Abstract
Background
Non-alcoholic fatty liver disease (NAFLD) is a common disorder that is known to be the leading cause of chronic liver disease worldwide. This study aims to systematically review and meta-analyze the association between PNPLA3 rs738409 polymorphism and non-alcoholic fatty liver.
Methods
Following a systematic review and meta-analysis method, articles without any time limitation, were extracted from SID, MagIran, IranDoc, Scopus, Embase, Web of Science (WoS), PubMed and ScienceDirect international databases. Random effects model was used for analysis, and heterogeneity of studies was investigated considering the I2 index and using Comprehensive Meta-Analysis software.
Results
The odds ratio of CC genotype in patients with non-alcoholic fatty liver demonstrates the protective effect of CC genotype with the ratio of 0.52, whereas CG genotype presents an increasing effect of CG genotype with the ratio of 0.19, and GG genotype also showed an increasing effect of GG genotype with the ratio of 1.05. Moreover, CG + GG genotypes as a single group demostrated an odds rartio of 0.88.
Conclusion
This meta-analysis highlights that people with CC genotype has 52% lower chance of developing non-alcoholic fatty liver disease, and those with CG genotype had 19% higher risk of developing non-alcoholic fatty liver. Those with GG genotype were 105% more likely to develop non-alcoholic fatty liver than others. Moreover, those present in a population with CG + GG genotypes were 88% more likely to have non-alcoholic fatty liver disease.
Keywords: Polymorphism, Gene, Non-alcoholic fatty liver, NAFLD, PNPLA3
Background
Non-alcoholic Fatty Liver Disease (NAFLD) is a common disorder that is known to be the leading cause of chronic liver disease worldwide. This disorder is caused by abnormal accumulation of fat in liver tissue cells and can eventually lead to liver cirrhosis [1, 2]. The disease was first diagnosed in 1980 and is subsequently being recognised as one of the major contributors to mortality from liver disorders [3].
In people with NAFLD, deaths from liver disease were reported to be 0.77 per thousand people per year, and cardiovascular deaths were 4.79 per thousand people per year [4]. The prevalence of NAFLD in East Asian countries is 15–45% and in the developed Western countries is found to be 20–30% [5]. According to a study in 2010, the prevalence of this disorder in the general population was 35% and in another study in 2019 the prevalence of NAFLD was 25% [6, 7].
The prevalence of NAFLD is associated with disorders such as obesity and insulin-resistant diabetes and metabolic syndrome [4, 8]. In the study of Yamamoto et al., it has been estimated that the number of obese and overweight people will rise to more than 2 billion by the year 2030, so the occurance of NAFLD is expected to increase with the rise in obesity levels in the general population. Moreover, with the prevalence of obesity in children in recent years, NAFLD has been recognised as the most common liver disorder in children [9, 10]. Other causes of non-alcoholic fatty liver disease include sedentary lifestyle, poor diet and genetic polymorphism of different genes [11].
Different types of genes may be involved in the pathogenesis of NAFLD. Genetic factors cause NAFLD in 27 to 39% of cases. One of the most important genetic factors for NAFLD is Single Nucleotide Polymorphism (SNP) (rs738409) in the patatin-like phospholipase domain containing protein 3 (PNPLA3). This SNP was first identified in 2008 by two independent studies on the independent genome.
The PNPLA3 rs738409 C > G SNP is a type of Missense that results in the replacement of cytosine with guanosine and, ultimately, the incorrect coding of methionine rather than isoleucine at position 148. This single nucleotide polymorphism is located in the third exon of the pnpla3 gene. The PNPLA3 gene is located on human chromosome 22 (chr22q13.31).
PNPLA3 encodes a protein known as adiponutrin (ADPN). This protein is expressed in adipocytes and hepatocytes. Moreover, this protein has lipolytic and lipogenic properties, however the exact function of adiponutrin is still unclear. PNPLA has also been reported to be highly expressed on human stellate cells. The encoded protein has retinol esterase activity and allows retinol secretion from hepatocytes while the mutation induces intracellular retention of this compound, therefore, PNPLA3 rs738409 is susceptible to NAFLD.
The function of PNPLA3 rs738409 is still unknown, however in vitro studies have shown that PNPLA3 protein has tricylglycerol (TG) hydrolase and lysophosphatidyl acyltransferase (LPAAT) and calcium independent phospholipase A2 activities. PNPLA3 also plays a critical role in homestasis of lipid metabolism. PNPLA3 eventually causes glycerolipid hydrolasis in the liver and inhibits lipid outflow into peripheral adipose tissue, thus contributing to hepatic steatosis and related disorders. NAFLD is characterised by the accumulation of lipids in hepatic steatosis.
The PNPLA3 gene is associated not only with liver fat content, but also with hepatic inflammation, hepatic steatohepatitis, fibrosis and cirrhosis, indicating that it plays a key role in the development of NAFLD. Inflammatory infiltration and liver damage are greater in patients carrying PNPLA3 I148M than in wild-type genotype individuals; this gene is thought to be closely linked to liver inflammation. Compared to non-carriers, homozygous carriers has 73% higher liver fat content, 3.2 times higher risk in high necroinflammatory scores and 3.2 times higher risk of developing fibrosis [4, 5, 7, 11–19]. rs738409 of the patatin-like phospholipase domain containing gene 3 (PNPLA3) is known to be the most common and most potent gene in the development of NAFLD [4].
Furthermore, the association of PNPLA3 gene polymorphisms with other liver disorders such as alcoholic fatty liver (ALD) has also been observed [20]..
Since non-alcoholic fatty liver disease is very common and can have adverse side effects, understanding the factors affecting its occurrence can play a key role in the prevention and control of this disease and the treatment of those affected. This study aims to systematically review and meta-analyse the association between PNPLA3 rs738409 polymorphism and non-alcoholic fatty liver.
Methods
Search method
This study was performed to determine the association between PNPLA3 rs73409 C > G polymorphism using a systematic review and meta-analysis. Data were collected from Iranian and international databases of Web of Science (WoS), Embase, Scopus, PubMed, science direct, ProQuest, Google Scholar, SID, Irandoc and other international databases. International databses were searched using the keywords (PNPLA3 gene or PNPLA3 polymorphism OR patatin-like phospholipase domain-containing protein3) and (Non-alcoholic Fatty Liver Disease or NAFLD or Nonalcoholic Steatohepatitis) and their possible combination; Persian equivalent of keywords were used for searches within the Persian databases. The Google Scholar search engine was also used with both English and Persian keywords. In order to assess gray literature review, sites related to the subject, as well as the references within the found sources were analysed.
