ABSTRACT
Intracellular bacterial pathogens spend portions of their life cycle both inside and outside host cells. While in these two distinct environments, they release or shed bacterial components, including virulence factors that promote their survival and replication. Some of these components are released through extracellular vesicles, which are either derived from the bacteria themselves or from the host cells. Bacteria- and host-derived vesicles have been studied almost exclusively in isolation from each other, with little discussion of the other type of secreted vesicles, despite the fact that both are generated during an in vivo infection and both are likely play a role in bacterial pathogenesis and host immunity. In this Review, we aim to bridge this gap and discuss what we know of bacterial membrane vesicles in their generation and composition. We will compare and contrast this with the composition of host-derived vesicles with regard to bacterial components. We will also compare host cell responses to the different vesicles, with a focus on how these vesicles modulate the immune response, using Mycobacterium, Listeria and Salmonella as specific examples for these comparisons.
KEY WORDS: Bacteria, Extracellular vesicles, Mycobacterium, Outer membrane vesicles
Summary: This Review discusses the content and function of extracellular vesicles released from bacteria and from host cells infected with bacteria, comparing Mycobacterium, Listeria and Salmonella pathogens.
Introduction
Cells communicate through various mechanisms, including the release of proteins, lipids and even nucleic acids. The origin and functions of these different molecules have been studied in the context of all aspects of cell biology, extending from bacterial communication to cell-to-cell communication within multicellular organisms. These factors are often released as single molecules; however, it has been become increasingly clear that some molecules released from cells are ‘packaged’ inside vesicles. The release of these extracellular vesicles (EVs) is a common process throughout evolution and provides a mechanism to deliver in one distinct unit a bolus of factors that can induce a plethora of effects in recipient cells. Therefore, in the context of a bacterial infection, both the pathogen and the host can release EVs that could influence the outcome of the infection. In this Review we discuss three intracellular pathogens, Mycobacterium, Listeria and Salmonella, as these important human pathogens have been studied in the context of both bacteria-derived and host-derived vesicles, allowing for a comparison of the two vesicle populations in their content and function.
A brief overview over EVs
Host-derived EVs
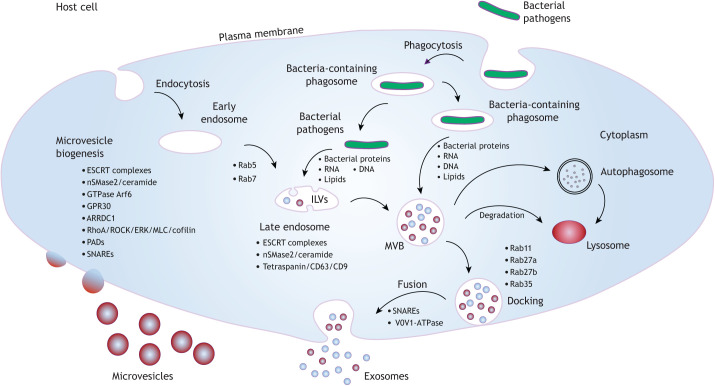
During an infection with an intracellular pathogen, host cells release EVs that carry not only host cellular content, including proteins, DNA, RNA and lipids, but also pathogen-derived macromolecules (Schorey et al., 2015). Thus far, studies of host-derived EVs (hEVs) and infectious diseases have focused on exosomes and microvesicles. As exosomes and microvesicles have overlapping sizes and some common markers, they can co-purify together and are often grouped under the common term of EVs. Nevertheless, these vesicles have different origins, content and function (D'Souza-Schorey and Schorey, 2018). Exosomes are 30–150 nm vesicles that are initially formed as intraluminal vesicles (ILVs) in multivesicular bodies (MVBs) within eukaryotic cells and are later released via the fusion of MVBs with the plasma membrane (Fig. 1). Initial studies indicated that MVBs fuse with lysosomes or autophagosomes, leading to the degradation of ILVs (Woodman and Futter, 2008), but later studies have found that MVBs can fuse with the plasma membrane to release ILVs, which upon extracellular release are called exosomes (Harding et al., 1984; Pan and Johnstone, 1983). It remains to be determined whether there are distinct MVB populations and, if so, how these populations are defined (Mobius et al., 2002). Exosome biogenesis is mediated by two general mechanisms: endosomal sorting complexes required for transport (ESCRT)-dependent and ESCRT-independent pathways (Trajkovic et al., 2008; Stuffers et al., 2009; Tamai et al., 2010) (Fig. 1). ESCRT is a multiprotein complex that enables membrane remodeling, resulting in membranes bending or budding away from the cytoplasm. The first member of this large complex, ESCRT-0, is recruited onto the endosomal surface and, through its ubiquitin-interacting domain, serves as a docking point for the monoubiquitylated proteins that will be packaged into ILVs (Raiborg and Stenmark, 2002). Subsequently, ESCRT-I, followed by ESCRT-II and ESCRT-III, protein complexes are recruited to the membrane and drive ILV and MVB formation (Babst et al., 2002; Katzmann et al., 2001; Wollert et al., 2009). Although ESCRT-dependent MVB biogenesis is common to many cell types, evidence suggests that some cells use an ESCRT-independent method for the generation of MVBs. For instance, in oligodendroglial cells, MVB and exosome biogenesis requires ceramide, produced through the activity of sphingomyelinase nSMase2 (also known as SMPD3). Ceramide is highly enriched in exosomes and has been shown to facilitate membrane curvature (Trajkovic et al., 2008). Tetraspanins, such as CD9 and CD81, have also been implicated in MVB and exosome biogenesis (Buschow et al., 2009). Trafficking of the MVBs to the plasma membrane and their fusion with the plasma membrane require a number of different small GTPases, including Rab11 (also known as Rab11a), Rab27a, Rab27b and Rab35 (Savina et al., 2002; Ostrowski et al., 2010; Hsu et al., 2010; Baietti et al., 2012) (Fig. 1).
Fig. 1.
Pathways and factors involved in EV biogenesis in host cells during an infection. Host factors required for microvesicle and exosome production and release, including the generation of MVBs and their transport to the plasma membrane for fusion and exosome release, are shown. For each type of EV, factors known to be involved in their biogenesis are indicated; these include ceramide and components of the ESCRT complex, which are involved in generating both exosomes and microvesicles. Also indicated are the types of macromolecules that are released from bacteria and are trafficked and packaged into EVs. These macromolecules can be released either from cytosolic bacteria or from bacteria within a phagosomal compartment. ERK, extracellular signal-regulated kinases; GPR30, G-protein-coupled receptor 30; MLC, myosin light chain; PADs, peptidylarginine deiminases.
Unlike exosomes, microvesicles are formed and released by outward budding of the plasma membrane (Fig. 1). In some cases, the microvesicle and exosome biogenesis pathways share common factors, including the ESCRT complex component TSG101 and vesicle-associated membrane protein-3 (VAMP3), both of which promote membrane fusion (Clancy et al., 2015). They also contain shared lipids, such as cholesterol and ceramide (Del Conde et al., 2005; Haraszti et al., 2016; Budnik et al., 2016), suggesting some commonalities in exosome and microvesicle biogenesis (Sedgwick and D'Souza-Schorey, 2018; D'Souza-Schorey and Schorey, 2018). Microvesicle formation also requires a reorganization of the cytoskeleton in parental cells, and therefore actin homeostasis is important. Rho family GTPases such as RhoA are required for microvesicle formation (Li et al., 2012; Sedgwick et al., 2015). Myosin light chain kinase, activated by RhoA, localizes to the microvesicle ‘neck’ and promotes microvesicle formation by regulating actomyosin contractility. (Sedgwick et al., 2015).
Bacteria-derived EVs
Gram-negative bacteria release vesicles through the budding of the outer membrane and therefore are referred to as outer membrane vesicles (OMVs) (reviewed in Schwechheimer and Kuehn, 2015; Kulp and Kuehn, 2010). Until recently, it was believed that the release of extracellular vesicles was specific to Gram-negative bacteria, as the presence of a complex cell wall with a peptidoglycan (PG) layer would restrict the release of membrane vesicles from Gram-positive bacteria (Briaud and Carroll, 2020). However, there is compelling evidence that both Gram-negative and Gram-positive bacteria release membrane vesicles that harbor a select catalog of molecules (Schwechheimer et al., 2014; Lee et al., 2009; Briaud and Carroll, 2020). These vesicles have a diverse set of functions, including delivery of virulence factors to modulate the host immune system, aiding in nutrient acquisition and ecological niche protection and helping to regulate the timing and generation of environmental responses such as the production of biofilms (Schwechheimer and Kuehn, 2015; Toyofuku et al., 2019).
