ABSTRACT
The mammalian central nervous system (CNS) exhibits limited regenerative capacity and the mechanisms that mediate its regeneration are not fully understood. Here, we present a novel experimental design to damage the CNS by using a contusion injury paradigm. The design of this protocol allows the study of long-term and short-term cellular responses, including those of the CNS and the immune system, and of any implications regarding functional recovery. We demonstrate for the first time that adult Drosophila melanogaster glial cells undergo spontaneous functional recovery following crush injury. This crush injury leads to an intermediate level of functional recovery after damage, which is ideal to screen for genes that facilitate or prevent the regeneration process. Here, we validate this model and analyse the immune responses of glial cells as a central regulator of functional regeneration. Additionally, we demonstrate that glial cells and macrophages contribute to functional regeneration through mechanisms involving the Jun N-terminal kinase (JNK) pathway and the Drosophila protein Draper (Drpr), characteristic of other neural injury paradigms. We show that macrophages are recruited to the injury site and are required for functional recovery. Further, we show that the proteins Grindelwald and Drpr in Drosophila glial cells mediate activation of JNK, and that expression of drpr is dependent on JNK activation. Finally, we link neuron-glial communication and the requirement of neuronal vesicular transport to regulation of the JNK pathway and functional recovery.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: JNK, Macrophages, CNS damage, Glia, Immune response, Regeneration
Summary: Central nervous system crush injury paradigm in adult Drosophila melanogaster is a suitable model to study the cellular events, and genetic pathways behind injury responses and functional regeneration. We describe the immune responses of glial cells, neurons and macrophages following injury, and the functional relevance of each response.
INTRODUCTION
In contrast to the peripheral nervous system (PNS), the mammalian central nervous system (CNS) has limited regenerative capacity (Curcio and Bradke, 2018). Consequently, brain stroke, spinal cord injury or neurodegenerative diseases result in permanent functional impairment. The limited regenerative capacity of neurons, myelin-associated inhibitory factors and the presence of glial scars, restrict regeneration of the mammalian CNS (Cregg et al., 2014; Silver and Miller, 2004). Other vertebrates, such as zebrafish, undergo spontaneous regeneration of the PNS and CNS following injury, thus facilitating functional recovery (Rasmussen and Sagasti, 2017). However, this regenerative capacity deteriorates with age; therefore, as animals age, functional recovery following spinal cord injury is limited (Becker et al., 1997). Invertebrate models, such as Caenorhabditis elegans and Drosophila melanogaster allow extensive genetic manipulations that contribute to deduce the mechanisms that mediate regeneration of the CNS. Other injury models, such as axon regeneration following laser ablation, have emerged as being suitable to investigate the cellular properties of regeneration (Stone et al., 2010; Yanik et al., 2004). This model has contributed to uncover the implication of microtubules (Chen et al., 2011), dendrite-axon interconversion (Stone et al., 2010) and the decline of spontaneous functional recovery with age (Byrne et al., 2014). However, although these mechanisms are conserved in mammals, they do not participate in CNS regeneration (Curcio and Bradke, 2018).
Neural regeneration studies are primarily focused on axon regeneration as a strategy to restore neural function. The stimulation of damaged neurons has been proposed to overcome deleterious signals originated in injured neurons and neighbouring cells (McQuarrie and Grafstein, 1973; Silver, 2009). Several mechanisms control axonal regeneration, including local cytoskeletal modifications that promote growth cone formation and axon extension (Bradke et al., 2012), axon-soma retrograde signalling (Rishal and Fainzilber, 2014), mitochondrial trafficking (Sheng, 2017), activation of specific regenerative transcriptional programs (Mahar and Cavalli, 2018) and epigenetic modification of selected genes (Weng et al., 2017).
The microenvironment also modulates regeneration following CNS injury. Damaged axons of adult rat CNS neurons regrow when placed in a permissive environment for regeneration (Curcio and Bradke, 2018), an axon laser ablation model in Drosophila demonstrated that the glial metabolic status can promote or inhibit axon regrowth (Li et al., 2020). These results suggest a different role of permissive and non-permissive environments for regeneration. Glial cells maintain those neural environments (Sawa et al., 2004). Glial cells proliferate, undergo morphological changes, enwrap axons and engulf cellular debris in response to injury. This process is known as the glial regenerative response (GRR) and it is conserved across species (Kato et al., 2018). GRR can be divided into two, the proliferative response and the immune response (Losada-Perez, 2018). Previous studies in Drosophila and mice described a genetic network that regulates the GRR to injury (Kato et al., 2011, 2015; Losada-Perez et al., 2016, 2017). This network includes the genes kon-tiki/Ng2, Notch/Notch1, pros/Prox1 and dorsal/NFkB, and controls glial cell division and differentiation. GRR has been described in brains of Drosophila larvae, although neural trauma in adults is the most common form of CNS injury in humans and, thus, its role during adult CNS damage needs to be investigated.
The GRR immune response involves gene expression of draper (drpr) and signalling via Jun N-terminal kinase (MAPK8, also known as and hereafter referred to as JNK). drpr is the Drosophila orthologue of mammalian Megf10 (CED-1 in C. elegans) (Chiu et al., 2018). Upon neural injury, engulfing cells, macrophages or neighbouring cells remove cellular debris. These cells respond to local cues known as ‘eat-me’ signals released from injured tissue via the Drpr/Megf10/CED-1 receptor (Awasaki et al., 2006; Chiu et al., 2018; Chung et al., 2000; Franc et al., 1996, 1999; Hamon et al., 2006; Kurant et al., 2008; MacDonald et al., 2006).
The engulfment of dying cells in the germ line or epithelial cells is mediated by the phagocytic receptor Drpr through activation of the JNK pathway (Casas-Tintó et al., 2015; Etchegaray et al., 2012). Likewise, mutations of JNK reduce axon regeneration and provoke accumulation of axon debris after injury (Chiu et al., 2018). In addition, knockdown of JNK-pathway components block CED-1 mediated axon regeneration and axon debris removal (Chiu et al., 2018), suggesting a role for JNK pathway in Drpr/CED-1-mediated axon regrowth. This immune system response operates in both Drosophila CNS and PNS (Gadani et al., 2015). Drosophila adult glial cells become reactive in response to axotomy through JNK-mediated upregulation of drpr (MacDonald et al., 2013). In addition to the glial immune response, the immune system also contributes to regeneration of the CNS following injury. Studies in vertebrates revealed the role of macrophages during wound healing, debris removal or through the recruitment of other cells to the injury site (Brancato and Albina, 2011; Koh and DiPietro, 2011; Werner and Grose, 2003). However, little is known about the role of macrophages after neural injury in Drosophila.
Some vertebrates recover locomotion following limb amputation by adapting the activity of the central pattern generator (CPG), a feature that illustrates the robustness of the locomotor system (Isakov et al., 2016; Kirpensteijn et al., 1999; Mendes et al., 2014). To identify bona fide mechanisms of neural regeneration it is, therefore, necessary to find a reliable method to investigate injured-limb movement instead of the locomotor capacity of the complete animal. Here, we present a novel paradigm of injury in adult flies, which reproduces the main features described in other animal models after CNS injury and allows the study of functional regeneration in a temporally restricted manner. We describe for the first-time the spontaneous functional regeneration following injury in adult Drosophila, the activation of immune components, such as Drpr or JNK in neural cells and the requirement of macrophages for functional recovery.
Therefore, application of crush injury is a suitable technique to study cellular events and genetic pathways to understand responses of the CNS to injury and their relevance for functional recovery.
RESULTS
A procedure for adult crush injury
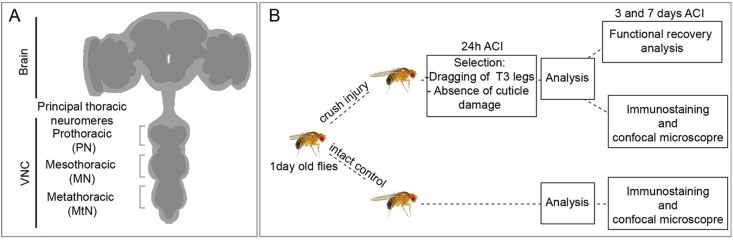
To study GRR in adult flies we used forceps to apply pressure from the exterior to damage the metathoracic segment of adult flies (1 day after eclosion), leading to impaired movement of T3 legs (innervated by the metathoracic ganglia) and locomotor dysfunction. To cause the injury, we first anesthetised the animal on a CO2 plate, ventral side up, and gently pinched the thorax with forceps pointing to the metathoracic segment (Fig. 1A). As the injury was performed manually, variability of the damage was expected to be broad. To reduce variability, we classified injured flies 24 h after crush injury (ACI) (unless otherwise indicated) and selected only individuals that were dragging the third pair of legs. In contrast to other injury methods, crush injury allows the study of functional recovery as the cuticle and legs remain intact (Fig. 1B).
Fig. 1.
Crush injury protocol. (A) Schematic representation of adult Drosophila CNS showing the main thoracic neuromeres. (B) Diagram of standard protocol followed in this work.
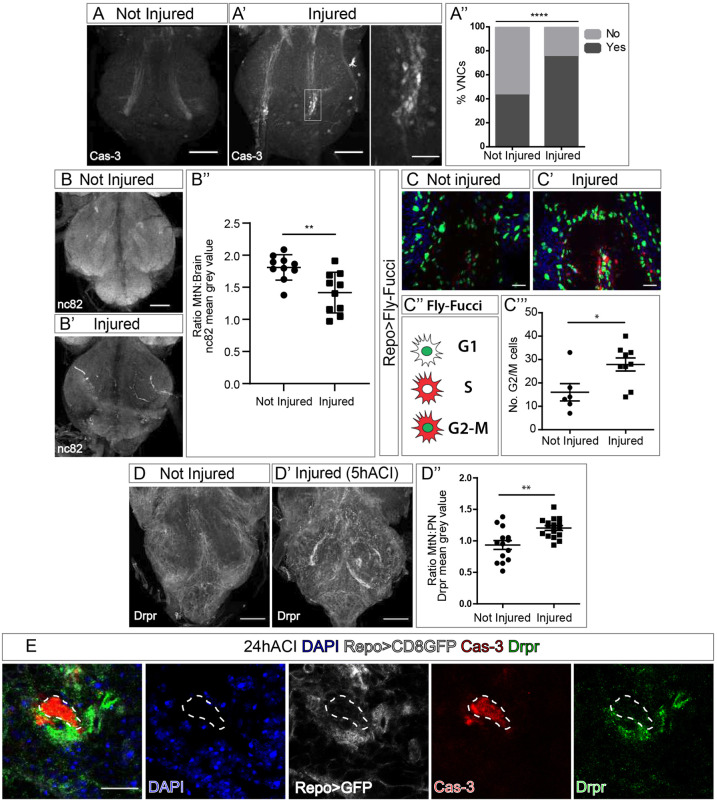
To validate the injury site, we determined cell damage in the CNS through staining for caspase-3 to detect cell death (Sarkissian et al., 2014) (Fig. 2A,A′). In most injured ventral nerve cords (VNCs) we found caspase-3-positive signals, as this feature is statistically less common in non-injured controls (Fig. 2A″). To ensure that the caspase-3 signal correlates with programmed cell death, we examined whether cells with caspase-3-positive signal correspond with condensed nuclei (Lu et al., 2005; Toné et al., 2007). We measured the DAPI signal in caspase-3-positive and caspase-3-negative nuclei of injured VNCs, and found that the average intensity of DAPI signals in caspase-3-positive nuclei is significantly higher compared to caspase-3-negative nuclei (Fig. S1A). Thus, we conclude that the caspase-3 signal we observed in injured VNCs corresponds to cell death.
