Dear Editor,
We identified hypermethylated PCDHGB7 as a novel cancer marker and applied it to early cervical cancer (CC) screening. It outperforms the widely implemented high‐risk human papillomavirus (hrHPV) test and ThinPrep cytologic test (TCT) and even can be used in the self‐sampled vaginal secretions, proving itself as a much more convenient yet highly effective screening method.
DNA methylation aberration occurs during cancer progression. DNA methylation has emerged as a promising diagnostic, prognostic, and predictive biomarker of various types of cancer. 1 However, the common biomarker of cancers has been rarely explored. Previously, we provided the concept of Universal Cancer Only Marker (UCOM) and identified hypermethylated HIST1H4F as the first UCOM marker. 2 In our genome‐wide methylation analysis, we found PCDH family genes were cancer cell‐differentially methylated genes (CC‐DMG). 2 In the current study, we focused on PCDHGB7, a member of the protocadherin gamma gene cluster, which plays critical roles in the establishment and function of specific neuronal connections, 3 and investigated whether it could be a novel UCOM marker. As CC is one of the most common female malignancies 4 and the widely implemented hrHPV and TCT yield a high false‐positive rate, 5 , 6 we aimed to applied PCDHGB7 in the early CC screening.
We compared the methylation status of PCDHGB7 in 17 cancer types with their corresponding normal tissues in TCGA and GEO database (n = 7114). It turned out PCDHGB7 was hypermethylated in all cancer types (Figure 1A). When analyzing FIGO staging, we found that PCDHGB7 was already hypermethylated in stage I of all cancer types analyzed (Figure S1), suggesting hypermethylated PCDHGB7 could be an early‐stage cancer indicator. Additionally, in different histological types, keratinizing squamous cell carcinoma, lymphovascular invasion, or histologic grades, there was no methylation difference of PCDHGB7 (Figure S2). To verify these analytical results, we collected 13 types of clinical cancer samples (n = 727), in which PCDHGB7 was hypermethylated accordingly (Figure 1B). Hypermethylation may account for the downregulated expression of PCDHGB7 (Figure S3) and the lower frequency of CTCF peaks located on PCDHGB7 promoter (Figure S4). Additionally, we assessed the performance of PCDHGB7 hypermethylation as a biomarker for distinguishing between cancer and normal samples. The area under the curve (AUC) values were obtained for distinguishing 13 types of clinical cancer and control tissues with pyrosequencing data (Figure 1C and Table S1). It showed that all the AUC was larger than 0.85 (Table S1), especially in biliary cancer (AUC = 0.98) and esophagus cancer (AUC = 0.99). These results highly suggested that hypermethylated PCDHGB7 can serve as a novel UCOM marker and play vital roles in CC progression.
FIGURE 1.
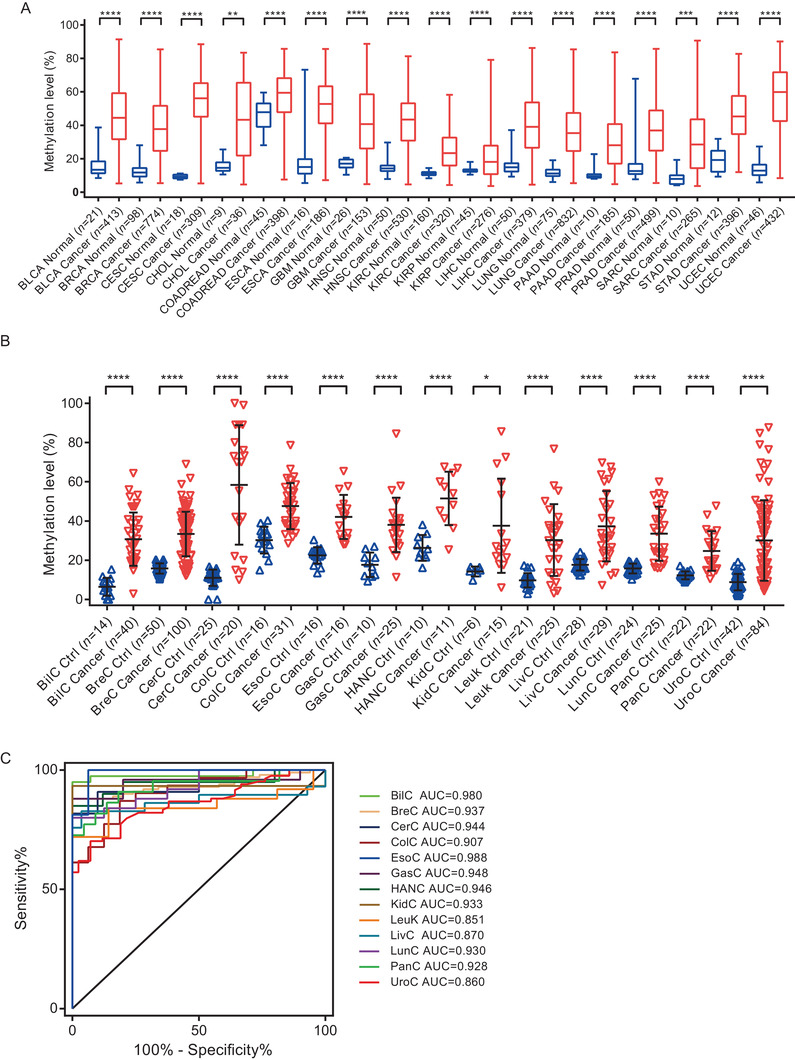
