Fig. 3A and B). 50-gene risk profiles were significantly predictive of mortality in the IPF-University of Chicago (HR:5·26, 95%CI:1·81–15·27, P = 0·0013) and IPF-Imperial College London (HR:4·31, 95%CI:1·81–10·23, P = 0·0016) cohorts (Fig. 3C and D). These results confirmed our previous findings [7,8] and indicated an overlapping outcome-associated transcriptomic signature between COVID-19 and IPF.
Fig. 3.
50-gene risk profile SAMS cutoffs predictive of COVID-19 outcomes are also predictive of poor IPF outcomes. (a) Clustering of IPF-University of Chicago and (b) IPF-Imperial College London cohort based on 50-gene risk profiles (high versus low) derived from the COVID-19 Discovery cohort (Up score >0.41 and Down Score <−0.41). Every column represents a subject and every row represents a gene. Log-based two-color scale is shown next to the heatmaps. Red denotes increased expression over the median of the sample and green denotes decrease expression. (c) Mortality differs between 50-gene risk profiles in the IPF-University of Chicago and (d) IPF-Imperial College London cohort. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
In this study, we show that a high-risk, 50-gene profile, previously shown to predict IPF mortality is also predictive of worse outcomes in COVID-19 patients. The transcriptomic overlap captured in different cohorts and experimental settings suggests a remarkably conserved systemic gene expression signature evoked by COVID-19 and IPF. Moreover, this overlapping profile combined with the observed pathological and radiological surrogates of pulmonary fibrosis shown by some severe COVID-19 patients suggests that both diseases share, to some extent, common host response features. The single-cell RNA-seq data in COVID-19 subjects points at the cells expressing the 50 genes predictive of poor disease outcomes. These data suggest that CD14+ monocytes, dendritic cells and neutrophils are critical regulators of the high-risk profile. In SARS-CoV-2 infected primates, increased circulating levels of classical and non-classical monocytes, and neutrophilic migration to the lungs [15] was associated with poor disease outcomes. In humans, reports have shown that severe COVID-19 is associated with elevated numbers of neutrophil precursors and circulating levels of CD14+monocytes with high expression of alarmins S100A8/9/12 and low expression of HLA-DR. [16]. The present analysis is consistent with that data. A recent report also indicates that serum calprotectin, which belongs to the S100 protein family, is associated with IPF diagnosis and correlates with diffusing capacity for carbon monoxide (DLCO) and the composite physiologic index (CPI) [17]. Moreover, previous evidence indicates that S100A9 is elevated in bronchoalveolar lavage fluid from IPF patients in comparison with healthy controls [18] and increased circulating levels of CD14+ monocytes were found to be predictive of mortality in IPF and other fibrotic lung diseases [19]. The single-cell RNA sequencing data shows increased proportion of CD4 and CD8 T lymphocytes and immunoglobulin-producing plasmablasts in individuals with a low-risk genomic profile, suggesting an association between a strong T cell response [20,21] and better disease outcomes [22]. This finding is consistent with recent data indicating that severe COVID-19 infection induces a distinct inflammatory program characterized by suppression of the innate immune system in the periphery and that milder cases evoke a more robust T cell response [23].
The biomarker and therapeutic implications of this discovery are significant since the identification of 50-gene risk profiles in COVID-19, in addition to clinical variables, can facilitate healthcare utilization such as triage of patients to the most appropriate location (home, ward, ICU), reduce hospital length of stay, allow for proper allocation of limited resources including mechanical ventilators and reduce the cost of inappropriate hospitalization. It could also allow the early identification of patients likely to deteriorate and resolve specific transcriptomic sub-phenotypes that are amenable to certain treatments. For example, while corticosteroids are currently recommended for hospitalized COVID-19 patients due to their positive effect in survival [24], the use of corticosteroids in IPF has been controversial due to increased risk of death and hospitalizations associated with immunosuppressive therapy [25]. Thus, the 50-gene, high-risk profile may facilitate the identification of patients that are more likely to respond to COVID-19 targeted therapies such as corticosteroids and others [26] or to identify a subgroup of IPF patients who may benefit from a limited course of corticosteroid therapy. The use of 50-gene risk profiles could also support the rationale to investigate the use of IPF-targeted antifibrotic medications [27,28] to prevent short- and long-term sequela of COVID-19. Another important aspect of our study that is worth mentioning is the remarkable ability of SAMS scores derived from the COVID-19 Discovery cohort, to identify 50-gene risk profiles predictive of poor outcomes across two additional COVID-19 and IPF cohorts despite using different genomic technologies and different starting material (bulk versus single-cell RNA).