Criteria for selection and evaluation of articles
Following the search process, all articles were collected in the EndNote software, and all duplicates were removed. Inclusion criteria were: 1- Case control studies, 2- Cohort, and 3- Studies examining the relationship between pnpla3 gene and non-alcoholic fatty liver disease and Exclusion criteria were: 1- Cross-sectional studies, 2- Case reports, 3- Intervention studies, 4- Letters to editor, 5- Studies where the full-text was not available, and 6- Studies in which individuals in the population under study have underlying disease.
Then a list of titles and abstracts was prepared and after hiding the full text of the articles they were provided to the reviewers. Each article was independently reviewed by two reviewers, and in case of disagreement between the two reviewers, the third reviewer’s judgement was considered as the criterion for approval of articles.
During the qualitative evaluation phase, the STROBE checklist was used to evaluate the studies qualitatively. This checklist consists of 22 criteria, of which 18 are used to assess all research papers, and 4 are specific to the type study. The checklist is used to evaluate the study objectives, determination of appropriateness of the sample size, type of study, sampling method, research population, data collection method(s), definition of variables and method of sampling, study data collection tools, study objectives, the statistical test used to assess the findings, and the maximum score derived from this checklist is 32. The articles with a score below 14 were excluded. Studies were then reviewed according to the PRISMA 2009 four-step process, including article identification, screening, eligibility criteria and finally meta-analysis.
Statistical analysis
In this study, heterogeneity of studies was investigated using I2 test, data were analyzed using Comprehensive Meta-analysis software (Biostat, Englewood, NJ, USA version 3), probability of publication bias results were evaluated using both funnel diagrams and Egger test; please note that the significance level was set at 0.05.
Results
This study investigated the association between PNPLA3 I148M rs738409 polymorphism and non-alcoholic fatty liver disease through systematic review and meta-analysis. Following searching various databases, a total of 1391 articles entered the study, of which 220 articles were from EMBASE database, 47 articles from ProQuest, 109 articles from PubMed, 84 articles from ScienceDirect, 243 articles from Scopus, 447 articles from Web of Science (WoS), 1 article from SID, 57 articles from Irandoc, 145 articles from Google Scholar, and 2 articles were selected following the reviews of other articles, and were found within the references.
Once the articles were collected, 360 duplicate articles were eliminated, and after reviewing the title and abstracts, 692 other articles were also removed and 339 articles were left subjected to secondary evaluation. After reviewing the full text of the articles in terms of thematic relevance as well as qualitative review of the articles, 308 additional articles were excluded and finally 31 articles entered the meta-analysis process (please see Tables 1 and 2).
Table 1.
Characteristics of studies entered into the meta-analysis
| Row | Author [References] | Publication year | Area | Age of case group | Age of control group | Case Group Size | Control group Size |
|---|---|---|---|---|---|---|---|
| 1 | Alam, S [21]. | 2017 | Bangladesh | 39.1 ± 8.6 | 29.64 ± 7.03 | 99 | 75 |
| 2 | Baclig, M. O [22]. | 2014 | Philippines | 20–70 | 20–70 | 32 | 36 |
| 3 | Bhatt, S. P [23]. | 2013 | India | 38.2 ± 7 | 37.1 ± 6.9 | 162 | 173 |
| 4 | Chen, L. Z [24]. | 2019 | China | 26.71 ± 2.81 | 22.48 ± 3.12 | 512 | 451 |
| 5 | Choobini, Neda [19] | 2016 | Iran | 47.9 ± 12.3 | 40 ± 13.9 | 95 | 183 |
| 6 | Di Costanzo, A [25]. | 2018 | Italy | 54 | 49.7 | 218 | 227 |
| 7 | Gorden, A [26]. | 2013 | America | 47 ± 10.6 | 46 ± 11.8 | 748 | 344 |
| 8 | Hotta, K [27]. | 2010 | Japan | 51.7 ± 15 | 47.2 ± 14.8 | 253 | 578 |
| 9 | Hudert, C. A [28]. | 2019 | Germany | 14.11 ± 2.15 | 46.73 ± 16.3 | 70 | 200 |
| 10 | Karoli, R [29]. | 2019 | India | 45 ± 8.2 | 46 ± 7 | 100 | 100 |
| 11 | Kawaguchi, Takahisa [30] | 2012 | Japan | 52.05 ± 14.85 | 48.8 ± 16.3 | 529 | 932 |
| 21 | Krishnasamy, N [31]. | 2020 | India | 43.15 ± 9.245 | 41.99 ± 12.7 | 105 | 102 |
| 13 | Lee, S. S [32]. | 2014 | Korea | 45.3 ± 15.5 | 45.3 ± 10.6 | 155 | 184 |
| 14 | Li, Y. L. [33] | 2012 | China | 46.7 ± 13.6 | 43.1 ± 13.4 | 203 | 202 |
| 15 | Liu, W. Y [34]. | 2019 | China | 40.2 ± 12.5 | 46.6 ± 9.2 | 349 | 58 |
| 16 | Niriella, M. A [35]. | 2017 | Sri Lanka | 42–71 | (42–71) | 1360 | 391 |
| 17 | Niu, T. H [36]. | 2014 | China | 49.7 ± 16.7 | 47.69 ± 15.68 | 390 | 409 |
| 18 | Oniki, Kentaro [37] | 2015 | Argentina | 61.2 ± 10.5 | 67.5 ± 6 | 393 | 740 |
| 19 | Park, J. H [38]. | 2015 | South Korea | 48.9 ± 7 | 49.1 ± 7.2 | 602 | 761 |
| 20 | Peng, X. E [39]. | 2012 | China | 45.33 ± 12.48 | 43.87 ± 13 | 553 | 553 |
| 21 | Rametta, R [40]. | 2014 | Italy | 49.7 ± 12.1 | 47.7 ± 12.1 | 137 | 260 |
| 22 | Shang, X. R [41]. | 2015 | China | 11.81 ± 2.20 | 11.44 ± 2.99 | 162 | 865 |
| 23 | Uygun, A [42]. | 2017 | Turkey | 42.1 ± 11.4 | 34.1 ± 12.8 | 216 | 150 |
| 24 | Valenti, L. [43] | 2012 | Italy | 49.5 ± 12 | 48(1 ± 2 | 144 | 257 |
| 25 | Valenti, L. [44] | 2010 | Italy | 46.4 ± 11 | 48.4 ± 13 | 253 | 179 |
| 26 | Vespasiani-Gentilucci, U [45]. | 2016 | Italy | 51.5 ± 12.3 | 40.1 ± 13.1 | 60 | 125 |
| 27 | Wang, C. W [18]. | 2011 | Taiwan | 48.11 ± 12.05 | 45.4 ± 15.93 | 156 | 723 |
| 28 | Wang, X. L. [46] | 2016 | China | 45 ± 13 | 45 ± 13 | 376 | 382 |
| 29 | Xia, M. F [16]. | 2016 | China | 60 | 61 | 1385 | 2915 |
| 30 | Yang, H. H [47]. | 2018 | China | 70.95 ± 4.73 | 72.53 ± 4.73 | 97 | 362 |
| 31 | Zhang, R. N [48]. | 2016 | China | 38.2 ± 13.78 | 42.64 ± 10.58 | 59 | 72 |
Table 2.