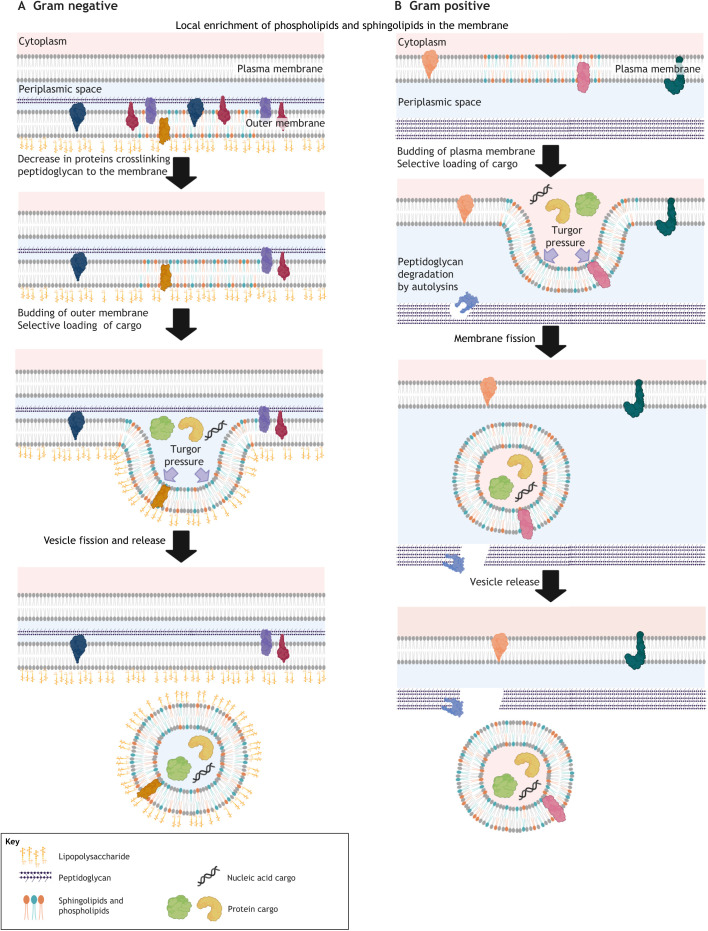
OMVs from Gram-negative bacteria are ∼20–200 nm in size (Fig. 2). Because OMVs are formed from the pinching of the outer membrane, proteins involved in crosslinking the membrane with the cell wall PG (Box 1) play a key role in regulating OMV generation. Mutations resulting in a loss of proteins such as OmpA, which functions to increase crosslinking between PG and the outer membrane, have been found to increase OMV generation in Escherichia coli, Salmonella and Acinetobacter (Moon et al., 2012; Deatherage et al., 2009; Sonntag et al., 1978). In addition to membrane–PG crosslinking, some lipids, such as sphingolipids, may facilitate vesicle budding owing to their ability to promote membrane curvature (Kulkarni et al., 2014). Gene function and biochemical pathway analyses in E. coli further support the close links between lipid composition and OMV formation (Liu et al., 2017a). The presence of unsaturated and branched-chain fatty acids in OMVs suggest that areas of membrane with increased fluidity are sites of OMV formation. However, it is important to note that there are species-specific differences in OMV lipid composition, and the generalizations indicated above should be used with caution.
Fig. 2.
Generation of EVs from Gram-negative and Gram-positive bacteria. The general processes required for release of bacteria-derived EVs are shown. (A) In Gram-negative bacteria, the budding from the outer membrane requires the removal of proteins that crosslink peptidoglycan to the lipid membrane. Membrane budding in both Gram-negative and Gram-positive bacteria requires generation of turgor pressure and is facilitated by membrane lipids that promote membrane curvature. (B) Gram-positive bacteria have the added requirement of remodeling of the peptidoglycan layer to allow transport of the EV through the cell wall. During this process, a selective set of proteins and nucleic acids are incorporated into the EVs. Figure created with BioRender.com
Box 1. Cell wall peptidoglycan.
Peptidoglycan (PG) is a polysaccharide made of two glucose derivatives, N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM), that alternate to form long linear chains. The chains are crosslinked to one another by a tetrapeptide – L-alanine, D-glutamine, L-lysine or meso-diaminopimelic acid (DPA), and D-alanine – that extends from the NAM sugar unit, allowing a lattice-like structure to form. Inhibition of PG biosynthesis is a common mode of action for various antibiotics. For example, penicillin interferes with the production of PG by binding to bacterial enzymes known as penicillin-binding proteins. By inhibiting these enzymes, the final step in cell wall biosynthesis, the crosslinking of the polysaccharide, is blocked. This causes the cell wall to weaken due to fewer crosslinks, allowing excess water to flow into the cell resulting in cell lysis and death.
Despite the requirement to traverse the PG layer, EVs from Gram-positive bacteria can be larger in size compared to OMVs and range from 20 to 400 nm (Briaud and Carroll, 2020). Similar to OMVs, EVs from Gram-positive bacteria are enriched in certain lipids, including sphingolipids, suggesting similar biophysical properties are involved in bacterial generation of EVs whether these vesicles are formed from the outer membrane or the cytoplasmic membrane (Toyofuku et al., 2019) (Fig. 2). An additional step in EV release from Gram-positive bacteria is their passage through the PG layer. The presence of transpeptidases and autolysins (involved in PG synthesis and degradation, respectively) inside these EVs suggest an important role for PG modification during EV release (Banzhaf et al., 2012). This notion is supported by drug studies, which have shown that sublethal exposure of Staphylococcus aureus to β-lactam antibiotics results in decreased PG crosslinking and a significant increase in EV release (Andreoni et al., 2019). Similarly, S. aureus mutants that are defective in autolysin production show decreased EV production (Wang et al., 2018). However, how this cell wall plasticity is regulated remains an open but critical question, as the release of EVs is an important part of bacterial adaptation to different surroundings, including the host microenvironment.
Mycobacteria and EVs
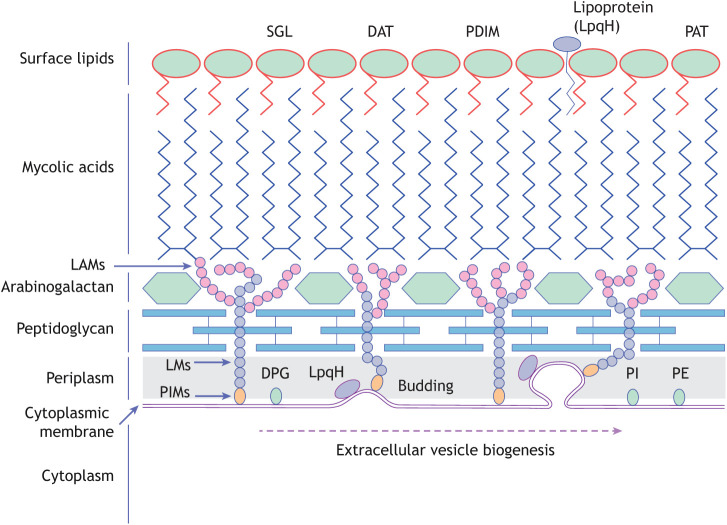
The genus Mycobacterium includes over 190 species, some of which are known animal or human pathogens, including Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis – a disease that is responsible for ∼1.5 million deaths annually (Schorey and Schlesinger, 2016). One of the most intriguing findings related to the release of EVs from Gram-positive bacteria is the observation that mycobacteria release EVs (Jurkoshek et al., 2016; Brown et al., 2015). This was particularly unexpected, because the EVs from mycobacteria must transverse not only the PG layer, but also an arabinogalactan layer and a thick, waxy mycolic acid layer (Fig. 3) (Hoffmann et al., 2008). EVs carrying mycobacterial components can also be generated from infected host cells, including macrophages, which are the primary host cell for Mtb and other pathogenic mycobacteria (Bhatnagar et al., 2007; Bhatnagar and Schorey, 2007; Singh et al., 2015). Addressing how mycobacteria-derived EVs (MycEVs) and hEVs work synergistically or antagonistically to modulate the host immune response during an infection is necessary for our understanding of mycobacterial pathogenesis.
Fig. 3.
Mycobacterium tuberculosis cell wall. Diagram of the mycobacterial cell wall, indicating the location of key lipid components and the site of membrane budding to form EVs. DAT, diacyltrehalose; DPG, diphosphatidylglycerol; LAMs, lipoarabinomannans; LM, lipomannan; PAT, polyacyltrehalose; PDIM, phthiocerol dimycocerosate; PE, phosphatidylethanolamine; PI, phosphatidyl-myo-inositol; PIMs, phosphoinositol mannosides; SGL, sulfoglycolipid.
Biogenesis of mycobacteria-derived EVs
The release of EVs by mycobacteria was first described for Mycobacterium ulcerans in 2007 (Marsollier et al., 2007), but EV release has subsequently been shown for all mycobacteria species tested (Brown et al., 2015; Layre, 2020). Production of EVs is increased in Mtb grown in low-iron medium, an environment that is present inside host macrophages (Prados-Rosales et al., 2014b). The presence of metal ions at concentrations believed to be similar to those in Mtb-containing phagosomes also induces EV biogenesis (Chiplunkar et al., 2019). This suggests that vesicle formation is regulated by the mycobacteria, and although the underlying mechanisms remains undefined, genetic studies suggest a role for VirR (also known as Rv0431), a protein of unknown function whose deletion results in elevated EV release (Rath et al., 2013). Factors that regulate cell wall and membrane biosynthesis have also been linked to EV formation. For instance, IniA and IniC, which are dynamin-like proteins that appear to regulate membrane fusion (Wang et al., 2019), have been implicated in EV release (Gupta et al., 2020 preprint). Although these EV biogenesis studies were conducted using in vitro growth conditions, they have given us valuable information on the regulation of EV biogenesis by mycobacteria.
