ABSTRACT
In Saccharomyces cerevisiae, the selective autophagic degradation of mitochondria, termed mitophagy, is critically regulated by the adapter protein Atg32. Despite our knowledge about the molecular mechanisms by which Atg32 controls mitophagy, its physiological roles in yeast survival and fitness remains less clear. Here, we demonstrate a requirement for Atg32 in promoting spermidine production during respiratory growth and heat-induced mitochondrial stress. During respiratory growth, mitophagy-deficient yeast exhibit profound heat-stress induced defects in growth and viability due to impaired biosynthesis of spermidine and its biosynthetic precursor S-adenosyl methionine. Moreover, spermidine production is crucial for the induction of cytoprotective nitric oxide (NO) during heat stress. Hence, the re-addition of spermidine to Atg32 mutant yeast is sufficient to both enhance NO production and restore respiratory growth during heat stress. Our findings uncover a previously unrecognized physiological role for yeast mitophagy in spermidine metabolism and illuminate new interconnections between mitophagy, polyamine biosynthesis and NO signaling.
KEY WORDS: ATG32, Autophagy, Mitophagy, Nitric oxide, S-adenosyl methionine, Spermidine
Summary: Impaired mitophagy leads to a decrease in cellular SAM and spermidine levels, which in turn causes an impaired cytoprotective nitric oxide response otherwise necessary for heat stress tolerance.
INTRODUCTION
Mitochondria are essential for the generation of energy and essential metabolites but are prone to aging and dysfunction (Guo et al., 2013; Spinelli and Haigis, 2018; Jamieson, 1998). Autophagy, a cellular degradation process, selectively prunes damaged and unnecessary mitochondria in eukaryotic cells. This selective process, termed ‘mitophagy’ functions to prevent the pathogenesis of several mitochondrial-related diseases (Um and Yun, 2017). Mammalian mitophagy is regulated by multiple putative cargo receptors, whereas in yeast, Atg32 is the principal cargo adaptor uniquely required for mitophagy (Kanki et al., 2009b; Okamoto et al., 2009). Unlike mammalian cells, mitophagy-deficient yeast lacking Atg32 fail to exhibit obvious physiological anomalies, thus, little is known regarding its physiological importance (Kanki and Klionsky, 2010; Kanki et al., 2009b). Yeast mitophagy is enhanced under conditions of respiratory (non-fermentative) growth followed by nitrogen or amino acid starvation, during stationary phase, or in stress conditions that induce mitochondrial dysfunction; however, the lack of short-term gross morphological effects suggests mitophagy plays minimal roles in maintaining essential cellular functions in these conditions (Priault et al., 2005; Okamoto et al., 2009; Kanki and Klionsky, 2008; Vlahakis et al., 2016). One study demonstrates that mitophagy-deficient yeast are prone to mitochondrial genome instability as they age due to chronic reactive oxygen species (ROS) production during exceptionally prolonged nitrogen starvation (Kurihara et al., 2012). Nevertheless, a detailed understanding of the physiological function of yeast mitophagy and its effect on cell metabolism during mitochondrial stress remains limited. Here, we used an unbiased metabolomics approach to clarify the physiological roles of mitophagy on yeast metabolism during respiratory growth and heat-induced mitochondrial stress in the yeast Saccharomyces cerevisiae.
RESULTS AND DISCUSSION
Atg32-dependent mitophagy is necessary for respiratory growth during heat stress
Heat stress induces general mitochondrial dysfunction and activates multiple mitochondrial stress responses, including the mitochondrial unfolded protein response, decreased oxidative phosphorylation and decreased mitochondrial translation (Koike et al., 2018; Itami et al., 2018; Wilkening et al., 2018). These connections between heat stress and a myriad of mitochondrially related stress responses led us to postulate that mitophagy contributes to heat-stress tolerance.
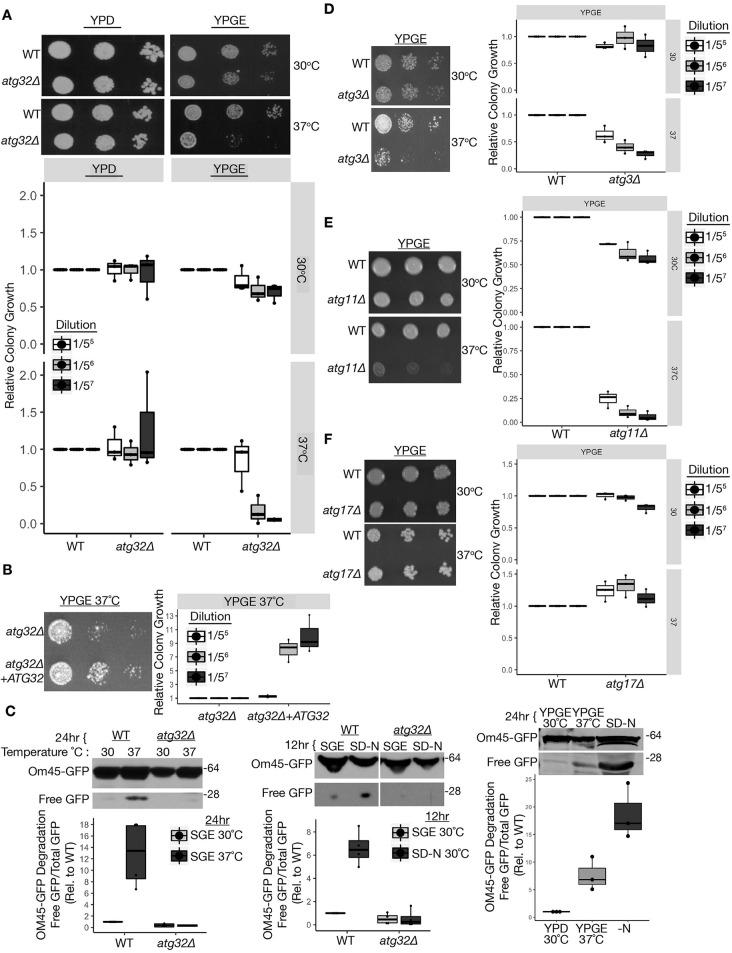
We first investigated the requirement for Atg32 in mediating fermentative and respiratory growth under conditions of heat-induced mitochondrial stress in S. cerevisiae. Glucose promotes fermentative growth and represses mitochondrial biogenesis and respiration, whereas non-fermentative carbon sources, such as glycerol/ethanol, upregulate mitochondrial biogenesis and respiration via oxidative phosphorylation to sustain growth (Roberts and Hudson, 2006). Growing yeast in these carbon sources allowed us to functionally interrogate the coordinate requirements for mitochondria and mitophagy in maintaining yeast cellular viability during heat stress. We compared the growth of wild-type and mitophagy (atg32Δ) mutant yeast undergoing fermentative (yeast peptone dextrose, YPD) versus non-fermentative (obligate) respiratory (yeast peptone glycerol ethanol, YPGE) growth at physiological (30°C) and heat-stress (37°C)-inducing temperatures. Respiratory growing atg32Δ yeast demonstrated robust growth defects compared to wild-type cells under heat stress (Fig. 1A). Re-introduction of the ATG32 wild-type gene into atg32Δ yeast augmented growth under these conditions (Fig. 1B). Hence, Atg32 is necessary to sustain the growth and viability of respiratory cells during heat-induced mitochondrial stress.