Fig. 2.
Crush injury can be used to study GRR. (A,A′) Representative metathoracic neuromeres (MtNs) showing anti-caspase-3 staining in not injured (A) and injured (24 h ACI) VNCs (A′). The boxed area in A′ is shown magnified in the most-right image. (A′′) Binomial test, comparing the percentage of injured and not injured VNCs (%VNCs) with caspase-3 (Cas-3)-positive signal (Yes), and VNCs without Cas-3-positive signal (No). ****P<0.0001. (B,B′) Representative images of not injured (B) and injured (B′, 24 h ACI) VNCs stained for Bruchpilot (nc82). (B″) Quantification of the MtN:Brain nc82 ratio mean intensity signal. Unpaired t-test, **P<0.005. (C,C′) Representative images showing Fly-FUCCI in glial cells of not injured (C) and injured (C′, 24 h ACI) VNCs. (C″) Schematic of Fly-FUCCI corresponding to phases of the cell-cycle. (C‴) Quantification of cells positive for GFP and RFP in G2/M phase. Unpaired t-test, *P<0.05. (D) Drpr staining of not injured (D) and injured (D′, 5 h ACI) VNCs. Quantification of the MtN:PN mean grey value ratio of the Drpr signal (D″). Unpaired t-test, **P<0.005. (E) Images of cell samples 24 h ACI stained for Drpr (green) colocalising with glial membrane (Repo>myrRFP, grey) and surrounding the cell debris (stained for Cas-3, red) 24 h ACI. Areas surrounded by dashed lines indicate the Cas-3 signal. Nuclei were stained with DAPI (blue). Genotypes: wild type (A,B,D); repoGal4>UAS-Fly-FUCCI (C); repoGal4>UAS-myrRFP (E). Scale bars: 50 μm (A,B,D); 15 μm (A′ right panel, C,E).
Next, to demonstrate that this novel crush injury produces cellular damage, we analysed the synapses in the VNC. Bruchpilot (Brp, also known as and hereafter referred to as nc82) is a presynaptic protein located in the active zones (Jordán-Álvarez et al., 2017), and one of the last proteins incorporated to synapses (Fouquet et al., 2009). Therefore, nc82 staining is a reliable marker for mature synapses, indicating the cellular state of the neuropil. nc82 immunostaining of VNCs 24 h ACI revealed a decrease of the nc82 signal in injured animals compared to non-injured controls of the same age (Fig. 2B,B′). We observed that the nc82 signal in the non-injured area of the VNC also decreased, probably due to synapse loss of dying neurons within the injured area that project to other areas. Therefore, to normalise the nc82 signal, we calculated the ratio of metathoracic neuromeres to brain (MtN:Brain) in each fly, corresponding to the ratio of injured to intact area. These results suggest that the injury provokes a reduction of the number of synapses (Fig. 2B″), consistent with the lack of movement of the third pair of legs observed after crush injury (Movie 1).
To determine the extent of the damage caused by crush injury, we analysed the cuticle and muscles in the area of damage. To avoid selecting flies with unwanted damage, e.g. bacterial infection, we discarded those flies that 24 h ACI showed any sign of undesirable external damage, such as deformation or change of pigmentation (Fig. S1B). To determine whether our injury paradigm results in muscle damage, we dissected the thorax of adult flies 24 h ACI. We damaged flies expressing green fluorescent protein (GFP) in muscle cells (mhc-Gal4) to visualise the ultrastructure of their metathoracic dorsoventral muscle, which are in the damaged area. Confocal images show that crush injury causes noticeable fibre disruption and damage to this muscle (Fig. S1C). We also investigated whether the integrity of injured VNCs remained intact. Although we found disruption of the blood brain barrier (Fig. S1D), we did not observe defects in the macrostructure of the VNC (Fig. S1E,E′). Therefore, we concluded that there is substantive damage in both the CNS and metathoracic dorsoventral muscles upon crush injury, comparable to what happens to humans with spinal cord injuries (Biering-Sørensen et al., 2009; Emery et al., 1998; Kakulas, 1999; Norenberg et al., 2004).
To elucidate whether this new paradigm produces the stereotypical responses of glial cells described in previous reports (Griffiths and Hidalgo, 2004; Kato et al., 2009, 2011; Losada-Perez et al., 2016; Purice et al., 2017; Smith et al., 1987; Treherne et al., 1984), we first determined whether crush injury induced proliferation of glial cells. To visualise cell-cycle progression and to quantify cell proliferation, we applied a 24-h pulse with the thymidine analog bromodeoxyuridine (BrdU). Quantification of BrdU-positive cells in the metathoracic region revealed significantly increased BrdU incorporation in glial cells (Repo+) upon injury (Fig. S1F,F″). To further confirm glial cell proliferation in response to crush injury, we visualised the fluorescent ubiquitination-based cell-cycle indicator (FUCCI) cell-cycle reporter (Zielke et al., 2014). This Fly-FUCCI reports accumulation of E2F (GFP) and CycB (RFP) protein, indicating phases G1, S or G2/M of the cell cycle. We restricted expression of this UAS-reporter to glial cells under the control of the repoGal4 promoter (Casas-Tintó et al., 2017) and quantified the number of RFP- and GFP-double-positive (RFP+GFP+) cells to determine mitotic events. Our analysis showed that crush injury induces glial cell proliferation (Fig. 2C,C″).
It is well-documented that expression of the engulfment factor drpr as well as engulfment of cellular debris are features of GRR (Doherty et al., 2009, 2014; Logan et al., 2012; MacDonald et al., 2006, 2013). To determine the amount of Drpr in glial cells after injury, we used antibody specifically recognising Drpr to immunostain dissected VNCs 5 h ACI. Compared to that of controls, the Drpr signal is significantly higher in the metathoracic ganglia of injured VNCs (Fig. 2D,D′, Fig. S2A). We calculated the ratio between MtN and prothoracic neuromere (MtN:PN), which corresponds to the ratio of injured to non-injured area. The results indicate that, upon crush injury, Drpr protein levels were higher in the injured area (Fig. 2D″). Subsequently, 24 h ACI the Drpr signal colocalised with glial membranes that surround caspase-3-positive bodies (Fig. 2E, Movie 2). These results indicate that crush injury can activate the phagocytic response in glial cells of the CNS.
Next, we evaluated functional recovery following crush injury. Previous work in Drosophila larvae demonstrated that GRR is a natural mechanism that promotes tissue repair (Kato et al., 2011). However, to date, there is no paradigm that uncovers functional recovery in adult flies. We, therefore, performed a functional assay to investigate whether this tissue repair can be translated into a functional recovery in Drosophila adults. We monitored locomotion of injured flies and analysed the movement of the damaged legs; i.e. we selected injured flies that dragged rear T3 legs 24 h ACI (T0) (Movie 1) and, subsequently, analysed the movement at 3 and 7 days ACI in comparison with T0.
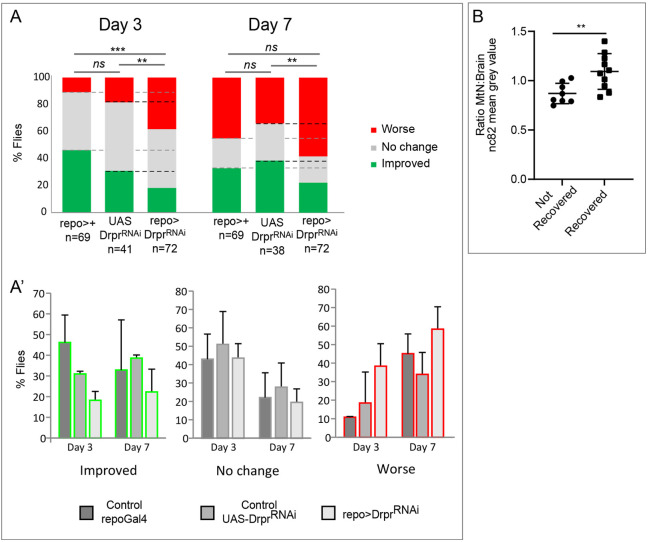
These results demonstrate that injured flies recover the movement of the legs at 3 and 7 days ACI (Fig. 3A,A′). To demonstrate that functional recovery implies CNS regeneration, we quantified the number of synapses within the injured area by staining for nc82 at 7 days ACI. We compared the ratio of the MtN:Brain nc82 signal in recovered and not recovered flies, and found it to be higher in recovered flies (Fig. 3B). These results demonstrate for the first time that functional recovery following CNS injury in Drosophila correlates with synapse number in the CNS. Additionally, we observed that the MtN:Brain ratio of the nc82 signal in recovered and non-recovered flies is lower than at 24 h ACI (Fig. 2B″), which suggests that the loss of synapses is most likely to occur 7 days ACI.
Fig. 3.
Adult Drosophila recover functionality after CNS crush injury and this requires glial Drpr. (A) Histograms representing the percentage of injured flies that correspond to each of the three movement categories Worse (red), No change (grey) or Improved (green), compared to T0 (24 h ACI). Functional recovery capacity of control flies (repoGal4 and UAS-drprRNAi alone) compared with drpr knockdown in glia. Chi-square test with two degrees of freedom, ***P<0.0001, **P<0.005. (A′) Histograms represent the percentage of flies corresponding to each category separately. (B) Quantification of the MtN:Brain nc82 mean intensity signal ratio of not recovered and recovered flies 7 days ACI. Unpaired t-test, **P<0.005. Genotypes: tubGal80ts, repoGal4/+, UAS DrprRNAi/+, tubGal80ts, repoGal4>UAS-DrprRNAi (A,A′); wild type (B).
Next, we aimed to study the mechanisms that govern neural repair. Previous work has focused on the proliferative component of GRR; however, we hypothesise that immune components play a central role in neural repair. We focused our initial efforts on Drpr, as this protein is one of the most-extensively studied components of the immune system in injury models (Doherty et al., 2014; Logan et al., 2012; MacDonald et al., 2013; Purice et al., 2017). We observed that drpr is consistently upregulated in glial cells within the injured area (Fig. 2D″) – in line with previous reports regarding other injury models (Doherty et al., 2009, 2014; Logan et al., 2012; MacDonald et al., 2006, 2013). Next, we tested whether expression of drpr in glial cells is required for functional recovery following crush injury. By using RNA interference (RNAi), we downregulated expression of drpr in glial cells (repo-Gal4) and analysed leg movement. The results show that downregulation drpr in glial cells (repo>drprRNAi) prevented the spontaneous functional recovery of the movement (Fig. 3A,A′). To further validate the contribution of Drpr, we tested the functional recovery in drpr mutant flies (draper Δ5 null allele) and obtained similar results (Fig. S3A,B). These results highlight that manipulation of gene expression in glial cells is sufficient to modify functional recovery, and that Drpr is required for functional recovery following CNS crush injury.