Content of mycobacteria-derived EVs
Composition defines function, and therefore it is critical to identify the molecules present in MycEVs. Analysis of their lipid content indicates that MycEVs are derived from the inner membrane of the mycobacteria, because they contain phospholipids characteristic of this membrane, such as phosphatidylglycine, phosphatidylethanolamine (PE) and phosphatidyl-myo-inositol (PI) (Prados-Rosales et al., 2011). Electron microscopy studies indicate that infected macrophages also release MycEVs containing lipoarabinomannan (LAM) and LpqH (Prados-Rosales et al., 2011). Proteomic studies have identified hundreds of mycobacterial proteins in MycEVs, such as antigen 85B, KatG and CFP10, (Prados-Rosales et al., 2011), which are known to be strong inducers of an acquired immune response (Lee et al., 2015). The vesicles also appear to be enriched in lipoproteins, such as LppX, PstS1 and LpqH (Prados-Rosales et al., 2011). Interestingly, MycEVs contain well-defined secreted proteins (for example, SodB, Apa and FbpA) as well as cytoplasmic proteins (for example, KatG, GlnA1 and AcpM) in approximately equal abundance (Table 1).
Table 1.
Comparison of mycobacterial proteins identified in mycobacteria-derived and host-derived EVs
Function of mycobacteria-derived EVs
Functionally, MycEVs have been shown to induce production of pro-inflammatory cytokines such as interleukin-1β and TNF by macrophages, which is dependent on their expression of toll-like receptor 2 (TLR2) (Prados-Rosales et al., 2011), a pattern recognition receptor (PRR) known to bind to various pathogen molecules (Kirschning and Schumann, 2002) including the Mtb lipoprotein LpqH (Drage et al., 2009). In addition, MycEVs also carry antigens that can be transferred to dendritic cells for presentation to T cells (Jurkoshek et al., 2016). In contrast, direct incubation of T cells with MycEVs has been found to inhibit T cell activation, likely through the transfer of LAM, which is known to inhibit T cell activation (Athman et al., 2017). The function of MycEVs in vivo is less clear. Intra-tracheal administration of EVs released from Mycobacterium bovis BCG, which is an attenuated strain of M. bovis and the only FDA-approved vaccine for Mtb (Foster et al., 2021), leads to an exacerbation of infection when EV-treated animals are subsequently infected with Mtb (Prados-Rosales et al., 2011). In contrast, subcutaneous administration of MycEVs results in activation of an antigen-specific acquired immune response and control of subsequent Mtb infection to a level comparable to that of M. bovis BCG-vaccinated mice (Prados-Rosales et al., 2014a). The different host response to the vesicles could stem from the difference in the route of administration, differing amounts of EVs used or the timing of treatment and/or infection.
Content of host-derived EVs carrying mycobacterial molecules
The mycobacterial content of hEVs has been characterized in various experimental conditions, including infected cells, infected mice and tuberculosis patients (Giri et al., 2010; Kruh-Garcia et al., 2014; Mehaffy et al., 2017; Singh et al., 2015). Early studies identified both mycobacterial lipids, such as phosphatidylinositol dimannoside (PIM2), LAM and lipomannan (LM) (Beatty et al., 2000; Rhoades et al., 2003; Beatty and Russell, 2001), and inner membrane lipids, such as PE and phosphatidylglycine, in hEVs, as well as the mycobacterial lipoproteins LpqH and LprG (Ullrich et al., 2000). More than 50 mycobacterial proteins have been identified in hEVs (Giri et al., 2010; Ullrich et al., 2000; Mehaffy et al., 2017; Wang et al., 2014). Support that the majority of these mycobacterial proteins are present in hEVs and not MycEVs comes from studies that looked at the eukaryotic cell ubiquitylation process. Treatment of Mtb-infected macrophages with the ubiquitylation inhibitor PYR-41 results in a loss of mycobacterial proteins in hEVs (Smith et al., 2015). As discussed above, monoubiquitylated proteins bind to ESCRT-I for their incorporation into MVBs, suggesting that mycobacterial proteins are transferred by a host trafficking pathway to MVBs and into exosomes. Further findings have shown that CD63-positive EVs released from infected macrophages also contain mycobacterial RNA (Singh et al., 2015). In addition, the mycobacterial SecA2 secretion system has been shown to be required for the RNA release from mycobacteria, but the mechanism by which the RNA is incorporated into hEVs is unknown (Cheng and Schorey, 2018).
Function of host-derived EVs
Initial experiments have determined that Mycobacterium avium-infected macrophages release EVs that can stimulate a pro-inflammatory response in non-infected or naïve macrophages (Bhatnagar and Schorey, 2007). A pro-inflammatory cytokine response has also been observed in naïve macrophages treated with hEVs released from macrophages infected with M. avium subspecies Paratuberculosis (Wang et al., 2014), as well as those infected with Mtb or M. bovis BCG (Bhatnagar et al., 2007). The ability of these hEVs to stimulate the pro-inflammatory response in naïve macrophages is dependent on the recipient cells expressing TLR2 (Bhatnagar et al., 2007). Subsequent work has suggested that the lipoprotein LpqH, which is present on hEVs released from Mtb-infected cells, is the TLR2 ligand and the primary driver of this inflammatory response (Schorey and Bhatnagar, 2008). Furthermore, exosomes isolated from the bronchoalveolar lavage (BAL) fluid of M. bovis BCG-infected mice contain mycobacterial components, including LpqH, and are pro-inflammatory ex vivo (Bhatnagar et al., 2007). In the same study, hEVs released from M. bovis BCG- or Mtb-infected macrophages were found to stimulate a pro-inflammatory response in vivo, with intranasal injection of hEVs into mice inducing TNF and IL-12 production, as well as recruitment of macrophages and neutrophils to the lung (Bhatnagar et al., 2007). Moreover, hEVs containing mycobacterial RNA activate the cytoplasmic RNA sensor RIG-I (also known as DDX58) in recipient cells and promote the maturation of Mtb–containing phagosomes, leading to increased bacterial killing (Cheng and Schorey, 2019). EVs isolated from Mtb-infected neutrophils have also been found to promote macrophage autophagy and antimicrobial activity (Alvarez-Jiménez et al., 2018). These results suggest that hEVs released from mycobacterial-infected cells can promote the recruitment and activation of immune cells both in vitro and in vivo and may play a role in facilitating the innate immune response to mycobacterial infection.
Host-cell-derived EVs released from Mtb- or M. bovis BCG-infected macrophages, or from macrophages treated with Mtb proteins isolated from the culture medium (CFP), activate antigen-specific CD4+ and CD8+ T cells in vivo and promote activation and maturation of bone marrow-derived dendritic cells (BMDCs) (Giri and Schorey, 2008). Moreover, hEVs released from CFP-treated macrophages, when administered intranasally, protect mice against a low-dose aerosolized infection with Mtb to a level equivalent to that observed in BCG-vaccinated mice (Cheng and Schorey, 2013). In addition, different EV populations, described as exosomes and microvesicles, released from Mtb- or M. bovis BCG-infected cells have been shown to stimulate an antigen-specific T cell response (Ramachandra et al., 2010). Taken together, these results suggest that hEVs are a source of antigen to stimulate the acquired immune response. However, other mechanisms of antigen delivery during a mycobacterial infection have been proposed and include infected dendritic cells and/or macrophages, necrotic cells (Sridharan and Upton, 2014), apoptotic bodies (Behar et al., 2011) and the release of free antigen (Srivastava and Ernst, 2014). The extent of antigen transfer through EVs during an in vivo infection remains an open question, but recent studies suggest that hEVs play at least some role in this process (Smith et al., 2017).
Collectively, these data suggest that EVs might be beneficial in controlling a mycobacterial infection. However, the mycobacterial components present on or in hEVs could also suppress the immune response. Indeed, hEVs from Mtb-infected cells partially suppress the ability of recipient macrophages to respond to interferon-γ (IFN-γ), and this inhibition is dependent on the expression of TLR2 in macrophages (Singh et al., 2011). Inhibition of a cellular response is not limited to macrophages, as hEVs from Mtb-infected cells can inhibit activation of CD4+ T cells, likely due to the transfer of LAM to the T cells, where it is known to induce a state of T cell anergy (Athman et al., 2017).
Comparison between mycobacteria- and host-derived EVs
Although there are some commonalities in the lipids and glycolipids present in MycEVs and hEVs, such as the presence of LAM and LpqH, there are clear differences in lipids between the EV types (Prados-Rosales et al., 2011; Beatty et al., 2000). The positioning of mycobacterial lipids in the bacterial cell and whether they are released from the mycobacteria during an infection will, in part, determine which lipids are present in hEVs. Moreover, the environmental conditions within the host will affect which lipids or glycolipids are released and to where they are trafficked. The dynamic nature of the host response, which will have an qualitative and quantitative effect on the mycobacterial cell wall, makes it difficult to obtain a clear answer with regard to which mycobacterial lipids are unique to different EV populations; however, MycEVs appear to have a higher concentration of polar lipids such as PI and PE compared to hEVs released from mycobacterial-infected macrophages (Prados-Rosales et al., 2011; Beatty et al., 2000).