Fig. 1.
Atg32-dependent mitophagy is necessary for respiratory growth during heat stress. (A-B) Yeast serial dilutions grown on YPD and YPGE media at 30°C and 37°C (n=3). (C) Immunoblot of Om45-GFP lysosomal turnover from strains cultured at 30°C in SGE or YPGE media, and shifted to heat stress [SGE (n=4) or YPGE (n=3), 37°C] or nitrogen starvation 30°C (SD-N) (n=4). (D-F) Serial dilutions of yeast grown on YPGE at 30°C and 37°C (n=3). WT, wild type. The center line denotes the median value (50th percentile), while the box contains the 25th to 75th percentiles of the dataset. The black whiskers with a dot at the end of a line mark the 5th and 95th percentiles, and values beyond these upper and lower bounds are considered outliers, marked with black dots.
Given the functional requirement for Atg32 during heat stress, we investigated whether heat stress induces mitophagy in respiratory yeast. We measured the degradation of the outer mitochondrial membrane protein OM45 fused to GFP as a method to quantify mitophagy flux, quantifying free GFP due to lysosome cleavage as detected by immunoblot analysis (Kanki et al., 2009a). Under conditions of respiratory growth and 37°C heat stress, wild-type cells exhibited increased ATG32-dependent Om45-GFP degradation compared to 30°C (Fig. 1C, left). Similar results were observed in these strains due to mitophagy induction following nitrogen starvation (Fig. 1C, middle); remarkably, nitrogen starvation elicited a more robust mitophagy response compared to heat stress (Fig. 1C, right) (Kanki et al., 2009a; Kanki and Klionsky, 2008; Kanki et al., 2009b). This observation in yeast is consistent with previous reports in mammalian cells demonstrating that short-term heat stress induces mitochondrial degradation and enhances mitochondrial quality in an autophagy-dependent manner, indicating that this is a conserved response (Itami et al., 2018; Ganesan et al., 2018). Similarly, mitophagy and Atg32 upregulation in yeast required prior respiration induced by non-fermentable carbon sources (Kanki and Klionsky, 2008; Kanki et al., 2009b; Okamoto et al., 2009).
We next investigated whether Atg32 supports viability during heat stress via its established role in mitophagy. Macro-autophagy and mitophagy both functionally require Atg3, which is necessary for autophagosome formation. Cells lacking Atg3 phenocopied the respiratory growth arrest of atg32Δ yeast during heat stress (Fig. 1D), corroborating a requirement for macro-autophagy for heat stress tolerance during respiratory growth. Although many core Atg proteins required for macro-autophagy are also necessary for mitophagy, there are certain key differences in the genetic requirements for mitophagy (Kanki and Klionsky, 2008; Kanki et al., 2009b; Okamoto et al., 2009). Importantly, the selective autophagy scaffold protein Atg11, which interacts with Atg32, is required for mitophagy but dispensable for macro-autophagy (Kim et al., 2001; Kanki and Klionsky, 2008). Moreover, Atg17 is a scaffold protein necessary for macro-autophagy but only partly needed for starvation-induced mitophagy, and it is completely dispensable for mitophagy induced by prolonged respiratory growth (Kanki and Klionsky, 2008; Okamoto et al., 2009; Kawamata et al., 2008). These context-dependent genetic requirements for mitophagy prompted us to assess the respiratory growth of atg11Δ and atg17Δ yeast during heat stress. Analogous to the genetic dependencies described for mitophagy during respiratory growth, atg11Δ exhibited atg32Δ-like respiratory growth defects during heat stress, whereas atg17Δ exhibited wild-type like growth under these conditions (Fig. 1E,F). These findings demonstrate a physiological requirement for Atg32-dependent mitophagy in promoting cellular viability during conditions of heat stress in cells in which mitochondrial respiration is obligatory.
Atg32 is necessary for S-adenosyl methionine and spermidine production during heat stress
We next sought to determine the metabolic impact of impaired mitophagy in respiratory growing cells under heat stress. We employed metabolomics analysis by gas chromatography-time of flight-mass spectrometry (GC-TOF-MS) to ascertain changes in metabolites of respiratory wild-type and atg32Δ yeast grown in YPEG at 30°C and 37°C.
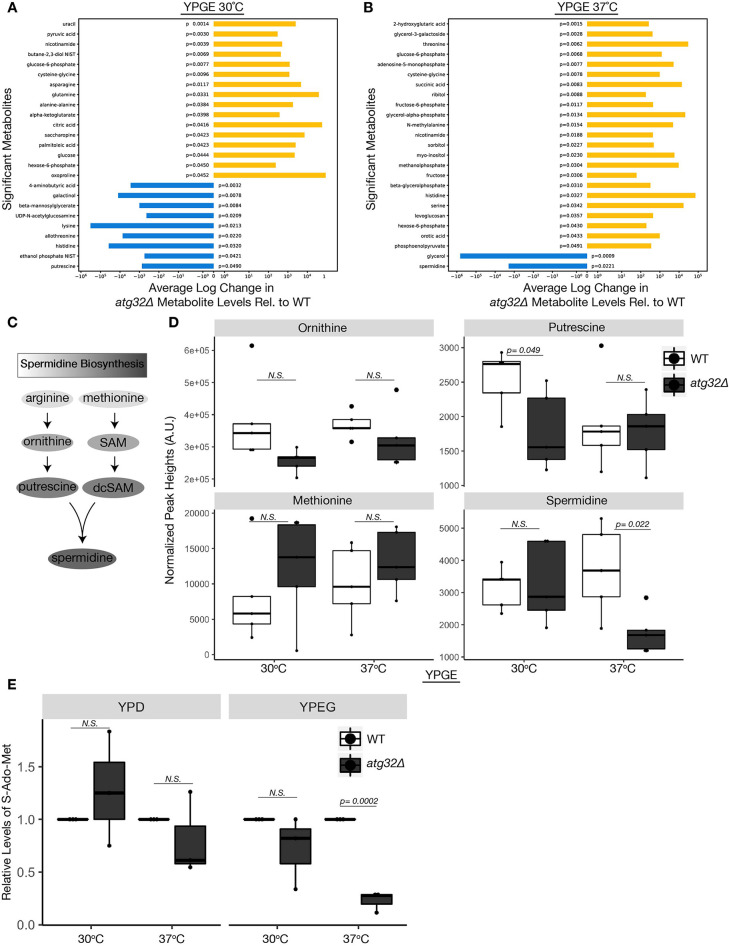