Overview of CC, CG, GG and CG + GG genotypes based on the obtained studies
| Row | Author [References] | Genotype | Dominant | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CG | GG | CG + GG | ||||||||||
| Case | Control | OR | Case | Control | OR | Case | Control | OR | Case | Control | OR | ||
| 1 | Alam, S [21]. | 45 | 37 | 0.398 | 27 | 43 | 1.365 | 3 | 19 | 5.700 | 30 | 62 | 2.514 |
| 2 | Baclig, M. O [22]. | 26 | 14 | 0.299 | 8 | 12 | 2.100 | 2 | 6 | 3.923 | 10 | 18 | 3.343 |
| 3 | Bhatt, S. P [23]. | 149 | 112 | 0.361 | 16 | 35 | 2.704 | 8 | 15 | 2.105 | 24 | 50 | 2.772 |
| 4 | Chen, L. Z [24]. | 196 | 114 | 0.373 | 194 | 236 | 1.133 | 61 | 162 | 2.959 | 255 | 398 | 2.683 |
| 5 | Choobini, Neda [19] | 15 | 13 | 1.776 | 14 | 17 | 2.631 | 154 | 65 | 0.408 | 168 | 82 | 0.563 |
| 6 | Di Costanzo, A [25]. | 123 | 92 | 0.617 | 56 | 91 | 2.188 | 48 | 35 | 0.713 | 104 | 126 | 1.620 |
| 7 | Gorden, A [26]. | 218 | 411 | 0.705 | 103 | 244 | 1.133 | 5 | 47 | 4.546 | 108 | 291 | 1.391 |
| 8 | Hotta, K [27]. | 175 | 45 | 0.498 | 296 | 111 | 0.745 | 104 | 97 | 2.834 | 400 | 208 | 2.057 |
| 9 | Hudert, C. A [28]. | 118 | 20 | 0.278 | 71 | 31 | 1.444 | 11 | 19 | 6.401 | 82 | 50 | 3.598 |
| 10 | Karoli, R [29]. | 51 | 20 | 0.240 | 32 | 55 | 2.597 | 17 | 25 | 1.627 | 59 | 80 | 2.780 |
| 11 | Kawaguchi, Takahisa [30] | 247 | 88 | 0.553 | 468 | 236 | 0.799 | 217 | 203 | 2.052 | 685 | 439 | 1.759 |
| 12 | Krishnasamy, N [31]. | 59 | 19 | 0.261 | 29 | 50 | 2.288 | 14 | 36 | 3.280 | 43 | 86 | 6.211 |
| 13 | Lee, S. S [32]. | 55 | 31 | 0.586 | 92 | 75 | 0.938 | 37 | 49 | 1.837 | 129 | 124 | 1.705 |
| 14 | Li, Y. L. [33] | 94 | 70 | 0.605 | 90 | 84 | 0.878 | 18 | 49 | 3.253 | 108 | 133 | 1.654 |
| 15 | Liu, W. Y [34]. | 24 | 94 | 0.522 | 24 | 152 | 1.093 | 8 | 85 | 2.012 | 32 | 237 | 1.719 |
| 16 | Niriella, M. A [35]. | 25 | 54 | 0.605 | 134 | 464 | 0.993 | 232 | 842 | 1.114 | 366 | 1306 | 1.652 |
| 17 | Niu, T. H [36]. | 183 | 48 | 0.173 | 176 | 153 | 0.855 | 50 | 189 | 6.751 | 226 | 342 | 5.769 |
| 18 | Oniki, Kentaro [37] | 223 | 38 | 0.248 | 394 | 111 | 0.346 | 121 | 45 | 0.662 | 515 | 156 | 0.288 |
| 19 | Park, J. H [38]. | 280 | 172 | 0.678 | 364 | 293 | 1.034 | 117 | 137 | 1.622 | 481 | 430 | 1.455 |
| 20 | Peng, X. E [39]. | 235 | 183 | 0.669 | 259 | 276 | 1.131 | 59 | 93 | 1.693 | 318 | 369 | 1.482 |
| 21 | Rametta, R [40]. | 150 | 51 | 0.435 | 95 | 67 | 1.662 | 15 | 19 | 2.630 | 110 | 86 | 2.299 |
| 22 | Shang, X. R [41]. | 338 | 60 | 0.917 | 418 | 74 | 0.899 | 109 | 28 | 1.449 | 527 | 102 | 1.090 |
| 23 | Uygun, A [42]. | 85 | 64 | 0.322 | 50 | 90 | 1.429 | 15 | 62 | 3.623 | 65 | 152 | 3.106 |
| 24 | Valenti, L.(2012) [43] | 146 | 55 | 0.470 | 95 | 68 | 1.526 | 16 | 21 | 2.572 | 111 | 89 | 2.128 |
| 25 | Valenti, L.(2010) [44] | 118 | 103 | 0.355 | 56 | 114 | 1.801 | 5 | 36 | 5.773 | 61 | 150 | 2.817 |
| 26 | Vespasiani-Gentilucci, U [45]. | 83 | 29 | 0.473 | 34 | 18 | 1.147 | 8 | 13 | 4.045 | 42 | 31 | 2.112 |
| 27 | Wang, C. W [18]. | 269 | 40 | 0.582 | 335 | 80 | 1.219 | 119 | 36 | 1.523 | 454 | 116 | 1.718 |
| 28 | Wang, X. L. [46] | 169 | 122 | 0.605 | 174 | 191 | 1.234 | 39 | 63 | 1.770 | 213 | 254 | 1.652 |
| 29 | Xia, M. F [16]. | 1200 | 486 | 0.773 | 1363 | 684 | 1.111 | 352 | 215 | 1.338 | 1715 | 899 | 1.294 |
| 30 | Yang, H. H [47]. | 110 | 27 | 0.884 | 123 | 40 | 1.364 | 129 | 30 | 0.809 | 252 | 70 | 1.132 |
| 31 | Zhang, R. N [48]. | 32 | 12 | 0.319 | 31 | 27 | 1.116 | 9 | 20 | 3.590 | 40 | 47 | 3.133 |
The PRISMA 4-step process highlighting the processes in obtaining the final articles for our meta-analysis is presented in Fig. 1.
