Abstract
Purpose:
The persistence of HIV-1-infected cells, despite the introduction of the combinatorial antiretroviral therapy (cART), is a major obstacle to HIV-1 eradication. Understanding the nature of HIV reservoir will lead to novel therapeutic approaches for the functional cure or eradication of the virus. In this review, we will update the recent development in imaging applications towards HIV-1/SIV viral reservoirs research and highlight some of their limitations.
Recent Findings:
CD4 T cells are the primary target of HIV-1/SIV and the predominant site for productive and latent reservoirs. This viral reservoir preferentially resides in lymphoid compartments that are difficult to access, which renders sampling and measurements problematical and a hurdle for understanding HIV-1 pathogenicity. Novel non-invasive technologies are needed to circumvent this and urgently help to find a cure for HIV-1. Recent technological advancements have had a significant impact on the development of imaging methodologies allowing the visualization of relevant biomarkers with high resolution and analytical capacity. Such methodologies have provided insights into our understanding of cellular and molecular interactions in health and disease.
Keywords: HIV-1, reservoir, lymphoid organs, TFH, tissue, imaging
Summary:
Imaging of the HIV-1 reservoir can provide significant insights for the nature (cell types), spatial distribution and the role of the tissue microenvironment for its in vivo dynamics and potentially lead to novel targets for the virus elimination.
Introduction
Although the introduction of cART has significantly improved the life expectancy and the quality of life of HIV-infected subjects, there is still need for novel strategies aiming at elimination of HIV. The existence of a pool of infected cells (viral reservoir) allowing the long-lasting preservation of replication-competent HIV-1 in cART donors poses a major obstacle for viral eradication. Understanding the nature of HIV-1 reservoir, where only a minority of the infected cells harbor replication-competent provirus (1), as well as the dynamic interplay between HIV-1 and host cells in the reservoir microenvironment is a prerequisite for the designing of novel strategies to eliminate the virus. Upon transmission, HIV-1 persists in the mucosal tissues, within days spreads to the lymphoid organs and to the whole body through circulation after 1–2 weeks (2). Lymphoid tissues (e.g., spleen, thymus, lymph nodes (LN), gut-associated lymphoid tissue (GALT)) represent the main anatomical sites of HIV-1 reservoir (2). LNs and gut mucosa contain the highest frequencies of infected cells and level of viral replication (3). HIV-infected cells have also been detected in the brain, bone marrow, lungs, kidney, liver, adipose tissue and genitourinary tract (4). Besides harboring HIV-1 reservoir, however, LNs and other secondary lymphoid organs represent the major sites for the development of the systemic adaptive immune response to the virus. Therefore, investigation of lymphoid organ microenvironment (tissue architecture, cellularity, cytokine/chemokine milieu and soluble mediators of cell-cell communication) will improve significantly our understanding of HIV-1 reservoir persistence and inform the development of novel strategies for the attack and elimination of infected cells.
LNs as a model for HIV-1 reservoir investigation
LNs are “ecosystems” characterized by a unique architecture, high cellular diversity and compartmentalization of cell types and soluble mediators ensuring the orchestrated function of specific cell types and signals ultimately leading to the optimal cellular and humoral responses to pathogens. Regarding the maintenance of the viral reservoir, follicles and germinal centers (GC) are considered to be immunologically privileged areas meaning they are protected from damage caused by local inflammation and recruitment of effector immune cells (5). The presence of NK (6) and CD8 T (7) cells within follicles, particularly germinal centers, and the expression of cytolytic proteins are limited during homeostasis. It is well established that CD8 T cells play a significant role in controlling viral replication within lymphoid tissues (8). Significant infiltration of activated, effector (GrzB positive) CD8+ T cells into the LN and follicles/GCs associated with FDC network disruption occurs in chronic infection (7, 9{Petrovas, 2017 #22). Increased numbers of follicular CD8 T cells were found even in ‘intact’ mature follicles in chronic SIV infection suggesting that this is not a passive phenomenon facilitated by the destruction of the T/B cell borders (10). The different expression profiles of perforin and granzyme B in circulating compared to LN HIV-specific CD8 T cells suggest that lymphoid tissue CD8+ T cells may employ alternative mechanisms to attach infected cells in LNs. Administration of engineered antibodies (e.g. bispecific antibodies recognizing TCR and broad neutralizing antibody) (11) could, however, take advantage of this extensive prevalence of follicular CD8 T cells and be used as alternative intervention for killing infected cells (7, 10). Therefore, the follicular/GC areas are not representing immunologically privileged areas anymore in chronic HIV infection, a process with direct consequences for the maintenance of the viral reservoir and possible immune interventions for its elimination. To this end, the understanding of the molecular mechanisms driving the aforementioned dynamics of effector CD8 T cells is of special interest.
TFH cells represent an important cellular reservoir.