Statistical analysis revealed significant changes in 25 metabolites between respiratory growing wild-type and atg32Δ yeast at 30°C, and a distinct set of 25 metabolites at 37°C (Fig. 2A,B). In conditions of heat stress, only two metabolites, glycerol and the polyamine spermidine, exhibited a significant decrease in atg32Δ yeast compared to wild type (Fig. 2B-D). Heat stress triggers glycerol efflux via the yeast glyceroporin FPS1 as a mechanism to stimulate Slt2 and high osmolarity response (HOG) MAPK signaling, which in turn is necessary for the induction of mitophagy. It is conceivable that cells with inhibited mitophagy hyperactivate this glycerol-driven cellular response as a result of heat-stress intolerance (Dunayevich et al., 2018; Mao et al., 2011). Spermidine is a polyamine necessary for a growing repertoire of cytoprotective processes due to its anti-inflammatory properties, ability to enhance mitochondrial function and respiration, condense and protect DNA from oxidative stress, promote translation of electron transport chain (ETC) complex proteins and improve proteostasis and chaperone activity (Puleston et al., 2019; Puleston and Simon, 2015; Lee et al., 2011; Madeo et al., 2018). Furthermore, deficiencies in spermidine are associated with diverse age-associated pathologies that are often attributed to mitochondrial dysregulation, including cardiovascular disease, neurodegeneration and cancer (Eisenberg et al., 2016; Madeo et al., 2018). Several studies attribute the cytoprotective and anti-aging capacity of spermidine to autophagy induction, which is observed across several organisms, including yeast, flies, worms and mammalian cells (Pietrocola et al., 2015; Morselli et al., 2011; Eisenberg et al., 2009; García-Prat et al., 2016). Furthermore, studies in cardiomyocytes and other cultured cell lines demonstrate that spermidine increases mitophagy to promote mitochondrial fitness (Eisenberg et al., 2016; García-Prat et al., 2016; Qi et al., 2016). Our findings reinforce these interconnections and uncover a previously unrecognized functional role for Atg32-mediated mitophagy in sustaining spermidine during conditions of heat stress in respiratory growing cells dependent on mitochondrial function.
Fig. 2.
Atg32 required for SAM and spermidine production during heat stress. (A-B) GC-TOF-MS metabolomics results depicting average change of metabolites exhibiting significant differences between wild-type (WT) and atg32Δ yeast grown in YPGE medium at 30°C (A) or 37°C (B) (n=5). (C) Spermidine biosynthesis metabolic pathway. In this polyamine biosynthesis pathway, arginine is converted into the polyamine ornithine, which is the precursor to putrescine. Separately, methionine is the metabolic precursor for S-adenosyl methionine (SAM), which becomes de-carboxylated to form dcSAM. Putrescine and dcSAM converge to create spermidine. (D) Steady state levels of spermidine and spermidine precursors from extracts of wild-type and atg32Δ yeast grown at 30°C and 37°C in YPGE medium. Metabolite peak heights were determined by GC-TOF-MS and normalized using the mTIC (sum of peak heights of known metabolites) (n=5). (E) Relative SAM levels between wild-type and atg32Δ yeast grown in fermentative YPD medium and respiratory YPGE medium at 30°C and 37°C (n=3). (A-E). P-values determined using a unpaired two-tailed Student's t-test. A.U., arbitrary units; N.S., not significant. The center line denotes the median value (50th percentile), while the box contains the 25th to 75th percentiles of the dataset. The black whiskers with a dot at the end of a line mark the 5th and 95th percentiles, and values beyond these upper and lower bounds are considered outliers, marked with black dots.
Spermidine biosynthesis requires two metabolic branches, including the conversion of the polyamine ornithine to putrescine, and the conversion of methionine to S-adenosyl methionine (SAM) and its decarboxylated (dc) form, which converge to form spermidine (Fig. 2C) (Pegg, 1969). Cellular levels of ornithine, putrescine and methionine were not significantly altered between strains at 37°C (Fig. 2D) and could not explain the observed spermidine deficiency.
We used a fluorogenic reporter assay to directly assess metabolic cellular levels of SAM (S-Ado-Met) in fermentative (YPD) and respiratory (YPGE) growing wild-type and atg32Δ yeast at both 30°C and 37°C. Consistent with the observed decrease in spermidine levels, respiratory growing atg32Δ yeast exhibited significantly decreased levels of SAM compared to wild type during of heat stress (37°C) (Fig. 2E). SAM is the rate-limiting step in spermidine production and often a point of strict metabolic regulation (Pegg, 1969). These data suggest that cells lacking Atg32 exhibit low spermidine levels as a result of decreased SAM production, despite having adequate levels of upstream metabolites methionine, ornithine and putrescine. Although our metabolomic analysis revealed putrescine levels to be significantly lower in atg32Δ cells at 30°C, this did not significantly impact spermidine levels, further supporting SAM as a rate-limiting step in spermidine biosynthesis (Fig. 2A,D). Based on these data, we conclude that the mitophagy adaptor protein Atg32 is necessary to sustain cellular levels of spermidine and its biosynthetic precursor SAM during heat stress under conditions of respiratory growth.