Despite the relevance and reproducibility of our findings, we need to acknowledge some limitations of our study. COVID-19 and IPF are diseases with different etiologies. COVID-19, the illness caused by SARS-CoV-2 infection, is characterized by diffuse [1] and extensive alveolar damage, dysmorphic pneumocytes and thrombosis of the lung micro, and macro-vasculature [29]. Poor outcomes in COVID-19 are predominantly driven by the host response to the infection [22]. IPF is a specific form of chronic fibrosing interstitial pneumonia of unknown etiology, limited to the lung and histologically characterized by usual interstitial pneumonia [30]. Accumulating evidence suggests that under genetic predisposition and environmental factors, the fibrotic response seen in IPF is driven by abnormally activated alveolar epithelial cells (AECs) leading to epithelial to mesenchymal transition and activation, proliferation, and differentiation of fibroblasts to myofibroblasts [31]. While our study focuses on predictive features of peripheral blood transcriptomic profiles in COVID-19 and IPF, we did not study the underlying mechanisms triggering this aberrant immune response and its potential relationship to alveolar epithelial cell injury or any other molecular mechanisms shared between COVID-19 and IPF. Future studies could be performed to characterize lung autopsy findings in deceased individuals with a 50-gene, high-risk profile or whether COVID-19 survivors with a high-risk profile are more likely to develop chronic ILD changes and a fibroproliferative phenotype. While this is to our knowledge the first systematic analysis of the overlapping gene expression signature of COVID-19 and IPF, we believe these data needs corroboration in large, prospective trials including more diverse patient populations to generalize our findings. That research could be complemented with an unbiased whole-exome analysis of circulating blood in both diseases, which could uncover other relevant genes associated with poor outcomes. Also, it would be important to determine whether the identification of 50-gene expressing cells in COVID-19 can be replicated in single-cell RNA-seq analyses of IPF patients, which could help define if the mentioned overlap is driven by similar cell type distributions in both diseases. Finally, given the retrospective nature of our study, we were limited by the lack of a comprehensive radiological assessment of COVID-19 subjects in both cohorts. Future studies should focus on comparing the radiological characteristics of subjects with a 50-gene high versus low risk profile.
In conclusion, peripheral blood, 50-gene risk profiles predict ICU admission, need for mechanical ventilation and in-hospital mortality in COVID-19 and overlaps a signature known to predict poor IPF outcomes. The cellular sources of these gene expression changes suggest common mechanisms implicating innate and adaptive immune responses in both diseases. A 50-gene, risk profile test in peripheral blood could be a potentially useful biomarker to predict COVID-19 mortality and morbidity.
5. Contributors
BJG, JS and JHM conceptualized, designed the study, collected data, carried out the initial analyses and drafted the initial manuscript. BJG, JS, TZ, BX, JB and JHM performed statistical analyses. JB, Y.H, SFM, PM, TM, IN, GM and AJ collected data, critically reviewed and drafted the revised manuscript. BJG and JS contributed equally to this work. All authors read and approved the final version of the manuscript. BJG, JS and JHM verified the underlying data.
Data sharing statement
Gene expression and clinical data has been previously deposited in the Gene Expression Omnibus (GEO) under the following accession numbers: GSE149689, GSE157103, GSE174818 GSE150728, GSE28221 and GSE93606.
Declaration of Competing Interest
JHM has a patent titled “52-gene signature in peripheral blood identifies a genomic profile associated with increased risk of mortality and poor disease outcomes in idiopathic pulmonary fibrosis” that relates to the work presented in this manuscript. IN receives consulting fees from Boehringer Ingelheim, Genentech and Parion Sciences.
Acknowledgments
This work is dedicated to those who lost their lives due to COVID-19. This work was supported by the Ubben Pulmonary Fibrosis Fund - University of South Florida (USF) Foundation (JHM). The work was also supported in part by the National Institute for Health Research Clinician Scientist Fellowship NIHR: CS-2013-13-017 (TMM); Action for Pulmonary Fibrosis Mike Bray fellowship (PLM); The National Heart, Lung, and Blood Institute (NHLBI) through award K01-HL-130704 (AJ), R01HL130796 and UG3HL145266 (IN).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103439.