Proteomics data indicate that EVs released from mycobacteria or from host cells infected with mycobacteria have an overlapping protein content (Table 1), but there are some proteins that are unique to each vesicle type. However, the observed compositions of the vesicles are only ‘snapshots’ and are specific to the experimental conditions used, and it is likely that there are additional, undefined proteins in the EVs. Again, this makes it difficult to determine what is truly unique to the different EV types. However, it is interesting that MycEVs contain both secreted and cytoplasmic proteins (Prados-Rosales et al., 2011), whereas hEVs appear to primarily contain secreted proteins (Giri et al., 2010; Kruh-Garcia et al., 2014). This likely stems from the difference in their biosynthesis, as MycEVs are produced from pinching of the inner membrane and therefore can incorporate cytoplasmic mycobacterial proteins during their production. In contrast, incorporation into hEVs requires mycobacterial proteins to first be released from the bacterial cell and either trafficked to MVBs for eventual release via exosomes, or to the plasma membrane to be included in microvesicles.
Functionally, as outlined above, both MycEVs and hEVs induce a number of host immune responses, including but not limited to promoting an inflammatory response and providing antigens to drive a T cell response. However, the presence and importance of mycobacterial RNA in MycEVs has not been addressed, so it is unclear whether MycEVs can activate a cytoplasmic RNA sensor such as RIG-I, as was found for hEVs from Mtb-infected macrophages (Cheng and Schorey, 2019). Of course, host molecules are also present in hEVs, and the presence of specific host factors adds another layer of complexity. Studies have shown that hEVs released from infected cells contain host mRNA and microRNA that promotes a more pro-inflammatory response in recipient macrophages (Singh et al., 2015).
Taken together, these findings suggest that Mtb components can spread beyond the infected cell via EVs to either activate or suppress the immune response. We hypothesize that these EVs will be involved in multiple steps during the infection process. Nevertheless, a sufficient concentration of MycEVs to induce a biological response may be limited to stages of the infection where extracellular mycobacteria are abundant. This is likely not observed during the initial weeks of infection, as the recruitment of macrophages provides sufficient host cells for the multiplying mycobacteria (Schorey and Schlesinger, 2016). However, as granulomas form and necrotic tissue develops, a higher concentration extracellular mycobacteria is observed (Lin and Flynn, 2018; Cadena et al., 2017), suggesting that it is during these later stages of infection where MycEVs may have the most biologically relevant effect. However, it is also possible that MycEVs are released directly from infected macrophages. Indeed, Athman et al. have shown that MycEVs released from Mtb-infected RAW264.7 cells contain the mycobacterial glycolipids LAM and LM (Athman et al., 2015) (Fig. 3). This conclusion is based on the detection of LAM-positive vesicles lacking the exosomal markers CD9 and CD63; however, recent data for exosomes and other hEVs indicate a significantly greater heterogeneity in vesicle composition than previously appreciated, with classic exosome markers such as CD9 and CD63 being present on only a subset of exosomes (Kowal et al., 2016). Therefore, a proteomic analysis of the LAM-positive CD9- and/or CD63-negative vesicles identified by Athman et al. (2015) may be needed to further confirm their bacterial origin.
Teasing out how the different EVs modulate the immune response and whether they induce responses at different stages of an infection would be aided by methods to selectively block the production of MycEVs and hEVs and to evaluate the outcome of the infection under these conditions. However, we presently lack targeted approaches to block production of MycEVs or hEVs. Alternatively, isolation of EVs from mycobacterial-infected mice or humans and characterization of their content, especially for markers specific to MycEVs and hEVs, would provide valuable information on when the different EV populations are produced.
EVs in the context of a Listeria monocytogenes infection
Listeria monocytogenes is a Gram-positive foodborne bacterial pathogen that causes listeriosis in humans (Swaminathan and Gerner-Smidt, 2007). Like pathogenic mycobacteria, L. monocytogenes can invade and proliferate in macrophages, but can also infect non-phagocytic cells. To successfully establish infection in host cells, Listeria are initially contained within the phagosome, but subsequently lyse the phagosomal membrane and enter the cytosol. The ability to escape the phagosome is mediated by multiple bacterial virulence factors, including listeriolysin O (LLO, encoded by the hly gene) and two phospholipases C (Gaillard et al., 1987). It has been reported that both Listeria and host cells release EVs during the course of L. monocytogenes infection (Kannan et al., 2020).
EVs released by L. monocytogenes
The mechanism for the generation of L. monocytogenes EVs remains unclear. Early studies indicate a role for the transcription factor σΒ, which is encoded by the sigB gene and is induced by environmental stress such as high salt concentration (Lee et al., 2013, 2018). In bacterial broth culture, ΔsigB L. monocytogenes produces fewer EVs than the wild-type parental strain, and σΒ likely functions in EV biogenesis by regulating the production of proteins involved in this process (Lee et al., 2013). Environmental stressors such as high salt increase σΒ levels, linking environmental factors to Listeria EV biogenesis (Lee et al., 2018). In addition to broth culture, L. monocytogenes also release EVs in infected macrophages, and they have a crucial role in bacterial survival in the host (Vdovikova et al., 2017; Coelho et al., 2019). Proteomic, lipidomic and metabolomic analyses of L. monocytogenes-derived EVs have found these EVs to carry the majority of known Listeria virulence proteins, including the pore-forming hemolysin LLO and phosphatidylinositol-specific phospholipase C (PI-PLC, encoded by plcA) (Lee et al., 2013; Coelho et al., 2019; Karthikeyan et al., 2020, 2019). L. monocytogenes-derived EVs induce significant cytotoxicity in host Caco-2 cells that is likely due to the presence of LLO and other toxins (Karthikeyan et al., 2020). Although LLO is a major virulence factor for Listeria pathogenesis, it can also activate antibacterial pathways that are detrimental for bacterial survival in host cells, such as autophagy (Meyer-Morse et al., 2010). In contrast, L. monocytogenes-released EVs containing oxidized or inactive LLO have been shown to inhibit native LLO-induced autophagy and cell death (Vdovikova et al., 2017).
The RNA profile in L. monocytogenes-released EVs has been determined using next-generation sequencing (Frantz et al., 2019), and the EVs were found to have all forms of bacterial RNAs, including mRNA, tRNA, rRNA and sRNA. RNAs isolated from L. monocytogenes-derived EVs can activate type I interferon production in recipient cells through the RIG-I cytosolic RNA-sensing pathway. Additional analysis indicates that ril32, a highly conserved L. monocytogenes sRNA, is packaged into EVs and activates RIG-I in recipient cells, and this activation promotes bacterial survival in macrophages (Frantz et al., 2019).
EVs released by L. monocytogene-infected host cells
During L. monocytogenes infection, host cells, such as mouse embryonic fibroblast cells and mouse macrophages, release exosomes carrying Listeria DNA. These may activate the production of interferon-β (IFN-β) in bystander cells through the cGAS–STING–TBK1 cytosolic DNA-sensing pathway (Nandakumar et al., 2019). Additionally, these hEVs promote the STING-dependent apoptosis of mouse splenic T lymphocytes that are stimulated with Fas ligand (FasL, also known as FASLG) or activated with anti-CD3 and anti-CD8, suggesting that they exert a detrimental effect on host defense mechanisms (Nandakumar et al., 2019). As described above, nSMase2 has been found to regulate exosome biogenesis in oligodendroglial cells. Treatment of L. monocytogene-infected macrophages with GW4869, an inhibitor of nSMase2, interferes with the exosome production. Interestingly, GW4869 treatment significantly decreases the bacterial load in L. monocytogene-infected mice, suggesting that EVs released from Listeria-infected cells promote bacterial survival (Nandakumar et al., 2019). Moreover, the host cGAS–STING–TBK1-dependent DNA-sensing pathway is also required for packaging Listeria DNA into exosomes via TBK1-mediated S222 phosphorylation of the MVB protein MVB12b, a key component of the ESCRT-I complex (Christ et al., 2017; Hansen et al., 2014). Furthermore, MVB12b has been found to be essential for the sorting of DNA into EVs and for stimulation of bystander cells (Nandakumar et al., 2019).
The evidence to date suggests that both Listeria-derived EVs and hEVs contain Listeria nucleic acids and that both vesicle types induce Type I interferon production and attenuate an effective immune response against Listeria infection. However, compared to mycobacterial infections, only a few studies have investigated the biological function of hEVs during L. monocytogenes infection (Zhang et al., 2012; Nandakumar et al., 2019), and the full protein content of the host cell-derived EVs remains to be defined. Performing a comparative proteomic analysis of host cell- and L. monocytogenes-derived EVs would help us better understand the roles of these distinct EV populations in the Listeria–host interactions. Additionally, a comparative study of EVs from Mtb-infected and L. monocytogenes-infected host cells would help us identify common or pathogen-specific factors that activate the EV-mediated pathways in host cells.