SAM and spermidine are necessary and sufficient to restore respiratory growth of atg32Δ cells during heat stress
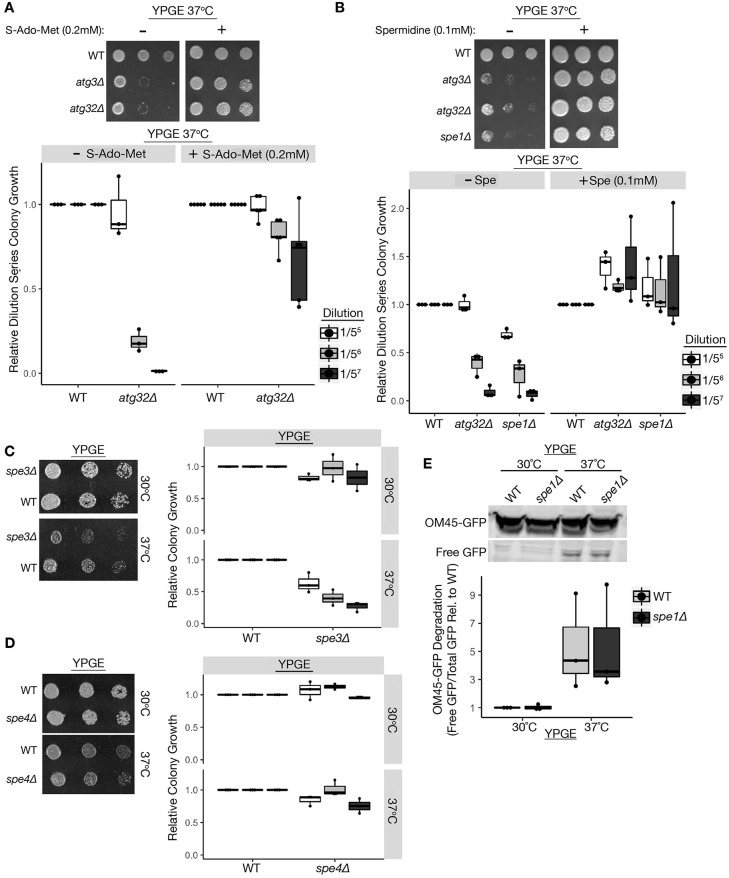
We next tested whether exogenous supplementation by SAM or spermidine is sufficient to restore the viability of respiratory growing atg32Δ cells under heat stress. Indeed, exogenous addition of 0.2 mM SAM or 0.1 mM spermidine was sufficient to significantly reverse the heat-stress-induced respiratory growth arrest of atg32Δ yeast and restore growth to wild-type-like levels (Fig. 3A,B). We further examined respiratory growth at 37°C in spermidine-deficient yeast lacking the ornithine decarboxylase Spe1 (spe1Δ), which catalyzes the first step in polyamine biosynthesis. Similar to atg32Δ yeast, spe1Δ mutants exhibited a severe heat stress-induced respiratory growth defect that was significantly restored to wild-type-like growth upon spermidine supplementation (Fig. 3A,B). Similarly, respiratory growing cells lacking the spermidine synthase Spe3 exhibited a significant heat-induced growth defect similar to atg32Δ and spe1Δ cells (Fig. 3C), whereas cells lacking the Spe4 enzyme that converts spermidine to spermine were unaffected (Fig. 3D). These findings further corroborate a unique role for spermidine in promoting viability during heat stress in respiratory growing cells. Interestingly, inhibiting spermidine biosynthesis did not significantly affect heat stress-induced mitophagy, as OM45-GFP degradation was equivalent between wild-type and spe1Δ strains (Fig. 3E). Overall, our data support Atg32-dependent mitophagy as an essential process for the maintenance of SAM and spermidine production, which is both necessary and sufficient for heat stress tolerance in respiratory growing yeast.
Fig. 3.
SAM and spermidine are necessary and sufficient to restore respiratory growth of atg32Δ cells during heat stress. (A-B) Serial dilutions of yeast spotted on YPD and YPGE plates with or without 0.2 mM SAM (A) or 0.1 mM spermidine (B) and grown at 37°C. (C-D) Serial dilutions of indicated yeast strains on YPGE plates at 30°C and 37°C. (E) Om45-GFP lysosomal turnover measured by immunoblot analysis from the indicated yeast cultured at 30°C in YPGE medium and shifted to 37°C. WT, wild type. The center line denotes the median value (50th percentile), while the box contains the 25th to 75th percentiles of the dataset. The black whiskers with a dot at the end of a line mark the 5th and 95th percentiles, and values beyond these upper and lower bounds are considered outliers, marked with black dots.
Mitophagy dependent spermidine production promotes cytoprotective nitric oxide response during heat stress
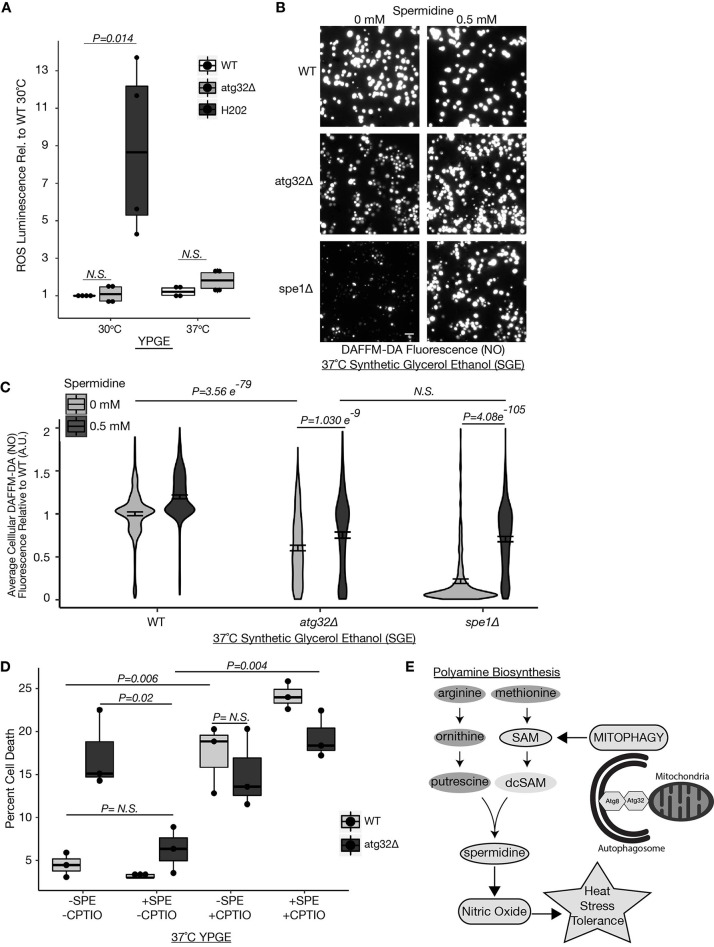
When respiratory yeast are shifted to glucose-containing nitrogen starvation medium, mitophagy prevents chronic ROS accumulation specifically in aging yeast (Kurihara et al., 2012). We investigated whether the sensitivity of respiratory growing atg32Δ yeast to heat stress is attributable to ROS using a luciferase-based luminescence assay (Ros-GLO, Promega). ROS production in respiratory growing atg32Δ cells was equivalent in all conditions (Fig. 4A). In contrast to that observed for extended nitrogen starvation, increased ROS is not a contributing factor to the respiratory growth defect of atg32Δ cells during heat stress.
Fig. 4.
Mitophagy-dependent spermidine production promotes cytoprotective NO during heat stress. (A) Relative ROS in wild-type (WT), atg32Δ, and WT plus H2O2 yeast grown at 30°C and 37°C in YPGE medium as determined by ROS-Glo luminescence assay (Promega). P-values determined using a unpaired two-tailed Student's t-test (n=4) (B,C) Fluorescence microscopy images (B) and quantification (C) of relative NO levels in yeast grown in SGE medium and subjected to 2.5 h of heat stress (37°C) with or without spermidine (0.5 mM) in the presence of DAF-FM-DA. (C) P-values determined using a unpaired two-tailed t-test. Violin plot representing all measurements from three bioreplicates. n=200 cells/condition/experiment (n=600 cells/condition). Data are mean±s.e.m. (D) Percentage of cell death in cells grown in YPGE medium with or without 1 mM NO-scavenger CPTIO and/or 0.5 mM spermidine. Cell death measured using Sytox Blue and fluorescence microscopy. ∼200 cells per condition (n=3). P-values determined using a unpaired two-tailed Student's t-test. (E) Schematic outlining the process by which mitophagy facilitates heat stress tolerance by promoting the biosynthesis of SAM and spermidine necessary for cytoprotective NO production during heat stress. Scale bar: 10 µm. A.U., arbitrary units; N.S., not significant. The center line denotes the median value (50th percentile), while the box contains the 25th to 75th percentiles of the dataset. The black whiskers with a dot at the end of a line mark the 5th and 95th percentiles, and values beyond these upper and lower bounds are considered outliers, marked with black dots.