Drpr and JNK in crush injury
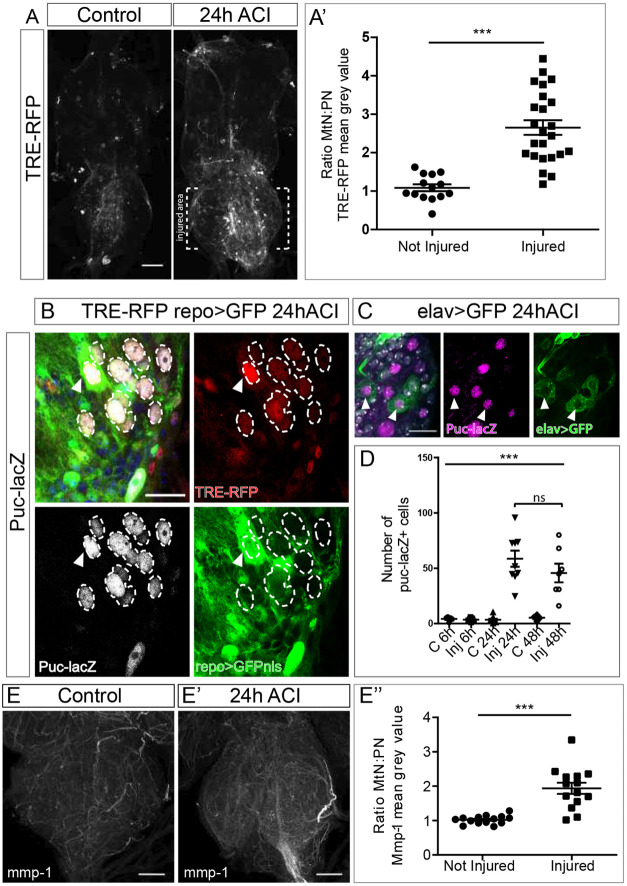
Drpr signalling has been studied in response to degenerating axons in alternative injury paradigms in Drosophila adult CNS; Drpr activates downstream pathways via Stat92E, MMP-1 or JNK (Doherty et al., 2014; MacDonald et al., 2013; Purice et al., 2017). Moreover, axotomy of legs or wings stimulate the activity of the AP-1/JNK pathway in the VNC (Purice et al., 2017). We analysed JNK activation in response to crush injury by using the AP1-recognition sequence TPA responsive element (TRE) conjugated to red fluorescent protein (RFP) (Chatterjee and Bohmann, 2012). This TRE-RFP signal was detected in the entire VNC in both injured as well as non-injured VNCs, with levels significantly increased in the injury area of damaged VNCs (Fig. 4A). To validate this observation, we calculated the MtN:PN ratio of RFP, which confirmed the increase of TRE-RFP reporter signal in injured VNCs (Fig. 4A′). To further confirm JNK activation, we used an additional JNK reporter, puckered-lacZ (puc-Z) (Martín-Blanco et al., 1998). To validate the reporter proteins, we first showed that all puc-Z-positive cells are also TRE-RFP positive (Fig. 4B). We also observed that JNK activation following injury occurs in both neurons and glial cells (Fig. 4B,C). Finally, we analysed the temporal activation of the JNK pathway in response to crush injury. We quantified JNK activation 6, 24 and 48 h ACI, and found a significant increase in the number of puc-Z-positive cells 24 h ACI, which is maintained up to 48 h ACI (Fig. 4D).
Fig. 4.
The JNK signalling pathway is activated in glial cells upon crush injury. (A) Representative images of not injured and injured VNCs showing the TRE-RFP reporter signal. (A′) Quantification of activated JNK signalling. Plotted is the metathoracic neuromere to prothoracic neuromere (MtN:PN) ratio of the TRE-RFP mean grey value of not injured to injured VNCs. Unpaired t-test, ***P<0.0001. (B) Images show a representative section of an injured MtN expressing the JNK reporters TRE-RFP and puc-Z, and the glial marker repo>GFP. puc-Z-positive cells, surrounded by dashed lines, that are also positive for TRE-RFP. Arrowheads indicate glial cells with active JNK. (C) Representative section of an injured MtN marked with the JNK reporter puc-Z and the neuronal marker elav>GFP. Arrowheads indicate neurons with active JNK. (D) Plotted is the number of puc-Z-positive cells in not injured and injured VNCs at different time points. One-way ANOVA, ***P<0.0001; Bonferoni's multiple comparison; n.s. not significant. (E-E′) Representative images of MtNs stained for Mmp-1 in not injured (E) and injured (E′; 24 h ACI) VNCs. (E′′) Quantification of the MtN:PN ratio of the Mmp-1 signal mean grey value. Unpaired t-test, ***P<0.0001. Genotypes: TRE-RFP; repoGal4>UAS-GFPnls (A,E). TRE-RFP; repoGal4>UAS-GFPnls/puckered-LacZ (B). elavLexA>lexAOP-GFP/+ (C). Scale bars: 50 μm (A,E), 15 μm (B,C).
In addition, we analysed the protein expression of matrix metalloproteinase 1 (MMP1), which is directly associated to JNK activity (Purice et al., 2017). We used a specific anti-MMP1 antibody and stained injured and non-injured VNCs. Quantification of the MMP1 signal showed an increase in the injured area (Fig. 4E-E″, Fig. S2B), in line with previous results. Thus, we conclude that crush injury activates JNK signalling in the CNS.
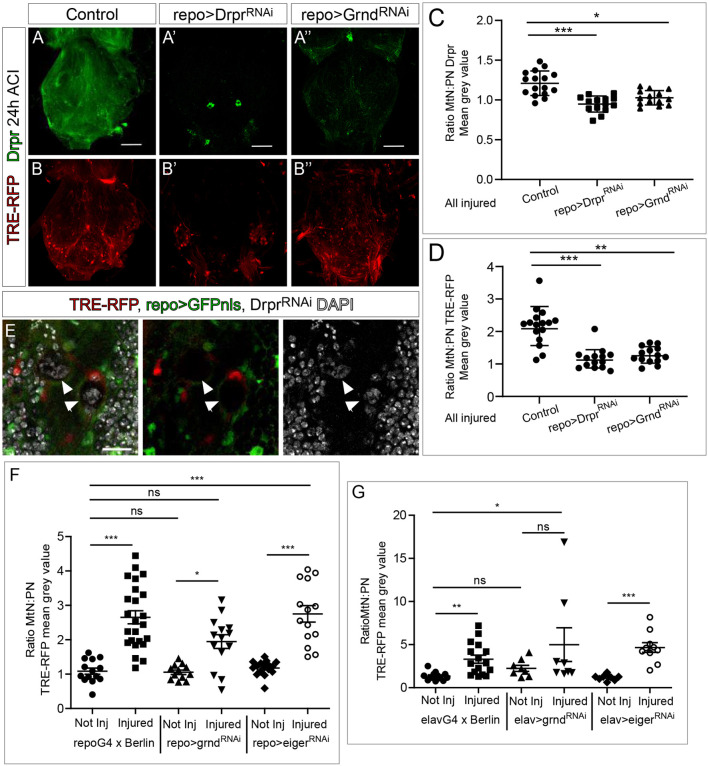
Next we asked whether activation of JNK signalling is triggered by Drpr, as described in other axotomy paradigms (MacDonald et al., 2013; Purice et al., 2017). Expression of drpr small interfering RNA (siRNA) reduced the Drpr protein signal in glial cells (we were unable to detect Drpr staining except in two neurosecretory neurons within the MtN) and prevented the increase of Drpr signalling (MtN:PN ratio) upon injury in the VNC (Fig. 5A,A′,C and Fig. S2C). Similarly, we analysed the activity of the JNK pathway – by using the TRE-RFP reporter – in injured animals upon drpr knockdown in glial cells. Quantification of the TRE-RFP signal showed a significant reduction of JNK activation upon injury when compared to VNCs of control injured flies (Fig. 5B,B′; Fig. S2D). In line with these results, the increase of TRE-RFP reporter signal ratio (MtN:PN ratio) upon injury was prevented (Fig. 5D). We also observed spots of DNA accumulation in the injured area, suggesting the presence of apoptotic cellular debris due to the absence of Drpr (Fig. 5E). These results indicate that Drpr is required for JNK activation, in line with previous reports in other injury models.
Fig. 5.
JNK and Drpr regulate each other and this requires Grnd. (A-B″) Representative metathoracic neuromeres (MtNs) stained for Drpr, showing the TRE-RFP reporter signal at 24 h ACI in control flies (A,B) and in flies with knocked down drpr (A′,B′) or knocked down grnd (A″,B″) in glial (repoGal4) cells. (C) Quantification of MtN:PN ratio of Drpr staining. One-Way ANOVA, *P<0.05; Bonferroni multiple comparison, ***P<0.0001. (D) Quantification of MtN:PN ratio of the TRE-RFP reporter signal. One-Way ANOVA, **P<0.001; Bonferroni multiple comparison, ***P<0.0001. (E) Images show the accumulation of nuclear debris (arrowheads, stained with DAPI, grey) due to downregulation of drpr expression in glial cells. The TRE-RFP JNK reporter is shown in red, glial cells (repo>GFP) are shown in green. (F,G) Quantification of the metathoracic neuromere to prothoracic neuromere (MtN:PN) ratios of the TRE-RFP mean grey value of not injured to injured VNCs after knocking down grnd or eiger in glial cells (repoGal4, F) or neurons (elavGal4, G). Dunn's multiple comparison test, *P<0.05, **P<0.001, ***P<0.0001. ns, not significant. Genotypes: TRE-RFP; repoGal4-UAS-GFPnls (A-D). TRE-RFP; repoGal4>UAS-GFPnls, UAS-drprRNAi (A′,B′,C-E). TRE-RFP; repoGal4>UAS-GFPnls, UAS-grndRNAi (A″,B″,C,D). TRE-RFP; repoGal4>UAS-GFPnls, TRE-RFP; repoGal4>UAS-GFPnls, UAS-grndRNAi, TRE-RFP; repoGal4>UAS-GFPnls, UAS-eigerRNAi (F). TRE-RFP; elavGal4>+, TRE-RFP; elavGal4>UAS-grndRNAi, TRE-RFP and elavGal4>UAS-eigerRNAi (G). Scale bars: 50 μm (A,B), 15 μm (E).
