ABSTRACT
Axons and dendrites are distinguished by microtubule polarity. In Drosophila, dendrites are dominated by minus-end-out microtubules, whereas axons contain plus-end-out microtubules. Local nucleation in dendrites generates microtubules in both orientations. To understand why dendritic nucleation does not disrupt polarity, we used live imaging to analyze the fate of microtubules generated at branch points. We found that they had different rates of success exiting the branch based on orientation: correctly oriented minus-end-out microtubules succeeded in leaving about twice as often as incorrectly oriented microtubules. Increased success relied on other microtubules in a parallel orientation. From a candidate screen, we identified Trim9 and kinesin-5 (Klp61F) as machinery that promoted growth of new microtubules. In S2 cells, Eb1 recruited Trim9 to microtubules. Klp61F promoted microtubule growth in vitro and in vivo, and could recruit Trim9 in S2 cells. In summary, the data argue that Trim9 and kinesin-5 act together at microtubule plus ends to help polymerizing microtubules parallel to pre-existing ones resist catastrophe.
KEY WORDS: Kinesin, Microtubule, Polarity
Summary: Trim9 and Klp61F contribute to a quality control mechanism near nucleation sites that promotes growth of new microtubules in parallel to existing ones.
INTRODUCTION
Neurons typically have highly branched dendrites that receive signals, and a single and long axon to send out signals. To establish and sustain such polarized structure and function, pre- and post-synaptic cargo, as well as other proteins and organelles, need to be sorted specifically to axons and dendrites. One type of directional cue that guides intracellular transport is the intrinsic polarity of the microtubule cytoskeleton (Kapitein et al., 2010; Maniar et al., 2011; Yu et al., 2000). Microtubules are made from α-/β-tubulin heterodimers in a head-to-tail formation. The α-tubulin exposed end is called the minus end and is relatively stable (Akhmanova and Steinmetz, 2019), whereas the β-tubulin exposed plus end is highly dynamic (Akhmanova and Steinmetz, 2010). Long-distance intracellular transport is accomplished by dynein and kinesin motors that read microtubule polarity and walk along microtubules, moving cargo towards minus ends or plus ends, respectively.
In neurons, microtubule polarity is coupled to compartment identity. Microtubules are oriented differently in axons and dendrites (Baas and Lin, 2011; Rolls and Jegla, 2015). Axonal microtubules in Caenorhabditis elegans, Drosophila, zebrafish, frog and mouse neurons, are organized with their plus ends distal to the cell body (plus-end-out) (Burton and Paige, 1981; Harterink et al., 2018; Heidemann et al., 1981; Lee et al., 2017; Stepanova et al., 2003; Stone et al., 2008), and plus-end-directed kinesin motors carry cargo into axons (Franker and Hoogenraad, 2013; Hirokawa et al., 2010; Maday et al., 2014; Nabb et al., 2020). Dendrites in rodent neurons have ∼50% of their microtubules oriented as minus-end-out and 50% as plus-end-out in vivo and in vitro (Baas et al., 1988; Yau et al., 2016). In most C. elegans and Drosophila neurons, microtubules are almost all minus-end-out in dendrites (Harterink et al., 2018; Stone et al., 2008). The presence of a large minus-end-out microtubule population in dendrites enables dynein to be a dendritic cargo transporter (Kapitein et al., 2010; Tempes et al., 2020). Failure to generate correctly polarized microtubules results in mislocalization of cargoes in neurons (Maniar et al., 2012; van Beuningen et al., 2015; Yan et al., 2013). Thus, microtubule polarity is a critical component of compartment identity and substrate for polarized transport in neurons. It is therefore important to understand how neuronal microtubule polarity is established and maintained.
Unlike many other cells, mature neurons do not use centrosomes as microtubule organizing centers (MTOCs) (Nguyen et al., 2011; Stiess et al., 2010). γ-Tubulin, together with associated proteins, forms the γ-Tubulin ring complex (γTuRC), from which new microtubules are generated (Lin et al., 2015; Oakley et al., 2015). In cells with centrosomal MTOCs, γTuRCs are concentrated around centrioles. In mature mammalian neurons, γTuRCs are much more broadly distributed throughout axons, dendrites and the soma (Cunha-Ferreira et al., 2018; Sánchez-Huertas et al., 2016; Yonezawa et al., 2015), likely in part by the Augmin/HAUS complex (Cunha-Ferreira et al., 2018; Sánchez-Huertas et al., 2016). In C. elegans and Drosophila dendrites, γTuRCs are organized around endosomes (Liang et al., 2020; Weiner et al., 2020). In developing C. elegans dendrites these microtubule organizing endosomes are positioned near the growing tip, nicely explaining how minus-end-out polarity can be established (Liang et al., 2020). However, in mature Drosophila sensory dendrites, microtubule organizing endosomes are present at dendrite branch points throughout the arbor (Weiner et al., 2020). The orientation of newly formed microtubules nucleated at branch points is not constrained by being positioned near the tip as in C. elegans, and so could disrupt minus-end-out polarity of mature Drosophila dendrites.
In addition to local nucleation, several independent mechanisms contribute to microtubule organization in Drosophila dendrites. Growth of minus ends into dendrites facilitated by Patronin helps establish polarity (Feng et al., 2019). Steering of microtubule plus ends through branch points along existing microtubules by kinesin-2 and Apc acts as a positive feedback loop to reinforce polarity (Mattie et al., 2010; Weiner et al., 2016). We wished to understand how local nucleation acts in this context.
To investigate the relationship between microtubule nucleation and dendritic microtubule polarity, we used Drosophila sensory neurons. The simplest class, or Class I, of branched sensory neurons in Drosophila helps coordinate body movement (Hughes and Thomas, 2007) by responding to folding of the larval cuticle (He et al., 2019; Vaadia et al., 2019). In the class I neuron ddaE, correct levels of nucleation are required for normal dendritic microtubule polarity (Nguyen et al., 2014), and nucleation sites are localized to dendrite branch points by endosomes containing Wnt signaling proteins (Weiner et al., 2020). We therefore monitored the fate of newly nucleated microtubules at dendrite branch points in ddaE neurons. We found that both plus-end-out and minus-end-out microtubules were nucleated in branch points. However, as they exited the branch point, plus-end-out microtubules depolymerized much more frequently than minus-end-out microtubules. We found that this biased growth promotion of correctly oriented microtubules was dependent on parallel interactions between new and pre-existing microtubules. From a small candidate screen, we identified Trim9 and Klp61F as proteins that promoted the growth of new minus-end-out microtubules from branch points. Trim9 could be recruited to microtubules by Eb1, and Klp61F enhanced microtubule growth in vitro. We propose that Trim9 and Klp61F reinforce dendritic microtubule polarity by promoting growth of new microtubules in parallel with existing microtubules.
RESULTS
Misdirected microtubules generated within branch points are selectively destabilized as they exit
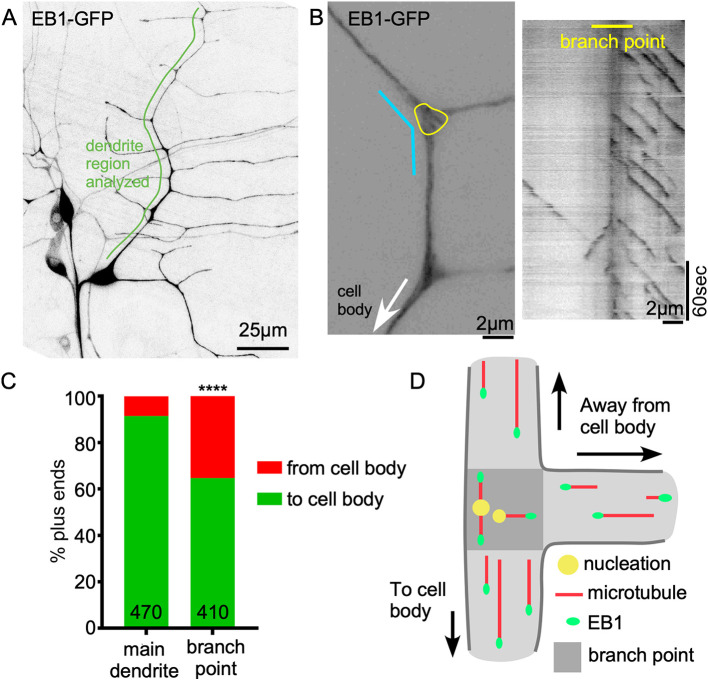
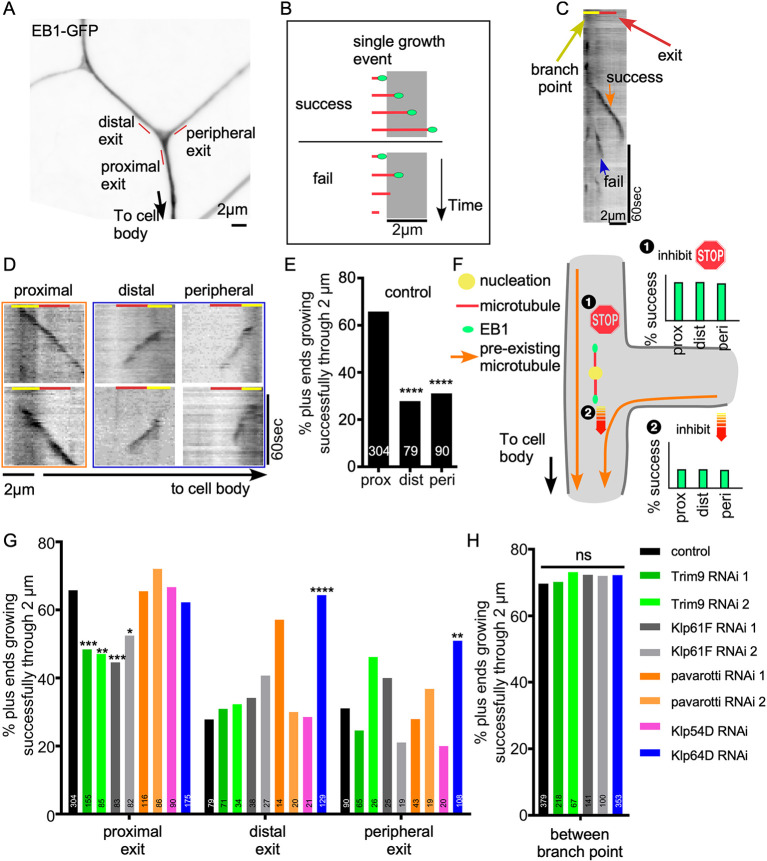
γ-Tubulin concentrates and functions at the branch points of Drosophila ddaE neurons (Nguyen et al., 2014; Weiner et al., 2020). To determine whether the generation of new microtubules in branch points might disrupt minus-end-out polarity, we monitored the behavior of microtubule plus ends that initiated growth in branch points. Growing ends of microtubules are recognized by EB proteins (Maurer et al., 2012), which can be used to track microtubule dynamics in neurons (Stepanova et al., 2003). The appearance of a new cluster, or comet, of tagged EB protein can represent either nucleation of a new microtubule or catastrophe rescue of an existing one. We focused on new Eb1-GFP comets initiating in branch points of the dorsal comb dendrite of ddaE (Fig. 1A,B; Movies 1, 2), as these have been associated with nucleation (Nguyen et al., 2014). For all analyses, we focused only on plus-end growth; growing minus ends are also labeled with Eb1-GFP, but can be distinguished from plus ends by their slow speed and typically smaller size (Feng et al., 2019). We compared the direction of new comet growth in branch points to comets moving in the regions between branch points. Between branch points ∼91% of comets moved towards the cell (Fig. 1C), similar to previous reports (Mattie et al., 2010; Stone et al., 2008). In contrast, within branch points only ∼65% of the newly emerged Eb1 comets traveled towards the cell body, and 16% grew towards distal dendrites and 19% towards peripheral dendrites (Fig. 1C,D). Because of the discrepancy between the behavior of microtubules within branch points and between them, we tracked the newly formed microtubules as they grew out of the branch point. We selected a 2-μm region at each branch point exit for analysis (Fig. 2A). We defined successful exit events as an Eb1-GFP comet initiated from within the branch point that traveled through this 2-μm dendrite segment without disappearing (Fig. 2B). Success rates were determined at proximal, distal and peripheral exits defined relative to the cell body. Although 66% of new microtubules exited the branch point when growing towards the cell body, only 30% exited successfully at the distal and proximal sites (Fig. 2C-E). Between branch points, 70% of comets were able to persistently grow for 2 μm (Fig. 2H). Thus, our data suggest that both plus-end-out and minus-end-out microtubules are nucleated at branch points, but plus-end-out microtubules are destabilized as they grow out of the branch point (Stone et al., 2008).