Although a high cellular level of oxidative free radicals is often attributed to cellular stress, membrane damage and cell death, recent reports have alternatively demonstrated cytoprotective roles for reactive nitrogen species in supporting cellular stress responses (Yoshikawa et al., 2016; Nishimura et al., 2013). Nishimura et al. (2013) demonstrated that heat-stress induced nitric oxide (NO) production in yeast is necessary for cellular viability and growth. Arginine is used to generate NO by nitric oxide synthases (NOS) and is also the first biosynthetic precursor in polyamine biosynthesis (Fig. 2C); thus, these pathways are linked in competition for arginine (Förstermann and Sessa, 2012; Nishimura et al., 2013; Yoshikawa et al., 2016; Whitney and Morris, 1978). Complex interconnections between autophagy and NO signaling that vary based on cellular context and metabolic state have been reported (He et al., 2014; Sarkar et al., 2011). To ascertain whether spermidine production impacts the NO-dependent heat stress response, we examined NO levels in wild-type, atg32Δ, and spe1Δ respiratory growing yeast during heat stress in the absence and presence of exogenous spermidine. NO levels were detected using the established NO-sensitive and specific fluorescence probe 4-amino-5-methylamino-2′,7′-di-fluorescein-diacetate (DAF-FM-DA) (Li et al., 2011; Balcerczyk et al., 2005). Consistent with a defect in cellular growth, respiratory atg32Δ yeast displayed significantly reduced NO compared to wild-type cells during heat stress (Fig. 4B,C). Further, spe1Δ cells exhibited severe defects in NO production under these conditions (Fig. 4B,C). These findings support that spermidine mediates the heat-stress-dependent induction of NO and demonstrate direct regulation between spermidine levels and NO production apart from the bioavailability of arginine.
As spermidine addition ameliorates the heat-stress induced respiratory growth arrest of atg32Δ and spe1Δ yeast, we tested whether exogenous spermidine addition impacted the heat-stress-induced production of cytoprotective NO in mutant cells. Consistent with a rescue in growth, spermidine supplementation significantly increased NO to wild-type-like levels in respiratory atg32Δ and spe1Δ yeast following heat stress (Fig. 4B,C). Thus, spermidine is essential for the NO production during heat stress and is sufficient to restore NO-dependent heat stress signaling in mitophagy mutants. We next investigated whether inhibiting NO ameliorates the benefits of spermidine addition on cellular viability during heat stress. Addition of the NO scavenger 2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (CPTIO) alone significantly increased cell death in respiratory growing wild-type cells to atg32Δ-like levels (Fig. 4D). Furthermore, quenching NO with CPTIO significantly reversed the protective effects of spermidine in both wild-type and atg32Δ yeast (Fig. 4D). Thus, spermidine functions through NO signaling to support heat stress tolerance. These findings corroborate numerous previous reports demonstrating a cytoprotective role for NO in promoting heat stress tolerance, and reveal an intimate reliance between mitophagy-dependent polyamine biosynthesis and NO production for heat stress tolerance in yeast cells in which mitochondrial respiration is obligatory. Our observations are consistent with a study in Arabidopsis thaliana demonstrating that addition of spermidine, but not upstream polyamines, augmented seedling NO release, suggesting a positive regulation between spermidine levels and NO production (Tun et al., 2006).
These findings provide a more comprehensive view into the physiological and metabolic roles of mitophagy during mitochondrial stress. In all, our findings support that Atg32-dependent mitophagy promotes heat stress tolerance in respiratory growing yeast by maintaining the production of SAM and its downstream polyamine metabolite spermidine, which are both functionally required to generate cytoprotective NO during heat stress (Fig. 4E). Given their individual roles in aging, our proposed model linking mitophagy, polyamine biosynthesis and NO signaling illuminates a potential functional network in age-related diseases.
MATERIALS AND METHODS
Yeast strains, plasmids and media
BY4741 wild-type and single deletion yeast strains used in this study were purchased from Thermo Fisher Scientific and verified by PCR. Yeast expressing chimeric Om45-GFP were generated by integrating a DNA fragment encoding GFP at the 3′ end of endogenous OM45 using a PCR-based integration method (Longtine et al., 1998). Parental plasmids pRS416 (Dr Peter Walter, UCSF, Dept of Biochemistry and Biophysics, CA, USA) and pRS416- prATG32-ATG32 (Kanki et al., 2009b) (Dr Daniel Klionsky, University of Michigan, Life Sciences Institute, Ann Arbor, MI , USA) were generously gifted.
Where indicated, yeast were grown in YPD medium [1% yeast extract (Difco), 2% peptone (Difco) and 2% dextrose (Difco)], YPGE medium [1% yeast extract (Difco), 2% peptone (Difco), 3% glycerol (Sigma-Aldrich) and 3% ethanol (Sigma-Aldrich)], synthetic dextrose (SD) medium [0.8% w/v yeast nitrogen base without amino acids (Difco), 0.79 g/l CSM powder (Sunrise Science) and 2% dextrose (Sigma-Aldrich)] or synthetic glycerol ethanol (SGE) medium [0.8% w/v yeast nitrogen base without amino acids (Difco), 0.79 g/l CSM powder (Sunrise Science), 3% glycerol (Sigma-Aldrich) and 3% ethanol (Sigma-Aldrich)] at 30°C (physiological) or 37°C (heat stress).
Yeast growth for heat stress experiments
Yeast cells were cultivated in liquid YPD medium with rapid agitation at 30°C overnight. Cells were passaged in fresh YPD medium at 30°C and grown to A600 of 0.8-1.2 followed by passage into both YPD and YPGE at 30°C and 37°C with a starting A600 of 0.2, and grown until A600 of 0.8-1.2 with rapid agitation. Cell pellets were then collected and harvested for experimentation. Where indicated, synthetic medium containing dextrose (SD) or glycerol (SGE) was used instead of YPD or YPGE.