The JNK pathway is evolutionarily conserved from fruit fly to humans (La Marca and Richardson, 2020; Pinal et al., 2019; Wu et al., 2019; Zeke et al., 2016). JNK signalling can be triggered by various extrinsic and intrinsic cues, and regulates a wide range of cellular process including proliferation, differentiation, migration and cell death (Weston and Davis, 2007). In Drosophila, binding of the tumor necrosis factor (TNF) orthologue Eiger (Egr) to its receptor Grindelwald (Grnd) activates JNK signalling pathway (Andersen et al., 2015; Glise et al., 1995; Igaki et al., 2002; Moreno et al., 2002; Portela et al., 2019a; Takatsu et al., 2000; Wu et al., 2019). To determine the contribution of the canonical ligand Eiger and the receptor Grnd in JNK pathway activation after crush injury, we downregulated expression of each, specifically in neurons or glia, and measured JNK pathway activation (assessed by measuring the MtN:PN ratio of TRE-RFP reporter). The results showed that grnd expression is required for JNK pathway activation in both neurons and glia, although the difference between non-injured and injured VNCs in repo>grndRNAi was still significant (Fig. 5F). Thus, to confirm the requirement of Grnd in glial cells, we expressed the dominant-negative form of Grnd, Grnd-Extra (Andersen et al., 2015). JNK activation (MtN:PN ratio) after injury was significantly decreased upon expression of Grnd-Extra in glial cells compared with controls (Fig. S2E). To further confirm the presence of Grnd in the injured area, we stained adult brains with a specific antibody against Grnd. Quantification of confocal images showed that Grnd is located in the adjacent region between the glial and neuron membrane (Fig. S2F,F′). By contrast, RNAi of eiger in glial cells or neurons did to not affect JNK activation in VNCs upon injury (Fig. 5F,G). To confirm the implication of Eiger in JNK pathway activation in response to crush injury, we analysed different eiger mutants that had recently been validated (Kodra et al., 2020). The results showed a similar degree of JNK activation compared to that in controls (Fig. S2G) and suggest that, in this context, Eiger is not required for the activation of JNK pathway. Activation of JNK signalling through the Grnd receptor – but in a way that is independent of its ligand Eiger – has been previously described in response to perturbation of cell polarity in epithelial tumor model (Andersen et al., 2015).
Moreover, the upregulation of drpr by JNK has previously been described in Drosophila (axotomy paradigms) and in C. elegans (Chiu et al., 2018; MacDonald et al., 2013). Therefore, we aimed to determine whether activity of the JNK pathway activates drpr expression through activation of the Grnd receptor. To reduce JNK pathway activity, we downregulated grnd by using RNAi specifically in glial cells (Fig. 5A,B″). We stained injured VNCs with anti-Drpr (Fig. 5A,A″), quantified the Drpr signal (Fig. S2C) and calculated the Drpr MtN:PN ratio (Fig. 5C). The results showed a reduction of Drpr signal in injured animals when grnd gene expression was downregulated, suggesting that JNK pathway activity in glial cells is required for upregulation of drpr expression upon injury in the VNC (Fig. 5A,A″,C). To validate these results, we reproduced similar experiments after overexpression of Grnd-Extra (Andersen et al., 2015) in glial cells, and obtained similar results (Fig. S2H). Together, our results indicated that drpr is upregulated in glial cells of the VNC upon crush injury, leading to activation of JNK signalling in glial cells and neurons via the Grnd receptor. We, therefore, concluded that JNK pathway activation in glial cells promotes drpr expression and vice versa.
Neuronal vesicular transport is required for regulation of JNK signalling and functional recovery upon injury
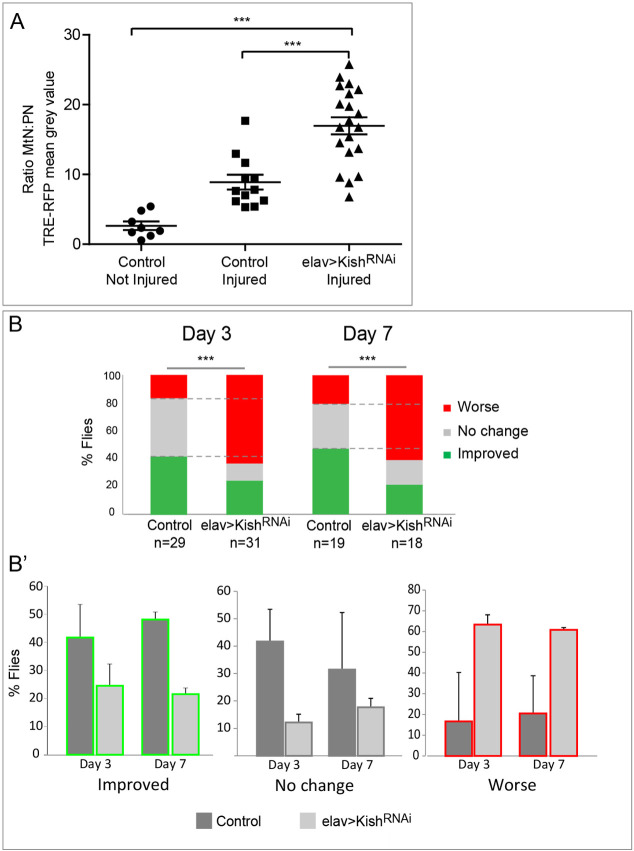
As glial cells receive signals from injured tissue, we hypothesised that affected neurons upon injury increase vesicular trafficking and secretory activity. To investigate whether the secretory nature of neurons implicated neuron-glial communication in this injury paradigm, we knocked down kish expression in neurons, which is reported to be involved in vesicle transport and secretion (Gershlick et al., 2016; Portela et al., 2019b; Wendler et al., 2010). We analysed JNK signalling following injury in these animals (elav>kishRNAi) and found that knockdown of kish stimulates JNK pathway signalling (Fig. 6A). However, kish knockdown in glial cells (repo>kishRNAi) did not affect activation of the JNK pathway in injured VNC (Fig. S4). These results suggested that secretion from neurons and, therefore, neuron-glial communication is required for JNK regulation in response to CNS injury.
Fig. 6.
Vesicular trafficking in neurons is required for JNK regulation and functional recovery. (A) Plotted is the metathoracic neuromere to prothoracic neuromere (MtN:PN) ratio of the TRE-RFP mean grey value of not injured and injured control flies, and injured flies whose vesicular transport in neurons is blocked. Dunn's multiple comparison, ***P<0.0001. (B) Percentage of functional recovery capacity of kish knockdown flies compared to that of controls. Chi-square test with two degrees of freedom, ***P<0.00001. (B′) Histograms represent percentage of flies in each category of movement, i.e. Worse (red), No change (grey) or Improved (green). Genotypes: TRE-RFP, tubGal80ts; UAS-KishRNAi/+ (Control) and TRE-RFP, tubGal80ts; elavGal4>UAS-KishRNAi (A,B).
To validate the relevance of neuron-glial communication in the activation of glial responses following injury, we next analysed functional recovery of injured flies after blocking vesicular transport in neurons. The results showed that functional recovery was compromised upon kish knockdown in neurons (Fig. 6B,B′). Hence, we concluded that, in neurons, the reduction of vesicular transport in response to kish RNAi is detrimental for functional recovery upon crush injury.
Taken together, these results highlight that neuron-glial communication involving vesicular transport in neurons is required for JNK pathway regulation and functional recovery in response to injury.
Macrophages are recruited to the crush injury site and are required for functional recovery
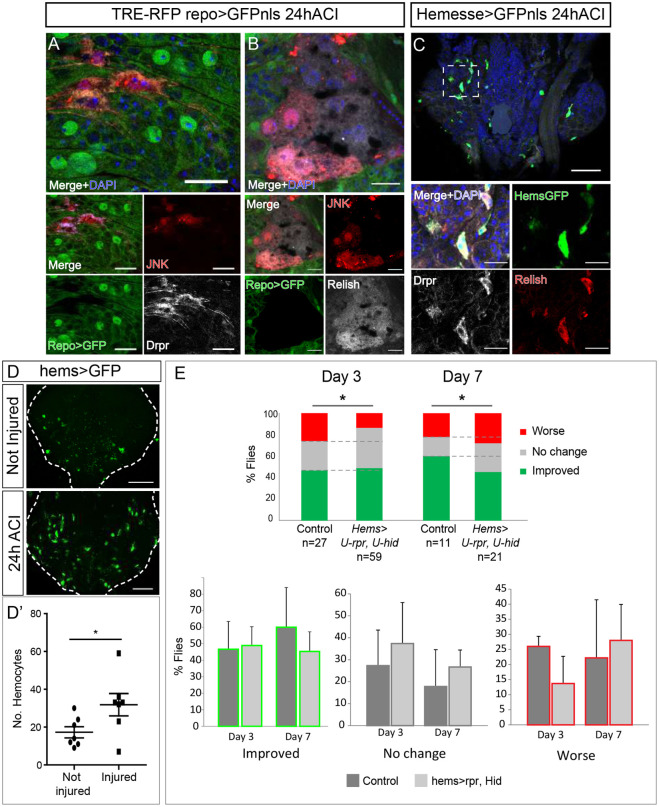
The immune response is crucial to facilitate the recovery process after injury in all animal systems. Previous studies have reported that the immune system plays a central role following brain injury (Gadani et al., 2015), and wound healing in epithelial tissues requires JNK signalling pathway activation (Galko and Krasnow, 2004). We report here the presence of non-glial cells [Repo negative (Repo−)] in injured VNCs, showing JNK pathway activation [positive for TRE and RFP (TRE-RFP+)] and Drpr at the injured site (Fig. 7A, Fig. S1D). These cells were located at the surface of the VNC, immediately above the outer-most glial layer. To investigate whether these cells express other genes of the immune system, we used specific antibody to analyse the amount of transcription factor Relish, the third Drosophila NFκB-related protein, which is homologous to mammalian NFκB1 (NFKB1) and activates the innate immune response (Tanji et al., 2010). We identified Repo−, TRE-RFP+, Relish+ cells, having a lamellocyte-like shape, above the outer-most glial cell layer (Fig. 7B). Thus, we asked whether non-glial cells that express the immune genes drpr and relish and whose JNK signalling pathway is activated are immune cells derived from the hemolymph, i.e. hemocytes, also known as macrophages in vertebrates. We used the specific hemocyte driver Hemesse-Gal4 to express membrane-targeted GFP tagged to CD8 (CD8-GFP) and stained injured VNCs for Drpr and Relish. We showed that GFP-positive cells (hemocytes) are recruited to the affected area within injured VNCs. These cells also stained positive for Drpr and Relish (Fig. 7C). Next, we quantified the number of hemocytes (Hems>RedStinger) in injured MtN compared to non-injured controls and found a significant increase within injured sites (Fig. 7D,D′). These results indicate that macrophages are recruited to the injury site in the VNC, where they activate JNK, Drpr and Relish.
Fig. 7.