It is know well established that TFH cells represent a potential sanctuary for HIV-1/SIV replication and viral persistence (12–17), making them an important target for interventions aiming to eliminate the virus. TFH cells are characterized by a unique biology, manifested by their distinct phenotype, transcriptome and molecular pathway signature (18–20) as well as their location in an area with limited access of effector antiviral mechanisms (e.g. HIV-specific CD8 T cells) (8, 21). The detection of HIV-1 mRNA as well as HIV-1 DNA positive TFH cells suggests that they can harbor both actively productive and latent virus and at frequencies higher to those found in non-TFH cells (17). Infection at a pre-TFH stage of differentiation, before their migration to the germinal center (22, 23) as well as in situ infection, facilitated by their continuous exposure to concentrated virus/virions trapped on FDC and GC B cells (24), could contribute to their establishment as a major reservoir site. Chronic HIV/SIV infection is associated with significant accumulation of TFH cells (17, 20) further increasing their importance for the reservoir maintenance. Unfortunately, the fate (distribution across the body) and phenotype of memory human TFH cells is not well known, therefore their contribution to overall viral reservoir is difficult to be estimated.
The role of tissue microenvironment
HIV infection is associated with dramatic changes in tissue stromal cells/structure, inflammation and innate immunity (25) and cytokine expression (26). Furthermore, TFH cells are exposed to a milieu of combined immunosuppression and antigenic stimulation by HIV-1. These are biological parameters with presumably significant impact in the TFH cell dynamics (differentiation, trafficking, survival). Cytokines expressed in the extra-follicular (IL10) and intrafollicular (IL10, IL4) areas can affect the survival of TFH cells (27), (28). The glycolytic background of TFH cells (29), at least for some of their subsets, associated with increased expression of Glut 1 (30) may facilitate their infection in a hypoxic environment like the GC (31) where access of some antiretroviral drugs is limited (32). On the other hand, the relatively low expression of CCR5, associated with high levels of CXCR4 in TFH cells, in combination with the reduced production of CXCL12, could contribute to the emergence of viruses using the CXCR4 receptor in chronic HIV disease (33). Therefore, better understanding of the tissue microenvironment surrounding TFH cells will reveal cellular and molecular biofactors with possible significant impact on the reservoir maintenance.
How do we investigate the tissue microenvironment?
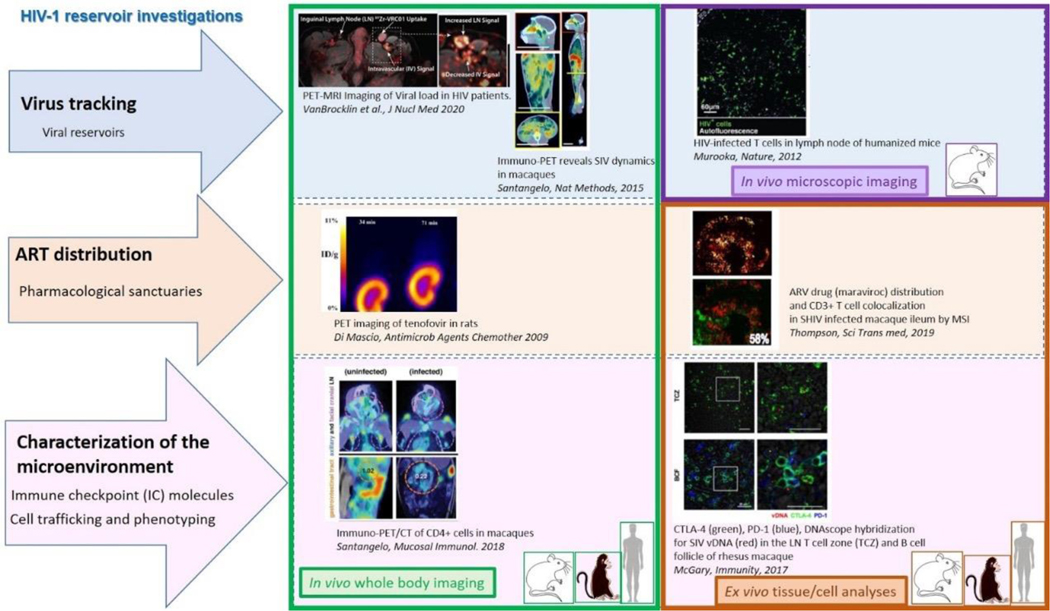
The comprehensive analysis of any tissue microenvironment (cells, soluble factors, structure elements, local operating signals/molecular pathways) requires the application of complementary methodologies that can inform for the phenotype, molecular profile, function and spatial positioning of relevant cells. To this end, imaging platforms are of great importance. An imaging platform should facilitate the simultaneous detection of several biomarkers (multiplexing) with high resolution and volume. Furthermore, the imaging/analysis of different types of molecules (mRNA, DNA, proteins, lipids, metabolites) should be components of a comprehensive analysis of the viral reservoir microenvironment. By allowing the simultaneous detection of biomarkers at a single focal plane, multiplexing reduces the possibility for interpretive errors, particularly for the analysis of rare cell populations like the HIV-1 infected cells, while preserves valuable tissue material for further analysis. High resolution/volume allows for the detailed analysis of cell-cell interactions, participating molecules, and intracellular events with respect to the expression of viral proteins or viral mRNA/DNA. The recent technological advancements provide opportunities for imaging methodologies allowing the tracking of specific cell types and virus across the body (e.g. PETscan based assays) combined with the in vivo recording of cellular dynamics (two-photon based assays) and their comprehensive illustration of their spatial topology (tissue section imaging, e.g. by scanning confocal microscopy) (Figure 1).