EVs in the context of a Salmonella enterica infection
As noted above, the EVs released from Gram-negative bacteria such as Salmonella spp. are referred to as OMVs. The OMVs from Salmonella have been studied extensively with regard to their composition and immune modulation, as well as in the context of vaccine development. Proteomic analyses have identified over 150 proteins in OMVs of S. enterica serovar Typhimurium, but the exact composition varies under different environmental conditions (Bai et al., 2014). Environmental conditions can also affect the RNA content of S. enterica serovar Typhimurium OMVs (Malabirade et al., 2018). OMVs of other Salmonella species have also been analyzed for their protein composition (Liu et al., 2017b). In addition to proteins, OMVs contain lipopolysaccharide (LPS), which can induce activation of macrophages and dendritic cells via TLR4 (Laughlin et al., 2015). Additionally, proteins that are regulated by PhoP, a two-component signaling system in Salmonella, also play a role in OMV-mediated dendritic cell activation, as vesicles from the pho-24 pmrA505 mutant strain induce a lower production of TNF, IL-12 p40 (also known as IL12B) and IL-6 compared to that induced by OMVs from wild-type bacteria (Laughlin et al., 2015). OMVs have also been shown to contain typhoid toxin, which can induce DNA damage in recipient cells (Guidi et al., 2013), as well as the virulence factors PagK1 (also known as PagK), PagK2 (also known as STM14_3167) and PagJ (also known as SarA), which can be transferred to non-infected cells during an infection (Yoon et al., 2011). A major research focus has been the development and testing of Salmonella OMVs as vaccine candidates. For example, initial characterization of OMVs derived from S. enterica serovar Enteritidis has revealed an enrichment in periplasmic and outer membrane proteins, and intranasal or intraperitoneal vaccination of mice with these OMVs has been found to elicit both a cell-mediated and humoral immune response (Liu et al., 2017a), with immunized mice having markedly higher rates of survival relative to control mice after challenge with a lethal dose of S. enterica serovar Enteritidis (Liu et al., 2017a). A similar vaccine efficacy for Salmonella OMVs has been observed in other studies (Pastor et al., 2021; Gasperini et al., 2020; Liu et al., 2016). A significant challenge for the use of Salmonella OMVs as vaccines is the presence of the endotoxin LPS in isolated vesicles. However, Lee et al. (2009) have observed a similar vaccine efficacy when using a low endotoxin-containing OMV from Salmonella enterica serovar Typhimurium compared to using OMVs with a normal level of LPS.
Information on hEVs in the context of a Salmonella infection is limited. Initial work has indicated that THP-1 cells infected with Salmonella enterica serovar Typhimurium release hEVs that contain Salmonella enterica serovar Typhimurium LPS, which stimulates TNF production in a TL4-dependent manner (Bhatnagar et al., 2007). More recently, EVs released from cells infected with Salmonella enterica serovar Typhimurium have been shown to induce an inflammatory response, primarily measured by TNF secretion (Hui et al., 2018). Furthermore, this effect appears to be macrophage specific, as EVs derived from infected macrophages induce TNF secretion in naïve macrophages and dendritic cells, whereas EVs from infected dendritic cells do not elicit such a response in either cell type. TNF secretion upon exposure to infection-derived EVs is partially reduced when a Salmonella enterica serovar Typhimurium mutant expressing an LPS variant with attenuated endotoxicity is used, suggesting that LPS may have a role in the observed effects. To dissect the importance of the different subsets of EVs, exosome populations from infected cells have been fractionated to isolate CD63+ and CD9+ EVs, and their secretory responses have been evaluated using cytokine arrays. Both EV populations induce similar cytokine profiles, with an increased secretion of pro-inflammatory cytokines. Treating macrophages that lack expression of TLR2 with EVs released from Salmonella enterica serovar Typhimurium-infected cells reveals a significant decrease in TNF secretion relative to treated wild-type macrophages, whereas TLR4-deficient macrophages show no TNF production upon treatment with the EVs (Hui et al., 2018).
Although hEVs from Salmonella-infected cells require additional characterization in regard to both their content and function, it is clear that both bacterial- and host-derived EVs contain known bacterial molecules, such as LPS, and can induce a pro-inflammatory response in recipient cells. Furthermore, the ability of Salmonella-released EVs to elicit a protective immune response indicates the presence of key antigens. Whether this extends to hEVs requires additional investigations, the results of which will help us define whether the two different EV populations are major suppliers of antigen during an in vivo infection.
Conclusions and next steps
We are still in our infancy in understanding the role of EVs in bacterial pathogenesis and host immunity, but it is clear that EVs carrying bacterial components are derived from both bacteria and host, especially in the case of intracellular pathogens. A common theme is the ability of the EVs generated during an infection to bind and activate PRRs. For example, activation of TLR2 or TLR4, as well as of nucleic acid sensors such as cGAS and RIG-I, has been observed for EVs released from various pathogenic bacteria and from host cells infected with these pathogens. However, the activation of these PRRs can induce different responses in different cells types. For instance, cGAS activation by EVs released from Listeria-infected macrophages induces IFN-β production in recipient macrophages, but leads to apoptosis in T cells (Nandakumar et al., 2019). Understanding the effects of EVs on different cell populations is essential if we are to define their role in immunity. Other important outstanding questions include how EV production is regulated during an in vivo infection, how EV composition and function change over time, and whether there are commonalities in host proteins between EVs released from cells infected with different pathogens. New techniques such as single-cell sequencing will help address the heterogeneity of the host response to different EV populations. Moreover, newly developed flow cytometry instruments and methods, which allow for isolation and characterization of individual EVs, will enable researchers to better define their EV population in terms of both content and function. Clearly there is much work to be done, but the results of such labor should provide new insight into host–pathogen interactions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Our work in this area is supported by the National Institutes of Health (AI 151206-01). Deposited in PMC for release after 12 months.
References
- Alvarez-Jiménez, V. D., Leyva-Paredes, K., García-Martínez, M., Vázquez-Flores, L., García-Paredes, V. G., Campillo-Navarro, M., Romo-Cruz, I., Rosales-García, V. H., Castañeda-Casimiro, J., González-Pozos, S.et al. (2018). Extracellular vesicles released from Mycobacterium tuberculosis-infected neutrophils promote macrophage autophagy and decrease intracellular mycobacterial survival. Front. Immunol. 9, 272. 10.3389/fimmu.2018.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoni, F., Toyofuku, M., Menzi, C., Kalawong, R., Mairpady Shambat, S., Francois, P., Zinkernagel, A. S. and Eberl, L. (2019). Antibiotics stimulate formation of vesicles in staphylococcus aureus in both phage-dependent and -independent fashions and via different routes. Antimicrob. Agents Chemother. 63, e01439-18. 10.1128/AAC.01439-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athman, J. J., Wang, Y., McDonald, D. J., Boom, W. H., Harding, C. V. and Wearsch, P. A. (2015). Bacterial membrane vesicles mediate the release of mycobacterium tuberculosis lipoglycans and lipoproteins from infected macrophages. J. Immunol. 195, 1044-1053. 10.4049/jimmunol.1402894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athman, J. J., Sande, O. J., Groft, S. G., Reba, S. M., Nagy, N., Wearsch, P. A., Richardson, E. T., Rojas, R., Boom, W. H., Shukla, S.et al. (2017). Mycobacterium tuberculosis membrane vesicles inhibit T cell activation. J. Immunol. 198, 2028-2037. 10.4049/jimmunol.1601199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., Katzmann, D. J., Snyder, W. B., Wendland, B. and Emr, S. D. (2002). Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3, 283-289. 10.1016/S1534-5807(02)00219-8 [DOI] [PubMed] [Google Scholar]
- Bai, J., Kim, S. I., Ryu, S. and Yoon, H. (2014). Identification and characterization of outer membrane vesicle-associated proteins in Salmonella enterica serovar Typhimurium. Infect. Immun. 82, 4001-4010. 10.1128/IAI.01416-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti, M. F., Zhang, Z., Mortier, E., Melchior, A., Degeest, G., Geeraerts, A., Ivarsson, Y., Depoortere, F., Coomans, C., Vermeiren, E.et al. (2012). Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14, 677-685. 10.1038/ncb2502 [DOI] [PubMed] [Google Scholar]