Fig. 1.
Both minus-end-out and plus-end-out microtubules are generated at dendrite branch points. (A) Overview image of the Drosophila Class I neuron ddaE expressing UAS-Eb1-GFP using 221-Gal4. (B) Left: example of a ddaE dendrite segment labeled by Eb1-GFP. The yellow line circles the branch point area in which nucleation direction was monitored. The cyan line indicates the region in which the kymograph on the right was generated. (C) Quantification of Eb1-GFP comet traveling direction in main dendrite (between branch points) and within dendrite branch points. Numbers on the graph are the total comets analyzed. ****P<0.0001 (Fisher's exact test). (D) Schematic diagram of nucleation direction and overall microtubule polarity in dendrites. Darker gray area indicates a branch point.
Fig. 2.
Quality control at dendrite branch point exits regulates microtubule plus-end polymerization. (A) A dendrite region of a ddaE neuron expressing Eb1-GFP under 221-Gal4 is shown. Distal, peripheral and proximal exits are indicated with red lines. (B) A schematic diagram of success and failure growth events is shown. Microtubules (red) growing plus ends are marked with Eb1-GFP (green dots). A success event is defined as an Eb1 comet traveling through a 2-μm segment without disappearing. A failure event is defined as a microtubule plus end that stops growing after it enters the 2-μm segment. (C) An example kymograph of Eb1-GFP at the proximal exit of a control neuron is shown. A failure and a success event are marked. The red line marks the 2-μm check point segment and the yellow line indicates the branch point region. (D) Example Eb1 traces at proximal, distal or peripheral exits of control neurons. An orange frame marks success events and a blue frame marks failure events. Yellow and red lines indicate the branch point and exit, respectively. (E) Quantification of success rates at proximal, distal or peripheral exits of branch points is shown. Numbers on the bars are the number of comets recorded from control animals. ****P<0.0001 (F-test). (F) Schematics of two hypotheses that could contribute to differential success at branch point exits. (G,H) Quantification of success rates at proximal, distal, peripheral exits and inbetween branch points is shown for neurons expressing control RNAi or RNAi against candidate genes. Numbers on the bars are number of comets analyzed for that condition. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; ns, not significant (F-test).
Trim9, Klp61F and Klp64D influence branch point exit success
We considered two basic mechanisms that could result in differential growth success of microtubules: growth in the ‘wrong’ direction away from the cell body could be inhibited, or growth in the ‘right’ direction towards the cell body could be promoted (Fig. 2F). In both cases, the new microtubule would need a cue to allow it to distinguish the ‘right’ direction from the ‘wrong’ one. We hypothesized that pre-existing stable microtubules could be this cue. In control neurons, most microtubules are oriented with plus ends towards the cell body, so they could be used to steer microtubules. If proteins associating with new growing microtubules promoted parallel microtubule arrangements, then they could help direct the microtubule tip towards the cell body. Alternatively, proteins that recognized anti-parallel orientation could be used to inhibit growth out of the branch away from the cell body (Fig. 2F). Based on this hypothesis, we selected candidate genes that promote microtubule interaction in parallel or counteract it in an anti-parallel orientation. Candidate genes were knocked down using cell type-specific RNAi (Dietzl et al., 2007), and we predicted that the reduction of a gene required to promote parallel interactions would result in low exit success in all directions, and reduction of an anti-parallel blocker would result in high exit success rates in all directions (example graphs in Fig. 2F). Kinesin-5 family member Klp61F was chosen because kinesin-5 motors form homotetramers that can slide anti-parallel microtubules apart (Kapitein et al., 2005), and thus could push a new incorrectly oriented microtubule back into the branch point. Kinesin-6 (pavarotti/pav is the somatic kinesin-6 in flies) can also slide anti-parallel microtubules (Nislow et al., 1992), so was included. Kinesin-12 family members can play a similar role to kinesin-5, but have also been shown to act on parallel microtubule bundles (Drechsler and McAinsh, 2016). Klp54D has been considered to be the Drosophila kinesin-12 (Radford et al., 2017), so was included. Kinesin-2 (a heterotrimeric complex of Klp64D, Klp68D and Kap3), in conjunction with Apc, Apc2 and Eb1, promotes growth of microtubule plus ends in parallel along pre-existing microtubules (Chen et al., 2014; Doodhi et al., 2014; Mattie et al., 2010; Weiner et al., 2016), so was a candidate for promoting branch point exit. Recently, the non-motor protein Trim46 was reported to facilitate parallel microtubule bundle formation at axon initial segments (Harterink et al., 2019; van Beuningen et al., 2015). We therefore also included Drosophila Trim9, the ortholog of mammalian Trim46, as a candidate. We used γ-Tubulin37C as a control because it is not expressed in somatic cells (Wiese, 2008), and we have not identified microtubule defects in cells in which it is targeted by RNAi (Weiner et al., 2020).
Transcripts encoding each of the candidate genes were targeted by RNAi in Class I dendritic arborization neurons using the Gal4-UAS system. Dicer2 and Eb1-GFP were also expressed under Gal4 control to facilitate RNA hairpin processing (Dietzl et al., 2007) and monitor microtubule growth (Rolls et al., 2007). Growing plus ends originating within branch points were tracked at proximal, distal and peripheral exits. We did not observe any change in microtubule behavior when pav or Klp54D were targeted (Fig. 2G). Knockdown of Trim9 and Klp61F significantly reduced successful exits at the proximal side (Fig. 2G) without affecting distal and peripheral exits. The same result was obtained with two independently generated RNAi lines for each gene. Knockdown of Klp64D, a subunit of kinesin-2, increased successful growth through distal and peripheral exits. Together, these data suggested that kinesin-2 functions as a negative regulator of microtubule growth, and Trim9 and Klp61F promote microtubule growth.
To determine whether the effects of kinesin-2, Trim9 and Klp61F were due to general changes in plus-end behavior, or were region specific, we assayed microtubule growth in regions between branch points. We saw no change in the rate of successful growth through a 2-μm region between branch points (Fig. 2H), suggesting that these proteins are most important when new microtubules exit branch points. We also examined overall microtubule polarity in dendrites with reduced Klp61F and Trim9 and found that they were no different from control neurons (Fig. S1). This robustness of overall polarity may be because other mechanisms are also controlling polarity in dendrites (see Discussion).
Successful growth out of branch points is facilitated by parallel microtubule tracks
The finding that kinesin-2 seemed to act as a negative regulator of microtubule growth (i.e. success rates increased when it was targeted by RNAi) was unexpected based on its known role in directing the growth of microtubule plus ends along parallel microtubules (Chen et al., 2014; Doodhi et al., 2014; Mattie et al., 2010; Weiner et al., 2016). We therefore considered that the phenotype could be due to a change in pre-existing microtubules rather than growing microtubule ends that were being tracked in the screen. In normal Drosophila dendrites, stable microtubules are oriented in parallel bundles with plus ends towards the cell body, and at branch points these bundles form a characteristic V shape (Fig. 3A,B; Stone et al., 2008). Loss of kinesin-2 results in mixed polarity of dendritic microtubules (Mattie et al., 2010), and at branch points mixed polarity bundles extended in all directions making a triangular shape (Fig. 3A,B). We hypothesized that the increase in successful exits at distal and peripheral branches in Klp64D RNAi neurons was due to the presence of stable microtubule tracks in parallel to the new microtubules in mixed polarity dendrites, rather than an effect of kinesin-2 on plus-end growth itself. To test this hypothesis, we generated mixed polarity dendrites by knocking down Patronin, a protein that functions only at microtubule minus ends.
Fig. 3.