Figure 1. Imaging approaches for HIV reservoir investigations.
HIV/SIV virus tracking can be performed by either the administration of fluorescent or radiolabeled probe (antibody or antibody fragment) for in vivo whole body PET imaging (40, 66) or in vivo microscopic imaging; or by tracking the expression of an imaging reporter gene within the viral construct (48). The distribution of ARV molecules is possible directly in vivo with the constraint of the development needed in radiochemistry to obtain a radiolabeled antiretroviral molecule (67). At the tissue scale, ex vivo quantitative imaging modalities such as the MSI-MALDI have showed their utility for evaluating the distribution of the ARV with the advantage of the possible combination with others imaging approaches allowing the multiplexing characterization of the microenvironment, with ISH for viral RNA or DNA detection, or IHF/IHC for the cell phenotyping (68) (69). At the whole-body scale, the characterization of the microenvironment (cell trafficking, expression of immune checkpoints) could also be performed by in vivo microscopic scale and also by PET imaging after administration of appropriate labeled probes (42).
Tissue Imaging Platforms
Major advancements have taken place in the field of imaging over the last decade. These technological breakthroughs have occurred at multiple levels; i) development of new hardware and software for image acquisition, processing and computational analysis, ii) characterization of numerous antibodies appropriate for multiplex staining and iii) invention of novel probes (e.g. fluorochrome- and metal ion-based probes). The existing imaging platforms, however, differ significantly in their spatial resolution as well as their depth penetration. The nature of the scientific question under investigation should set the criteria for resolution, type of probe(s) and level of multiplexing needed and guide the choice of the most relevant platform or combination of complementary technologies. Here, we discuss imaging tools with application in the HIV/SIV field that could provide unprecedented information for the characterization of viral reservoir.
Real time Imaging
Whole body level:
Positron Emission Tomography (PET) often associated with Computed Tomography (CT scan) or MRI is an imaging modality allowing whole-body exploration using positron emitter probes injected intravenously. Gamma rays induced by the radiotracer will be detected by PET detectors without major tissue attenuation and high sensitivity (34). PET imaging has been broadly used in clinics notably for tumor diagnosis and following diseases evolution over time. [18F]-FDG-PET (Fluorodesoxyglucose-PET) imaging brought crucial insights regarding inflammation-related diseases – notably in HIV-infected patients and preclinical SIV macaque models (35, 36). FDG-PET allowed the detection of the hypermetabolism linked to inflammation in lymphoid organs (e.g. spleen, lymph nodes, tonsils) but also in a broader panel of organs (brain, testicles, gut, glands, etc.) in HIV-infected patients. Several studies indeed revealed strong correlations between HIV-related (or SIV in macaques) organ hypermetabolism and disease or cART stage (37–39). However, FDG-PET imaging lacks specificity and may be inefficient to track latent disease or viral HIV-1/SIV reservoirs. Other PET probes –antibodies or associated fragments targeting envelop viral proteins- were described in macaques in the past few years to target more specifically virus particles (40). These anti-gp120 probes allowed the monitoring of whole-body SIV dynamics over the course of the disease and ART treatment actually in Non-Human Primates. Some pilot studies using anti-gp120 mAb VRC01 and other mAbs were also reported in HIV-infected (viremic or under ART) patients (41). These immune-PET tracers may then be crucial tools to study viral reservoirs in HIV-infected patients in the future. Tracers targeting cellular populations of interest – like CD4 T cells- were also reported to study longitudinally HIV-1/SIV related disease progression (42) and some radio-labelled antiretroviral drugs were also reported to be promising tools to study correlations between viral reservoirs and cART bio distribution (43).
In vivo tissue level.
The 2-photon microscopy is a non-linear optical imaging technique involving the simultaneously absorption of two photons in the near infrared. Unlike conventional linear microscopy, the excitation is strictly restricted to the focal plane of the objective, which significantly reduces photo damage in out-of-focus regions and tissue is thus kept intact allowing long-term imaging. Moreover, the use of longer excitation wavelengths strongly reduces scattering and absorption by biological tissues, allowing imaging to be performed at several hundred microns deep in various organs of living animals (44). The 2-photon microscopy is therefore perfectly suited for the 3D, real time visualization of cell migration and the dynamics cell interactions in a native environment at single-cell resolution (45). This imaging modality was used to show the role of perivascular macrophages in neutrophil recruitment during bacterial infection in mice skin (46). Moreover, it is possible to track in real-time the physical interactions between cells such as fluorescent-labeled CD8 T cells and dendritic cells (47). The dissemination of HIV virus expressing GFP and its impact on the behavior of infected T cells in lymph nodes of humanized mice were studied by 2-photon microscopy (48).