- Banzhaf, M., van den Berg van Saparoea, B., Terrak, M., Fraipont, C., Egan, A., Philippe, J., Zapun, A., Breukink, E., Nguyen-Disteche, M., den Blaauwen, T.et al. (2012). Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol. Microbiol. 85, 179-194. 10.1111/j.1365-2958.2012.08103.x [DOI] [PubMed] [Google Scholar]
- Beatty, W. and Russell, D. G. (2001). Analysis of mycobacterium-infected macrophages by immunoelectron microscopy and cell fractionation. Methods Mol. Med. 54, 281-293. 10.1385/1-59259-147-7:281 [DOI] [PubMed] [Google Scholar]
- Beatty, W. L., Rhoades, E. R., Ullrich, H. J., Chatterjee, D., Heuser, J. E. and Russell, D. G. (2000). Trafficking and release of mycobacterial lipids from infected macrophages. Traffic 1, 235-247. 10.1034/j.1600-0854.2000.010306.x [DOI] [PubMed] [Google Scholar]
- Behar, S. M., Martin, C. J., Nunes-Alves, C., Divangahi, M. and Remold, H. G. (2011). Lipids, apoptosis, and cross-presentation: links in the chain of host defense against Mycobacterium tuberculosis. Microbes Infect. 13, 749-756. 10.1016/j.micinf.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar, S. and Schorey, J. S. (2007). Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. J. Biol. Chem. 282, 25779-25789. 10.1074/jbc.M702277200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar, S., Shinagawa, K., Castellino, F. J. and Schorey, J. S. (2007). Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood 110, 3234-3244. 10.1182/blood-2007-03-079152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briaud, P. and Carroll, R. K. (2020). Extracellular vesicle biogenesis and functions in gram-positive bacteria. Infect. Immun. 88, e00433-20. 10.1128/IAI.00433-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, L., Wolf, J. M., Prados-Rosales, R. and Casadevall, A. (2015). Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 13, 620-630. 10.1038/nrmicro3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik, V., Ruiz-Cañada, C. and Wendler, F. (2016). Extracellular vesicles round off communication in the nervous system. Nat. Rev. Neurosci. 17, 160-172. 10.1038/nrn.2015.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschow, S. I., Nolte-'t Hoen, E. N., van Niel, G., Pols, M. S., ten Broeke, T., Lauwen, M., Ossendorp, F., Melief, C. J., Raposo, G., Wubbolts, R.et al. (2009). MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic 10, 1528-1542. 10.1111/j.1600-0854.2009.00963.x [DOI] [PubMed] [Google Scholar]
- Cadena, A. M., Fortune, S. M. and Flynn, J. L. (2017). Heterogeneity in tuberculosis. Nat. Rev. Immunol. 17, 691-702. 10.1038/nri.2017.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. and Schorey, J. S. (2013). Exosomes carrying mycobacterial antigens can protect mice against Mycobacterium tuberculosis infection. Eur. J. Immunol. 43, 3279-3290. 10.1002/eji.201343727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. and Schorey, J. S. (2018). Mycobacterium tuberculosis-induced IFN-beta production requires cytosolic DNA and RNA sensing pathways. J. Exp. Med. 215, 2919-2935. 10.1084/jem.20180508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. and Schorey, J. S. (2019). Extracellular vesicles deliver Mycobacterium RNA to promote host immunity and bacterial killing. EMBO Rep. 20, e46613. 10.15252/embr.201846613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiplunkar, S. S., Silva, C. A., Bermudez, L. E. and Danelishvili, L. (2019). Characterization of membrane vesicles released by Mycobacterium avium in response to environment mimicking the macrophage phagosome. Future Microbiol. 14, 293-313. 10.2217/fmb-2018-0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ, L., Raiborg, C., Wenzel, E. M., Campsteijn, C. and Stenmark, H. (2017). Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem. Sci. 42, 42-56. 10.1016/j.tibs.2016.08.016 [DOI] [PubMed] [Google Scholar]
- Clancy, J. W., Sedgwick, A., Rosse, C., Muralidharan-Chari, V., Raposo, G., Method, M., Chavrier, P. and D'Souza-Schorey, C. (2015). Regulated delivery of molecular cargo to invasive tumour-derived microvesicles. Nat. Commun. 6, 6919. 10.1038/ncomms7919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho, C., Brown, L., Maryam, M., Vij, R., Smith, D. F. Q., Burnet, M. C., Kyle, J. E., Heyman, H. M., Ramirez, J., Prados-Rosales, R.et al. (2019). Listeria monocytogenes virulence factors, including listeriolysin O, are secreted in biologically active extracellular vesicles. J. Biol. Chem. 294, 1202-1217. 10.1074/jbc.RA118.006472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage, B. L., Lara, J. C., Bergsbaken, T., Rassoulian Barrett, S. L., Lara, S. and Cookson, B. T. (2009). Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 72, 1395-1407. 10.1111/j.1365-2958.2009.06731.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Conde, I., Shrimpton, C. N., Thiagarajan, P. and Lopez, J. A. (2005). Tissue-factor-bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation. Blood 106, 1604-1611. 10.1182/blood-2004-03-1095 [DOI] [PubMed] [Google Scholar]
- Drage, M. G., Pecora, N. D., Hise, A. G., Febbraio, M., Silverstein, R. L., Golenbock, D. T., Boom, W. H. and Harding, C. V. (2009). TLR2 and its co-receptors determine responses of macrophages and dendritic cells to lipoproteins of Mycobacterium tuberculosis. Cell. Immunol. 258, 29-37. 10.1016/j.cellimm.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza-Schorey, C. and Schorey, J. S. (2018). Regulation and mechanisms of extracellular vesicle biogenesis and secretion. Essays Biochem. 62, 125-133. 10.1042/EBC20170078 [DOI] [PubMed] [Google Scholar]
- Foster, M., Hill, P. C., Setiabudiawan, T. P., Koeken, V., Alisjahbana, B. and van Crevel, R. (2021). BCG-induced protection against Mycobacterium tuberculosis infection: Evidence, mechanisms, and implications for next-generation vaccines. Immunol. Rev. 00, 1-23. 10.1111/imr.12965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz, R., Teubner, L., Schultze, T., La Pietra, L., Muller, C., Gwozdzinski, K., Pillich, H., Hain, T., Weber-Gerlach, M., Panagiotidis, G. D.et al. (2019). The secRNome of Listeria monocytogenes Harbors Small Noncoding RNAs That Are Potent Inducers of Beta Interferon. mBio 10, e01223-19. 10.1128/mBio.01223-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard, J. L., Berche, P., Mounier, J., Richard, S. and Sansonetti, P. (1987). In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect. Immun. 55, 2822-2829. 10.1128/IAI.55.11.2822-2829.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini, G., Alfini, R., Arato, V., Mancini, F., Aruta, M. G., Kanvatirth, P., Pickard, D., Necchi, F., Saul, A., Rossi, O.et al. (2020). Salmonella Paratyphi A outer membrane vesicles displaying Vi polysaccharide as multivalent vaccine against enteric fever. Infect. Immun. 89, e00699-20. 10.1128/IAI.00699-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri, P. K. and Schorey, J. S. (2008). Exosomes derived from M. Bovis BCG infected macrophages activate antigen-specific CD4+ and CD8+ T cells in vitro and in vivo. PLoS ONE 3, e2461. 10.1371/journal.pone.0002461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri, P. K., Kruh, N. A., Dobos, K. M. and Schorey, J. S. (2010). Proteomic analysis identifies highly antigenic proteins in exosomes from M. tuberculosis-infected and culture filtrate protein-treated macrophages. Proteomics 10, 3190-3202. 10.1002/pmic.200900840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi, R., Levi, L., Rouf, S. F., Puiac, S., Rhen, M. and Frisan, T. (2013). Salmonella enterica delivers its genotoxin through outer membrane vesicles secreted from infected cells. Cell. Microbiol. 15, 2034-2050. 10.1111/cmi.12172 [DOI] [PubMed] [Google Scholar]
- Gupta, S., Palacious, A., Khataokar, A., Wienrick, B., Lavin, J. L. and Sampedro, L. (2020). Dynamin-like proteins are essential for vesicle biogenesis in Mycobacterium tuberculosis. bioRxiv. 10.1101/2020.01.14.906362 [DOI] [Google Scholar]
- Hansen, K., Prabakaran, T., Laustsen, A., Jørgensen, S. E., Rahbaek, S. H., Jensen, S. B., Nielsen, R., Leber, J. H., Decker, T., Horan, K. A.et al. (2014). Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 33, 1654-1666. 10.15252/embj.201488029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraszti, R. A., Didiot, M. C., Sapp, E., Leszyk, J., Shaffer, S. A., Rockwell, H. E., Gao, F., Narain, N. R., DiFiglia, M., Kiebish, M. A.et al. (2016). High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell Vesicles 5, 32570. 10.3402/jev.v5.32570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, C., Heuser, J. and Stahl, P. (1984). Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: demonstration of a pathway for receptor shedding. Eur. J. Cell Biol. 35, 256-263. [PubMed] [Google Scholar]