Phagocytes are recruited to injured VNCs and required for functional recovery. (A) Surface of metathoracic neuromere (MtN) stained for Drpr (grey) and active JNK signaling (TRE-RFP, red) in non glial cells. Glial cells (repo>GFP) are in green; nuclei were stained with DAPI (blue). The top image is a magnified version of the merged image below. (B) Surface of MtN showing non glial cells stained for Relish (grey) and active JNK signalling (red); nuclei were stained with DAPI (blue). The top image is a magnified version of the merged image below. (C) MtN hemocytes stained for Drpr (grey) and Relish (red); nuclei were stained with DAPI (blue). The boxed area is shown magnified in the four images at the bottom in A-C. (D) Representative images and quantification of hemocytes in not injured and injured MtNs. (D′) Plotted are the number of hemocytes in not injured and injured MtNs. Unpaired t-test, *P<0.05 (E) Quantification of the functional recovery. Percentage of flies whose hemocytes had been genetically ablated compared to that of control flies. Data were quantified for each category of movement, i.e. Worse (red), No change (grey) or Improved (green). Chi-square test with two degrees of freedom, *P<0.1. Genotypes: TRE-RFP; repoGal4>UAS-GFPnls (A,B). HemesseGal4>UAS-CD8GFP (C,D), CD8 is a transmembrane protein, therefore the cell surface is labeled with GFP. HemesseGal4>UAS-RedStinger (D′). UAS-reaper, UAS-hid (Control) and tubGal80ts; HemesseGal4>UAS-reaper, UAS-hid (E). Scale bars: 15 μm (A, B, C bottom image); 50 μm (C top image, D).
Finally, we asked whether hemocytes contribute to functional recovery. We performed a functional recovery assay of injured flies and compared control flies with flies comprising genetically depleted hemocytes. To eliminate hemocytes from the organism, we overexpressed the pro-apoptotic genes hid and reaper under the control of a specific driver (Hems-Gal4>UAS-reaper, hid) – a strategy reported to eliminate all hemocytes (Lolo et al., 2012). Our results show that 3 days ACI, elimination of hemocytes does not impact recovery; on the contrary, it even improves as the percentage of worsening flies is reduced (Fig. 7E). Later, 7 days ACI, it is clear that the percentage of flies that improve locomotion is reduced when hemocytes are eliminated (Fig. 7E). So, all together the results suggest that, even though the initial response of the immune system can accelerate the worsening of flies, 7 days ACI elimination of hemocytes contributes to the worsening of flies in response to crush injury. This implies that hemocytes are required for functional recovery after crush injury.
DISCUSSION
Glial responses to CNS injury are conserved from Drosophila to mammals. Here, we show that the processes that occur following CNS injury in humans (Biering-Sørensen et al., 2009; Emery et al., 1998; Kakulas, 1999; Norenberg et al., 2004) were reproduced in Drosophila. However, the aggressiveness of the damage in this novel paradigm is less sever compared to other models – i.e. torn-off leg, needle stuck into the brain – as this crush injury model does not imply mutilation. We present a novel paradigm that allows to study cellular events upon CNS injury in Drosophila, recapitulating loss of synapses, apoptosis, activation of glial immune response and the JNK pathway as observed in other injury paradigms, and providing a novel tool to study the functional regeneration of the CNS. We focused on injury and cellular responses that occur in the CNS but cannot discard that other tissues might be affected in this paradigm.
We observed that glial cells divide in response to CNS crush injury, thereby reproducing the features of Drosophila and other species upon injury. In the cockroach, toxin-induced glial ablation produces compensatory glial proliferation (Smith et al., 1987; Treherne et al., 1984). In vertebrates, rodents and zebrafish, spinal cord injury or toxin ablation of oligodendrocytes or oligodendrocyte progenitor cells (OPCs) induce proliferation of the remaining OPCs (Kato et al., 2018). Finally, CNS injury throughout different developmental stages (embryo and larva) also produces glial proliferation in Drosophila (Griffiths and Hidalgo, 2004; Kato et al., 2009, 2011; Losada-Perez et al., 2016). These studies demonstrate the high conservation of this response and validate our model to study the glial proliferative response.
The glial phagocytic response to injury of CNS and PNS is also conserved across species. CNS injury of adult Drosophila stimulates phagocytic glial cells that engulf cell debris and apoptotic cells mediated by the upregulation of the engulfment receptor drpr/MEGF10 (Doherty et al., 2009; MacDonald et al., 2006; Morizawa et al., 2017). We showed here that drpr is upregulated in response to crush injury, which is in line with previous studies. Furthermore, in glial cells, Drpr activated the JNK signalling pathway that, in turn, is required to upregulate drpr gene expression – also in line with previous results (MacDonald et al., 2013). We demonstrated that JNK activation requires the receptor Grnd in both neurons and glia cells. Downregulation of grnd in neurons also reduced JNK activation following injury, highlighting the importance of neuron-glial communication. Moreover, vesicular transport in neurons is required to prevent further JNK activation in response to crush injury, supporting the significance of neuron-glial communication. These results demonstrate that, in our crush injury model, the glial immune response was activated; therefore, it can also be used to study the immune components of the GRR. We also showed the implication of circulating immune cells regarding the repair of CNS injury. Hemocytes were recruited to the injured site and are required for functional recovery. Thus, our crush injury paradigm allows the study of systemic immune responses upon CNS injury (Fig. 8).
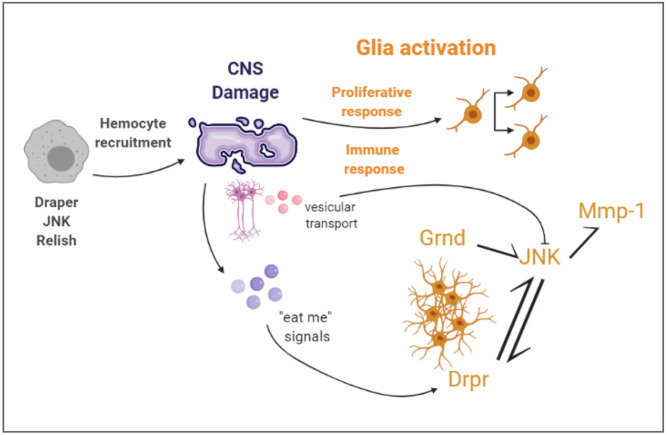
Fig. 8.
Schematic, summarizing the cellular events described in this article. Crush injury causes damage in the CNS, and triggers a proliferative and immune response in glial cells. The glial immune response includes activation of the JNK pathway via Grnd and Drpr. Mmp-1 activation, most probably downstream of JNK, also occurs upon crush injury. Vesicular transport in neurons is also required to modulate JNK activation upon injury. Hemocytes, containing Drpr and Relish and with activated JNK, are recruited to the injury site. Diagram created with BioRender.com.
Moreover, it enables the analysis of functional recovery and how expression of specific genes in different cell types contribute to it. Mammals and insects recover locomotion following limb amputation (Isakov et al., 2016; Kirpensteijn et al., 1999), showing that CPGs and locomotor controls are adapt to new requirements. After limb amputation flies learn to move normally (Isakov et al., 2016). Therefore, to study locomotion recovery it is required to focus on injured limbs. Our novel paradigm focussed on affected limbs and, therefore, we concentrated on the regeneration of damaged tissue. We analysed the rear legs during different time points (1, 3 and 7 days ACI), instead of following other standardised methods that measure the movement of the animal as a whole, such as during trikinetics or open-field-arena assays (Valente et al., 2007; Chiu et al., 2010; Soto-Padilla et al., 2018).
We show for the first time that wild-type flies spontaneously recover from the loss of synapses, and improve the movement of impaired legs to a certain extent. We found that, in glial cells, Drpr is required for functional recovery, linking the immune glial response to functional regeneration. kish knockdown in neurons reduces regenerative capacity upon injury, suggesting that vesicular transport is relevant for neuronal physiology or that neuron-glial communication is required for functional recovery itself. Previous publications (Portela et al., 2019b) suggest that RNAi of kish does not affect neuronal features; thus, we propose, in line with previous observations (Shklover et al., 2015), that neuron-glial communication is required for functional recovery. Finally, the fact that ablation of hemocytes – i.e. the Drosophila equivalent of macrophages in vertebrates – impedes functional recovery in response to crush injury, suggests that macrophages are required for functional regeneration. Together, our results demonstrate that functional recovery can be altered by genetic modifications, such as RNAi of grnd or drpr in glial cells or RNAi of kish neurons. It is of relevance to mention the intermediate phenotypes shown by some upstream activation sequence (UAS) Drosophila lines in the functional recovery results. The leakiness of the UAS system has been previously discussed (Akmammedov et al., 2017) and is associated to the residual activity of the Hsp70 minimal promoter. Therefore, even though results are consistent, some results based on the use of UAS fly lines as controls show a tendency consistent with the proposed residual activity.
A key challenge for the field is to determine the specific molecular and cellular causes of motor activity impairment and recovery; any information will facilitate the understanding of mechanisms that underlie functional regeneration. Physical damage to the nervous system causes a plethora of signals that need to be studied in-depth as a grouped response. The contribution of PI3K-GSK3 signalling pathway has been proposed to be a valid strategy to promote synaptogenesis, mitochondrial restoration and axon regeneration (Huang et al., 2019; Saijilafu et al., 2013). However, these studies mainly focused on the re-myelination process, an event crucial for the full regeneration of the nervous system but – on its own – insufficient to regenerate function upon damage. Here, we provide a new model to study physical damage of the nervous system, and determine the cellular and molecular events that are relevant for the final outcome of the organism upon injury. This paradigm of adult CNS injury brings new perspectives to this area of research, as it focuses on functional recovery, the most-relevant feature of regeneration. The advantages of the UAS/Gal4 system, the collection of specific Gal4 lines for each glial cell type, together with the ability to be able to manipulate functional recovery, make the crush-injury paradigm an ideal model in the search for new targets in order to improve CNS regeneration.
MATERIALS AND METHODS
Fly stocks and genetics
Experiments were performed in adult Drosophila melanogaster flies raised at 25°C or combining 17°C with 29°C for temporal control of UAS/Gal-4 using the thermo-sensitive repression system Gal80TS. Stocks were generated by conventional genetics. Fly stocks used were: UAS-CD8 GFP (BL-79626), UAS-myr-RFP (BL-7119), repo-Gal4 (BL-7415), tub Gal80ts (BL-7019), UAS-eiger RNAi (BL-58993), UAS-GFP.E2f, UAS-mRFP.CycB (Fly- FUCCI, BL-55100), UAS-RedStinger (BL 8546), elavLexA (BL-52676), LexOP-CD8GFP (BL-66545), eiger MIMIC (BL 59754), mhc-Gal4 (BL 55133) form Bloomington Drosophila Stock Center; UAS-kish RNAi (v40884) from Vienna Drosophila Resource Centre; UAS-Drpr RNAi (BL-67034) from M. Freeman; Berlin, UAS-puckered-LacZ, elav-Gal4 from A. Ferrus; UAS-hid, UAS-reaper from Carlos Estella (CBM-CSIC, Madrid, Spain); UAS-Grnd RNAi (v43454), UAS-Grnd-Extra, TRE-RFP (BL-59011) from Jean-Paul Vincent (The Francis Crick Institute, London, UK); Hemese-Gal4 (BL-8699) from Dan Hultmark (Tampere University, Tampere, Sweden); Draper Δ5 from Eduardo Moreno (Champalimaud Centre for the Unknown, Lisbon, Portugal); eiger1AG, eiger3AG from Laura Johnston (Columbia University, New York, NY).