Despite these advantages, this technique has however some limitations: the penetration depth still remains insufficient in optically dense samples (such as skin, lymph nodes) and the applications are limited to preclinical studies especially in small animal models like mice. The 3D visualization of the interactions between the virus and fluorescent-labeled immune effector cells would permit a better understanding of the structure of reservoirs and the organization of the microenvironment.
Ex vivo tissue level spatial-omics: microscopy technologies
Laser scanning confocal microscopy (LSCM) is a versatile single-cell, 3D imaging platform offering axial (X, Y) resolutions 180–250nm and an imaging depth up to 1000 um (49). Tissue samples for confocal imaging are prepared with traditional fluorescent immunocytochemistry (IHC) methods with antibodies conjugated to specific fluorochromes or oligonucleotide probes depending on the target of interest. Compared to traditional chromogenic IHC, LSCM modalities facilitate multiplexing allowing for >65 markers to be visualized when cyclic staining protocols, such as IBEX are adopted (50). Immune cell frequencies, cell-to-cell interactions and local protein expression patterns can be determined using commercial software with pipelines such as HistoCytometry (51) and Chrysalis (52) or open-source programs such as ImageJ (53). One disadvantage of LSCM pipelines however is that they are more time-consuming compared with 2D methodologies due to their volumetric nature. As such they are better suited for research applications requiring fine mechanistic spatial interpolations and less suited for batch analysis of clinical specimens.
Among the novel 2-dimension (2D) imaging techniques, which promise improved diagnostic benefit, are based on cyclic immunofluorescence, tyramide-based mIHC/IF and epitope-targeted mass spectrometry. CO-Detection by indexing (CODEX) is a multi-parametric imaging modality using antibodies conjugated to barcodes comprised of unique oligonucleotide sequences. The assay targets specific barcodes with dye-labelled reporters through iterative cycles of imaging and removal. The analysis relies on a bundled software. Concerning multiplexing, one can image 40+ markers, and at the resolution of 260 nm. One of the main limitations is that the scanning capacity is for one slide and the whole slide imaging is possible; however is very costly and time consuming (54, 55). Recently, the use of high-dimensional time-of-flight mass cytometry (CyTOF) to identify 40+ parameters simultaneously has emerged as a technique for broad-scale immune profiling and biomarker discovery (56). The method combines high-resolution laser ablation with mass cytometry for the simultaneous evaluation of theoretically 100 biomarkers, labelled with metal-tagged antibodies, with single cell resolution of 0.5 μm and spatial resolution. With regards to the analysis, with the recommended software HistoCat, the cell segmentation remains the most challenging process in multiplexed image analysis as the cell borders are not always clearly defined and individual pixels may contain information from more than one cell. The number of slides that can be imaged might also be limited (57, 58). Acquisition of imaging data from sequential labeled tissue sections is a way to overcome the limitation of volumetric analysis using a 2D platform. Although several programs facilitate the alignment and registration of sequential sections one should keep in mind that this a challenging process, depending on the required resolution (e.g. pixel-by-pixel alignment).
Volumetric detection of pathogen RNA/DNA or cell-specific transcripts is also possible through fluorescent in-situ hybridization (FISH) (Figure 2). One such methodology, RNAscope (59) pairs oligonucleotide probes with a custom hybridization-based signal amplification system for iterative detection of mRNA transcripts. Multiplexing LSCM with FISH can therefore increase significantly the dimensionality of parameters derived from a single tissue section. The Nanostring GeoMx digital spatial profiling (DSP) combines spatial and molecular profiling technologies, as it allows high-plex (30+) spatial detection of proteins and RNA using oligonucleotides detection, through barcodes that are conjugated to antibodies using a photocleavable UV light–sensitive linker. The oligo barcodes undergo quantitative analysis and are mapped back to tissue to allow spatial profiling at the defined regions of interest. There are limitations of the DSP platform, for instance, profiling every cell at single –cell resolution of 10–20μm, may be impractical, as there is not a whole image reconstruction (60–62).
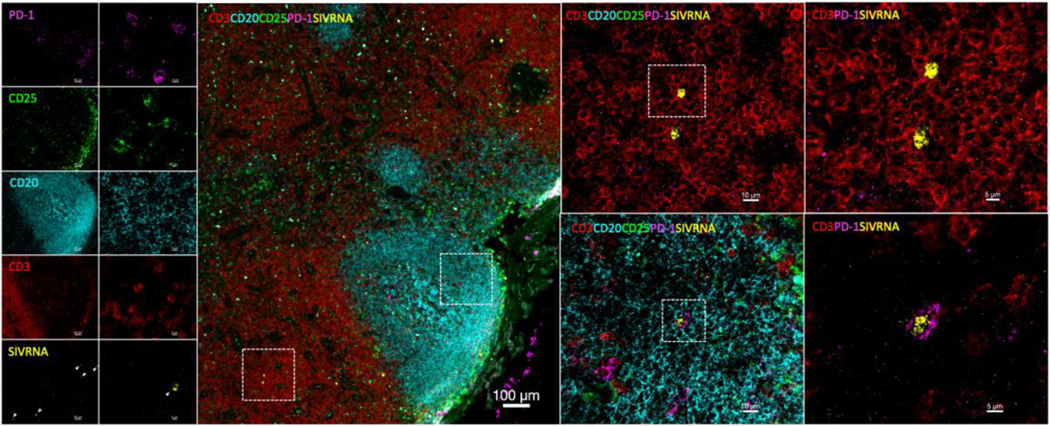
Figure 2. Detection of cell harboring actively transcribed virus using the RNAscope platform.