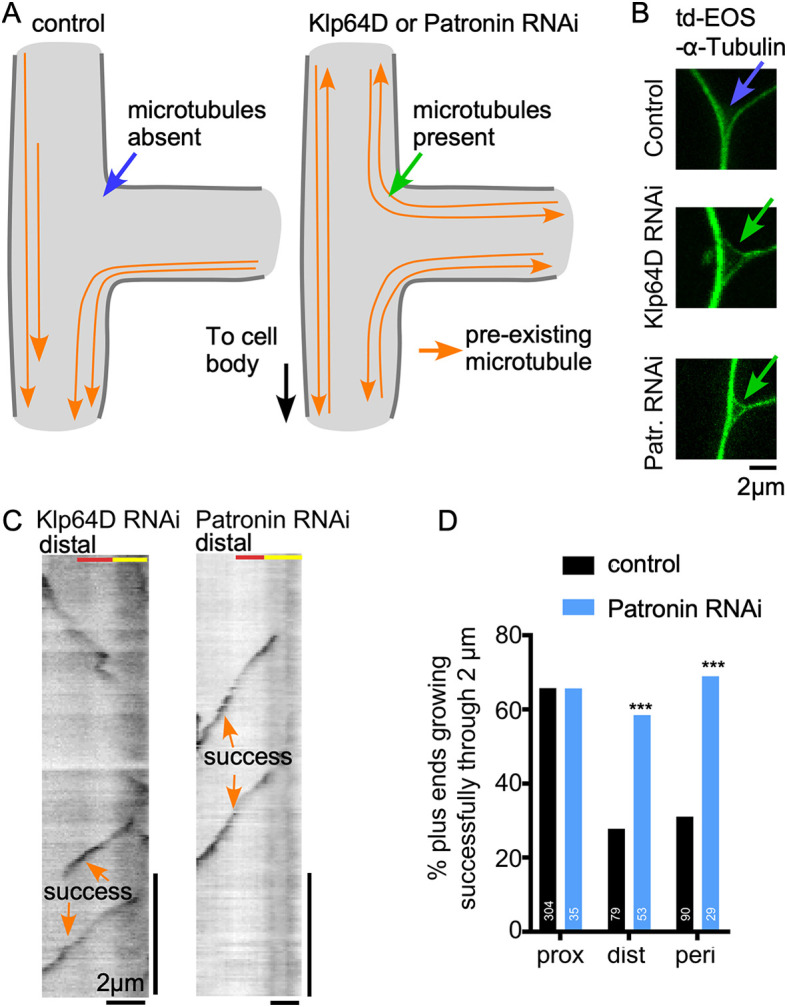
Parallel microtubule tracks positively regulate new microtubule growth. (A) Diagrams of microtubule layout in control, Klp64D or Patronin knockdown neurons are shown for a dendrite branch point. (B) Example images of ddaE dendrite branch points from neurons co-expressing td-EOS-α-tubulin with control, Klp64D or Patronin RNAi. (C) Example kymographs of ddaE neurons co-expressing Eb1-GFP with Klp64D or Patronin RNAi. Kymographs were generated at distal exits of dendrite branch points. (D) Quantification of success rates at branch point exits of neurons expressing control or Patronin RNAi. Data from control neurons from Fig. 2G are included for comparison. ***P<0.001 (Fisher's exact test).
Like Klp64D RNAi, Patronin RNAi causes mixed microtubule polarity in ddaE dendrites (Fig. 3A; Feng et al., 2019), and this was confirmed by the presence of a triangular arrangement of microtubule bundles at dendrite branch points (Fig. 3B). In Patronin knockdown neurons new microtubules behaved in the same way that they did in Klp64D knockdown cells: new microtubules had an equally high probability of successfully exiting branch points through proximal, distal and peripheral sites (Fig. 3C,D). As Patronin specifically recognizes minus ends (Goodwin and Vale, 2010; Hendershott and Vale, 2014), change in plus-end behavior is likely secondary to mixed microtubule polarity. This result suggests that the successful exit of growing microtubules depends on the presence of parallel microtubule tracks (Fig. 4A), and that anti-parallel microtubules, which are also present at all exits in Klp64D and Patronin RNAi dendrites, do not reduce exit success.
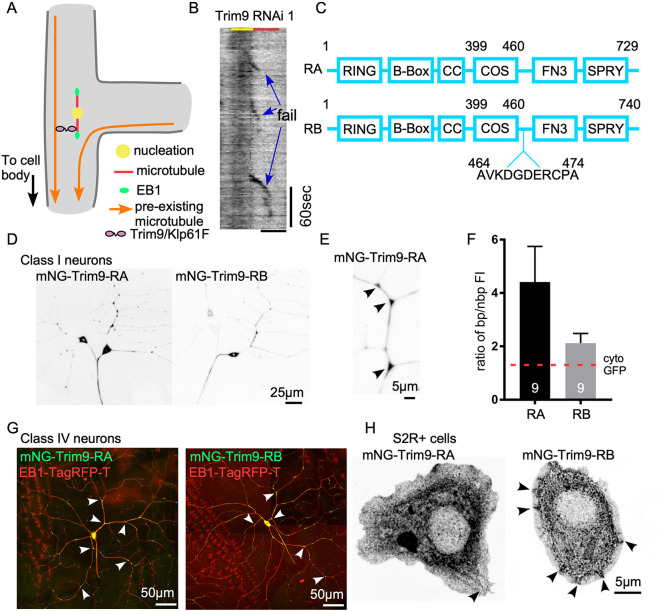
Fig. 4.
Trim9 concentrates at dendrite branch points. (A) Schematic diagram of Trim9/Klp61F promoting growth of new microtubules along parallel microtubule bundles. (B) Example kymograph of Eb1-GFP at the proximal exit of ddaE neuron expressing Trim9 RNAi 1. Three failure events are marked. Scale bar: 2 μm. (C) Schematic diagram of Trim9 RA and RB isoforms. Numbers refer to the amino acid sequence of each isoform. Lengths of boxes and lines are not scaled to the real sizes of each domain. (D) Example images of Class I neurons expressing UAS-mNG-Trim9-RA or UAS-mNG-Trim9-RB transgenes under 221-Gal4. (E) Zoomed-in image of a ddaE dendrite segment labeled with mNG-Trim9-RA under 221-Gal4. Arrowheads indicate branch points. (F) Quantification of the ratio of Trim9 RA and RB isoforms at branch point (bp) compared to non-branch point (nbp) regions. Data are presented as mean±s.d. The red dashed line on the graph marks the reported ratio of cytoplasmic GFP in the same neuron type. (G) Example images of Class IV ddaC neuron expressing mNG-Trim9-RA or mNG-Trim9-RB transgenes under ppk-Gal4. Eb1-TagRFP-T was co-expressed as a cell shape marker. Arrowheads indicate branch points. (H) UAS-mNG-Trim9-RA or UAS-mNG-Trim9-RB was expressed in Drosophila S2R+ cells with Actin-Gal4 (example images shown). The arrowhead in the RA panel points to weak microtubule localization. Arrowheads in RB panel point to patches at the cell peripheral.
Trim9-RA concentrates at dendrite branch points
Trim9 (also known as Asap, Anomalies in sensory axon patterning) is the only class I Trim protein subfamily member in Drosophila (Short and Cox, 2006). Trim9 mutants have axonal defects (Akin and Zipursky, 2016; Morikawa et al., 2011; Song et al., 2011; Yang et al., 2014), but dendrites of sensory neurons arborize normally (Morikawa et al., 2011). The axonal defects have been linked to actin regulation downstream of the Netrin pathway (Akin and Zipursky, 2016; Morikawa et al., 2011; Song et al., 2011; Yang et al., 2014). Dendritic or microtubule-related functions have not been reported for Drosophila Trim9, although it does contain the C-terminal subgroup one signature (COS) box that functions as a microtubule-binding domain (Short and Cox, 2006). To understand how Trim9 prevents failure of microtubule growth during exit from dendrite branch points (Fig. 4B), we tagged Trim9 with mNeonGreen (mNG), a very bright green fluorescent protein (Shaner et al., 2013). There are two splice isoforms of Trim9, RA and RB, in Drosophila, and we generated tagged versions of both. The RB isoform has an 11-amino acid insertion adjacent to the COS box (Fig. 4C). mNG-Trim9-RA and RB were expressed in Drosophila sensory neurons with the Gal4-UAS system. Both splice forms were found throughout Class I and Class IV neurons (Fig. 4D,E,G). Although both isoforms were present at dendrite branch points (Fig. 4D,G), the RA isoform accumulated at branch points at higher levels than RB (Fig. 4F). We have previously shown that cytoplasmic fluorescent proteins have a 1.3-fold higher fluorescence intensity at branch points compared to between them (Nguyen et al., 2014), presumably due to volume of the branch point. The two-fold and four-fold increases in intensity of the RB and RA isoforms at branch points suggests they are actively localized to this site. Beyond concentration at the branch point, the localization was largely diffuse with occasional brighter patches in the cell body and branch points. Higher resolution images did not reveal additional patterning, including to microtubules.
To determine whether either isoform could be associated with the cytoskeleton, we expressed mNG-Trim9-RA and RB in cultured Drosophila S2R+ cells. The pattern of RA hinted that it might associate at low levels with microtubules, and accumulation of RB near the cell periphery was more suggestive of recruitment by actin (Fig. 4H). However, neither pattern was sufficient by itself to make conclusions about cytoskeletal interaction, and this motivated us to probe cytoskeletal interactions further by co-expressing Trim9 with putative binding partners.
Drosophila Trim9 can be recruited to microtubules by Eb1 in S2 cells
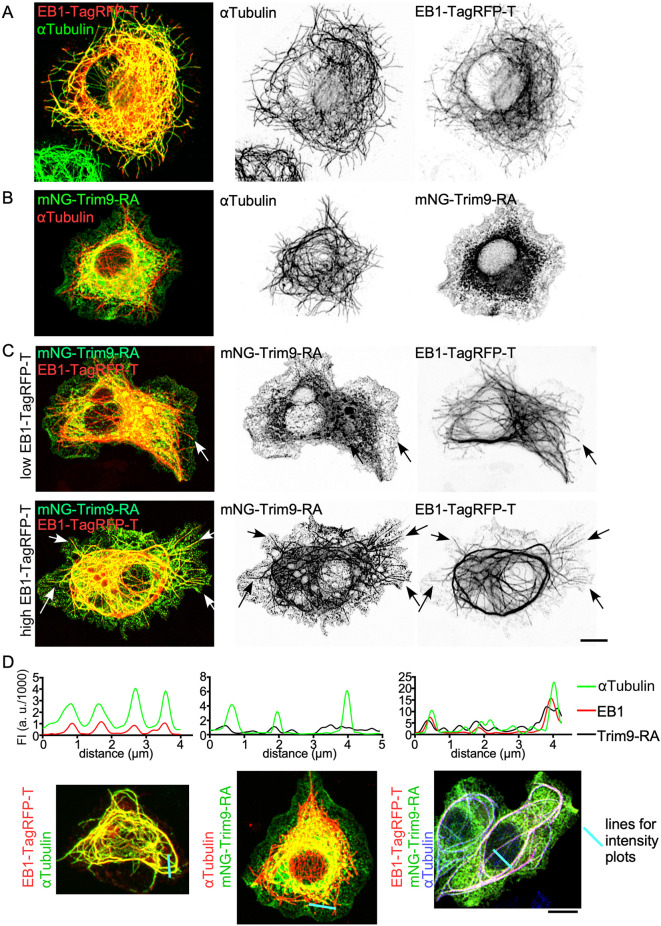
Because the function of Trim9 at branch point exits was to promote plus-end growth, and Trim9 did not seem to be robustly recruited to microtubules by itself, we hypothesized that the core plus-end-tracking protein (+TIP) Eb1 might recruit Trim9 to growing microtubules. To test this idea, we co-expressed Eb1-TagRFP-T with mNG-Trim9-RA in S2R+ cells. At the relatively high expression levels in these cells, Eb1-TagRFP-T associated with the microtubule lattice, not just the plus end (Fig. 5A). In contrast to Eb1, mNG-Trim9-RA showed little colocalization with microtubules on its own (Fig. 5B). When Eb1-TagRFP-T and mNG-Trim9-RA were co-transfected, Trim9-RA could be seen at microtubule plus ends with Eb1 (Fig. 5C). At higher expression levels of Eb1, Trim9-RA colocalized with Eb1 along the microtubule (Fig. 5C). This visual colocalization was confirmed by the co-alignment of peaks in line intensity plots (Fig. 5D). Although Trim9-RB did not show signs of microtubule localization when expressed in S2 cells alone, it could also be recruited to microtubules by Eb1, although perhaps to a lesser extent than Trim9-RA (Fig. S2). Overall, our data suggest that Eb1 can recruit Trim9 to microtubules, at least when overexpressed.