Fig. 1.

PRISMA flow diagram for the processes followed for includign studies in this systematic review and meta-analysis
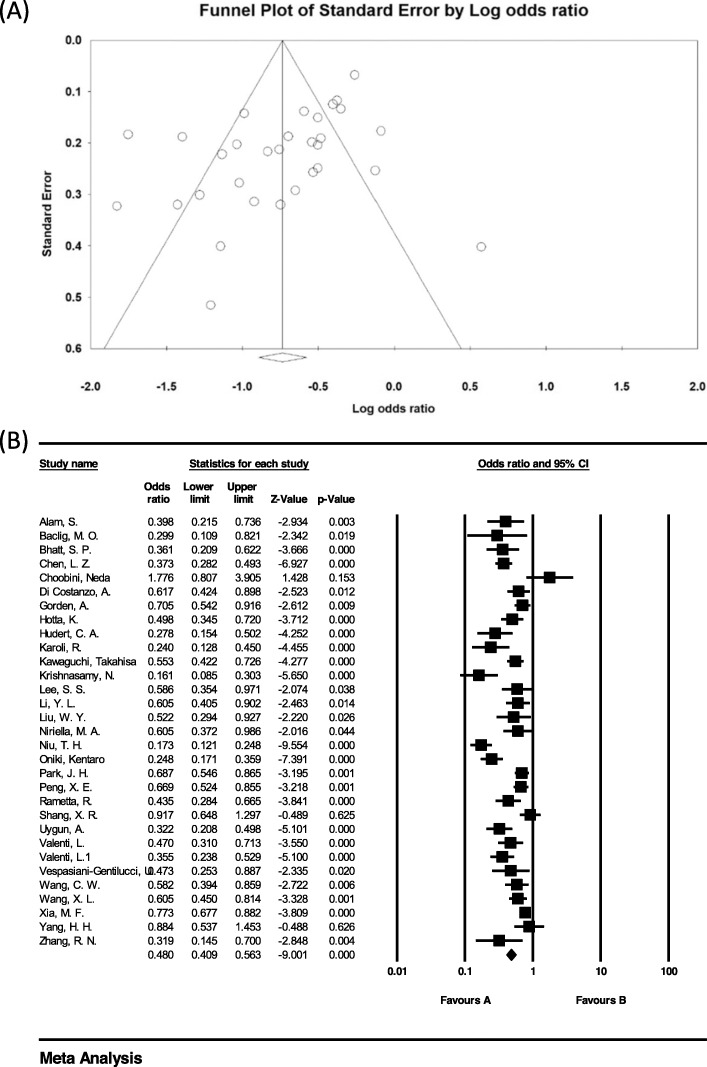
Investigation of heterogeneity and publication bias (CC genotype)
The heterogeneity of the studies was evaluated using the I2 test. Based on this test, I2 = 82.2% was obtained, which indicates high heterogeneity in the included studies. Moreover, the results of the publication bias study were compared with the Egger test (please see Fig. 2 A), which was not statistically significant (P = 0.052).
Fig. 2.
Funnel plot (A) and Overall forest plot of CC Genotype in Patients with Non-Alcoholic Fatty Liver Based on Random Model (B)
The total number of samples included in the case group and in the control group were 9973 and 13,048 respectively. The odds ratio of CC genotype in patients with non-alcoholic fatty liver was 0.48 based on meta-analysis (95% CI: 0.40–056), indicating an protective effect of CC genotype with 0.52, meaning that those with this genotype are 52% less likely to develop non-alcoholic fatty liver than others. In Fig. 2 B, the odds ratio based on the random effects model is shown where the black small rectanlges has the odds ratio and the rectangle length indicates the 95% confidence interval; the diamond shape represents the odds ratio for the entire study (Fig. 2 B).
Investigation of heterogeneity and publication bias (CG genotype)
The heterogeneity of the studies was evaluated using the I2 test. Based on this test, I2 = 80.3% was obtained, which indicates high heterogeneity in the included studies. Moreover, the results of the publication bias study were compared with the Egger test (please see Fig. 3 A), which was not statistically significant (P = 0.072).
Fig. 3.
Funnel plot (A) and Overall forest plot of CG Genotype in Patients with Non-Alcoholic Fatty Liver Based on Random Model (B)
The total number of samples included in the case group and in the control group were 9973 and 13,048 respectively. The odds ratio of CG genotype in patients with non-alcoholic fatty liver was 1.19 based on meta-analysis (95% CI: 1–1.33), indicating an increasing effect of CG genotype with 0.19, meaning that those with this genotype are 19% more likely to develop non-alcoholic fatty liver than others. In Fig. 3 B, the odds ratio based on the random effects model is shown where the black small rectanlges has the odds ratio and the rectangle length indicates the 95% confidence interval; the diamond shape represents the odds ratio for the entire study (Fig. 3 B).
Investigation of heterogeneity and publication bias (GG genotype)
The heterogeneity of the studies was evaluated using the I2 test. Based on this test, I2 = 86.3% was obtained, which indicates heterogeneity in the included studies. Moreover, the results of the publication bias study were compared with the Egger test (please see Fig. 4 A), which was not statistically significant (P = 0.064).
Fig. 4.
Funnel plot (A) and Overall forest plot of GG Genotype in Patients with Non-Alcoholic Fatty Liver Based on Random Model (B)
The total number of samples included in the case group and in the control group were 9973 and 13,048 respectively. The odds ratio of GG genotype in patients with non-alcoholic fatty liver was 2.05 based on meta-analysis (95% CI: 1.64–2.56), indicating an increasing effect of GG genotype with 1.05, meaning that those with this genotype are 105% more likely to develop non-alcoholic fatty liver than others. In Fig. 4 B, the odds ratio based on the random effects model is shown where the black small rectanlges has the odds ratio and the rectangle length indicates the 95% confidence interval; the diamond shape represents the odds ratio for the entire study (Fig. 4 B).
Investigation of heterogeneity and publication bias (CG + GG genotype)
The heterogeneity of the studies was evaluated using the I2 test. Based on this test, I2 = 90.7% was obtained, which indicates heterogeneity in the included studies. Moreover, the results of the publication bias study were compared with the Egger test (please see Fig. 5 A), which was not statistically significant (P = 0.054).
Fig. 5.