Representative example of vRNA+ staining in a viremic NHP LN tissue section following RNAscope FISH and immunofluorescent histochemistry. T cells harboring actively transcribed virus and B-cell follicles are detected and delineated by staining for the lineage specific markers CD3 (red) and CD20 (cyan). The markers CD25 (green) and PD-1 (magenta) are also used to track activated T-cells and Tfh (CD3hiPD-1hi cells within CD20hi/dim areas) irrespective of potential CD4 downregulation. Examples of infected T-cells in the T-cell area (CD3hi PD-1lo) and an infected Tfh (CD3hiPD-1hi) in the CD20hi/dim area are given in the zoomed in panels. Images were acquired on a Leica TCS SP8 confocal microscope using a 63x objective (NA 1.4). Scale bars are 100um (main merged figure), 10um and 5um.
The aforementioned methodologies rely on the availability of relevant probes for the molecules under investigation (either antibodies or mRNA/DNA probes). For the comprehensive analysis of the reservoir microenvironment, however, the investigation of other type of molecules, in a ‘non-hypothesis driven’ mode should be considered too. Imaging Mass-Spectrometry (IMS) methodologies support higher levels of multiplexing compared with microscopy-based methodologies as they are not limited by optics but have a lower overall sensitivity for proteins (63),(64). Preparation of tissues for IMS, such as MALDI-TOF IMS requires the application of an organic chemical matrix that aids in analyte desorption and ionization for visualization of a diverse range of biomarkers with broad mass ranges most notably lipids, drugs and metabolites at resolutions of 5–20um (65). Following acquisition detected mass signatures can be mapped to specific tissue pixels using commercially available software such as SCiLS Lab to create 2D maps rich in analyte information that facilitate the discovery of novel tissue-specific spatial patterns.
Future directions
Ideally, the characterization of HIV-1 reservoir at tissue level should include information related to cells harboring the virus (e.g. operation of particular intracellular signaling pathways, expression of specific transcription factors) and their surrounding microenvironment. This type of characterization sets the requirement for high resolution, volume and ability for imaging of a wide spectrum of molecules. Imaging of latent vs actively transcribed virus is a challenging process. The development/application of sophisticated probes targeting spicing isoforms of viral protein mRNAs can increase the confidence for detection of actively transcribed virus. The combined detection of mRNA, DNA and viral proteins is an alternative way for more accurate characterization of infected cells. In addition to acquisition of high dimensional data, there is need for the development of computational tools allowing i) the detailed characterization of the reservoir topology (frequencies, numbers and relative positioning/distances of particular cellular subsets) and ii) the integration of data generated by different platforms (e.g. imaging and sequencing data sets). This type of analysis will provide specific, personalized HIV-1 reservoir tissue signatures. Given the limited access to relevant human tissues, the non-human primate SIV model would be of special interest in this regard.
Key Points:
- HIV-1 reservoir preferentially resides in lymphoid compartments.
- Understanding the nature and topology of HIV reservoir is critical for the development of novel strategies to eliminate the virus.
- Comprehensive understanding of viral reservoir requires imaging methodologies allowing the acquisition of data with high resolution, dimension and volume.
- Current imaging technologies allow the tracking of virus and relevant cells at whole body, tissue and cellular level.
- Imaging technologies can inform for the nature of the reservoir as well as the possible role of the tissue microenvironment.
Acknowledgements
The authors would like to thank Dr Roger Le Grand and Dr Nabila Seddiki for their critical review of the current manuscript and the insightful suggestions. Part of this research was supported by CHUV institutional funds, the Intramural Research Program of the Vaccine Research Center, NIAID, National Institutes of Health, the European Union IMI2 program Immune Image (grant agreement No 831514), the National Research Agency (ANR; ImaCovPrim) and the Program ANRS RHIVIERA WP2. The Infectious Disease Models and Innovative Therapies (IDMIT) research infrastructure is supported by the ‘Programme Investissements d’Avenir’, managed by the ANR under reference ANR-11-INBS-0008.
Footnotes
Conflict of interest:
The authors declare no competing financial interests.