To downregulate gene expression, we used specific UAS-RNAi lines to target the genes of interest, under the control of pan-neuronal (elav>) or pan-glial (repo>) Gal4 drivers. To control the downregulation of different genes we combined the UAS/Gal4 system with the Tubulin-Gal80TS (temperature sensitive) tool. We maintained the desired genotypes at non-permissive temperature (i.e. 17°C, at which the Gal80TS repressor protein is active), to block UAS constructs overexpression. Then, we collected 1-2 day-old adult flies after eclosion (equivalent to 0-1 day-old flies grown at 25°C), and performed the crush injury, keeping them at permissive temperature (i.e. 29°C, at which Gal80TS is inactive and the Gal4/UAS system is active). Injured flies dragging the third pair of legs and did not show signs of cuticle damage were selected 24 h later (24 h ACI).
Crush injury
Flies were anesthetised on a CO2 plate and placed ventral side up. Each fly was immobilised using forceps. Finer forceps were used to gently apply pressure between the second and third pair of legs, aiming to injure the metathoracic VNC neuromere. Once injured, flies were placed in a fresh food vial. 24 h ACI (or 5 h ACI for experiments described in Fig. 2D); flies that dragged the third pair of legs were selected. Of those selected, flies with any sign of cuticle damage were discarded.
BrdU incorporation
For incorporation of the thymidine analog bromodeoxyuridine (BrdU), injured flies (immediately following crush injury) and not-injured flies were placed in Eppendorf tubes containing food and BrdU (5% sugar, 1% food colorant and 1 mg/ml BrdU, Roche). Not-injured and injured flies with colorant in the abdomen were selected 24 h after crush injury.
Immunohistochemistry and BrdU detection
Immunohistochemistry was performed following the protocol for adult Drosophila immunolabelling according to Purice et al. (2017). For BrdU detection, dissected VNCs were treated with 2 M HCl for 20 min (RT) prior to incubation with anti-BrdU.
We used the following antibodies: mouse anti-Repo (DSHB, 8D12 ID: AB_528448, 1:200), rat anti-BrdU (Abcam 1:500, ab6326), mouse anti-nc82 (anti-Bruchpilot; DSHB 1:30, 2ea ID: AB_2314866), rabbit anti-cleaved caspase-3 (Asp175) (5A1E) (Cell Signaling Technology, 1:50, #9664), mouse anti-Mmp1 (DSHB, 3A6B4 ID: AB_579780; 3B8D12 ID: AB_579781; 5H7B11 ID: AB_579779 used at 1:1:1, 1:50), mouse anti-Relish-C (DSHB, 21F3 ID: AB_1553772, 1:50), guinea pig anti-Repo [gift from B. Altenhein (von Hilchen et al., 2013)], rabbit anti-Drpr [gift from M. Freeman (Freeman et al., 2003)], guinea pig anti-grnd [gift from P. Leopold (Andersen et al., 2015)].
Caspase 3 quantification
We stained adult Drosophila VNCs with anti-cleaved-caspase 3 (Cell Signaling Technology 1:50, #9664 lot number 18). All experimental groups were treated in parallel and analysed in blind experiments. Samples showing a bright caspase-3 signal were considered to be positive, all else as negative. The number of samples considered to be caspase-3 positive is represented as a percentage of the total number of samples.
To determine whether caspase-3 signals colocalised with condensed nuclei, we analysed the intensity of the DAPI signal by using the ‘mean grey value’ in FIJI software; this measurement takes into account both intensity and area of the signal. We measure five caspase-3-positive and five caspase-3-negative nuclei in nine samples of injured flies (i.e. 90 cells were analysed). For ROI selection of each nucleus we used ‘Oval selection’ to select the whole signal. In these confocal stacks, nuclei normally occupy two to three 1 µm slides, so we analysed the ‘mean grey intensity value’ in the slice that was closer to the centre of the nucleus, i.e. where the DAPI signal is more intense.
Functional recovery
24 h ACI, flies were selected by the reduction of movement in the third pair of legs. We also considered the lack of external visible damage, and introduced each fly into individual food vials. Motor capacity of the third pair of legs was characterised 1, 3 and 7 days ACI. We took into account locomotion capacity, grooming and trembling phenotypes, and established distinct categories. The following categories we established from zero movement to full functionality: leg dragging without any movement (i), leg dragging with movement, flexion (ii), flexion with movement (iii), movement intention (iv), tremor or grooming intention (v), clumsy movement (vi) and complete movement (vii). For functional recovery experiments motor capacity between days 3 and 7 was compared to day 1 ACI, and registered as ‘improved’ (increased motor capacity), ‘no change’ or ‘worse’ (decreased motor capacity).
Quantification, statistical analysis and imaging
Images were acquired by confocal microscopy (LEICA TCS SP5) and processed using Fiji (ImageJ 1.50e) software. Images were assembled using Adobe Photoshop CS4 and Adobe Illustrator CS4.
The ‘mean grey value’ was measured (using the measurement tool) by quantifying equivalent regions of interest (ROIs) of complete z-stack projections by using the Fiji plugin ‘sum slices’. Images were then rotated, so all VNCs had the same orientation. ROIs in different experimental groups were identical in area size and shape. To determine the ROI, we draw a rectangle around most of the hemineuromere to measure the intensity of the signal, but without any area outside the neuromere.
We calculated the ratio of metathoracic to prothoracic ganglia to normalise individual variances. In the case of anti-nc82, we found a decrease of signal in prothoracic segment in the injured samples; thus we normalised the results with the signal of anti-nc82 in the central brain.
Cells were counted using the cell counter plugin from Fiji or Imaris (Imaris 6.3.1 software- Bitplane). Quantification of Grnd protein within the membrane of glial cells and neurons was done by using the ‘Analyze→ plot profile’ tool in Fiji software.
Data were analysed and plotted using GraphPad Prism v7.0.0 or v9.0.0. Qualitative data were analysed only in Fig. 2A, where we performed a binomial test (****P<0.0001). For quantitative data, we used a D'Agostino–Pearson normality test and analysed data with normal distributions by using two-tailed t-test with Welch’s unequal variances t-test. When data had multiple comparisons, we used one-way ANOVA with Bonferroni post hoc-test; we used two-tailed Mann–Whitney U-test or Kruskal–Wallis test and Dunn’s post hoc-test for data that did not pass normality testing. The Chi-square test with two degrees of freedom was used for functional recovery experiments. Error bars represent standard deviation (+s.d.).
Supplementary Material
Acknowledgements
We thank Professor Alberto Ferrús, Dr Paco Martín and the anonymous reviewers for critiques of the manuscript and helpful discussions, Clemencia Cuadrado for fly stocks maintenance and Niki Anthoney for English supervision. We are grateful to Marc Freeman (Vollum Institute, Oregon Health and Science University, Portland, OR), Jean-Paul Vincent (The Francis Crick Institute, London, UK), Benjamin Altenhein (Institute of Zoology, University of Cologne, Germany), Laura Johnston, Columbia University, New York, NY), the Vienna Drosophila Resource Centre, the Bloomington Drosophila stock Centre and the Developmental Studies Hydridoma Bank for supplying fly stocks and antibodies, and FlyBase for its wealth of information. We acknowledge the support of the Confocal Microscopy unit at the Cajal Institute.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Author contributions metadata Conceptualization: M.L.-P., S.C.-T.; Methodology: M.L.-P., S.C.-T.; Validation: M.L.-P., S.C.-T.; Formal analysis: M.L.-P., N.G.-G., S.C.-T.; Investigation: M.L.-P., N.G.-G., S.C.-T.; Resources: S.C.-T.; Writing - original draft: M.L.-P., S.C.-T.; Writing - review & editing: M.L.-P., S.C.-T.; Visualization: M.L.-P., N.G.-G.; Supervision: M.L.-P., S.C.-T.; Project administration: S.C.-T.; Funding acquisition: S.C.-T.
Funding
This work was supported by the Ministerio de Ciencia y Tecnología (grant number: PID2019-110116GB-100, MICINN to S.C.T.), the Consejo Superior de Investigaciones Científicas (CSIC, grant number: LINKA 20268 to S.C.T.) and Fellowship from the Comunidad de Madrid (grant number: 2016-T2-BMD-1295 to M.L.-P.)