Fig. 5.
Eb1-TagRFP-T recruits Trim9 to microtubules in S2R+ cells. (A) UAS-Eb1-TagRFP-T was expressed in S2R+ cells under Actin-Gal4. αTubulin was visualized by antibody staining. (B) UAS-mNG-Trim9-RA was expressed in S2R+ cells with Actin-Gal4, and αTubulin was visualized with an antibody. (C) Cells with mildly overexpressed or highly overexpressed Eb1-TagRFP-T. UAS-mNG-Trim9-RA and UAS-Eb1-TagRFP-T were expressed in S2R+ cells with Actin-Gal4. Arrows indicate colocalization of mNG-Trim9-RA and Eb1-TagRFP-T. (D) Line intensity traces of Eb1-TagRFP-T, αTubulin antibody staining or mNG-Trim9-RA were generated from the images below. The blue lines on the images indicate the lines used to generate fluorescent intensity (FI) in the graphs. Scale bars: 5 μm.
Klp61F promotes microtubule growth in vivo and in vitro
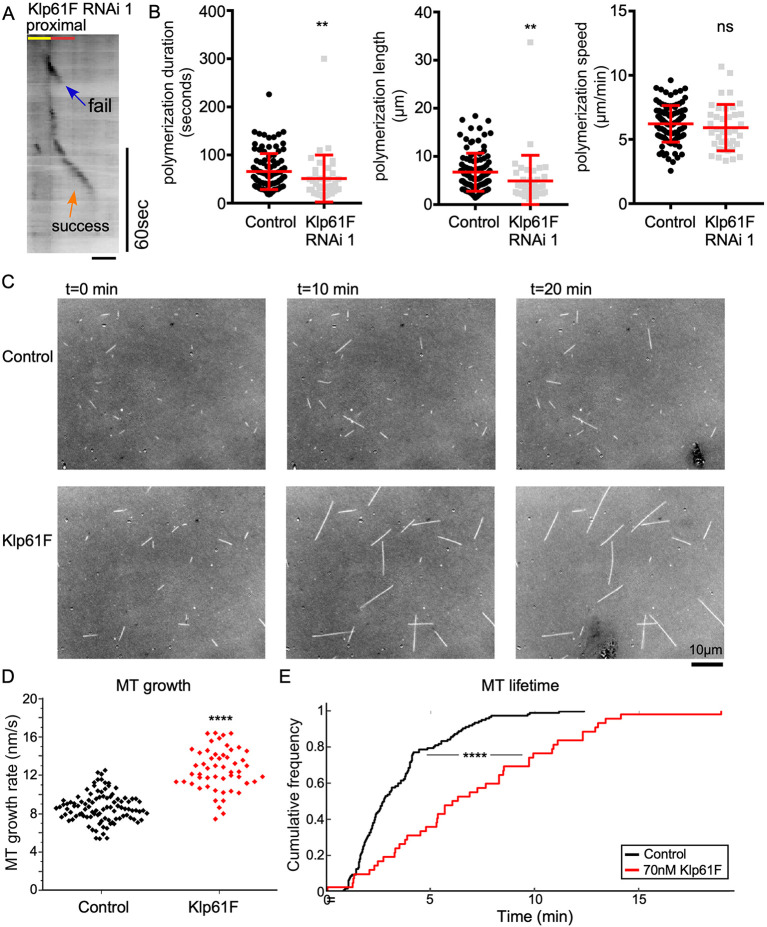
The other player identified as a positive regulator of microtubule growth out of branch points in the candidate screen (Fig. 2G, Fig. 6A), Klp61F, is a kinesin-5 family member. Like Trim9, tagged Klp61F concentrated at dendrite branch points (Fig. S3), where it seems to function. Indeed, the overall pattern of Klp61F distribution in dendrites matched that of Trim9-RA and Trim9-RB well, with the exception that Klp61F-mScarlet accumulated on additional bright puncta (Fig. S3). Canonically, kinesin-5 motors act during mitosis to establish a bipolar spindle and slide antiparallel microtubules apart (Mann and Wadsworth, 2019). However, kinesin-5 has also been shown to function in mammalian neurons. Reduction of kinesin-5 function results in faster growing axons in cultured sympathetic neurons (Haque et al., 2004; Myers and Baas, 2007), perhaps because its normal function is to reduce microtubule sliding (Myers and Baas, 2007). Kinesin-5 also functions in axon growth cones, in which it reduces the entry of microtubules in areas where it is active (Nadar et al., 2008, 2012). Dendrites of cultured mammalian neurons in which kinesin-5 is reduced are shorter and thinner (Kahn et al., 2015). None of these effects or functions seemed to explain how Klp61F promotes the exit of microtubules from dendritic branch points. We therefore considered other activities proposed for kinesin-5 based on in vitro studies. Purified kinesin-5 from Xenopus functions as a microtubule polymerase and inhibitor of catastrophe (Chen and Hancock, 2015), likely through a change in tubulin conformation induced by motor binding (Chen et al., 2019). To test whether this activity might be relevant in neurons in vivo, we examined microtubule plus-end behavior in dendrites in more detail. We analyzed the length over which individual plus ends grew, as well as the duration and speed of growth. Both length and duration of growth were slightly reduced in Klp61F RNAi neurons (Fig. 6B), consistent with the motor playing a role as a microtubule growth promoter. To test whether Drosophila kinesin-5 has the same polymerase activity as Xenopus kinesin-5 in vitro (Chen and Hancock, 2015), we generated a chimeric dimer consisting of the head and neck-linker domain of kinesin-5 and coiled-coil domain from kinesin-1. It was previously shown that the polymerase activity of Xenopus kinesin-5 resides in the motor domain itself (Chen et al., 2019), validating the use of a chimeric motor. When we added this purified Klp61F dimer to an in vitro microtubule polymerization assay, longer microtubules were observed over a 20-min period compared to a control condition without added kinesin (Fig. 6C; Movie 3). The Klp61F dimer also increased the speed of microtubule polymerization (Fig. 6D) and generated microtubules that persisted longer (Fig. 6E). These results are consistent with Klp61F promoting polymerization and likely also inhibiting catastrophe. The ability to promote polymerization of plus ends and reduce catastrophe frequency likely contributes to successful exit of new plus ends from dendrite branch points.
Fig. 6.
Klp61F promotes microtubule polymerization and inhibits catastrophe in vivo and in vitro. (A) Example kymograph of Eb1-GFP at the proximal exit of a ddaE neuron expressing Klp61F RNAi 1. Scale bar: 2 μm. (B) Quantification of microtubule plus-end polymerization under control or Klp61F RNAi conditions. Horizontal lines show the mean, error bars indicate s.d. (C) Snapshots of microtubules generated from seeds incubated with purified tubulin on slides. (D) Quantification of microtubule (MT) polymerization speed of microtubule growth in vitro (nMT=100 for control and nMT=53 for 70 nM Klp61F). An unpaired two-tailed t-test was used to compare the polymerization speeds. (E) Microtubule lifetime was also generated from movies of in vitro polymerization (nMT=123 for Control and nMT=43 for 70 nM Klp61F). **P<0.01; ****P<0.0001; ns, not significant [Kruskal–Wallis test (B); Kolmogorov–Smirnov test was used to compare the two lifetime distributions (E)].