References
- 1.Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, Swanstrom R. HIV reservoirs: what, where and how to target them. Nat Rev Microbiol. 2016;14(1):55–60. [DOI] [PubMed] [Google Scholar]
- 2.Chaillon A, Gianella S, Dellicour S, Rawlings SA, Schlub TE, De Oliveira MF, et al. HIV persists throughout deep tissues with repopulation from multiple anatomical sources. J Clin Invest. 2020;130(4):1699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohn LB, Chomont N, Deeks SG. The Biology of the HIV-1 Latent Reservoir and Implications for Cure Strategies. Cell Host Microbe. 2020;27(4):519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton K, Winckelmann A, Palmer S. HIV-1 Reservoirs During Suppressive Therapy. Trends Microbiol. 2016;24(5):345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong S, Van Kaer L. Immune privilege: keeping an eye on natural killer T cells. J Exp Med. 1999;190(9):1197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferlazzo G, Pack M, Thomas D, Paludan C, Schmid D, Strowig T, et al. Distinct roles of IL-12 and IL-15 in human natural killer cell activation by dendritic cells from secondary lymphoid organs. Proc Natl Acad Sci U S A. 2004;101(47):16606–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrovas C, Ferrando-Martinez S, Gerner MY, Casazza JP, Pegu A, Deleage C, et al. Follicular CD8 T cells accumulate in HIV infection and can kill infected cells in vitro via bispecific antibodies. Sci Transl Med. 2017;9(373).** This is the first study to show a more efficient ex vivo bispecific antibody mediated killing of infected targets by follicular compared to non-follicular CD8 T cells in chronic HIV infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med. 2015;21(2):132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tenner-Racz K, Racz P, Thome C, Meyer CG, Anderson PJ, Schlossman SF, et al. Cytotoxic effector cell granules recognized by the monoclonal antibody TIA-1 are present in CD8+ lymphocytes in lymph nodes of human immunodeficiency virus-1-infected patients. Am J Pathol. 1993;142(6):1750–8. [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrando-Martinez S, Moysi E, Pegu A, Andrews S, Nganou Makamdop K, Ambrozak D, et al. Accumulation of follicular CD8+ T cells in pathogenic SIV infection. J Clin Invest. 2018;128(5):2089–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabozzi G, Pegu A, Koup RA, Petrovas C. Bispecific antibodies: Potential immunotherapies for HIV treatment. Methods. 2019;154:118–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aid M, Dupuy FP, Moysi E, Moir S, Haddad EK, Estes JD, et al. Follicular CD4 T Helper Cells As a Major HIV Reservoir Compartment: A Molecular Perspective. Front Immunol. 2018;9:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, et al. PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med. 2016;22(7):754–61. [DOI] [PubMed] [Google Scholar]
- 14.Lindqvist M, van Lunzen J, Soghoian DZ, Kuhl BD, Ranasinghe S, Kranias G, et al. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J Clin Invest. 2012;122(9):3271–80.** The first study to show the accumulation of TFH cells in chronic HIV infection, a profile associated with increased germinal center reactivity and dyregulated production of IgG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Pond SLK, et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature. 2016;530(7588):51–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pantaleo G, Graziosi C, Demarest JF, Butini L, Montroni M, Fox CH, et al. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature. 1993;362(6418):355–8. [DOI] [PubMed] [Google Scholar]
- 17.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. 2013;210(1):143–56.** One of the first studies to show that TFH cells are a major reservoir for replication competent HIV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crotty S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity. 2019;50(5):1132–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular Helper T Cells. Annu Rev Immunol. 2016;34:335–68. [DOI] [PubMed] [Google Scholar]
- 20.Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, et al. CD4 T follicular helper cell dynamics during SIV infection. J Clin Invest. 2012;122(9):3281–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connick E, Folkvord JM, Lind KT, Rakasz EG, Miles B, Wilson NA, et al. Compartmentalization of simian immunodeficiency virus replication within secondary lymphoid tissues of rhesus macaques is linked to disease stage and inversely related to localization of virus-specific CTL. J Immunol. 2014;193(11):5613–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Weatherall C, Bailey M, Alcantara S, De Rose R, Estaquier J, et al. Simian immunodeficiency virus infects follicular helper CD4 T cells in lymphoid tissues during pathogenic infection of pigtail macaques. J Virol. 2013;87(7):3760–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H, Wang X, Malam N, Aye PP, Alvarez X, Lackner AA, et al. Persistent Simian Immunodeficiency Virus Infection Drives Differentiation, Aberrant Accumulation, and Latent Infection of Germinal Center Follicular T Helper Cells. J Virol. 2016;90(3):1578–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heesters BA, Lindqvist M, Vagefi PA, Scully EP, Schildberg FA, Altfeld M, et al. Follicular Dendritic Cells Retain Infectious HIV in Cycling Endosomes. PLoS Pathog. 