References
- Akmammedov, A., Geigges, M. and Paro, R. (2017). Single vector non-leaky gene expression system for Drosophila melanogaster. Sci. Rep. 7, 6899. 10.1038/s41598-017-07282-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, D. S., Colombani, J., Palmerini, V., Chakrabandhu, K., Boone, E., Röthlisberger, M., Toggweiler, J., Basler, K., Mapelli, M., Hueber, A.-O.et al. (2015). The Drosophila TNF receptor Grindelwald couples loss of cell polarity and neoplastic growth. Nature 522, 482-486. 10.1038/nature14298 [DOI] [PubMed] [Google Scholar]
- Awasaki, T., Tatsumi, R., Takahashi, K., Arai, K., Nakanishi, Y., Ueda, R. and Ito, K. (2006). Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron 50, 855-867. 10.1016/j.neuron.2006.04.027 [DOI] [PubMed] [Google Scholar]
- Becker, T., Wullimann, M. F., Becker, C. G., Bernhardt, R. R. and Schachner, M. (1997). Axonal regrowth after spinal cord transection in adult zebrafish. J. Comp. Neurol. 377, 577-595. 10.1002/(SICI)1096-9861(19970127)377:4<577::AID-CNE8>3.0.CO;2-# [DOI] [PubMed] [Google Scholar]
- Biering-Sørensen, B., Kristensen, I. B., Kjær, M. and Biering-Sørensen, F. (2009). Muscle after spinal cord injury. Muscle Nerve 40, 499-519. 10.1002/mus.21391 [DOI] [PubMed] [Google Scholar]
- Bradke, F., Fawcett, J. W. and Spira, M. E. (2012). Assembly of a new growth cone after axotomy: the precursor to axon regeneration. Nat. Rev. Neurosci. 13, 183-193. 10.1038/nrn3176 [DOI] [PubMed] [Google Scholar]
- Brancato, S. K. and Albina, J. E. (2011). Wound macrophages as key regulators of repair: origin, phenotype, and function. Am. J. Pathol. 178, 19-25. 10.1016/j.ajpath.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, A. B., Walradt, T., Gardner, K. E., Hubbert, A., Reinke, V. and Hammarlund, M. (2014). Insulin/IGF1 signaling inhibits age-dependent axon regeneration. Neuron 81, 561-573. 10.1016/j.neuron.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Tintó, S., Lolo, F.-N. and Moreno, E. (2015). Active JNK-dependent secretion of Drosophila Tyrosyl-tRNA synthetase by loser cells recruits haemocytes during cell competition. Nat. Commun. 6, 10022. 10.1038/ncomms10022 [DOI] [PubMed] [Google Scholar]
- Casas-Tintó, S., Arnés, M. and Ferrús, A. (2017). Drosophila enhancer-Gal4 lines show ectopic expression during development. R. Soc. Open Sci. 4, 170039. 10.1098/rsos.170039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, N. and Bohmann, D. (2012). A versatile ϕC31 based reporter system for measuring AP-1 and NRF2 signaling in Drosophila and in tissue culture. PLoS ONE 7, e34063. 10.1371/journal.pone.0034063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Wang, Z., Ghosh-Roy, A., Hubert, T., Yan, D., O'Rourke, S., Bowerman, B., Wu, Z., Jin, Y. and Chisholm, A. D. (2011). Axon regeneration pathways identified by systematic genetic screening in C. elegans. Neuron 71, 1043-1057. 10.1016/j.neuron.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, J. C., Low, K. H., Pike, D. H., Yildirim, E. and Edery, I. (2010). Assaying locomotor activity to study circadian rhythms and sleep parameters in Drosophila. J. Vis. Exp. 43, 2157. 10.3791/2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, H., Zou, Y., Suzuki, N., Hsieh, Y.-W., Chuang, C.-F., Wu, Y.-C. and Chang, C. (2018). Engulfing cells promote neuronal regeneration and remove neuronal debris through distinct biochemical functions of CED-1. Nat. Commun. 9, 4842. 10.1038/s41467-018-07291-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, S., Gumienny, T. L., Hengartner, M. O. and Driscoll, M. (2000). A common set of engulfment genes mediates removal of both apoptotic and necrotic cell corpses in C. elegans. Nat. Cell Biol. 2, 931-937. 10.1038/35046585 [DOI] [PubMed] [Google Scholar]
- Cregg, J. M., DePaul, M. A., Filous, A. R., Lang, B. T., Tran, A. and Silver, J. (2014). Functional regeneration beyond the glial scar. Exp. Neurol. 253, 197-207. 10.1016/j.expneurol.2013.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio, M. and Bradke, F. (2018). Axon regeneration in the central nervous system: facing the challenges from the inside. Annu. Rev. Cell Dev. Biol. 34, 495-521. 10.1146/annurev-cellbio-100617-062508 [DOI] [PubMed] [Google Scholar]
- Doherty, J., Logan, M. A., Taşdemir, Ö. E. and Freeman, M. R. (2009). Ensheathing glia function as phagocytes in the adult Drosophila brain. J. Neurosci. 29, 4768-4781. 10.1523/JNEUROSCI.5951-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, J., Sheehan, A. E., Bradshaw, R., Fox, A. N., Lu, T.-Y. and Freeman, M. R. (2014). PI3K signaling and Stat92E converge to modulate glial responsiveness to axonal injury. PLoS Biol. 12, e1001985. 10.1371/journal.pbio.1001985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, E., Aldana, P., Bunge, M. B., Puckett, W., Srinivasan, A., Keane, R. W., Bethea, J. and Levi, A. D. O. (1998). Apoptosis after traumatic human spinal cord injury. J. Neurosurg. 5, 911-920. 10.3171/foc.1998.5.4.1 [DOI] [PubMed] [Google Scholar]
- Etchegaray, J. I., Timmons, A. K., Klein, A. P., Pritchett, T. L., Welch, E., Meehan, T. L., Li, C. and McCall, K. (2012). Draper acts through the JNK pathway to control synchronous engulfment of dying germline cells by follicular epithelial cells. Development 139, 4029-4039. 10.1242/dev.082776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet, W., Owald, D., Wichmann, C., Mertel, S., Depner, H., Dyba, M., Hallermann, S., Kittel, R. J., Eimer, S. and Sigrist, S. J. (2009). Maturation of active zone assembly by Drosophila Bruchpilot. J. Cell Biol. 186, 129-145. 10.1083/jcb.200812150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc, N. C., Dimarcq, J.-L., Lagueux, M., Hoffmann, J. and Ezekowitz, R. A. B. (1996). Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity 4, 431-443. 10.1016/S1074-7613(00)80410-0 [DOI] [PubMed] [Google Scholar]
- Franc, N. C., Heitzler, P., Ezekowitz, R. A. B. and White, K. (1999). Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science 284, 1991-1994. 10.1126/science.284.5422.1991 [DOI] [PubMed] [Google Scholar]
- Freeman, M. R., Delrow, J., Kim, J., Johnson, E. and Doe, C. Q. (2003). Unwrapping glial biology:Gcm target genes regulating glial development, diversification, and function. Neuron 38, 567-580. 10.1016/S0896-6273(03)00289-7 [DOI] [PubMed] [Google Scholar]
- Gadani, S. P., Walsh, J. T., Lukens, J. R. and Kipnis, J. (2015). Dealing with danger in the CNS: the response of the immune system to injury. Neuron 87, 47-62. 10.1016/j.neuron.2015.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galko, M. J. and Krasnow, M. A. (2004). Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol. 2, e239. 10.1371/journal.pbio.0020239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershlick, D. C., Schindler, C., Chen, Y. and Bonifacino, J. S. (2016). TSSC1 is novel component of the endosomal retrieval machinery. Mol. Biol. Cell. 27, 2803-2883. 10.1091/mbc.E16-04-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glise, B., Bourbon, H. and Noselli, S. (1995). hemipterous encodes a novel drosophila MAP kinase kinase, required for epithelial cell sheet movement. Cell 83, 451-461. 10.1016/0092-8674(95)90123-x [DOI] [PubMed] [Google Scholar]
- Griffiths, R. L. and Hidalgo, A. (2004). Prospero maintains the mitotic potential of glial precursors enabling them to respond to neurons. EMBO J. 23, 2440-2450. 10.1038/sj.emboj.7600258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon, Y., Trompier, D., Ma, Z., Venegas, V., Pophillat, M., Mignotte, V., Zhou, Z. and Chimini, G. (2006). Cooperation between engulfment receptors: the case of ABCA1 and MEGF10. PLoS ONE 1, e120. 10.1371/journal.pone.0000120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., Miao, L., Yang, L., Liang, F., Wang, Q., Zhuang, P., Sun, Y. and Hu, Y. (2019). AKT-dependent and -independent pathways mediate PTEN deletion-induced CNS axon regeneration. Cell Death Dis. 10, 203. 10.1038/s41419-018-1289-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki, T., Kanda, H., Yamamoto-Goto, Y., Kanuka, H., Kuranaga, E., Aigaki, T. and Miura, M. (2002). Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 21, 3009-3018. 10.1093/emboj/cdf306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakov, A., Buchanan, S. M., Sullivan, B., Ramachandran, A., Chapman, J. K. S., Lu, E. S., Mahadevan, L. and De Bivort, B. (2016). Recovery of locomotion after injury in Drosophila melanogaster depends on proprioception. J. Exp. Biol. 219, 1760-1771. 10.1242/jeb.133652 [DOI] [PubMed] [Google Scholar]
- Jordán-Álvarez, S., Santana, E., Casas-Tintó, S., Acebes, Á. and Ferrús, A. (2017). The equilibrium between antagonistic signaling pathways determines the number of synapses in Drosophila. PLoS ONE 12, e0184238. 10.1371/journal.pone.0184238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakulas, B. A. (1999). A review of the neuropathology of human spinal cord injury with emphasis on special features. J. Spinal Cord Med. 22, 119-124. 10.1080/10790268.1999.11719557 [DOI] [PubMed] [Google Scholar]
- Kato, K., Awasaki, T. and Ito, K. (2009). Neuronal programmed cell death induces glial cell division in the adult Drosophila brain. Development 136, 51-59. 10.1242/dev.023366 [DOI] [PubMed] [Google Scholar]
- Kato, K., Forero, M. G., Fenton, J. C. and Hidalgo, A. (2011). The glial regenerative response to central nervous system injury is enabled by pros-notch and pros-NFκB feedback. PLoS Biol. 9, e1001133. 10.1371/journal.pbio.1001133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, K., Konno, D., Berry, M., Matsuzaki, F., Logan, A. and Hidalgo, A. (2015). Prox1 inhibits proliferation and is required for differentiation of the oligodendrocyte cell lineage in the mouse. PLoS ONE 10, e0145334. 10.1371/journal.pone.0145334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, K., Losada-Perez, M. and Hidalgo, A. (2018). Gene network underlying the glial regenerative response to central nervous system injury. Dev. Dyn. 247, 85-93. 10.1002/dvdy.24565 [DOI] [PubMed] [Google Scholar]
- Kirpensteijn, J., van den Bos, R. and Endenburg, N. (1999). Adaptation of dogs to the amputation of a limb and their owners’ satisfaction with the procedure. Vet. Rec. 144, 115-118. 10.1136/vr.144.5.115 [DOI] [PubMed] [Google Scholar]
- Kodra, A., de la Cova, C., Gerhold, A. R. and Johnston, L. A. (2020). Widely used mutants of eiger, encoding the Drosophila Tumor Necrosis factor, carry additional mutations in the NimrodC1 phagocytosis receptor. G3 10, 4707-4712. 10.1534/g3.120.401800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, T. J. and DiPietro, L. A. (2011). Inflammation and wound healing: the role of the macrophage. Expert Rev. Mol. Med. 13, e23. 10.1017/S1462399411001943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurant, E., Axelrod, S., Leaman, D. and Gaul, U. (2008). Six-microns-under acts upstream of Draper in the glial phagocytosis of apoptotic neurons. Cell 133, 498-509. 10.1016/j.cell.2008.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Marca, J. E. and Richardson, H. E. (2020). Two-faced: roles of JNK signalling during tumourigenesis in the Drosophila model. Front. Cell Dev. Biol. 8, 42. 10.3389/fcell.2020.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Sami, A., Noristani, H. N., Slattery, K., Qiu, J., Groves, T., Wang, S., Veerasammy, K., Chen, Y. X., Morales, J.et al. (2020). Glial metabolic rewiring promotes axon regeneration and functional recovery in the central nervous system. Cell Metab. 32, 767-785.E7. 10.1016/j.cmet.2020.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan, M. A., Hackett, R., Doherty, J., Sheehan, A., Speese, S. D. and Freeman, M. R. (2012). Negative regulation of glial engulfment activity by Draper terminates glial responses to axon injury. Nat. Neurosci. 15, 722-730. 10.1038/nn.3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolo, F.-N., Casas-Tintó, S. and Moreno, E. (2012). Cell competition time line: winners kill losers, which are extruded and engulfed by hemocytes. Cell Rep. 2, 526-539. 10.1016/j.celrep.2012.08.012 [DOI] [PubMed] [Google Scholar]