Funnel plot (A) and Overall forest plot of CG + GG Genotype in Patients with Non-Alcoholic Fatty Liver Based on Random Model (B)
The total number of samples included in the case group and in the control group were 9973 and 13,048 respectively. The odds ratio of CG + GG genotype in patients with non-alcoholic fatty liver was 1.88 based on meta-analysis (95% CI: 1.5–2.3), indicating an increasing effect of CG + GG genotype with 0.88, meaning that those with this genotype are 88% more likely to develop non-alcoholic fatty liver than others. In Fig. 5 B, the odds ratio based on the random effects model is shown where the black small rectanlges has the odds ratio and the rectangle length indicates the 95% confidence interval; the diamond shape represents the odds ratio for the entire study (Fig. 5 B).
Discussion
In this study, after investigating the association between different genotypes of PNPLA3 rs738409 polymorphism and non-alcoholic fatty liver disease, we highlighted that people with CC genotype with the odds ratio of 0.48, have 52% lower risk of developing non-alcoholic fatty liver, while this ratio in CG and GG genotypes were 1.19 and 2.05 respectively, and therefore the probability of developing the disease in those with these genotypes were 19% (CG) and 105% (GG) higher. On the other hand, considering the CG + GG groups as a single population/group, and following a statistical analysis, it was concluded that the odds ratio of this group in relation to occurance of Non-alcoholic fatty liver was 1.88, meaning that this group were 88% more likely to develop the disorder than others. The effect of the G allele on non-alcoholic fatty liver disease can also be emphasized. A study in India in 2020 also found that the G allele plays a key role in the development of NAFLD [31].
NAFLD is recognised as one of the most common liver diseases in the world with unknown etiology and pathogenesis. However, several factors including genetics, diet and inactivity, have been presented as some of the key reasons for the development of the disease. It has also been found that a good diet and regular exercise can reduce the risk of developing insulin resistance and can boost glucose homeostasis. Other SNPs such as rs2896019 and rs3810322 have also been reported to increase the risk of non-alcoholic fatty liver disease [1]. Past genomic studies have identified two genes PNPLA3 I148M and TM6SF2 E167K as the most likely genetic factors in the development of NAFLD [49].
According to meta-analysis by Zhang et al. (2015) on some studies undertaken in Asian countries, when comparing people having the G allele with a population with the C allele, the probability of non-alcoholic fatty liver disease was reported to be 1.92, and therefore it was concluded that the G allele is likely to increase the development of non-alcoholic fatty liver to the liver in people with G allele by 92%; Moreover, it can increase the risk of renal fibrosis and ALT serum levels. Development of NAFLD in the dominant phenotype (CG + GG) was 110% higher than the recessive phenotype. On the other hand, comparing the CG + GG populations with the CG genotype, it was concluded that the risk of NAFLD was higher in the homozygous GG population than in other populations [5].
Another meta-analysis conducted in 2019 stated that this polymorphism had a major impact on the development of tissue damage in liver and that the G allele was considered as a risk factor for NAFLD in such a way that the ratio of development of the disease in those with one G allele to those without it was 1.88, and 4.01 in those where both alleles were G. It has also been suggested that this gene increases alanine aminotransferase levels in serum [50].
According to a meta-analysis by Jiaying et al. (2020), this gene is involved in the development of non-alcoholic osteopathy (NASH) in children and adolescents; it is also accosiated with factors such as serum alanine transaminase, aspartate transaminase, gamma glutamyl transferase, that are indicators of liver damage [51].
Another meta-analysis in 2015, it was reported that all genetic variations in the rs738409 polymorphism in the pnpla3 gene was strongly associated with the incidence of NAFLD and NASH, especially in Asian and Spanish populations. In this study, however, no association was found between rs738409 polymorphism and hepatic steatosis. It was also reported that the GG genotype had a high impact on the development of NAFLD as well as renal fibrosis. The ratio of this genotype over inflammation occurance was reported as 3.13 [14].
According to a study by Chobin et al., the CG genotype was identified as a predisposing genotype to a 2.63-fold increase in the likelihood of developing the disease. Moreover, it was reported that the GG genotypes possess a protective effect, meaning that existence of such gentype results in a 59% decrease in developing NAFLD. Furtheremore, the odds of developing the disease in the CC genotype was 1.78 [19].
According to another study by Sood et al. (2016) in Japan, the odds ratio of the GG genotype was 36.5% in obese people and 47.8% in the non-obese population who had a fatty liver. Moreover, the modified odds ratio of non-alcoholic fatty liver disease in GG genotype was reported to be 4.15 in non-obese individuals, and 2.76 in obese pupulation. This genotype also increases the chances of developing steatosis and liver fibrosis [52]. A family history of NAFLD may result in higher levels of ALT and cholesterol among children. Moreover, it was reported that for every 10 unit increase in ALT (in IU / L) there will be approximately 1.5 times and for every 20 unit (mg / L) increase in body cholesterol, there will be approximately 2 times the risk of developing NAFLD in children [53, 54].
Limitation
The limitation of this study was the lack of access to the full-text of some of the sources.
Conclusion
This meta-analysis study demonstrated that people with the CC genotype were 52% less likely to develop non-alcoholic fatty liver disease, and people with CG genotype were 19% more likely to develop non-alcoholic fatty liver. Moreover, population with the GG genotype, had 105% more chance of developing a non-alcoholic fatty liver. Moreover, population with CG + GG genetypes demonstrate 88% more chance of developing the disease, and this is suggesting the effect of G allele on non-alcoholic fatty liver disease. In future, the effects of genetic and environmental factors on the level fo tissue damage, and also the effect of this gene on fibrosis and liver cirrhosis can be studied.
Acknowledgements
We hereby express our gratitude and appreciation to the Student Research Committee of Kermanshah University of Medical Sciences.
Abbreviations
- NAFLD
Non-alcoholic fatty liver disease
- SNP
Single Nucleotide Polymorphism
- TG
Tricylglycerol
- WoS
Web of Science
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analysis.
- STROBE
Strengthening the Reporting of Observational Studies in Epidemiology for cross- sectional Study
Authors’ contributions
ND and KM and HGH contributed to the design, MM statistical analysis, participated in most of the study steps. ND and FD and MHF prepared the manuscript. NS and ND and HGH assisted in designing the study, and helped in the, interpretation of the study. All authors have read and approved the content of the manuscript.
Funding
By Student Research Committee of Kermanshah University of Medical Sciences, Deputy for Research and Technology, Kermanshah University of Medical Sciences (IR) (990582).
Availability of data and materials
Datasets are available through the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
Ethics approval was received from the ethics committee of deputy of research and technology, Kermanshah University of Medical Sciences (IR.KUMS.REC.1399.205).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nader Salari, Email: n_s_514@yahoo.com.
Niloufar Darvishi, Email: darvishiniloofar@gmail.com.
Kamran Mansouri, Email: kamranmansouri@gmail.com.