2015;11(12):e1005285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Estes JD. Pathobiology of HIV/SIV-associated changes in secondary lymphoid tissues. Immunol Rev. 2013;254(1):65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biancotto A, Grivel JC, Iglehart SJ, Vanpouille C, Lisco A, Sieg SF, et al. Abnormal activation and cytokine spectra in lymph nodes of people chronically infected with HIV-1. Blood. 2007;109(10):4272–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vella AT, Dow S, Potter TA, Kappler J, Marrack P. Cytokine-induced survival of activated T cells in vitro and in vivo. Proc Natl Acad Sci U S A. 1998;95(7):3810–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribeiro SP, Aid M, Dupuy FP, Chan CN, Hultquist J, Delage C, et al. IL-10 driven memory T cell survival and Tfh differentiation promote HIV persistence. bioRxiv. 2021:2021.02.26.432955. [Google Scholar]
- 29.Zeng H, Cohen S, Guy C, Shrestha S, Neale G, Brown SA, et al. mTORC1 and mTORC2 Kinase Signaling and Glucose Metabolism Drive Follicular Helper T Cell Differentiation. Immunity. 2016;45(3):540–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padhan K, Moysi E, Noto A, Chassiakos A, Ghneim K, Shah S, et al. Human follicular CD4 T cell function is defined by specific molecular, positional and TCR dynamic signatures. bioRxiv. 2020:2020.05.12.089706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loisel-Meyer S, Swainson L, Craveiro M, Oburoglu L, Mongellaz C, Costa C, et al. Glut1-mediated glucose transport regulates HIV infection. Proc Natl Acad Sci U S A. 2012;109(7):2549–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A. 2014;111(6):2307–12.* This study demonstrates compromised concentration of antiretroviral drugs in the lymph nodes which is associated with persistent replication of HIV and raised the need for novel therapeutics that could penetrate such anatomical sites with higher efficiency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez N, Bermejo M, Calonge E, Jolly C, Arenzana-Seisdedos F, Pablos JL, et al. SDF-1/CXCL12 production by mature dendritic cells inhibits the propagation of X4-tropic HIV-1 isolates at the dendritic cell-T-cell infectious synapse. J Virol. 2010;84(9):4341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slomka PJ, Pan T, Germano G. Recent Advances and Future Progress in PET Instrumentation. Semin Nucl Med. 2016;46(1):5–19. [DOI] [PubMed] [Google Scholar]
- 35.Scharko AM, Perlman SB, Pyzalski RW, Graziano FM, Sosman J, Pauza CD. Whole-body positron emission tomography in patients with HIV-1 infection. Lancet. 2003;362(9388):959–61. [DOI] [PubMed] [Google Scholar]
- 36.Scharko AM, Perlman SB, Hinds PWn, Hanson JM, Uno H, Pauza CD. Whole body positron emission tomography imaging of simian immunodeficiency virus-infected rhesus macaques. Proc Natl Acad Sci U S A. 1996;93(13):6425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brust D, Polis M, Davey R, Hahn B, Bacharach S, Whatley M, et al. Fluorodeoxyglucose imaging in healthy subjects with HIV infection: impact of disease stage and therapy on pattern of nodal activation. AIDS. 2006;20(7):985–93. [DOI] [PubMed] [Google Scholar]
- 38.Sathekge M, Maes A, Kgomo M, Van de Wiele C. Fluorodeoxyglucose uptake by lymph nodes of HIV patients is inversely related to CD4 cell count. Nucl Med Commun. 2010;31(2):137–40. [DOI] [PubMed] [Google Scholar]
- 39.Henrich TJ, Hsue PY, VanBrocklin H. Seeing Is Believing: Nuclear Imaging of HIV Persistence. Front Immunol. 2019;10:2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santangelo PJ, Rogers KA, Zurla C, Blanchard EL, Gumber S, Strait K, et al. Whole-body immunoPET reveals active SIV dynamics in viremic and antiretroviral therapy-treated macaques. Nat Methods. 2015;12(5):427–32.** The first report regarding the use of a SIV anti-Gp120 antibody as a tracer for the real time, iv vivo imaging of viral dynamics in non-human primates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMahon JH, Zerbato JM, Lau JSY, Lange JL, Roche M, Tumpach C, et al. A clinical trial of non-invasive imaging with an anti-HIV antibody labelled with copper-64 in people living with HIV and uninfected controls. EBioMedicine. 2021;65:103252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santangelo PJ, Cicala C, Byrareddy SN, Ortiz KT, Little D, Lindsay KE, et al. Early treatment of SIV+ macaques with an alpha4beta7 mAb alters virus distribution and preserves CD4(+) T cells in later stages of infection. Mucosal Immunol. 2018;11(3):932–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bray M, Di Mascio M, de Kok-Mercado F, Mollura DJ, Jagoda E. Radiolabeled antiviral drugs and antibodies as virus-specific imaging probes. Antiviral Res. 2010;88(2):129–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller MJ, Wei SH, Parker I, Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 2002;296(5574):1869–73. [DOI] [PubMed] [Google Scholar]
- 45.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2(12):932–40. [DOI] [PubMed] [Google Scholar]
- 46.Abtin A, Jain R, Mitchell AJ, Roediger B, Brzoska AJ, Tikoo S, et al. Perivascular macrophages mediate neutrophil recruitment during bacterial skin infection. Nat Immunol. 2014;15(1):45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bousso P, Robey E. Dynamics of CD8+ T cell priming by dendritic cells in intact lymph nodes. Nat Immunol. 2003;4(6):579–85. [DOI] [PubMed] [Google Scholar]