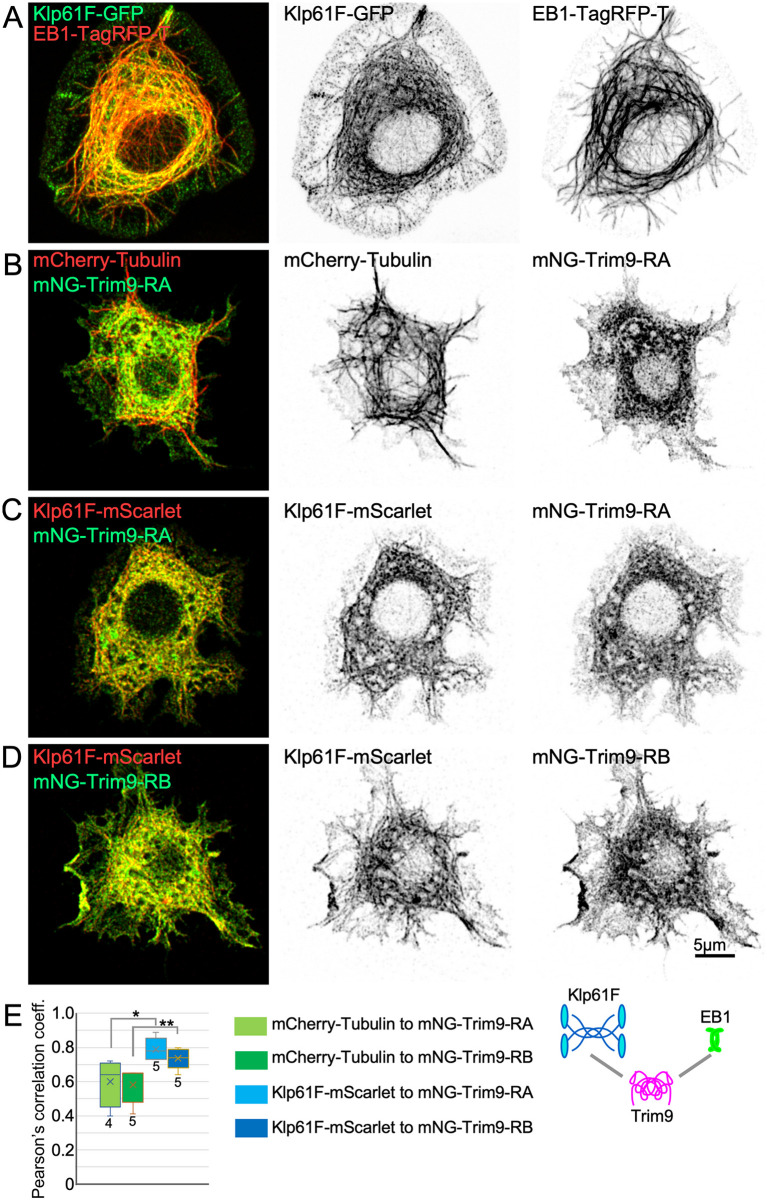
Klp61F and Trim9 colocalization in S2 cells suggests they interact
So far, we have shown that Klp61F promoted plus-end polymerization in vitro and in vivo, and that Trim9 can be recruited by Eb1. Thus, both Klp61F and Trim9 may function in dendrites at microtubule plus ends. To test whether they might be able to interact with one another at plus ends, we used an approach similar to that used for Eb1 and Trim9 (Fig. 5). The association of Klp61F-GFP and Klp61F-mScarlet with microtubules could be seen clearly when cold methanol was used to extract and fix cells (Fig. S4; Fig. 7A); microtubule association was not clearly detectable with paraformaldehyde fixation. Unlike Trim9, Klp61F recruitment to microtubules did not require overexpression of Eb1 (Fig. S4). Very little Trim9 colocalized with microtubules even when cells were extracted with methanol (Fig. 7B; Fig. S4). However, co-transfection of Klp61F and Trim9 resulted in the appearance of a more filamentous pattern in Trim9, and even more striking than this was the overall similarity between the distribution of Klp61F and Trim9 (Fig. 7C,D). To quantify this pattern similarity, we performed a Pearson's correlation analysis. The similarity between Trim9 and Klp61F was substantially higher than that between Trim9 and microtubules (Fig. 7E), indicating that Klp61F and Trim9 likely interact.
Fig. 7.
Colocalization of Klp61F and Trim9 in S2 cells. (A-D) Example images of S2 cells expressing the indicated tagged proteins. Cells were fixed with cold methanol to extract cytosolic background and allow microtubule-associated Klp61F to be visualized. (E) Pearson's correlation coefficients were calculated for the red and green channels from cytoplasmic regions. Boxes show the first to third quartiles, with a line representing the median and ‘x’ indicating the mean. Whiskers show the minimum and maximum values. An unpaired two-tailed t-test was used to compare across the conditions indicated. *P<0.05, **P<0.01. Four cells were analyzed for mCherry-Tubulin/mNG-Trim9-RA and five cells were analyzed for each of the other conditions.
DISCUSSION
The almost uniform minus-end-out microtubule polarity in Drosophila dendrites has proven a useful system for the identification of mechanisms that control microtubule organization and polarity. One surprise is that multiple mechanisms operate in parallel even in this very confined space with simple microtubule layout. Two basic types of polarity control mechanisms have been identified: those that can establish microtubule polarity independently of existing microtubules, and those that act as positive feedback loops to reinforce the predominant polarity.
Two mechanisms have been identified that can autonomously contribute to dendritic minus-end-out microtubule polarity: local nucleation and minus-end growth. Very early in the development of dendritic arborization neurons in the Drosophila embryo, new dendrites are populated by plus-end-out microtubules that grow in from the cell body (Feng et al., 2019). The next step is for slower-growing minus ends to enter dendrites from the cell body adding minus-end-out microtubules (Feng et al., 2019). Dendrites then remain with mixed polarity for the rest of embryogenesis, and eventually this resolves to minus-end-out in larvae (Hill et al., 2012). Minus-end-out microtubules can also be generated locally in dendrites by nucleation. Like microtubule growth from the cell body, nucleation can contribute plus-end-out and minus-end-out microtubules. In mature dendritic arborization neurons, nucleation at branch points is biased towards generating minus-end-out microtubules (Fig. 1). Several mechanisms to bias nucleation have been shown to operate in neurons, but they have not been shown to act in mature dendritic arborization neurons. In developing C. elegans sensory dendrites, nucleation sites cluster close to the tip, resulting in a short region of plus-end-out microtubules beyond the cluster and the proximal dendrite dominated by minus-end-out microtubules (Liang et al., 2020). However, in ddaE dendrites, nucleation sites are found not just at the dendrite tip but throughout the arbor at branch points (Nguyen et al., 2014). In ddaE dendrites, nucleation has been proposed to be biased by recruitment to only one side of Golgi outposts (Yalgin et al., 2015). However, nucleation sites have recently been shown to be recruited to endosomes rather than Golgi in ddaE dendrites (Weiner et al., 2020), making the earlier findings difficult to interpret. In summary, both growth from microtubule ends and nucleation of new microtubules create minus-end-out, as well as plus-end-out, microtubules in dendrites.
In addition to the de novo mechanisms that add minus-end-out microtubules to dendrites described above, feedback mechanisms reinforce the dominant polarity. One of these controls the direction of microtubule growth at branch points. As microtubules grow from distal regions of the dendrite into branch points, they encounter a choice to grow towards the cell body or away from the cell body. Kinesin-2, together with Eb1, Apc and Apc2, allows the growing plus end to track existing microtubules, reinforcing polarity (Mattie et al., 2010; Weiner et al., 2016). If this mechanism is eliminated, polarity in dendrites remains mixed. The final percentage of minus-end-out microtubules seems to depend on the angle of branches such that in ddaE dendrites, in which branch angles are close to 90° and microtubules can easily turn either direction at branch points, ∼50% of microtubules are minus-end-out (Mattie et al., 2010). In dendrites with more acute branch angles that help direct growing plus ends towards the cell body, 70% of microtubules remain minus-end-out, even when steering is eliminated (Mattie et al., 2010). The mechanism we describe here, that selectively promotes growth of new microtubules out of branch points along parallel microtubules, seems to act as an additional feedback loop that helps align newly nucleated microtubules with pre-existing ones. Impairing this mechanism seems to have little impact on overall polarity in dendrites (Fig. S1), unlike eliminating steering. Quality control of newly nucleated microtubules may have a small effect on overall polarity because nucleation itself is somewhat biased and nucleation may contribute at relatively low levels to the overall microtubule population at steady state in mature neurons. Quality control of new microtubules may exist as a backup mechanism to help maintain microtubule organization under stressful conditions like axon injury, which can upregulate nucleation (Chen et al., 2012).
Our data suggest that Trim9 and Klp61F are both needed for new microtubules to grow in parallel bundles with pre-existing microtubules. The fact that reduction of either results in a phenotype suggests that they act together, rather than in parallel, to control plus-end behavior. A vertebrate Trim9 family member, Trim46, organizes parallel microtubules at the axon initial segment (Harterink et al., 2019; van Beuningen et al., 2015) and has autonomous parallel bundling activity in vitro (Fréal et al., 2019). A role for the single Drosophila Trim9 family member in parallel orientation of microtubules suggests that this may be an ancestral function of this family, rather than a new function that evolved with the expansion of the family in vertebrates. If this is the case, then the other vertebrate family members in the C-1 subfamily containing a microtubule-binding COS box (Short and Cox, 2006) may all engage parallel microtubules, perhaps in different cell types or subcellular regions. In vitro, Trim46 has a strong preference for interacting with parallel bundles of microtubules over individual microtubules, and so tends to accumulate some distance behind dynamic plus ends (Fréal et al., 2019). When a depolymerizing plus end encounters a bundled region decorated with Trim46, catastrophe is strongly inhibited (Fréal et al., 2019). Although the ability of Trim46 to rescue catastrophes would be expected to promote growth like Drosophila Trim9, the site of action seems to be somewhat different. Trim46 acts at bundled regions of microtubules behind the dynamic plus end (Fréal et al., 2019), and Drosophila Trim9 prevents catastrophes from happening, likely at the plus end, with Eb1 and Klp61F.
The in vivo role of Klp61F in promoting continued growth of plus ends out of the branch point is consistent with previous in vitro studies showing that Eg5 dimers can accumulate at the plus end and promote polymerization (Chen and Hancock, 2015). Klp61F tetramers form rods ∼95 nm in length (Kashina et al., 1996) that crosslink microtubules with spacing of at least 60 nm (Sharp et al., 1999). The spacing of microtubules bundled with Trim46 is ∼37 nm (Harterink et al., 2019). Based on these general size considerations, as well as in vitro activities of Klp61F and Trim46, we propose the following model. Klp61F could be constantly traveling along stable bundles of microtubules. If a new microtubule approaches within 60 nm, then it could be captured. As Klp61F interacts with the growing plus end it could promote polymerization. At the same time, Trim9 could be recruited to the growing plus end by Eb1, perhaps through [S/T]-x-[I/L]-P motifs, which interact with Eb1 (Honnappa et al., 2009). Both Drosophila Trim9 proteins contain two potential motifs at amino acids; the first is at 42-SALP-46 in both, and the second is 479-TILP-483 in the RB isoform and 468-TILP-472 in the RA isoform. After Klp61F grabs the growing microtubule with Eb1 and Trim9 at its tip, Trim9 could reinforce the parallel interaction. Although this model provides an initial framework for the function of Klp61F and Trim9 in dendrites, it is quite speculative and raises many additional questions. For example, if Klp61F is in constant flux towards the plus end of microtubules, how is it transported into dendrites? And do these proteins really act sequentially? If so, perhaps family members collaborate in a similar way in other cellular scenarios; for example, at the axon initial segment.
MATERIALS AND METHODS
Drosophila strain maintenance and larva collection
Fly stocks were raised on cornmeal-agar-yeast food in humidified 25 or 20°C rooms. To generate Drosophila larvae for experiments, fly crosses were set up in bottles that had holes punctured with a syringe needle, and embryos were collected on 35 mm caps filled with normal fly food. After collecting embryos for 24 h, caps were removed to a Petri dish and aged for 3 days at 25°C. Only one neuron was sampled per larva in this study. Sex of larvae was not differentiated.