Hooman Ghasemi, Email: humanghasemi2010@gmail.com.
Melika Hosseinian-Far, Email: mel.hosseinian@gmail.com.
Fateme Darvishi, Email: darvishy.fateme@gmail.com.
Masoud Mohammadi, Email: Masoud.mohammadi1989@yahoo.com.
References
- 1.Song GH, et al. Association of patatin-like phospholipase domain-containing protein 3 gene polymorphisms with susceptibility of nonalcoholic fatty liver disease in a Han Chinese population. Medicine. 2016;95(33):e4569. [DOI] [PMC free article] [PubMed]
- 2.Seko Y, Yamaguchi K, Mizuno N, Okuda K, Takemura M, Taketani H, Hara T, Umemura A, Nishikawa T, Moriguchi M, Yasui K, Kamaguchi M, Nishioji K, Mochizuki N, Kobayashi M, Mori K, Tanaka S, Matsuura K, Tanaka Y, Itoh Y. Combination of PNPLA3 and TLL1 polymorphism can predict advanced fibrosis in Japanese patients with nonalcoholic fatty liver disease. J Gastroenterol. 2018;53(3):438–448. doi: 10.1007/s00535-017-1372-8. [DOI] [PubMed] [Google Scholar]
- 3.Tan HL, Zain SM, Mohamed R, Rampal S, Chin KF, Basu RC, Cheah PL, Mahadeva S, Mohamed Z. Association of glucokinase regulatory gene polymorphisms with risk and severity of non-alcoholic fatty liver disease: an interaction study with adiponutrin gene. J Gastroenterol. 2014;49(6):1056–1064. doi: 10.1007/s00535-013-0850-x. [DOI] [PubMed] [Google Scholar]
- 4.Xia MF, Lin HD, Chen LY, Wu L, Ma H, Li Q, Aleteng Q, Hu Y, He WY, Gao J, Bian H, Li XY, Gao X. The PNPLA3 rs738409 C>G variant interacts with changes in body weight over time to aggravate liver steatosis, but reduces the risk of incident type 2 diabetes. Diabetologia. 2019;62(4):644–654. doi: 10.1007/s00125-018-4805-x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L, You W, Zhang H, Peng R, Zhu Q, Yao A, Li X, Zhou Y, Wang X, Pu L, Wu J. PNPLA3 polymorphisms (rs738409) and non-alcoholic fatty liver disease risk and related phenotypes: a meta-analysis. J Gastroenterol Hepatol (Australia) 2015;30(5):821–829. doi: 10.1111/jgh.12889. [DOI] [PubMed] [Google Scholar]
- 6.Xia MF, Chen LY, Wu L, Ma H, Li Q, Aleteng Q, Hu Y, He WY, Gao J, Lin HD, Gao X. The PNPLA3 rs738409 C>G variant influences the association between low skeletal muscle mass and NAFLD: the Shanghai Changfeng study. Aliment Pharmacol Ther. 2019;50(6):684–695. doi: 10.1111/apt.15372. [DOI] [PubMed] [Google Scholar]
- 7.Zain SM, Mohamed R, Mahadeva S, Cheah PL, Rampal S, Basu RC, Mohamed Z. A multi-ethnic study of a PNPLA3 gene variant and its association with disease severity in non-alcoholic fatty liver disease. Hum Genet. 2012;131(7):1145–1152. doi: 10.1007/s00439-012-1141-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tai CM, Huang CK, Tu HP, Hwang JC, Chang CY, Yu ML. PNPLA3 genotype increases susceptibility of nonalcoholic steatohepatitis among obese patients with nonalcoholic fatty liver disease. Surg Obes Relat Dis. 2015;11(4):888–894. doi: 10.1016/j.soard.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 9.Marzuillo P, Grandone A, Perrone L, Miraglia del Giudice E. Understanding the pathophysiological mechanisms in the pediatric non-alcoholic fatty liver disease: the role of genetics. World J Hepatol. 2015;7(11):1439–1443. doi: 10.4254/wjh.v7.i11.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamamoto K, Kogiso T, Taniai M, Hashimoto E, Tokushige K. Differences in the genetic backgrounds of patients with alcoholic liver disease and non-alcoholic fatty liver disease. Jgh Open. 2019;3(1):17–24. doi: 10.1002/jgh3.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JZ, Cao HX, Chen JN, Pan Q. PNPLA3 rs738409 underlies treatment response in nonalcoholic fatty liver disease. World J Clin Cases. 2018;6(8):167–175. doi: 10.12998/wjcc.v6.i8.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan S, Liu H, Yuan D, Xu J, Chen Y, Xu X, Xu F, Liang H. PNPLA3 I148M mediates the regulatory effect of NF-kB on inflammation in PA-treated HepG2 cells. J Cell Mol Med. 2020;24(2):1541–1552. doi: 10.1111/jcmm.14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J, Marsh S, Hu J, Feng W, Wu C. The pathogenesis of nonalcoholic fatty liver disease: interplay between diet, gut microbiota, and genetic background. Gastroenterol Res Pract. 2016;2016:1–13. doi: 10.1155/2016/2862173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu R, et al. Association between Patatin-Like Phospholipase Domain Containing 3 Gene (PNPLA3) Polymorphisms and Nonalcoholic Fatty Liver Disease: a HuGE Review and Meta-Analysis. Scientific Reports. 2015;5:9284. [DOI] [PMC free article] [PubMed]
- 15.Xu M, Li Y, Zhang S, Wang X, Shen J, Zhang S. Interaction of TM6SF2 E167K and PNPLA3 I148M variants in NAFLD in Northeast China. Ann Hepatol. 2019;18(3):456–460. doi: 10.1016/j.aohep.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Xia MF, Ling Y, Bian H, Lin HD, Yan HM, Chang XX, Li XM, Ma H, Wang D, Zhang LS, Wang SS, Wu BJ, He WY, Zhao NQ, Gao X. I148M variant of PNPLA3 increases the susceptibility to non-alcoholic fatty liver disease caused by obesity and metabolic disorders. Aliment Pharmacol Ther. 2016;43(5):631–642. doi: 10.1111/apt.13521. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Song J, Shang X, Chawla N, Yang Y, Meng X, Wang H, Ma J. Physical activity and sedentary behavior can modulate the effect of the PNPLA3 variant on childhood NAFLD: a case-control study in a Chinese population. Bmc Medical Genetics. 2016;17(1):90. doi: 10.1186/s12881-016-0352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang CW, Lin HY, Shin SJ, Yu ML, Lin ZY, Dai CY, Huang JF, Chen SC, Li SSL, Chuang WL. The PNPLA3 I148M polymorphismis associated with insulin resistance and nonalcoholic fatty liver disease in a normoglycaemic population. Liver Int. 2011;31(v9):1326–1331. doi: 10.1111/j.1478-3231.2011.02526.x. [DOI] [PubMed] [Google Scholar]