Om45-GFP processing mitophagy assay
To monitor mitophagy, the Om45-GFP processing assay was adapted from Kanki et al. (2009a) (Kanki and Klionsky, 2008). Briefly, indicated yeast strains expressing endogenously GFP-tagged OM45 were grown in SGE medium (A600 0.8-1.2) at 30°C, and were then shifted to SD-N media or SGE media and cultured for the indicated time at 37°C. Cell aliquots equivalent to 1.0 A600 were harvested, and TCA-precipitated whole-cell proteins were run on a 4-12% PAGE gel, and western blot analysis was performed using primary antibody against GFP (1:1000, Abcam, AB6556, and Antibodies Incorporated, 75-131), Pgk1 (loading control, 1:5000, Abcam, AB113687) and horseradish peroxidase (HRP) secondary antibody (1:5000, HRP donkey anti-rabbit IgG, Thermo Fisher Scientific, 45-000-682). GFP antibody was validated with western blot analysis using lysates from yeast strains without GFP. Western blots were quantified using Fiji.
Yeast dilution series growth plates
Cells were first grown in YPD medium at 30°C to A600 of 0.8-1.2 log phase growth. Serial dilutions were performed in corresponding medium starting with a concentration of 1 OD/ml and serially diluted 1:5 for seven iterations, then ∼2 μl of yeast suspension was plated with a multichannel pipette onto YPD or YPGE agar growth plates. Cells were allowed to grow on plates at 30°C or 37°C for 4 days. For genetic rescue experiments, atg32Δ yeast expressing either control empty vector pRS416 or pRS416- prATG32-ATG32 were grown in -URA SD dropout medium (CSM –URA, 1004-010, Sunrise Science). Serial dilutions were then plated on YPD or YPGE agar plates and grown at 37°C for 4 days. In cases in which dilutions were plated on SGE plates, growth was allowed for 7 days. YPD or YPGE plates containing 0.1 mM spermidine or 0.5 mM SAM were created by diluting S-(5′Adenodyl)-L-methionine chloride (Cayman Chemical, 13956) or spermidine (Cayman Chemical, 14918) in water and adding directly to warm medium containing agar to the indicated final concentration.
Quantification of colony spot growth was performed using Fiji to determine the density/intensity of each patch of yeast for each dilution. Images were converted to 8-bit tiffs and the rectangle selection was used to select the regions of interest. The Analyze> Gels>Plot Lanes function was then used to generate a profile plot of intensity peaks. Relevant peaks were isolated from background using the line tool, and the relative density/intensity for each dilution patch was quantified using the wand selection tool. Quantifications represent fold change relative to wild-type yeast at each respective dilution.
Untargeted metabolic profiling by GC-TOF-MS
Yeast cells were cultivated in liquid YPD medium at 30°C and grown to A600 of 0.8-1.2 followed by passage into either YPD or YPGE with a starting A600 of 0.2 and grown at both 30°C and 37°C until an A600 of 0.8-1.2. Yeast were harvested to obtain a pellet of 1.5 OD per sample, and samples were submitted to the Fiehn Laboratory at the National Institutes of Health West Coast Metabolomics Center where metabolites were analyzed by GC-TOF-MS using the LECO Pegasus IV time of flight mass spectrometer. Samples were prepared for analysis by MS by adding 5 ml of cold quenching buffer (3:2 methanol:water) to 1.5 OD yeast pellet and centrifuging (1000 g, 5 min) to separate the cell pellet. The cell pellet was mechanically ground in 1 ml of extraction solvent (3:3:2 acetonitrile:isopropanol:water) and centrifuged at 14,000 g for 5 min. The supernatant was then collected. This was repeated with remaining cell pellet to obtain 2 ml of supernatant. Extracted supernatant (50 μl) was dried down for GC-TOF-MS analysis. Quantification and statistical analysis of metabolomic results was performed using RStudio, Jupyter, SciPy and Pandas+Matplotlib to calculate and plot average differences of metabolites that exhibit a significant change between wild-type and atg32Δ yeast grown in YPGE medium at 30°C or 37°C. Associated P-values were also calculated using an unpaired two-tailed t-test assuming equal variance (scipy.stats.ttest_ind). Metabolite peak heights were normalized using the metabolites total ion chromatogram, also known as ‘mTIC’, which is the sum of the peak heights of the known metabolites
SAM fluorescence bridge-it assay
Yeast cells were cultivated in liquid YPD medium at 30°C and grown to OD A600 of 0.8-1.2, followed by passage into either YPD or YPGE medium with a starting A600 of 0.2 and grown at both 30°C and 37°C until A600 of 0.8-1. Relative cellular SAM levels were determined using the SAM Bridge-It fluorometric assay kit (Mediomics, NC1099716) as per the manufacturer's recommendations. Briefly, 1 OD A600 of yeast was pelleted/condition, and cellular SAM was extracted using the provided CM assay buffer by mechanical cell lysis with glass beads at ambient temperature for 1 h, and supernatant was clarified by centrifugation (15,000 g for 15 min). The resulting supernatant (2 µl) and 18 µl of provided SAM assay solution was mixed and incubated at ambient temperature for 30 min. Subsequent SAM-dependent fluorescence was determined using a SpectraMax M2e (Molecular Devices) microplate reader with excitation of 485 nm and emission of 665 nm.
Detecting ROS levels using a ROS-Glo H2O2 luminescence assay
ROS-dependent cellular luminescence was determined as per the manufacturer's instructions (Promega, G8820) using the ROS-Glo H2O2 kit. Yeast cells were cultivated in liquid YPGE medium with rapid agitation at 30°C overnight starting at an A600 of 0.2 and grown until A600 of 0.8. Next, 80 μl of cells were placed into a 96-well plate and 20 μl of 125 μM H2O2 substrate (25 μM final concentration), in the provided dilution buffer, was added. Positive control wells contained 80 μl of yeast plus 1 mM H2O2 at 30°C. Cells were then placed in a 30°C or 37°C incubator for 2 h before 100 μl of provided ROS-GLO detection reagent was added, which included the recommended addition of 1 μl each of D-cysteine and signal enhancer solution. Plates were incubated for 20 min at room temperature. Relative luminescence was recorded using a SpectraMax L Microplate Reader luminometer (Molecular Devices).
Determining NO levels using the NO-specific fluorogenic probe DAF-FM-DA and fluorescent microscopy
Yeast cells were cultivated in liquid SD medium at 30°C and grown to A600 of 0.8-1.2, followed by passage into SGE medium with or without 0.5 mM spermidine starting at A600 of 0.2 and grown at 30°C until an OD A600 of 0.5. Fresh SGE medium with or without spermidine was replenished, and yeast were allowed to continue growing until an OD of 1 at 30°C. Subsequently, 15 μM DAF-FM-DA (Cayman Chemical, 18767; diluted in DMSO) was added and cells were incubated in SGE medium with or without spermidine for an additional 2.5 h at 37°C. Cells were next rinsed with respective medium lacking DAF-FM-DA and were imaged by fluorescence microscopy using the Delta Vision Elite microscope (Applied Precision GE Healthcare Life Sciences) fitted with a 1.4 -NA 100× objective and PCO Edge cMOS camera, and run by softWoRx Resolve3D software. Capture and post-capture image processing were performed using softWoRx software (Applied Precision GE Healthcare Life Sciences) and Fiji (Schindelin et al., 2012). The average fluorescence across each cell was quantified using the line tool and Analyze-Measure functions in Fiji, and results were normalized for each experiment by dividing all values by the average for wild type at 30°C. For each experiment, 200 cells per condition were quantified and three biological replicates (n=600 cells/condition) were represented as a violin plot with s.e.m. generated in RStudio.