- 48.Murooka TT, Deruaz M, Marangoni F, Vrbanac VD, Seung E, von Andrian UH, et al. HIV-infected T cells are migratory vehicles for viral dissemination. Nature. 2012;490(7419):283–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moysi E, Estes JD, Petrovas C. Novel Imaging Methods for Analysis of Tissue Resident Cells in HIV/SIV. Curr HIV/AIDS Rep. 2016;13(1):38–43. [DOI] [PubMed] [Google Scholar]
- 50.Radtke AJ, Kandov E, Lowekamp B, Speranza E, Chu CJ, Gola A, et al. IBEX: A versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc Natl Acad Sci U S A. 2020;117(52):33455–65.** This study describes a high dimension, repetitive staining and chemical bleaching method that allows the detection of more than >65 biomarkers in the same tissue section without physical degradation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37(2):364–76.** The first description of Histo-cytometry, a method for quantitative analysis of imaging data using flow cytometry platform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kotov DI, Pengo T, Mitchell JS, Gastinger MJ, Jenkins MK. Chrysalis: A New Method for High-Throughput Histo-Cytometry Analysis of Images and Movies. J Immunol. 2019;202(1):300–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18(1):529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parra ER, Francisco-Cruz A, Wistuba II State-of-the-Art of Profiling Immune Contexture in the Era of Multiplexed Staining and Digital Analysis to Study Paraffin Tumor Tissues. Cancers (Basel). 2019;11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan WCC, Nerurkar SN, Cai HY, Ng HHM, Wu D, Wee YTF, et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun (Lond). 2020;40(4):135–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hartmann FJ, Bendall SC. Immune monitoring using mass cytometry and related high-dimensional imaging approaches. Nat Rev Rheumatol. 2020;16(2):87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gadalla R, Noamani B, MacLeod BL, Dickson RJ, Guo M, Xu W, et al. Validation of CyTOF Against Flow Cytometry for Immunological Studies and Monitoring of Human Cancer Clinical Trials. Front Oncol. 2019;9:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ijsselsteijn ME, van der Breggen R, Farina Sarasqueta A, Koning F, de Miranda N. A 40-Marker Panel for High Dimensional Characterization of Cancer Immune Microenvironments by Imaging Mass Cytometry. Front Immunol. 2019;10:2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deleage C, Wietgrefe SW, Del Prete G, Morcock DR, Hao XP, Piatak M Jr.,, et al. Defining HIV and SIV Reservoirs in Lymphoid Tissues. Pathog Immun. 2016;1(1):68–106.* The first study to describe the use of RNAscope for the detection of HIV and SIV at tissue level. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee HJ, Lee JJ, Song IH, Park IA, Kang J, Yu JH, et al. Prognostic and predictive value of NanoString-based immune-related gene signatures in a neoadjuvant setting of triple-negative breast cancer: relationship to tumor-infiltrating lymphocytes. Breast Cancer Res Treat. 2015;151(3):619–27. [DOI] [PubMed] [Google Scholar]
- 61.Danaher P, Warren S, Dennis L, D’Amico L, White A, Disis ML, et al. Gene expression markers of Tumor Infiltrating Leukocytes. J Immunother Cancer. 2017;5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Millar E, Browne L, Slapetova I, Shang F, Ren Y, Bradshaw R, et al. TILs Immunophenotype in Breast Cancer Predicts Local Failure and Overall Survival: Analysis in a Large Radiotherapy Trial with Long-Term Follow-Up. Cancers (Basel). 2020;12(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caprioli RM. Imaging Mass Spectrometry: A Perspective. J Biomol Tech. 2019;30(1):7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yagnik G, Liu Z, Rothschild KJ, Lim MJ. Highly Multiplexed Immunohistochemical MALDI-MS Imaging of Biomarkers in Tissues. J Am Soc Mass Spectrom. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neumann EK, Djambazova KV, Caprioli RM, Spraggins JM. Multimodal Imaging Mass Spectrometry: Next Generation Molecular Mapping in Biology and Medicine. J Am Soc Mass Spectrom. 2020;31(12):2401–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.VanBrocklin H, Beckford Vera D, Schulte B, Flavell R, Seo Y, Levi J, et al. <strong>Imaging viral load and T cell activation in HIV: Tools for cure development</strong>. Journal of Nuclear Medicine. 2020;61(supplement 1):540-.31562222 [Google Scholar]
- 67.Di Mascio M, Srinivasula S, Bhattacharjee A, Cheng L, Martiniova L, Herscovitch P, et al. Antiretroviral tissue kinetics: in vivo imaging using positron emission tomography. Antimicrob Agents Chemother. 2009;53(10):4086–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson CG, Rosen EP, Prince HMA, White N, Sykes C, de la Cruz G, et al. Heterogeneous antiretroviral drug distribution and HIV/SHIV detection in the gut of three species. Sci Transl Med. 2019;11(499). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGary CS, Deleage C, Harper J, Micci L, Ribeiro SP, Paganini S, et al. CTLA-4(+)PD-1(−) Memory CD4(+) T Cells Critically Contribute to Viral Persistence in Antiretroviral Therapy-Suppressed, SIV-Infected Rhesus Macaques. Immunity. 2017;47(4):776–88 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]