- 19.Choobini N, Azarpira N, Mashayekhi MR. Association of PNPLA3 gene polymorphism (rs738409) and nonalcoholic fatty liver disease in southern Iranian population. JFUMS. 2016;6(1):60–68. [Google Scholar]
- 20.Salameh H, Hanayneh MA, Masadeh M, Naseemuddin M, Matin T, Erwin A, Singal AK. PNPLA3 as a genetic determinant of risk for and severity of non-alcoholic fatty liver disease Spectrum. J Clin Transl Hepatol. 2016;4(3):175–191. doi: 10.14218/JCTH.2016.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alam S, Islam MS, Islam S, Mustafa G, Saleh AA, Ahmad N. Association of single nucleotide polymorphism at PNPLA3 with fatty liver, steatohepatitis, and cirrhosis of liver. Indian J Gastroenterol. 2017;36(5):366–372. doi: 10.1007/s12664-017-0784-y. [DOI] [PubMed] [Google Scholar]
- 22.Baclig MO, Lozano-Kühne JP, Mapua CA, Gopez-Cervantes J, Natividad FF, St Luke’s Liver Diseases Study Group Genetic variation I148M in patatin-like phospholipase 3 gene and risk of non-alcoholic fatty liver disease among Filipinos. Int J Clin Exp Med. 2014;7(8):2129–2136. [PMC free article] [PubMed] [Google Scholar]
- 23.Bhatt SP, Nigam P, Misra A, Guleria R, Pandey RM, Pasha MAQ. Genetic variation in the patatin-like phospholipase domain-containing protein-3 (PNPLA-3) gene in Asian Indians with nonalcoholic fatty liver disease. Metab Syndr Relat Disord. 2013;11(5):329–335. doi: 10.1089/met.2012.0064. [DOI] [PubMed] [Google Scholar]
- 24.Chen LZ, et al. Combining I148M and E167K variants to improve risk prediction for nonalcoholic fatty liver disease in Qingdao Han population, China. Lipids in Health and Disease. 2019;18(1):45. [DOI] [PMC free article] [PubMed]
- 25.Di Costanzo A, et al. Evaluation of polygenic determinants of non-alcoholic fatty liver disease (NAFLD) by a candidate genes resequencing strategy. Sci Rep. 2018;8(1):3702. doi: 10.1038/s41598-018-21939-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorden A, Yang R, Yerges-Armstrong LM, Ryan KA, Speliotes E, Borecki IB, Harris TB, Chu X, Wood GC, Still CD, Shuldiner AR, Gerhard GS, the GOLD Consortium<sup>1</sup> Genetic variation at NCAN locus is associated with inflammation and fibrosis in non-alcoholic fatty liver disease in morbid obesity. Hum Hered. 2013;75(1):34–43. doi: 10.1159/000346195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hotta K, Yoneda M, Hyogo H, Ochi H, Mizusawa S, Ueno T, et al. Association of the rs738409 polymorphism in PNPLA3 with liver damage and the development of nonalcoholic fatty liver disease. Bmc Medical Genetics. 2010;11(1). 10.1186/1471-2350-11-172. [DOI] [PMC free article] [PubMed]
- 28.Hudert CA, Selinski S, Rudolph B, Bläker H, Loddenkemper C, Thielhorn R, Berndt N, Golka K, Cadenas C, Reinders J, Henning S, Bufler P, Jansen PLM, Holzhütter HG, Meierhofer D, Hengstler JG, Wiegand S. Genetic determinants of steatosis and fibrosis progression in paediatric non-alcoholic fatty liver disease. Liver Int. 2019;39(3):540–556. doi: 10.1111/liv.14006. [DOI] [PubMed] [Google Scholar]
- 29.Karoli R, et al. Association of genetic non-alcoholic fatty liver disease with insulin resistance-are we different? J Assoc Physicians India. 2019;67(March):34–38. [PubMed] [Google Scholar]
- 30.Kawaguchi T, et al. Genetic Polymorphisms of the Human PNPLA3 Gene Are Strongly Associated with Severity of Non-Alcoholic Fatty Liver Disease in Japanese. PLoS One. 2012;7(6):e38322. [DOI] [PMC free article] [PubMed]
- 31.Krishnasamy N, et al. Association of metabolic syndrome and patatin-like phospholipase 3 - rs738409 gene variant in non-alcoholic fatty liver disease among a Chennai-based south Indian population. J Gene Med. 2020;22(4):e3160. [DOI] [PubMed]
- 32.Lee SS, Byoun YS, Jeong SH, Woo BH, Jang ES, Kim JW, Kim HY. Role of the PNPLA3 I148M polymorphism in nonalcoholic fatty liver disease and fibrosis in Korea. Dig Dis Sci. 2014;59(12):2967–2974. doi: 10.1007/s10620-014-3279-z. [DOI] [PubMed] [Google Scholar]
- 33.Li YL, et al. Genetic variant I148M in PNPLA3 is associated with the ultrasonography-determined steatosis degree in a Chinese population. Bmc Medical Genetics. 2012;13(1). 10.1186/1471-2350-13-113. [DOI] [PMC free article] [PubMed]
- 34.Liu WY, Zheng KI, Pan XY, Ma HL, Zhu PW, Wu XX, Rios RS, Targher G, Byrne CD, Wang XD, Chen YP, Zheng MH. Effect of PNPLA3 polymorphism on diagnostic performance of various noninvasive markers for diagnosing and staging nonalcoholic fatty liver disease. J Gastroenterol Hepatol. 2020;35(6):1057-64. [DOI] [PubMed]
- 35.Niriella MA, Pathmeswaran A, de Silva ST, Kasturiratna A, Perera R, Subasinghe CE, Kodisinghe K, Piyaratna C, Rishikesawan V, Dassanayaka AS, de Silva AP, Wickramasinghe R, Takeuchi F, Kato N, de Silva HJ. Incidence and risk factors for non-alcoholic fatty liver disease: a 7-year follow-up study among urban, adult Sri Lankans. Liver Int. 2017;37(11):1715–1722. doi: 10.1111/liv.13478. [DOI] [PubMed] [Google Scholar]