Determining heat-stress-induced yeast cell death with Sytox blue nucleic acid stain following addition of spermidine and NO scavenger CPTIO
Yeast cells were cultivated in liquid YPD medium at 30°C and grown to A600 of 0.8-1.2, followed by passage into YPEG medium with or without 0.5 mM spermidine and 1 mM CPTIO (Cayman Chemical, 81540) starting at A600 of 0.2 and grown at 37°C until an OD of A600 of 0.5. Fresh medium with or without spermidine and CPTIO was replenished, and yeast were allowed to continue growing until an OD of 1 at 37°C (∼24 h total at 37°C). Subsequently, 1 μM Sytox blue (Thermo Fisher Scientific, S34857) nucleic acid stain was added, and cells were incubated for 10 min. Cells were next rinsed with PBS and were imaged in the DAPI channel by fluorescence microscopy using an Echo Revolve fluorescence microscope. Images were quantified using Fiji. n=∼200 cells/condition for a total of three bioreplicates quantified for percentage of cell death.
Supplementary Material
Acknowledgements
We thank Dr Peter Walter and Dr Daniel Klionsky for providing plasmids.
Footnotes
Competing interests
J.D. is a Scientific Advisory Board Member for Vescor Therapeutics, LLC.
Funding
This work was supported by the National Institutes of Health (CA126792 and AG057462 to J.D.; T32CA108462 to A.V.; and U2C-DK119886 to West Coast Metabolomics) and a National Science Foundation Student Fellowship (1144247 to J.G.). Deposited in PMC for release after 12 months.
Data availability
The GC-TOF-MS metabolomics dataset is available at the National Institutes of Health Common Fund National Metabolomics Data Repository Metabolomics Workbench (https://www.metabolomicsworkbench.org) (Study ID: ST001318; Project ID: PR000895; DOI: 10.21228/M80Q38). Associated source code for metabolomics analysis and manuscript graphs is available at the following GitHub repository: www.github.com/arivlahakis/vlahakis-kaur-source-code. The dataset for Fig. 4C is available at Dryad (https://doi.org/10.7272/Q6348HMP).
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/article-lookup/doi/10.1242/jcs.253781
References
- Balcerczyk, A., Soszynski, M. and Bartosz, G. (2005). On the specificity of 4-amino-5-methylamino-2′,7′-difluorofluorescein as a probe for nitric oxide. Free Radic. Biol. Med. 39, 327-335. 10.1016/j.freeradbiomed.2005.03.017 [DOI] [PubMed] [Google Scholar]
- Dunayevich, P., Baltanás, R., Clemente, J. A., Couto, A., Sapochnik, D., Vasen, G. and Colman-Lerner, A. (2018). Heat-stress triggers MAPK crosstalk to turn on the hyperosmotic response pathway. Sci. Rep. 8, 15168. 10.1038/s41598-018-33203-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg, T., Knauer, H., Schauer, A., Buttner, S., Ruckenstuhl, C., Carmona-Gutierrez, D., Ring, J., Schroeder, S., Magnes, C., Antonacci, L.et al. (2009). Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 11, 1305-1314. 10.1038/ncb1975 [DOI] [PubMed] [Google Scholar]
- Eisenberg, T., Abdellatif, M., Schroeder, S., Primessnig, U., Stekovic, S., Pendl, T., Harger, A., Schipke, J., Zimmermann, A., Schmidt, A.et al. (2016). Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med. 22, 1428-1438. 10.1038/nm.4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstermann, U. and Sessa, W. C. (2012). Nitric oxide synthases: regulation and function. Eur. Heart J. 33, 829-837. 10.1093/eurheartj/ehr304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan, S., Pearce, S. C., Gabler, N. K., Baumgard, L. H., Rhoads, R. P. and Selsby, J. T. (2018). Short-term heat stress results in increased apoptotic signaling and autophagy in oxidative skeletal muscle in Sus scrofa. J. Therm. Biol. 72, 73-80. 10.1016/j.jtherbio.2018.01.003 [DOI] [PubMed] [Google Scholar]
- García-Prat, L., Martínez-Vicente, M., Perdiguero, E., Ortet, L., Rodríguez-Ubreva, J., Rebollo, E., Ruiz-Bonilla, V., Gutarra, S., Ballestar, E., Serrano, A. L.et al. (2016). Autophagy maintains stemness by preventing senescence. Nature 529, 37-42. 10.1038/nature16187 [DOI] [PubMed] [Google Scholar]
- Guo, C., Sun, L., Chen, X. and Zhang, D. (2013). Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural Regen. Res. 8, 2003-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, H., Feng, Y.-S., Zang, L.-H., Liu, W.-W., Ding, L.-Q., Chen, L.-X., Kang, N., Hayashi, T., Tashiro, S.-I., Onodera, S.et al. (2014). Nitric oxide induces apoptosis and autophagy; autophagy down-regulates NO synthesis in physalin A-treated A375-S2 human melanoma cells. Food Chem. Toxicol. 71, 128-135. 10.1016/j.fct.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Itami, N., Shirasuna, K., Kuwayama, T. and Iwata, H. (2018). Short-term heat stress induces mitochondrial degradation and biogenesis and enhances mitochondrial quality in porcine oocytes. J. Therm. Biol. 74, 256-263. 10.1016/j.jtherbio.2018.04.010 [DOI] [PubMed] [Google Scholar]
- Jamieson, D. J. (1998). Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14, 1511-1527. [DOI] [PubMed] [Google Scholar]
- Kanki, T. and Klionsky, D. J. (2008). Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 283, 32386-32393. 10.1074/jbc.M802403200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki, T. and Klionsky, D. J. (2010). The molecular mechanism of mitochondria autophagy in yeast. Mol. Microbiol. 75, 795-800. 10.1111/j.1365-2958.2009.07035.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki, T., Kang, D. and Klionsky, D. J. (2009a). Monitoring mitophagy in yeast: the Om45-GFP processing assay. Autophagy 5, 1186-1189. 10.4161/auto.5.8.9854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki, T., Wang, K., Cao, Y., Baba, M. and Klionsky, D. J. (2009b). Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17, 98-109. 10.1016/j.devcel.2009.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata, T., Kamada, Y., Kabeya, Y., Sekito, T. and Ohsumi, Y. (2008). Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell 19, 2039-2050. 10.1091/mbc.e07-10-1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Kamada, Y., Stromhaug, P. E., Guan, J., Hefner-Gravink, A., Baba, M., Scott, S. V., Ohsumi, Y., Dunn, W. A., Jr. and Klionsky, D. J. (2001). Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153, 381-396. 10.1083/jcb.153.2.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike, N., Hatano, Y. and Ushimaru, T. (2018). Heat shock transcriptional factor mediates mitochondrial unfolded protein response. Curr. Genet. 64, 907-917. 10.1007/s00294-018-0809-9 [DOI] [PubMed] [Google Scholar]