- Hoffmann, C., Leis, A., Niederweis, M., Plitzko, J. M. and Engelhardt, H. (2008). Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. USA 105, 3963-3967. 10.1073/pnas.0709530105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, C., Morohashi, Y., Yoshimura, S., Manrique-Hoyos, N., Jung, S., Lauterbach, M. A., Bakhti, M., Grønborg, M., Møbius, W., Rhee, J.et al. (2010). Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J. Cell Biol. 189, 223-232. 10.1083/jcb.200911018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, W. W., Hercik, K., Belsare, S., Alugubelly, N., Clapp, B., Rinaldi, C. and Edelmann, M. J. (2018). Salmonella enterica Serovar typhimurium alters the extracellular proteome of macrophages and leads to the production of proinflammatory exosomes. Infect. Immun. 86, e00386-17. 10.1128/IAI.00386-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkoshek, K. S., Wang, Y., Athman, J. J., Barton, M. R. and Wearsch, P. A. (2016). Interspecies communication between pathogens and immune cells via bacterial membrane vesicles. Front Cell Dev Biol 4, 125. 10.3389/fcell.2016.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan, S., Balakrishnan, J. and Govindasamy, A. (2020). Listeria monocytogens - Amended understanding of its pathogenesis with a complete picture of its membrane vesicles, quorum sensing, biofilm and invasion. Microb. Pathog. 149, 104575. 10.1016/j.micpath.2020.104575 [DOI] [PubMed] [Google Scholar]
- Karthikeyan, R., Gayathri, P., Gunasekaran, P., Jagannadham, M. V. and Rajendhran, J. (2019). Comprehensive proteomic analysis and pathogenic role of membrane vesicles of Listeria monocytogenes serotype 4b reveals proteins associated with virulence and their possible interaction with host. Int. J. Med. Microbiol. 309, 199-212. 10.1016/j.ijmm.2019.03.008 [DOI] [PubMed] [Google Scholar]
- Karthikeyan, R., Gayathri, P., Gunasekaran, P., Jagannadham, M. V. and Rajendhran, J. (2020). Functional analysis of membrane vesicles of Listeria monocytogenes suggests a possible role in virulence and physiological stress response. Microb. Pathog. 142, 104076. 10.1016/j.micpath.2020.104076 [DOI] [PubMed] [Google Scholar]
- Katzmann, D. J., Babst, M. and Emr, S. D. (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145-155. 10.1016/S0092-8674(01)00434-2 [DOI] [PubMed] [Google Scholar]
- Kirschning, C. J. and Schumann, R. R. (2002). TLR2: cellular sensor for microbial and endogenous molecular patterns. Curr. Top. Microbiol. Immunol. 270, 121-144. 10.1007/978-3-642-59430-4_8 [DOI] [PubMed] [Google Scholar]
- Kowal, J., Arras, G., Colombo, M., Jouve, M., Morath, J. P., Primdal-Bengtson, B., Dingli, F., Loew, D., Tkach, M. and Thery, C. (2016). Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 113, E968-E977. 10.1073/pnas.1521230113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruh-Garcia, N. A., Wolfe, L. M., Chaisson, L. H., Worodria, W. O., Nahid, P., Schorey, J. S., Davis, J. L. and Dobos, K. M. (2014). Detection of Mycobacterium tuberculosis peptides in the exosomes of patients with active and latent M. tuberculosis infection using MRM-MS. PLoS ONE 9, e103811. 10.1371/journal.pone.0103811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, H. M., Swamy Ch, V. and Jagannadham, M. V. (2014). Molecular characterization and functional analysis of outer membrane vesicles from the antarctic bacterium Pseudomonas syringae suggest a possible response to environmental conditions. J. Proteome Res. 13, 1345-1358. 10.1021/pr4009223 [DOI] [PubMed] [Google Scholar]
- Kulp, A. and Kuehn, M. J. (2010). Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64, 163-184. 10.1146/annurev.micro.091208.073413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin, R. C., Mickum, M., Rowin, K., Adams, L. G. and Alaniz, R. C. (2015). Altered host immune responses to membrane vesicles from Salmonella and Gram-negative pathogens. Vaccine 33, 5012-5019. 10.1016/j.vaccine.2015.05.014 [DOI] [PubMed] [Google Scholar]
- Layre, E. (2020). Trafficking of mycobacterium tuberculosis envelope components and release within extracellular vesicles: host-pathogen interactions beyond the wall. Front. Immunol. 11, 1230. 10.3389/fimmu.2020.01230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. R., Kim, S. H., Jeong, K. J., Kim, K. S., Kim, Y. H., Kim, S. J., Kim, E., Kim, J. W. and Chang, K. T. (2009). Multi-immunogenic outer membrane vesicles derived from an MsbB-deficient Salmonella enterica serovar typhimurium mutant. J. Microbiol. Biotechnol. 19, 1271-1279. [PubMed] [Google Scholar]
- Lee, J. H., Choi, C. W., Lee, T., Kim, S. I., Lee, J. C. and Shin, J. H. (2013). Transcription factor sigmaB plays an important role in the production of extracellular membrane-derived vesicles in Listeria monocytogenes. PLoS ONE 8, e73196. 10.1371/journal.pone.0073196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Kim, S. H., Choi, D. S., Lee, J. S., Kim, D. K., Go, G., Park, S. M., Kim, S. H., Shin, J. H., Chang, C. L.et al. (2015). Proteomic analysis of extracellular vesicles derived from Mycobacterium tuberculosis. Proteomics 15, 3331-3337. 10.1002/pmic.201500037 [DOI] [PubMed] [Google Scholar]
- Lee, T., Jun, S. H., Choi, C. W., Kim, S. I., Lee, J. C. and Shin, J. H. (2018). Salt stress affects global protein expression profiles of extracellular membrane-derived vesicles of Listeria monocytogenes. Microb. Pathog. 115, 272-279. 10.1016/j.micpath.2017.12.071 [DOI] [PubMed] [Google Scholar]
- Li, B., Antonyak, M. A., Zhang, J. and Cerione, R. A. (2012). RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene 31, 4740-4749. 10.1038/onc.2011.636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Liu, Q., Yi, J., Liang, K., Hu, B., Zhang, X., Curtiss, R., III and Kong, Q. (2016). Outer membrane vesicles from flagellin-deficient Salmonella enterica serovar Typhimurium induce cross-reactive immunity and provide cross-protection against heterologous Salmonella challenge. Sci. Rep. 6, 34776. 10.1038/srep34776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, P. L. and Flynn, J. L. (2018). The end of the binary era: revisiting the spectrum of tuberculosis. J. Immunol. 201, 2541-2548. 10.4049/jimmunol.1800993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Yi, J., Liang, K., Zhang, X. and Liu, Q. (2017a). Outer membrane vesicles derived from salmonella enteritidis protect against the virulent wild-type strain infection in a mouse model. J. Microbiol. Biotechnol. 27, 1519-1528. 10.4014/jmb.1705.05028 [DOI] [PubMed] [Google Scholar]
- Liu, Q., Yi, J., Liang, K., Zhang, X. and Liu, Q. (2017b). Salmonella Choleraesuis outer membrane vesicles: proteomics and immunogenicity. J. Basic Microbiol. 57, 852-861. 10.1002/jobm.201700153 [DOI] [PubMed] [Google Scholar]
- Malabirade, A., Habier, J., Heintz-Buschart, A., May, P., Godet, J., Halder, R., Etheridge, A., Galas, D., Wilmes, P. and Fritz, J. V. (2018). The RNA complement of outer membrane vesicles from salmonella enterica serovar typhimurium under distinct culture conditions. Front. Microbiol. 9, 2015. 10.3389/fmicb.2018.02015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsollier, L., Brodin, P., Jackson, M., Kordulakova, J., Tafelmeyer, P., Carbonnelle, E., Aubry, J., Milon, G., Legras, P., Andre, J. P.et al. (2007). Impact of Mycobacterium ulcerans biofilm on transmissibility to ecological niches and Buruli ulcer pathogenesis. PLoS Pathog. 3, e62. 10.1371/journal.ppat.0030062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehaffy, C., Dobos, K. M., Nahid, P. and Kruh-Garcia, N. A. (2017). Second generation multiple reaction monitoring assays for enhanced detection of ultra-low abundance Mycobacterium tuberculosis peptides in human serum. Clin Proteomics 14, 21. 10.1186/s12014-017-9156-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Morse, N., Robbins, J. R., Rae, C. S., Mochegova, S. N., Swanson, M. S., Zhao, Z., Virgin, H. W. and Portnoy, D. (2010). Listeriolysin O is necessary and sufficient to induce autophagy during Listeria monocytogenes infection. PLoS ONE 5, e8610. 10.1371/journal.pone.0008610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobius, W., Ohno-Iwashita, Y., van Donselaar, E. G., Oorschot, V. M., Shimada, Y., Fujimoto, T., Heijnen, H. F., Geuze, H. J. and Slot, J. W. (2002). Immunoelectron microscopic localization of cholesterol using biotinylated and non-cytolytic perfringolysin O. J. Histochem. Cytochem. 50, 43-55. 10.1177/002215540205000105 [DOI] [PubMed] [Google Scholar]
- Moon, D. C., Choi, C. H., Lee, J. H., Choi, C. W., Kim, H. Y., Park, J. S., Kim, S. I. and Lee, J. C. (2012). Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles. J. Microbiol. 50, 155-160. 10.1007/s12275-012-1589-4 [DOI] [PubMed] [Google Scholar]
- Nandakumar, R., Tschismarov, R., Meissner, F., Prabakaran, T., Krissanaprasit, A., Farahani, E., Zhang, B. C., Assil, S., Martin, A., Bertrams, W.et al. (2019). Intracellular bacteria engage a STING-TBK1-MVB12b pathway to enable paracrine cGAS-STING signalling. Nat Microbiol 4, 701-713. 10.1038/s41564-019-0367-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski, M., Carmo, N. B., Krumeich, S., Fanget, I., Raposo, G., Savina, A., Moita, C. F., Schauer, K., Hume, A. N., Freitas, R. P.et al. (2010). Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19-30; sup pp 1-13. 10.1038/ncb2000 [DOI] [PubMed] [Google Scholar]
- Pan, B. T. and Johnstone, R. M. (1983). Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell 33, 967-978. 10.1016/0092-8674(83)90040-5 [DOI] [PubMed] [Google Scholar]