- 36.Niu TH, Jiang M, Xin YN, Jiang XJ, Lin ZH, Xuan SY. Lack of association between apolipoprotein C3 gene polymorphisms and risk of nonalcoholic fatty liver disease in a Chinese Han population vs. World J Gastroenterol. 2014;20(13):3655–3662. doi: 10.3748/wjg.v20.i13.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oniki K, et al. Influence of the PNPLA3 rs738409 polymorphism on non-alcoholic fatty liver disease and renal function among normal weight subjects. PloS one. 2015;10(7):e0132640. [DOI] [PMC free article] [PubMed]
- 38.Park JH, Cho BL, Kwon H, Prilutsky D, Yun JM, Choi HC, Hwang KB, Lee IH, Kim JI, Kong SW. I148M variant in PNPLA3 reduces central adiposity and metabolic disease risks while increasing nonalcoholic fatty liver disease. Liver Int. 2015;35(12):2537–2546. doi: 10.1111/liv.12909. [DOI] [PubMed] [Google Scholar]
- 39.Peng XE, et al. Genetic Variants in PNPLA3 and Risk of Non-Alcoholic Fatty Liver Disease in a Han Chinese Population. Plos One. 2012;7(11):e50256. [DOI] [PMC free article] [PubMed]
- 40.Rametta R, Ruscica M, Dongiovanni P, Macchi C, Fracanzani AL, Steffani L, Fargion S, Magni P, Valenti L. Hepatic steatosis and PNPLA3 I148M variant are associated with serum Fetuin-a independently of insulin resistance. Eur J Clin Investig. 2014;44(7):627–633. doi: 10.1111/eci.12280. [DOI] [PubMed] [Google Scholar]
- 41.Shang XR, Song JY, Liu FH, Ma J, Wang HJ. GWAS-identified common variants with nonalcoholic fatty liver disease in Chinese children. J Pediatr Gastroenterol Nutr. 2015;60(5):669–674. doi: 10.1097/MPG.0000000000000662. [DOI] [PubMed] [Google Scholar]
- 42.Uygun A, Ozturk K, Demirci H, Oztuna A, Eren F, Kozan S, Yilmaz Y, Kurt O, Turker T, Vatansever S, Alper E, Unsal B. The association of nonalcoholic fatty liver disease with genetic polymorphisms: a multicenter study. Eur J Gastroenterol Hepatol. 2017;29(4):441–447. doi: 10.1097/MEG.0000000000000813. [DOI] [PubMed] [Google Scholar]
- 43.Valenti L, Rametta R, Ruscica M, Dongiovanni P, Steffani L, Motta BM, et al. The i148m Pnpla3 polymorphism influences serum adiponectin in patients with fatty liver and healthy controls. BMC Gastroenterol. 2012;12(1). 10.1186/1471-230X-12-111. [DOI] [PMC free article] [PubMed]
- 44.Valenti L, al-Serri A, Daly AK, Galmozzi E, Rametta R, Dongiovanni P, Nobili V, Mozzi E, Roviaro G, Vanni E, Bugianesi E, Maggioni M, Fracanzani AL, Fargion S, Day CP. Homozygosity for the Patatin-like Phospholipase-3/Adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51(4):1209–1217. doi: 10.1002/hep.23622. [DOI] [PubMed] [Google Scholar]
- 45.Vespasiani-Gentilucci U, Gallo P, Porcari A, Carotti S, Galati G, Piccioni L, de Vincentis A, Dell’Unto C, Vorini F, Morini S, Riva E, Picardi A. The PNPLA3 rs738409 C > G polymorphism is associated with the risk of progression to cirrhosis in NAFLD patients. Scand J Gastroenterol. 2016;51(8):967–973. doi: 10.3109/00365521.2016.1161066. [DOI] [PubMed] [Google Scholar]
- 46.Wang XL, et al. Additive effects of the risk alleles of PNPLA3 and TM6SF2 on non-alcoholic fatty liver disease (NAFLD) in a Chinese population. Front Genet. 2016;7. 10.3389/fgene.2016.00140. [DOI] [PMC free article] [PubMed]
- 47.Yang HH, et al. A novel index including SNPs for the screening of nonalcoholic fatty liver disease among elder Chinese: a population-based study. Medicine. 2018;97(13):e0272. [DOI] [PMC free article] [PubMed]
- 48.Zhang RN, Zheng RD, Mi YQ, Zhou D, Shen F, Chen GY, Zhu CY, Pan Q, Fan JG. APOC3 rs2070666 is associated with the hepatic steatosis independently of PNPLA3 rs738409 in Chinese Han patients with nonalcoholic fatty liver diseases. Dig Dis Sci. 2016;61(8):2284–2293. doi: 10.1007/s10620-016-4120-7. [DOI] [PubMed] [Google Scholar]
- 49.Liu ZT, Chen TC, Lu XX, Cheng J, Xie HY, Zhou L, Zheng SS. PNPLA3 I148M variant affects non-alcoholic fatty liver disease in liver transplant recipients. World J Gastroenterol. 2015;21(34):10054–10056. doi: 10.3748/wjg.v21.i34.10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai G, et al. Association between PNPLA3 rs738409 polymorphism and nonalcoholic fatty liver disease (NAFLD) susceptibility and severity: a meta-analysis. Medicine (United States). 2019;98(7):e14324. [DOI] [PMC free article] [PubMed]
- 51.Li J, et al. Effect of the patatin‐like phospholipase domain containing 3 gene (PNPLA3) I148M polymorphism on the risk and severity of nonalcoholic fatty liver disease and metabolic syndromes: A meta-analysis of paediatric and adolescent individuals. Pediatric Obesity. 2020;15(6):e12615. [DOI] [PubMed]
- 52.Honda Y, Yoneda M, Kessoku T, Ogawa Y, Tomeno W, Imajo K, Mawatari H, Fujita K, Hyogo H, Ueno T, Chayama K, Saito S, Nakajima A, Hotta K. Characteristics of non-obese non-alcoholic fatty liver disease: effect of genetic and environmental factors. Hepatol Res. 2016;46(10):1011–1018. doi: 10.1111/hepr.12648. [DOI] [PubMed] [Google Scholar]
- 53.Sood V, Khanna R, Rawat D, Sharma S, Alam S, Sarin SK. Study of family clustering and PNPLA3 gene polymorphism in pediatric non alcoholic fatty liver disease. Indian Pediatr. 2018;55(7):561–567. doi: 10.1007/s13312-018-1297-1. [DOI] [PubMed] [Google Scholar]
- 54.Chen LZ, Xin YN, Geng N, Jiang M, Zhang DD, Xuan SY. PNPLA3 I148M variant in nonalcoholic fatty liver disease: demographic and ethnic characteristics and the role of the variant in nonalcoholic fatty liver fibrosis. World J Gastroenterol. 2015;21(3):794–802. doi: 10.3748/wjg.v21.i3.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets are available through the corresponding author upon reasonable request.