- Kurihara, Y., Kanki, T., Aoki, Y., Hirota, Y., Saigusa, T., Uchiumi, T. and Kang, D. (2012). Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J. Biol. Chem. 287, 3265-3272. 10.1074/jbc.M111.280156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. B., Park, J. H., Folk, J. E., Deck, J. A., Pegg, A. E., Sokabe, M., Fraser, C. S. and Park, M. H. (2011). Inactivation of eukaryotic initiation factor 5A (eIF5A) by specific acetylation of its hypusine residue by spermidine/spermine acetyltransferase 1 (SSAT1). Biochem. J. 433, 205-213. 10.1042/BJ20101322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., Skinner, C., Castello, P. R., Kato, M., Easlon, E., Xie, L., Li, T., Lu, S.-P., Wang, C., Tsang, F.et al. (2011). Identification of potential calorie restriction-mimicking yeast mutants with increased mitochondrial respiratory chain and nitric oxide levels. J. Aging Res. 2011, 673185. 10.4061/2011/673185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., 3rd, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P. and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Madeo, F., Eisenberg, T., Pietrocola, F. and Kroemer, G. (2018). Spermidine in health and disease. Science 359, eaan2788. 10.1126/science.aan2788 [DOI] [PubMed] [Google Scholar]
- Mao, K., Wang, K., Zhao, M., Xu, T. and Klionsky, D. J. (2011). Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J. Cell Biol. 193, 755-767. 10.1083/jcb.201102092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli, E., Mariño, G., Bennetzen, M. V., Eisenberg, T., Megalou, E., Schroeder, S., Cabrera, S., Bénit, P., Rustin, P., Criollo, A.et al. (2011). Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 192, 615-629. 10.1083/jcb.201008167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, A., Kawahara, N. and Takagi, H. (2013). The flavoprotein Tah18-dependent NO synthesis confers high-temperature stress tolerance on yeast cells. Biochem. Biophys. Res. Commun. 430, 137-143. 10.1016/j.bbrc.2012.11.023 [DOI] [PubMed] [Google Scholar]
- Okamoto, K., Kondo-Okamoto, N. and Ohsumi, Y. (2009). Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87-97. 10.1016/j.devcel.2009.06.013 [DOI] [PubMed] [Google Scholar]
- Pegg, A. E. (1969). Reactivity of analogues of S-adenosylmethionine in the enzymic synthesis of spermidine by mammalian tissues. Biochim. Biophys. Acta (BBA) Gen. Subj. 177, 361-364. 10.1016/0304-4165(69)90151-2 [DOI] [PubMed] [Google Scholar]
- Pietrocola, F., Lachkar, S., Enot, D. P., Niso-Santano, M., Bravo-San Pedro, J. M., Sica, V., Izzo, V., Maiuri, M. C., Madeo, F., Mariño, G.et al. (2015). Spermidine induces autophagy by inhibiting the acetyltransferase EP300. Cell Death Differ. 22, 509-516. 10.1038/cdd.2014.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priault, M., Salin, B., Schaeffer, J., Vallette, F. M., di Rago, J.-P. and Martinou, J.-C. (2005). Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 12, 1613-1621. 10.1038/sj.cdd.4401697 [DOI] [PubMed] [Google Scholar]
- Puleston, D. J. and Simon, A. K. (2015). New roles for autophagy and spermidine in T cells. Microb. Cell 2 91-93. 10.15698/mic2015.03.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleston, D. J., Buck, M. D., Klein Geltink, R. I., Kyle, R. L., Caputa, G., O'sullivan, D., Cameron, A. M., Castoldi, A., Musa, Y., Kabat, A. M.et al. (2019). Polyamines and eIF5A Hypusination Modulate Mitochondrial Respiration and Macrophage Activation. Cell Metab. 30, 352-363.e8. 10.1016/j.cmet.2019.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Y., Qiu, Q., Gu, X., Tian, Y. and Zhang, Y. (2016). ATM mediates spermidine-induced mitophagy via PINK1 and Parkin regulation in human fibroblasts. Sci. Rep. 6, 24700. 10.1038/srep24700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, G. G. and Hudson, A. P. (2006). Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Mol. Genet. Genomics 276, 170-186. 10.1007/s00438-006-0133-9 [DOI] [PubMed] [Google Scholar]
- Sarkar, S., Korolchuk, V. I., Renna, M., Imarisio, S., Fleming, A., Williams, A., Garcia-Arencibia, M., Rose, C., Luo, S., Underwood, B. R.et al. (2011). Complex inhibitory effects of nitric oxide on autophagy. Mol. Cell 43, 19-32. 10.1016/j.molcel.2011.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B.et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli, J. B. and Haigis, M. C. (2018). The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 20, 745-754. 10.1038/s41556-018-0124-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun, N. N., Santa-Catarina, C., Begum, T., Silveira, V., Handro, W., Floh, E. I. and Scherer, G. F. (2006). Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant Cell Physiol. 47, 346-354. 10.1093/pcp/pci252 [DOI] [PubMed] [Google Scholar]
- Um, J.-H. and Yun, J. (2017). Emerging role of mitophagy in human diseases and physiology. BMB Rep. 50, 299-307. 10.5483/BMBRep.2017.50.6.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahakis, A., Lopez Muniozguren, N. and Powers, T. (2016). Calcium channel regulator Mid1 links TORC2-mediated changes in mitochondrial respiration to autophagy. J. Cell Biol. 215, 779-788. 10.1083/jcb.201605030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney, P. A. and Morris, D. R. (1978). Polyamine auxotrophs of Saccharomyces cerevisiae. J. Bacteriol. 134, 214-220. 10.1128/JB.134.1.214-220.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkening, A., Rüb, C., Sylvester, M. and Voos, W. (2018). Analysis of heat-induced protein aggregation in human mitochondria. J. Biol. Chem. 293, 11537-11552. 10.1074/jbc.RA118.002122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa, Y., Nasuno, R., Kawahara, N., Nishimura, A., Watanabe, D. and Takagi, H. (2016). Regulatory mechanism of the flavoprotein Tah18-dependent nitric oxide synthesis and cell death in yeast. Nitric Oxide 57, 85-91. 10.1016/j.niox.2016.04.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.