- Pastor, Y., Ting, I., Berzosa, M., Irache, J. M. and Gamazo, C. (2021). Vaccine based on outer membrane vesicles using hydrogels as vaccine delivery system. Methods Mol. Biol. 2182, 153-160. 10.1007/978-1-0716-0791-6_14 [DOI] [PubMed] [Google Scholar]
- Prados-Rosales, R., Baena, A., Martinez, L. R., Luque-Garcia, J., Kalscheuer, R., Veeraraghavan, U., Camara, C., Nosanchuk, J. D., Besra, G. S., Chen, B.et al. (2011). Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J. Clin. Invest. 121, 1471-1483. 10.1172/JCI44261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados-Rosales, R., Carreño, L. J., Batista-Gonzalez, A., Baena, A., Venkataswamy, M. M., Xu, J., Yu, X., Wallstrom, G., Magee, D. M., LaBaer, J.et al. (2014a). Mycobacterial membrane vesicles administered systemically in mice induce a protective immune response to surface compartments of Mycobacterium tuberculosis. mBio 5, e01921-e01914. 10.1128/mBio.01921-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados-Rosales, R., Weinrick, B. C., Pique, D. G., Jacobs, W. R., Jr, Casadevall, A. and Rodriguez, G. M. (2014b). Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J. Bacteriol. 196, 1250-1256. 10.1128/JB.01090-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg, C. and Stenmark, H. (2002). Hrs and endocytic sorting of ubiquitinated membrane proteins. Cell Struct. Funct. 27, 403-408. 10.1247/csf.27.403 [DOI] [PubMed] [Google Scholar]
- Ramachandra, L., Qu, Y., Wang, Y., Lewis, C. J., Cobb, B. A., Takatsu, K., Boom, W. H., Dubyak, G. R. and Harding, C. V. (2010). Mycobacterium tuberculosis synergizes with ATP to induce release of microvesicles and exosomes containing major histocompatibility complex class II molecules capable of antigen presentation. Infect. Immun. 78, 5116-5125. 10.1128/IAI.01089-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath, P., Huang, C., Wang, T., Wang, T., Li, H., Prados-Rosales, R., Elemento, O., Casadevall, A. and Nathan, C. F. (2013). Genetic regulation of vesiculogenesis and immunomodulation in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 110, E4790-E4797. 10.1073/pnas.1320118110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, E., Hsu, F., Torrelles, J. B., Turk, J., Chatterjee, D. and Russell, D. G. (2003). Identification and macrophage-activating activity of glycolipids released from intracellular Mycobacterium bovis BCG. Mol. Microbiol. 48, 875-888. 10.1046/j.1365-2958.2003.03473.x [DOI] [PubMed] [Google Scholar]
- Savina, A., Vidal, M. and Colombo, M. I. (2002). The exosome pathway in K562 cells is regulated by Rab11. J. Cell Sci. 115, 2505-2515. [DOI] [PubMed] [Google Scholar]
- Schorey, J. S. and Bhatnagar, S. (2008). Exosome function: from tumor immunology to pathogen biology. Traffic 9, 871-881. 10.1111/j.1600-0854.2008.00734.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorey, J. S. and Schlesinger, L. S. (2016). Innate immune responses to tuberculosis. Microbiol Spectr 4, TBTB2-0010-2016. 10.1128/microbiolspec.TBTB2-0010-2016 [DOI] [PubMed] [Google Scholar]
- Schorey, J. S., Cheng, Y., Singh, P. P. and Smith, V. L. (2015). Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 16, 24-43. 10.15252/embr.201439363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C. and Kuehn, M. J. (2015). Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 13, 605-619. 10.1038/nrmicro3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C., Kulp, A. and Kuehn, M. J. (2014). Modulation of bacterial outer membrane vesicle production by envelope structure and content. BMC Microbiol. 14, 324. 10.1186/s12866-014-0324-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick, A. E. and D'Souza-Schorey, C. (2018). The biology of extracellular microvesicles. Traffic 19, 319-327. 10.1111/tra.12558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick, A. E., Clancy, J. W., Olivia Balmert, M. and D'Souza-Schorey, C. (2015). Extracellular microvesicles and invadopodia mediate non-overlapping modes of tumor cell invasion. Sci. Rep. 5, 14748. 10.1038/srep14748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P. P., LeMaire, C., Tan, J. C., Zeng, E. and Schorey, J. S. (2011). Exosomes released from M. tuberculosis infected cells can suppress IFN-gamma mediated activation of naive macrophages. PLoS One 6, e18564. 10.1371/journal.pone.0018564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P. P., Li, L. and Schorey, J. S. (2015). Exosomal RNA from mycobacterium tuberculosis-infected cells is functional in recipient macrophages. Traffic 16, 555-571. 10.1111/tra.12278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, V. L., Jackson, L. and Schorey, J. S. (2015). Ubiquitination as a mechanism to transport soluble mycobacterial and eukaryotic proteins to exosomes. J. Immunol. 195, 2722-2730. 10.4049/jimmunol.1403186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, V. L., Cheng, Y., Bryant, B. R. and Schorey, J. S. (2017). Exosomes function in antigen presentation during an in vivo Mycobacterium tuberculosis infection. Sci. Rep. 7, 43578. 10.1038/srep43578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag, I., Schwarz, H., Hirota, Y. and Henning, U. (1978). Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J. Bacteriol. 136, 280-285. 10.1128/JB.136.1.280-285.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan, H. and Upton, J. W. (2014). Programmed necrosis in microbial pathogenesis. Trends Microbiol. 22, 199-207. 10.1016/j.tim.2014.01.005 [DOI] [PubMed] [Google Scholar]
- Srivastava, S. and Ernst, J. D. (2014). Cell-to-cell transfer of M. tuberculosis antigens optimizes CD4 T cell priming. Cell Host Microbe 15, 741-752. 10.1016/j.chom.2014.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuffers, S., Sem Wegner, C., Stenmark, H. and Brech, A. (2009). Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 10, 925-937. 10.1111/j.1600-0854.2009.00920.x [DOI] [PubMed] [Google Scholar]
- Swaminathan, B. and Gerner-Smidt, P. (2007). The epidemiology of human listeriosis. Microbes Infect. 9, 1236-1243. 10.1016/j.micinf.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Tamai, K., Tanaka, N., Nakano, T., Kakazu, E., Kondo, Y., Inoue, J., Shiina, M., Fukushima, K., Hoshino, T., Sano, K.et al. (2010). Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem. Biophys. Res. Commun. 399, 384-390. 10.1016/j.bbrc.2010.07.083 [DOI] [PubMed] [Google Scholar]
- Toyofuku, M., Nomura, N. and Eberl, L. (2019). Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 17, 13-24. 10.1038/s41579-018-0112-2 [DOI] [PubMed] [Google Scholar]
- Trajkovic, K., Hsu, C., Chiantia, S., Rajendran, L., Wenzel, D., Wieland, F., Schwille, P., Brugger, B. and Simons, M. (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244-1247. 10.1126/science.1153124 [DOI] [PubMed] [Google Scholar]
- Ullrich, H. J., Beatty, W. L. and Russell, D. G. (2000). Interaction of Mycobacterium avium-containing phagosomes with the antigen presentation pathway. J. Immunol. 165, 6073-6080. 10.4049/jimmunol.165.11.6073 [DOI] [PubMed] [Google Scholar]
- Vdovikova, S., Luhr, M., Szalai, P., Nygåard Skalman, L., Francis, M. K., Lundmark, R., Engedal, N., Johansson, J. and Wai, S. N. (2017). A novel role of listeria monocytogenes membrane vesicles in inhibition of autophagy and cell death. Front. Cell Infect. Microbiol. 7, 154. 10.3389/fcimb.2017.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.-J., Chen, C., Xie, P.-F., Pan, Y., Tan, Y.-H. and Tang, L.-J. (2014). Proteomic analysis and immune properties of exosomes released by macrophages infected with Mycobacterium avium. Microbes Infect. 16, 283-291. 10.1016/j.micinf.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Wang, X., Thompson, C. D., Weidenmaier, C. and Lee, J. C. (2018). Release of Staphylococcus aureus extracellular vesicles and their application as a vaccine platform. Nat. Commun. 9, 1379. 10.1038/s41467-018-03847-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Guo, X., Yang, X., Zhang, B., Ren, J., Liu, A., Ran, Y., Yan, B., Chen, F., Guddat, L. W.et al. (2019). Mycobacterial dynamin-like protein IniA mediates membrane fission. Nat. Commun. 10, 3906. 10.1038/s41467-019-11860-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert, T., Wunder, C., Lippincott-Schwartz, J. and Hurley, J. H. (2009). Membrane scission by the ESCRT-III complex. Nature 458, 172-177. 10.1038/nature07836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman, P. G. and Futter, C. E. (2008). Multivesicular bodies: co-ordinated progression to maturity. Curr. Opin. Cell Biol. 20, 408-414. 10.1016/j.ceb.2008.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, H., Ansong, C., Adkins, J. N. and Heffron, F. (2011). Discovery of Salmonella virulence factors translocated via outer membrane vesicles to murine macrophages. Infect. Immun. 79, 2182-2192. 10.1128/IAI.01277-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Zhang, R., Zhang, H., Liu, J., Yang, Z., Xu, P., Cai, W., Lu, G., Cui, M., Schwendener, R. A.et al. (2012). Microparticles released by Listeria monocytogenes-infected macrophages are required for dendritic cell-elicited protective immunity. Cell. Mol. Immunol. 9, 489-496. 10.1038/cmi.2012.33 [DOI] [PMC free article] [PubMed] [Google Scholar]