Trim9 plasmid construction
To create mNeonGreen (mNG) tagged Trim9, we first amplified the mNeonGreen coding region from pNCS-mNeonGreen plasmid (Allele Biotechnology and Pharmaceuticals) and inserted it into the pUAST vector (Brand and Perrimon, 1993) using EcoR1 and Asc1 restriction sites. The Trim9-RA coding region was cloned from cDNA plasmid RE22018 (Drosophila Genomics Resource Center collection) and cloned into pUAST-mNG using Spe1 and Kpn1 sites (primers are included in Table S1). The Trim9-RB coding region was made by adding the additional 33 nucleotides using a Q5 mutagenesis kit (New England Biolabs). Then Trim9-RB was subcloned into pUAST-mNG in the same way as the RA isoform. The pUAST-mNG-Trim9-RA and pUAS-mNG-Trim9-RB plasmids were injected into Drosophila embryos by BestGene Inc. Transgene insertions were mapped and balanced using standard Drosophila genetics protocols. Fly lines that generated the best signal-to-noise ratio with least aggregation when crossed to 221-Gal4 were used in this study. These two plasmids were also used in Drosophila S2R+ cells.
Klp61F plasmid construction
To generate pUAST-Klp61F-GFP, the Klp61F coding sequence was amplified from cDNA plasmid LD15641 (Drosophila Genomics Resource Center collection) and inserted into pUAST-GFP between EcoRI and AscI sites using In-Fusion cloning kit (Takara). To generate pUAST-Klp61F-mScarlet, we digested the pUAST-Klp61F-GFP construct with NheI and XbaI (New England Biolabs) to remove the GFP sequence. The mScarlet coding sequence was amplified from pUAST-mScarlet and subcloned into the pUAST-Klp61F backbone using In-Fusion cloning. The pUAST-mScarlet plasmid was used to make transgenic flies by BestGene Inc. Insertion sites were mapped to chromosomes using standard Drosophila genetics techniques.
Transgenic fly strains
Drosophila tester lines used in this study included (1) UAS-Dicer2; 221-Gal4, UAS-Eb1-GFP/TM6; (2) UAS-Dicer2; 221-Gal4, UAS-tdEOS-α-Tubulin/TM6; (3) 221-Gal4; and (4) ppk-Gal4, UAS-Eb1-TagRFP-T. RNAi lines and other transgenic lines were crossed with tester lines. Dicer2 was included to generate better RNAi knockdown (Dietzl et al., 2007). 221-Gal4 and ppk-Gal4 were used to express transgenes in class I or class IV neurons, respectively. γ-Tubulin37C RNAi was used as control RNAi because γ-Tubulin37C is only maternally expressed (Wiese, 2008). Fly lines that were acquired from the Bloomington Drosophila Stock Center (BDSC, BL) or the Vienna Drosophila Resource Center (VDRC) included γ-Tubulin37C RNAi line VDRC25271; Klp64D RNAi VDRC45373; Patronin RNAi line BL36659; Trim9 RNAi line 1 VDRC100767; Trim9 RNAi line12 VDRC21405; Klp61F RNAi line 1 VDRC52549; Klp61F RNAi line 2 BL33685; pavarotti RNAi line 1 BL42573; pavarotti RNAi line 2 VDRC46137; and Klp54D RNAi VDRC100140 (see Table S1).
Drosophila live imaging
Three-day-old larvae were mounted between a glass slide with a drop of dried agarose and a 22×40 mm coverslip; the ends of the coverslip were taped to the slide to immobilize larvae. Live imaging of whole larvae was performed on a widefield Zeiss Imager M2 microscope equipped with a Colibri2 LED system with AxioCam M2 or AxioCam 506 camera under 63×1.4NA Plan APOCHROMAT oil immersion objective. Movies of Eb1 comets were recorded at 1 frame/s for 300 frames. Overview images of neurons expressing Eb1-GFP, tdEOS-αTubulin or mNG-Trim9 were acquired with a Zeiss LSM800 confocal microscope using a 63×1.4 NA oil objective. All movies were aligned and analyzed using ImageJ.
For assessment of nucleation direction, new Eb1 comets that emerged within branch points and traveled towards one of the three exits were counted. Comets that initiated outside branch points and obvious rescue events were excluded. The area of a branch point was defined as the enlarged dendrite linking site between a side branch and the comb dendrite. A 2-μm dendrite segment immediately next to a branch point was defined as the exit or checkpoint. At least ten neurons were analyzed for each condition. Success and failure events were determined from Eb1 movies by way of kymographs. Kymographs were generated for figures to capture events across time in a single frame.
Cell culture and immunostaining
Drosophila S2R+ cells (Drosophila Genomics Resource Center) were maintained in Schneider's Drosophila medium (Gibco) supplied with 10% fetal bovine serum (Gibco) and kept at 25°C. Cells were passed every 4 days, and frozen and recovered as needed following standard protocols. All transfections were performed in six-well plates using an Effectene Transfection Reagent Kit (Qiagen). Twenty-four hours after transfection cells were replated into six-well plates containing coverslips treated with concanavalin-A to increase adhesion. After 30 min, cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 10 min (experiments in Figs 4, 5) or −20°C methanol for 10 min (Fig. 7). Plasmids used were pUAST-Eb1-TagRFP-T, pUAST-mNG-Trim9-RA, pUAST-mNG-Trim9-RB, pUAST-Klp61F-GFP, pUAST-Klp61F-mScarlet and pAc-mCherry-αTubulin (see Table S1 for additional details of plasmids); pAC-Gal4 was used to drive expression. For immunostaining (Fig. 5), cells were permeabilized using PBST (PBS with 0.2% Triton X-100) for 20 min at room temperature. Cells were then incubated in blocking buffer, PBST with 5% normal goal serum, for 1 h at room temperature. Next, primary antibody against α-tubulin (Abcam, 18251) diluted 1:2000 in blocking buffer was incubated with samples overnight at 4°C. After several washes with blocking buffer, secondary antibody (goat anti-rabbit IgG Alexa Fluor 488, Abcam for Fig. 5A; goat anti-rabbit IgG Rhodamine Red-X, Jackson ImmunoResearch, for Fig. 5B; and goat anti-rabbit Cy5, Invitrogen, for Fig. 5D) diluted at 1:1000 was applied at room temperature for 1 h. Cells were washed with blocking buffer, and coverslips were mounted in HardSet antifade mounding medium (Vectashield; Figs 4, 5), or 85% glycerol and 50 mM Tris (pH 8) (Fig. 7). Cells that were not stained (Figs 4, 7) were washed with PBS and mounted for imaging immediately after fixation. Images were acquired using a Zeiss LSM800 microscope with a 63×1.4 NA Oil objective using the Airyscan detector in optimal image acquisition mode. Processed Airyscan images are shown. Image assembly and analysis was performed using ImageJ. Pearson's correlation analysis was performed using a single z slice. Rectangular regions fully within the cell and away from the nucleus were selected, with only one region per cell used. Coloc 2 within ImageJ was used to generate the Pearson's R value.
Klp61F and tubulin purification
Klp61F dimers were generated by fusing the motor and neck linker domains (residues 1-368) of Drosophila kinesin-5 to the neck-coil and coil-1 of KHC (345-406), to generate Klp61F-406. Plasmids were transformed into Escherichia coli BL21 (DE3) for protein expression and grown in 2 l cultures. Protein expression was induced by adding 1 mM IPTG and grown at 18°C overnight. The collected pellets were collected and suspended in 25A200 buffer, and lysed by sonication. After Ni-column extraction, motors were exchanged into 0.5 µM mantADP with BRB80 buffer using a GE HiTrap Desalting column, followed by the addition of 10 vol% sucrose as a cryoprotectant for flash freezing in liquid nitrogen for storage at −80°C.
Phosphocellulose grade tubulin was obtained from freshly harvested cow brains. The brains were homogenized and clarified to make the tubulin readily available in solution. Three cycles of polymerization at 37°C, centrifugation at 37°C, depolymerization at 4°C and centrifugation at 4°C were performed to purify soluble and active tubulin while getting rid of excess protein that may have been packed with the tubulin. A final two cycles of the polymerization and depolymerization were carried out on the phosphocellulose grade tubulin, yielding tubulin with a purity of >99%. The tubulin concentration was recorded using an absorbance at 280 nm with an extinction coefficient of 11500 M−1 cm−1. The tubulin samples were then flash frozen in liquid nitrogen and stored at −80°C.
Microtubule dynamics assay
Coverslips were made by an overnight bath in 6 M HCl and subsequently rinsed with ddH20. Slides were then plasma-cleaned for 10 min and incubated overnight in a vacuum-based desiccator with silane-vapor. Microscopes slides were washed with 70% ethanol and rinsed using ddH20 and dried using nitrogen. The coverslips were fixed to the slides with two pieces of double-sided tape making a silanized flow chamber.
Double-cycled GMPCPP seeds were used as the nucleating templates to observe microtubule dynamics in vitro. They were formed by the addition of 2 µM tubulin (0.6 µM biotinylated plus 1.4 µM unlabeled tubulin) to a solution of 1 mM GMPCPP and 4 mM MgCl2 for 2 h at 37°C. Seeds were then spun in an airfuge at 30 psi for 5 min to remove free tubulin. Solutions were suspended in an equal volume of BRB80 and kept on ice at 4°C for 30-40 min to depolymerize the seeds. This solution was then polymerized again as described earlier. The seeds were then spun and suspended in a 50% Glycerol BRB80 mixture and flash frozen in liquid nitrogen and stored at −80°C. Seed preparation on the day of the experiment required a quick thaw at 37°C and another spin on the airfuge to remove the glycerol from solution. The preparation was then suspended in a buffer containing 0.1 mM Mg-GMPCPP.
The silanized flow chamber went through sequential 5-min washes of 600 nM Neutravidin, 5 wt% Pluronic-127, 2 mg/ml of casein, double cycled-GMPCPP seeds and biotinylated-bovine serum albumin to bind any open Neutravidin. Lastly, polymerization solution was made containing 70 nM KLP61F-560, 12.5 µM unlabeled bovine brain tubulin, 1 mM Mg-ATP and 1 mM Mg-GTP (0.05% methylcellulose) (Chen et al., 2019). The slide was placed on top of an objective heater that was set to 26°C. The slide was allowed to equilibrate to 26°C for ∼5 min before imaging occurred. The polymerization dynamics were imaged using interference reflection microscopy on a Nikon TE-2000 TIRF microscope with a blue and green LED from the pE-300 white used to produce a white light. The microtubule signal recorded was the interference pattern between the signal reflecting from the surface of the glass and the signal from the scattered light of the microtubule (Mahamdeh et al., 2018). Images were corrected for uneven illumination by taking a background image in solution. The dynamics were imaged at 2 frames/s for 30 min. Microtubule growth was analyzed by making kymographs in ImageJ and fitting a line through the growth phases of the microtubules. The microtubule lifetimes were recorded as the time from which a growth phase began until a catastrophe would occur. All images shown are inverted images of the signal recorded.