- Losada-Perez, M. (2018). Glia: from ‘just glue’ to essential players in complex nervous systems: a comparative view from flies to mammals. J. Neurogenet. 32, 78-91. 10.1080/01677063.2018.1464568 [DOI] [PubMed] [Google Scholar]
- Losada-Perez, M., Harrison, N. and Hidalgo, A. (2016). Molecular mechanism of central nervous system repair by the Drosophila NG2 homologue kon-tiki. J. Cell Biol. 214, 587-601. 10.1083/jcb.201603054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada-Perez, M., Harrison, N. and Hidalgo, A. (2017). Glial kon/NG2 gene network for central nervous system repair. Neural Regen. Res. 1, 31-34, doi:10.4103/1673-5374.198969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Z., Zhang, C. and Zhai, Z. (2005). Nucleoplasmin regulates chromatin condensation during apoptosis. Proc. Natl. Acad. Sci. USA 102, 2778-2783. 10.1073/pnas.0405374102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, J. M., Beach, M. G., Porpiglia, E., Sheehan, A. E., Watts, R. J. and Freeman, M. R. (2006). The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron 50, 869-881. 10.1016/j.neuron.2006.04.028 [DOI] [PubMed] [Google Scholar]
- MacDonald, J. M., Doherty, J., Hackett, R. and Freeman, M. R. (2013). The c-Jun kinase signaling cascade promotes glial engulfment activity through activation of draper and phagocytic function. Cell Death Differ. 20, 1140-1148. 10.1038/cdd.2013.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahar, M. and Cavalli, V. (2018). Intrinsic mechanisms of neuronal axon regeneration. Nat. Rev. Neurosci. 19, 323-337. 10.1038/s41583-018-0001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Blanco, E., Gampel, A., Ring, J., Virdee, K., Kirov, N., Tolkovsky, A. M. and Martinez-Arias, A. (1998). puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 12, 557-570. 10.1101/gad.12.4.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuarrie, I. G. and Grafstein, B. (1973). Axon outgrowth enhanced by a previous nerve injury. Arch. Neurol. 29, 53-55. 10.1001/archneur.1973.00490250071008 [DOI] [PubMed] [Google Scholar]
- Mendes, C. S., Rajendren, S. V., Bartos, I., Márka, S. and Mann, R. S. (2014). Kinematic responses to changes in walking orientation and gravitational load in Drosophila melanogaster. PLoS ONE 9, e109204. 10.1371/journal.pone.0109204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, E., Yan, M. and Basler, K. (2002). Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr. Biol. 12, 1263-1268. 10.1016/S0960-9822(02)00954-5 [DOI] [PubMed] [Google Scholar]
- Morizawa, Y. M., Hirayama, Y., Ohno, N., Shibata, S., Shigetomi, E., Sui, Y., Nabekura, J., Sato, K., Okajima, F., Takebayashi, H.et al. (2017). Reactive astrocytes function as phagocytes after brain ischemia via ABCA1-mediated pathway. Nat. Commun. 8, 1598. 10.1038/s41467-017-01594-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norenberg, M. D., Smith, J. and Marcillo, A. (2004). The pathology of human spinal cord injury: defining the problems. J. Neurotrauma 21, 429-4440. 10.1089/089771504323004575 [DOI] [PubMed] [Google Scholar]
- Pinal, N., Calleja, M. and Morata, G. (2019). Pro-apoptotic and pro-proliferation functions of the JNK pathway of Drosophila: roles in cell competition, tumorigenesis and regeneration. Open Biol. 9, 180256. 10.1098/rsob.180256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela, M., Venkataramani, V., Fahey-Lozano, N., Seco, E., Losada-Perez, M., Winkler, F. and Casas-Tintó, S. (2019a). Glioblastoma cells vampirize WNT from neurons and trigger a JNK/MMP signaling loop that enhances glioblastoma progression and neurodegeneration. PLoS Biol. 17, e3000545. 10.1371/journal.pbio.3000545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela, M., Segura-Collar, B., Argudo, I., Sáiz, A., Gargini, R., Sánchez-Gómez, P. and Casas-Tintó, S. (2019b). Oncogenic dependence of glioma cells on kish/TMEM167A regulation of vesicular trafficking. Glia 67, 404-417. 10.1002/glia.23551 [DOI] [PubMed] [Google Scholar]
- Purice, M. D., Ray, A., Münzel, E. J., Pope, B. J., Park, D. J., Speese, S. D. and Logan, M. A. (2017). A novel Drosophila injury model reveals severed axons are cleared through a draper/MMP-1 signaling cascade. eLife 6, e23611. 10.7554/eLife.23611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, J. P. and Sagasti, A. (2017). Learning to swim, again: axon regeneration in fish. Exp. Neurol. 287, 318-330. 10.1016/j.expneurol.2016.02.022 [DOI] [PubMed] [Google Scholar]
- Rishal, I. and Fainzilber, M. (2014). Axon-soma communication in neuronal injury. Nat. Rev. Neurosci. 15, 32-42. 10.1038/nrn3609 [DOI] [PubMed] [Google Scholar]
- Saijilafu, Hur, E.-M., Liu, C.-M., Jiao, Z., Xu, W.-L. and Zhou, F.-Q. (2013). PI3K-GSK3 signalling regulates mammalian axon regeneration by inducing the expression of Smad1. Nat. Commun. 4, 2690. 10.1038/ncomms3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkissian, T., Timmons, A., Arya, R., Abdelwahid, E. and White, K. (2014). Detecting apoptosis in Drosophila tissues and cells. Methods. 68, 89-96. 10.1016/j.ymeth.2014.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, A., Pletnikov, M. V. and Kamiya, A. (2004). Neuron-glia interactions clarify genetic-environmental links in mental illness. Trends Neurosci. 27, 294-297. 10.1016/j.tins.2004.03.012 [DOI] [PubMed] [Google Scholar]
- Sheng, Z.-H. (2017). The interplay of axonal energy homeostasis and mitochondrial trafficking and anchoring. Trends Cell Biol. 27, 403-416. 10.1016/j.tcb.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shklover, J., Mishnaevski, K., Levy-Adam, F. and Kurant, E. (2015). JNK pathway activation is able to synchronize neuronal death and glial phagocytosis in Drosophila. Cell Death Dis. 6, e1649. 10.1038/cddis.2015.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver, J. (2009). CNS regeneration: only on one condition. Curr. Biol. 19, R444-R446. 10.1016/j.cub.2009.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver, J. and Miller, J. H. (2004). Regeneration beyond the glial scar. Nat. Rev. Neurosci. 5, 146-156. 10.1038/nrn1326 [DOI] [PubMed] [Google Scholar]
- Smith, P. J., Howes, E. A. and Treherne, J. E. (1987). Mechanisms of glial regeneration in an insect central nervous system. J. Exp. Biol. 132, 59-78. https://pubmed.ncbi.nlm.nih.gov/3323407/ [DOI] [PubMed] [Google Scholar]
- Soto-Padilla, A., Ruijsink, R., Span, M., van Rijn, H. and Billeter, J.-C. (2018). An automated method to determine the performance of Drosophila in response to temperature changes in space and time. J. Vis. Exp. 140, 10.3791/58350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, M. C., Nguyen, M. M., Tao, J., Allender, D. L. and Rolls, M. M. (2010). Global up-regulation of microtubule dynamics and polarity reversal during regeneration of an axon from a dendrite. Mol. Biol. Cell 21, 767-777. 10.1091/mbc.e09-11-0967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsu, Y., Nakamura, M., Stapleton, M., Danos, M. C., Matsumoto, K., O'Connor, M. B., Shibuya, H. and Ueno, N. (2000). TAK1 Participates in c-Jun N-terminal Kinase signaling during Drosophila development. Mol. Cell. Biol. 20, 3015-3026. 10.1128/mcb.20.9.3015-3026.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji, T., Yun, E.-Y. and Ip, Y. T. (2010). Heterodimers of NF-κB transcription factors DIF and Relish regulate antimicrobial peptide genes in Drosophila. Proc. Natl. Acad. Sci. USA 107, 14715-14720. 10.1073/pnas.1009473107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toné, S., Sugimoto, K., Tanda, K., Suda, T., Uehira, K., Kanouchi, H., Samejima, K., Minatogawa, Y. and Earnshaw, W. C. (2007). Three distinct stages of apoptotic nuclear condensation revealed by time-lapse imaging, biochemical and electron microscopy analysis of cell-free apoptosis. Exp. Cell Res. 313, 3635-3644. 10.1016/j.yexcr.2007.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treherne, J. E., Harrison, J. B., Treherne, J. M. and Lane, N. J. (1984). Glial repair in an insect central nervous system: Effects of surgical lesioning. J. Neurosci 4, 2689-2697. 10.1523/JNEUROSCI.04-11-02689.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente D., Golani I. and Mitra P. P. (2007). Analysis of the Trajectory of Drosophila melanogaster in a Circular Open Field Arena. PloS ONE 2, e1083. doi:10.1371/journal.pone.0001083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hilchen, C. M., Bustos, Á. E., Giangrande, A., Technau, G. M. and Altenhein, B. (2013). Predetermined embryonic glial cells form the distinct glial sheaths of the Drosophila peripheral nervous system. Development 140, 3657-3668. 10.1242/dev.093245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler, F., Gillingham, A. K., Sinka, R., Rosa-Ferreira, C., Gordon, D. E., Franch-Marro, X., Peden, A. A., Vincent, J.-P. and Munro, S. (2010). A genome-wide RNA interference screen identifies two novel components of the metazoan secretory pathway. EMBO J. 29, 304-314. 10.1038/emboj.2009.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, Y.-L., An, R., Cassin, J., Joseph, J., Mi, R., Wang, C., Zhong, C., Jin, S.-G., Pfeifer, G. P., Bellacosa, A.et al. (2017). An intrinsic epigenetic barrier for functional axon regeneration. Neuron 94, 337-346.e6. 10.1016/j.neuron.2017.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, S. and Grose, R. (2003). Regulation of wound healing by growth factors and cytokines. Physiol. Rev. 83, 835-870. 10.1152/physrev.2003.83.3.835 [DOI] [PubMed] [Google Scholar]
- Weston, C. R. and Davis, R. J. (2007). The JNK signal transduction pathway. Curr. Opin. Cell Biol. 19, 142-149. 10.1016/j.ceb.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Wu, C., Li, Z., Ding, X., Guo, X., Sun, Y., Wang, X., Hu, Y., Li, T., La, X., Li, J.et al. (2019). Snail modulates JNK-mediated cell death in Drosophila. Cell Death Dis. 10, 893. 10.1038/s41419-019-2135-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanik, M. F., Cinar, H., Cinar, H. N., Chisholm, A. D., Jin, Y. and Ben-Yakar, A. (2004). Neurosurgery: functional regeneration after laser axotomy. Nature 432, 822. 10.1038/432822a [DOI] [PubMed] [Google Scholar]
- Zeke, A., Misheva, M., Reményi, A. and Bogoyevitch, M. A. (2016). JNK signaling: regulation and functions based on complex protein-protein partnerships. Microbiol. Mol. Biol. Rev. 80, 793-835. 10.1128/MMBR.00043-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke, N., Korzelius, J., vanStraaten, M., Bender, K., Schuhknecht, G. F. P., Dutta, D., Xiang, J. and Edgar, B. A. (2014). Fly-FUCCI: a versatile tool for studying cell proliferation in complex tissues. Cell Rep. 7, 588-598. 10.1016/j.celrep.2014.03.020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.