Appendix. Supplementary materials
References
- 1.Carsana L., Sonzogni A., Nasr A. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-center descriptive study. Lancet Infect Dis. 2020;20(10):1135–1140. doi: 10.1016/S1473-3099(20)30434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann M., Verleden S.E., Kuehnel M. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020;383(2):120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grillo F., Barisione E., Ball L., Mastracci L., Fiocca R. Lung fibrosis: an undervalued finding in COVID-19 pathological series. Lancet Infect Dis. 2020;21(4) doi: 10.1016/S1473-3099(20)30582-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian S., Xiong Y., Liu H. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Mod Pathol. 2020;33(6):1007–1014. doi: 10.1038/s41379-020-0536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou S., Wang Y., Zhu T., Xia L. CT Features of Coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. AJR Am J Roentgenol. 2020;214(6):1287–1294. doi: 10.2214/AJR.20.22975. [DOI] [PubMed] [Google Scholar]
- 6.Spagnolo P., Balestro E., Aliberti S. Pulmonary fibrosis secondary to COVID-19: a call to arms? Lancet Respir Med. 2020;8(8):750–752. doi: 10.1016/S2213-2600(20)30222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herazo-Maya J.D., Noth I., Duncan S.R. Peripheral blood mononuclear cell gene expression profiles predict poor outcome in idiopathic pulmonary fibrosis. Sci Transl Med. 2013;5(205) doi: 10.1126/scitranslmed.3005964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herazo-Maya J.D., Sun J., Molyneaux P.L. Validation of a 52-gene risk profile for outcome prediction in patients with idiopathic pulmonary fibrosis: an international, multicentre, cohort study. Lancet Respir Med. 2017;5(11):857–868. doi: 10.1016/S2213-2600(17)30349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith G.B., Prytherch D.R., Meredith P., Schmidt P.E., Featherstone P.I. The ability of the National Early Warning Score (NEWS) to discriminate patients at risk of early cardiac arrest, unanticipated intensive care unit admission, and death. Resuscitation. 2013;84(4):465–470. doi: 10.1016/j.resuscitation.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Lee J.S., Park S., Jeong H.W. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferon’s in development of severe COVID-19. Sci Immunol. 2020;5(49) doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overmyer K.A., Shishkova E., Miller I.J. Large-Scale multi-omic analysis of COVID-19 severity. Cell Syst. 2020;12(2):23–40. doi: 10.1016/j.cels.2020.10.003. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilk A.J., Rustagi A., Zhao N.Q. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26(7):1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molyneaux P.L., Willis-Owen S.A.G., Cox M.J. Host-microbial interactions in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2017;195(12):1640–1650. doi: 10.1164/rccm.201607-1408OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R Core Team R: a language and environment for statistical computing. R Found Stat Comput. 2020 https://www.R-project.org/ Vienna, Austria. [Google Scholar]
- 15.Fahlberg M.D., Blair R.V., Doyle-Meyers L.A. Cellular events of acute, resolving or progressive COVID-19 in SARS-CoV-2 infected non-human primates. Nat Commun. 2020;11(1):6078. doi: 10.1038/s41467-020-19967-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulte-Schrepping J., Reusch N., Paclik D. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182(6):1419–1440. doi: 10.1016/j.cell.2020.08.001. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machahua C., Guler S.A., Horn M.P. Serum calprotectin as new biomarker for disease severity in idiopathic pulmonary fibrosis: a cross-sectional study in two independent cohorts. BMJ Open Respir Res. 2021;8(1) doi: 10.1136/bmjresp-2020-000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara A., Sakamoto N., Ishimatsu Y. S100A9 in BALF is a candidate biomarker of idiopathic pulmonary fibrosis. Respir Med. 2012;106(4):571–580. doi: 10.1016/j.rmed.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 19.Scott M.K.D., Quinn K., Li Q. Increased monocyte count as a cellular biomarker for poor outcomes in fibrotic diseases: a retrospective, multicenter cohort study. Lancet Respir Med. 2019;7(6):497–508. doi: 10.1016/S2213-2600(18)30508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z., John Wherry E. T cell responses in patients with COVID-19. Nat Rev Immunol. 2020;20(9):529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atyeo C., Fischinger S., Zohar T. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53(3):524–532. doi: 10.1016/j.immuni.2020.07.020. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X., Tan Y., Ling Y. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583(7816):437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 23.Arunachalam P.S., Wimmers F., Mok C.K.P. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369(6508):1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Group R.C., Horby P., Lim W.S., Emberson J.R, The RECOVERY Collaborative Group Dexamethasone in hospitalized patients with COVID-19. N Engl J Med. 2021;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Idiopathic Pulmonary Fibrosis Clinical Research N. Raghu G., Anstrom K.J., King T.E., Lasky J.A., Martinez F.J. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med. 2012;366(21):1968–1977. doi: 10.1056/NEJMoa1113354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shrestha G.S., Paneru H.R., Vincent J.L. Precision medicine for COVID-19: a call for better clinical trials. Critical Care. 2020;24(1):282. doi: 10.1186/s13054-020-03002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King T.E., Bradford W.Z., Castro-Bernardini S. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 28.Richeldi L., du Bois R.M., Raghu G. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2071–2082. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- 29.Bussani R., Schneider E., Zentilin L. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine. 2020;61 doi: 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.American Thoracic S, European Respiratory S American Thoracic Society/European Respiratory Society International multidisciplinary consensus classification of the idiopathic interstitial pneumonias. This joint statement of the American Thoracic Society (ATS), and the European Respiratory Society (ERS) was adopted by the ATS board of directors, June 2001 and by the ERS Executive Committee, June 2001. Am J Respir Crit Care Med. 2002;165(2):277–304. doi: 10.1164/ajrccm.165.2.ats01. [DOI] [PubMed] [Google Scholar]
- 31.Selman M., Pardo A. The leading role of epithelial cells in the pathogenesis of idiopathic pulmonary fibrosis. Cell Signal. 2020;66 doi: 10.1016/j.cellsig.2019.109482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.