Supplementary Material
Acknowledgements
We thank the Bloomington Drosophila Stock Center (National Institutes of Health, P40OD018537) and the Vienna Drosophila Resource Center for maintaining invaluable fly stocks and providing the stocks used in this study. We are also grateful to Rolls and Hancock lab members for discussions and ideas.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: C.F., J.M.C., W.O.H., M.M.R.; Investigation: C.F., J.M.C., G.O.K., M.C.S., A.T.W., J.I.H., M.M.R.; Writing - original draft: C.F., M.M.R.; Writing - review & editing: J.M.C., J.I.H., W.O.H.; Visualization: C.F.; Supervision: W.O.H., M.M.R.; Funding acquisition: W.O.H., M.M.R.
Funding
This work was supported by the National Institutes of Health (NIH; GM085115 to M.M.R. and GM076476 to W.O.H.). J.M.C. was supported by NIH T32 training grant GM108563. Deposited in PMC for release after 12 months.
References
- Akhmanova, A. and Steinmetz, M. O. (2010). Microtubule +TIPs at a glance. J. Cell Sci. 123, 3415-3419. 10.1242/jcs.062414 [DOI] [PubMed] [Google Scholar]
- Akhmanova, A. and Steinmetz, M. O. (2019). Microtubule minus-end regulation at a glance. J. Cell Sci. 132, jcs227850. 10.1242/jcs.227850 [DOI] [PubMed] [Google Scholar]
- Akin, O. and Zipursky, S. L. (2016). Frazzled promotes growth cone attachment at the source of a Netrin gradient in the Drosophila visual system. Elife 5, e20762. 10.7554/eLife.20762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas, P. W. and Lin, S. (2011). Hooks and comets: the story of microtubule polarity orientation in the neuron. Dev. Neurobiol. 71, 403-418. 10.1002/dneu.20818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas, P. W., Deitch, J. S., Black, M. M. and Banker, G. A. (1988). Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc. Natl. Acad. Sci. USA 85, 8335-8339. 10.1073/pnas.85.21.8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H. and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415. 10.1242/dev.118.2.401 [DOI] [PubMed] [Google Scholar]
- Burton, P. R. and Paige, J. L. (1981). Polarity of axoplasmic microtubules in the olfactory nerve of the frog. Proc. Natl. Acad. Sci. USA 78, 3269-3273. 10.1073/pnas.78.5.3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. and Hancock, W. O. (2015). Kinesin-5 is a microtubule polymerase. Nat. Commun. 6, 8160. 10.1038/ncomms9160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Stone, M. C., Tao, J. and Rolls, M. M. (2012). Axon injury and stress trigger a microtubule-based neuroprotective pathway. Proc. Natl. Acad. Sci. USA 109, 11842-11847. 10.1073/pnas.1121180109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Rolls, M. M. and Hancock, W. O. (2014). An EB1-kinesin complex is sufficient to steer microtubule growth in vitro. Curr. Biol. 24, 316-321. 10.1016/j.cub.2013.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G.-Y., Cleary, J. M., Asenjo, A. B., Chen, Y., Mascaro, J. A., Arginteanu, D. F. J., Sosa, H. and Hancock, W. O. (2019). Kinesin-5 promotes microtubule nucleation and assembly by stabilizing a lattice-competent conformation of tubulin. Curr. Biol. 29, 2259-2269.e4. 10.1016/j.cub.2019.05.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira, I., Chazeau, A., Buijs, R. R., Stucchi, R., Will, L., Pan, X., Adolfs, Y., van der Meer, C., Wolthuis, J. C., Kahn, O. I.et al. (2018). The HAUS complex is a key regulator of non-centrosomal microtubule organization during neuronal development. Cell Rep 24, 791-800. 10.1016/j.celrep.2018.06.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl, G., Chen, D., Schnorrer, F., Su, K.-C., Barinova, Y., Fellner, M., Gasser, B., Kinsey, K., Oppel, S., Scheiblauer, S.et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151-156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Doodhi, H., Katrukha, E. A., Kapitein, L. C. and Akhmanova, A. (2014). Mechanical and geometrical constraints control kinesin-based microtubule guidance. Curr. Biol. 24, 322-328. 10.1016/j.cub.2014.01.005 [DOI] [PubMed] [Google Scholar]
- Drechsler, H. and McAinsh, A. D. (2016). Kinesin-12 motors cooperate to suppress microtubule catastrophes and drive the formation of parallel microtubule bundles. Proc. Natl. Acad. Sci. USA 113, E1635-E1644. 10.1073/pnas.1516370113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, C., Thyagarajan, P., Shorey, M., Seebold, D. Y., Weiner, A. T., Albertson, R. M., Rao, K. S., Sagasti, A., Goetschius, D. J. and Rolls, M. M. (2019). Patronin-mediated minus end growth is required for dendritic microtubule polarity. J. Cell Biol. 218, 2309-2328. 10.1083/jcb.201810155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franker, M. A. M. and Hoogenraad, C. C. (2013). Microtubule-based transport - basic mechanisms, traffic rules and role in neurological pathogenesis. J. Cell Sci. 126, 2319-2329. 10.1242/jcs.115030 [DOI] [PubMed] [Google Scholar]
- Fréal, A., Rai, D., Tas, R. P., Pan, X., Katrukha, E. A., van de Willige, D., Stucchi, R., Aher, A., Yang, C., Altelaar, A. F. M.et al. (2019). Feedback-driven assembly of the axon initial segment. Neuron 104, 305-321.e8. 10.1016/j.neuron.2019.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, S. S. and Vale, R. D. (2010). Patronin regulates the microtubule network by protecting microtubule minus ends. Cell 143, 263-274. 10.1016/j.cell.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque, S. A., Hasaka, T. P., Brooks, A. D., Lobanov, P. V. and Baas, P. W. (2004). Monastrol, a prototype anti-cancer drug that inhibits a mitotic kinesin, induces rapid bursts of axonal outgrowth from cultured postmitotic neurons. Cell Motil. Cytoskeleton 58, 10-16. 10.1002/cm.10176 [DOI] [PubMed] [Google Scholar]
- Harterink, M., Edwards, S. L., de Haan, B., Yau, K. W., van den Heuvel, S., Kapitein, L. C., Miller, K. G. and Hoogenraad, C. C. (2018). Local microtubule organization promotes cargo transport in C. elegans dendrites. J. Cell Sci. 131, jcs223107. 10.1242/jcs.223107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harterink, M., Vocking, K., Pan, X., Soriano Jerez, E. M., Slenders, L., Fréal, A., Tas, R. P., van de Wetering, W. J., Timmer, K., Motshagen, J.et al. (2019). TRIM46 Organizes microtubule fasciculation in the axon initial segment. J. Neurosci. 39, 4864-4873. 10.1523/JNEUROSCI.3105-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L., Gulyanon, S., Mihovilovic Skanata, M., Karagyozov, D., Heckscher, E. S., Krieg, M., Tsechpenakis, G., Gershow, M. and Tracey, W. D.Jr (2019). Direction selectivity in drosophila proprioceptors requires the mechanosensory channel Tmc. Curr. Biol. 29, 945-956.e3. 10.1016/j.cub.2019.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann, S. R., Landers, J. M. and Hamborg, M. A. (1981). Polarity orientation of axonal microtubules. J. Cell Biol. 91, 661-665. 10.1083/jcb.91.3.661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershott, M. C. and Vale, R. D. (2014). Regulation of microtubule minus-end dynamics by CAMSAPs and Patronin. Proc. Natl. Acad. Sci. USA 111, 5860-5865. 10.1073/pnas.1404133111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, S. E., Parmar, M., Gheres, K. W., Guignet, M. A., Huang, Y., Jackson, F. R. and Rolls, M. M. (2012). Development of dendrite polarity in Drosophila neurons. Neural Dev. 7, 34. 10.1186/1749-8104-7-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa, N., Niwa, S. and Tanaka, Y. (2010). Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610-638. 10.1016/j.neuron.2010.09.039 [DOI] [PubMed] [Google Scholar]
- Honnappa, S., Gouveia, S. M., Weisbrich, A., Damberger, F. F., Bhavesh, N. S., Jawhari, H., Grigoriev, I., van Rijssel, F. J., Buey, R. M., Lawera, A.et al. (2009). An EB1-binding motif acts as a microtubule tip localization signal. Cell 138, 366-376. 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Hughes, C. L. and Thomas, J. B. (2007). A sensory feedback circuit coordinates muscle activity in Drosophila. Mol. Cell. Neurosci. 35, 383-396. 10.1016/j.mcn.2007.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn, O. I., Sharma, V., González-Billault, C. and Baas, P. W. (2015). Effects of kinesin-5 inhibition on dendritic architecture and microtubule organization. Mol. Biol. Cell 26, 66-77. 10.1091/mbc.e14-08-1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein, L. C., Peterman, E. J., Kwok, B. H., Kim, J. H., Kapoor, T. M. and Schmidt, C. F. (2005). The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435, 114-118. 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Kapitein, L. C., Schlager, M. A., Kuijpers, M., Wulf, P. S., van Spronsen, M., MacKintosh, F. C. and Hoogenraad, C. C. (2010). Mixed microtubules steer dynein-driven cargo transport into dendrites. Curr. Biol. 20, 290-299. 10.1016/j.cub.2009.12.052 [DOI] [PubMed] [Google Scholar]
- Kashina, A. S., Baskin, R. J., Cole, D. G., Wedaman, K. P., Saxton, W. M. and Scholey, J. M. (1996). A bipolar kinesin. Nature 379, 270-272. 10.1038/379270a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T. J., Lee, J. W., Haynes, E. M., Eliceiri, K. W. and Halloran, M. C. (2017). The kinesin adaptor calsyntenin-1 organizes microtubule polarity and regulates dynamics during sensory axon arbor development. Front. Cell Neurosci. 11, 107. 10.3389/fncel.2017.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X., Kokes, M., Fetter, R. D., Sallee, M. D., Moore, A. W., Feldman, J. L. and Shen, K. (2020). Growth cone-localized microtubule organizing center establishes microtubule orientation in dendrites. Elife 9, e56547. 10.7554/eLife.56547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, T.-C., Neuner, A. and Schiebel, E. (2015). Targeting of γ-tubulin complexes to microtubule organizing centers: conservation and divergence. Trends Cell Biol. 25, 296-307. 10.1016/j.tcb.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Maday, S., Twelvetrees, A. E., Moughamian, A. J. and Holzbaur, E. L. (2014). Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron 84, 292-309. 10.1016/j.neuron.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamdeh, M., Simmert, S., Luchniak, A., Schaffer, E. and Howard, J. (2018). Label-free high-speed wide-field imaging of single microtubules using interference reflection microscopy. J. Microsc. 272, 60-66. 10.1111/jmi.12744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniar, T. A., Kaplan, M., Wang, G. J., Shen, K., Wei, L., Shaw, J. E., Koushika, S. P. and Bargmann, C. I. (2011). UNC-33 (CRMP) and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting. Nat. Neurosci. 15, 48-56. 10.1038/nn.2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniar, T. A., Kaplan, M., Wang, G. J., Shen, K., Wei, L., Shaw, J. E., Koushika, S. P. and Bargmann, C. I. (2012). UNC-33 (CRMP) and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting. Nat. Neurosci. 15, 48-56. 10.1038/nn.2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, B. J. and Wadsworth, P. (2019). Kinesin-5 regulation and function in mitosis. Trends Cell Biol. 29, 66-79. 10.1016/j.tcb.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Mattie, F. J., Stackpole, M. M., Stone, M. C., Clippard, J. R., Rudnick, D. A., Qiu, Y., Tao, J., Allender, D. L., Parmar, M. and Rolls, M. M. (2010). Directed microtubule growth, +TIPs, and Kinesin-2 are required for uniform microtubule polarity in dendrites. Curr. Biol. 20, 2169-2177. 10.1016/j.cub.2010.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer, S. P., Fourniol, F. J., Bohner, G., Moores, C. A. and Surrey, T. (2012). EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell 149, 371-382. 10.1016/j.cell.2012.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa, R. K., Kanamori, T., Yasunaga, K.-i. and Emoto, K. (2011). Different levels of the Tripartite motif protein, Anomalies in sensory axon patterning (Asap), regulate distinct axonal projections of Drosophila sensory neurons. Proc. Natl. Acad. Sci. USA 108, 19389-19394. 10.1073/pnas.1109843108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, K. A. and Baas, P. W. (2007). Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J. Cell Biol. 178, 1081-1091. 10.1083/jcb.200702074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabb, A. T., Frank, M. and Bentley, M. (2020). Smart motors and cargo steering drive kinesin-mediated selective transport. Mol. Cell. Neurosci. 103, 103464. 10.1016/j.mcn.2019.103464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadar, V. C., Ketschek, A., Myers, K. A., Gallo, G. and Baas, P. W. (2008). Kinesin-5 is essential for growth-cone turning. Curr. Biol. 18, 1972-1977. 10.1016/j.cub.2008.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadar, V. C., Lin, S. and Baas, P. W. (2012). Microtubule redistribution in growth cones elicited by focal inactivation of kinesin-5. J. Neurosci. 32, 5783-5794. 10.1523/JNEUROSCI.0144-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, M. M., Stone, M. C. and Rolls, M. M. (2011). Microtubules are organized independently of the centrosome in Drosophila neurons. Neural Dev. 6, 38. 10.1186/1749-8104-6-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, M. M., McCracken, C. J., Milner, E. S., Goetschius, D. J., Weiner, A. T., Long, M. K., Michael, N. L., Munro, S. and Rolls, M. M. (2014). Gamma-tubulin controls neuronal microtubule polarity independently of Golgi outposts. Mol. Biol. Cell 25, 2039-2050. 10.1091/mbc.e13-09-0515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow, C., Lombillo, V. A., Kuriyama, R. and McIntosh, J. R. (1992). A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature 359, 543-547. 10.1038/359543a0 [DOI] [PubMed] [Google Scholar]
- Oakley, B. R., Paolillo, V. and Zheng, Y. (2015). γ-Tubulin complexes in microtubule nucleation and beyond. Mol. Biol. Cell 26, 2957-2962. 10.1091/mbc.E14-11-1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford, S. J., Go, A. M. and McKim, K. S. (2017). Cooperation between kinesin motors promotes spindle symmetry and chromosome organization in oocytes. Genetics 205, 517-527. 10.1534/genetics.116.194647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls, M. M. and Jegla, T. J. (2015). Neuronal polarity: an evolutionary perspective. J. Exp. Biol. 218, 572-580. 10.1242/jeb.112359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls, M. M., Satoh, D., Clyne, P. J., Henner, A. L., Uemura, T. and Doe, C. Q. (2007). Polarity and compartmentalization of Drosophila neurons. Neural Dev. 2, 7. 10.1186/1749-8104-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Huertas, C., Freixo, F., Viais, R., Lacasa, C., Soriano, E. and Luders, J. (2016). Non-centrosomal nucleation mediated by augmin organizes microtubules in post-mitotic neurons and controls axonal microtubule polarity. Nat. Commun. 7, 12187. 10.1038/ncomms12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner, N. C., Lambert, G. G., Chammas, A., Ni, Y., Cranfill, P. J., Baird, M. A., Sell, B. R., Allen, J. R., Day, R. N., Israelsson, M.et al. (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407-409. 10.1038/nmeth.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D. J., McDonald, K. L., Brown, H. M., Matthies, H. J., Walczak, C., Vale, R. D., Mitchison, T. J. and Scholey, J. M. (1999). The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 144, 125-138. 10.1083/jcb.144.1.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short, K. M. and Cox, T. C. (2006). Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J. Biol. Chem. 281, 8970-8980. 10.1074/jbc.M512755200 [DOI] [PubMed] [Google Scholar]
- Song, S., Ge, Q., Wang, J., Chen, H., Tang, S., Bi, J., Li, X., Xie, Q. and Huang, X. (2011). TRIM-9 functions in the UNC-6/UNC-40 pathway to regulate ventral guidance. J. Genet. Genomics 38, 1-11. 10.1016/j.jcg.2010.12.004 [DOI] [PubMed] [Google Scholar]
- Stepanova, T., Slemmer, J., Hoogenraad, C. C., Lansbergen, G., Dortland, B., De Zeeuw, C. I., Grosveld, F., van Cappellen, G., Akhmanova, A. and Galjart, N. (2003). Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 23, 2655-2664. 10.1523/JNEUROSCI.23-07-02655.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiess, M., Maghelli, N., Kapitein, L. C., Gomis-Rüth, S., Wilsch-Bräuninger, M., Hoogenraad, C. C., Tolić-Nørrelykke, I. M. and Bradke, F. (2010). Axon extension occurs independently of centrosomal microtubule nucleation. Science 327, 704. 10.1126/science.1182179 [DOI] [PubMed] [Google Scholar]
- Stone, M. C., Roegiers, F. and Rolls, M. M. (2008). Microtubules have opposite orientation in axons and dendrites of Drosophila neurons. Mol. Biol. Cell 19, 4122-4129. 10.1091/mbc.e07-10-1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempes, A., Weslawski, J., Brzozowska, A. and Jaworski, J. (2020). Role of dynein-dynactin complex, kinesins, motor adaptors, and their phosphorylation in dendritogenesis. J. Neurochem. 155, 10-28. 10.1111/jnc.15010 [DOI] [PubMed] [Google Scholar]
- Vaadia, R. D., Li, W., Voleti, V., Singhania, A., Hillman, E. M. C. and Grueber, W. B. (2019). characterization of proprioceptive system dynamics in behaving drosophila larvae using high-speed volumetric microscopy. Curr. Biol. 29, 935-944.e4. 10.1016/j.cub.2019.01.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beuningen, S. F. B., Will, L., Harterink, M., Chazeau, A., van Battum, E. Y., Frias, C. P., Franker, M. A. M., Katrukha, E. A., Stucchi, R., Vocking, K.et al. (2015). TRIM46 Controls neuronal polarity and axon specification by driving the formation of parallel microtubule arrays. Neuron 88, 1208-1226. 10.1016/j.neuron.2015.11.012 [DOI] [PubMed] [Google Scholar]
- Weiner, A. T., Lanz, M. C., Goetschius, D. J., Hancock, W. O. and Rolls, M. M. (2016). Kinesin-2 and Apc function at dendrite branch points to resolve microtubule collisions. Cytoskeleton (Hoboken) 73, 35-44. 10.1002/cm.21270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner, A. T., Seebold, D. Y., Torres-Gutierrez, P., Folker, C., Swope, R. D., Kothe, G. O., Stoltz, J. G., Zalenski, M. K., Kozlowski, K., Barbera, D. J.et al. (2020). Endosomal Wnt signaling proteins control microtubule nucleation in dendrites. PLoS Biol. 18, e3000647. 10.1371/journal.pbio.3000647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese, C. (2008). Distinct Dgrip84 isoforms correlate with distinct gamma-tubulins in Drosophila. Mol. Biol. Cell 19, 368-377. 10.1091/mbc.e07-08-0801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalgin, C., Ebrahimi, S., Delandre, C., Yoong, L. F., Akimoto, S., Tran, H., Amikura, R., Spokony, R., Torben-Nielsen, B., White, K. P.et al. (2015). Centrosomin represses dendrite branching by orienting microtubule nucleation. Nat. Neurosci. 18, 1437-1445. 10.1038/nn.4099 [DOI] [PubMed] [Google Scholar]
- Yan, J., Chao, D. L., Toba, S., Koyasako, K., Yasunaga, T., Hirotsune, S. and Shen, K. (2013). Kinesin-1 regulates dendrite microtubule polarity in Caenorhabditis elegans. eLIFE 2, e00133. 10.7554/eLife.00133.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Li, R., Kaneko, T., Takle, K., Morikawa, R. K., Essex, L., Wang, X., Zhou, J., Emoto, K., Xiang, Y.et al. (2014). Trim9 regulates activity-dependent fine-scale topography in Drosophila. Curr. Biol. 24, 1024-1030. 10.1016/j.cub.2014.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau, K. W., Schatzle, P., Tortosa, E., Pages, S., Holtmaat, A., Kapitein, L. C. and Hoogenraad, C. C. (2016). Dendrites In Vitro and In Vivo contain microtubules of opposite polarity and axon formation correlates with uniform plus-end-out microtubule orientation. J. Neurosci. 36, 1071-1085. 10.1523/JNEUROSCI.2430-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa, S., Shigematsu, M., Hirata, K. and Hayashi, K. (2015). Loss of gamma-tubulin, GCP-WD/NEDD1 and CDK5RAP2 from the centrosome of neurons in developing mouse cerebral and cerebellar cortex. Acta Histochem. Cytochem. 48, 145-152. 10.1267/ahc.15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, W., Cook, C., Sauter, C., Kuriyama, R., Kaplan, P. L. and Baas, P. W. (2000). Depletion of a microtubule-associated motor protein induces the loss of dendritic identity. J. Neurosci. 20, 5782-5791. 10.1523/JNEUROSCI.20-15-05782.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.