Hypermethylated PCDHGB7 is identified as a UCOM marker. (A) PCDHGB7 was hypermethylated in 17 cancer types compared with their normal tissues in TCGA databases. Box and whiskers plots were plotted; box represents the upper quartile, lower quartile, and median; whiskers represent minimum to maximum. BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COADREAD, colon adenocarcinoma and rectal adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD‐LUSC, lung adenocarcinoma and lung squamous cell carcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; SARC, sarcoma; STAD, stomach adenocarcinoma; UCEC, uterine corpus endometrial carcinoma. (B) PCDHGB7 hypermethylated was confirmed in 13 types of cancers compared with their normal tissues in clinical samples. Error bar represents upper quartile, lower quartile, and median. (C) The AUC values for distinguishing cancer from control tissues in 13 cancer types. BilC, biliary cancer; BreC, breast cancer; CerC, cervical cancer; ColC, colorectal cancer; EsoC, esophagus cancer; GasC, gastric cancer; HANC, head and neck cancer; KidC, kidney cancer; Leuk, leukemia; LivC, liver cancer; LunC, lung cancer; PanC, pancreatic cancer; UroC, urothelial cancer. In both (A) and (B), P values were calculated using the two‐tailed unpaired parametric test by GraphPad Prism 7.0. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001
The management strategies for high‐ and low‐grade squamous intraepithelial lesion (HSIL, LSIL) are distinct; hence, there is an urgent demand for distinguishing HSIL from LSIL. We found the methylation level of PCDHGB7 in HSIL or CC (defined as “≥HSIL”) was significantly higher than that in LSIL and normal samples (defined as “≤LSIL”) (Figure 2A), implying it could act as a stage divider to classify ≥HSIL from ≤LSIL stage and an early cervical precancerous lesion biomarker. To avoid bisulfite treatment in bisulfite‐PCR pyrosequencing, we modified methylation‐sensitive restriction enzyme combined real‐time fluorescent quantitative PCR (MSRE‐qPCR) to quantify methylation status. In samples with lower methylation levels (10%–20%), the value of ΔCt dropped dramatically (Figure 2B), indicating MSRE‐qPCR was superior for early cancer screening since less cancerous DNA existed alongside relatively lower methylation level. In 404 cervical smears, ΔCt for quantified PCDHGB7 methylation was significantly lower in ≥HSIL compared with that in ≤LSIL (Figure 2C). Furthermore, the ROC curve showed that MSRE‐qPCR quantification of PCDHGB7 methylation could be used for classifying CC and distinguishing HSIL from ≤LSIL samples. The AUC was 0.97 for CC, 0.87 for HSIL, and 0.88 for ≥HSIL (Figure 2D). With the methylation cutoff ΔCt = 4.0 when the Youden index is maximized (ΔCt ≤ 4.0 indicates ≥HSIL; ΔCt > 4.0 indicates ≤ LSIL), the specificity was 94.3%, and the sensitivity was 96.0% for CC (Figure 2E).
FIGURE 2.
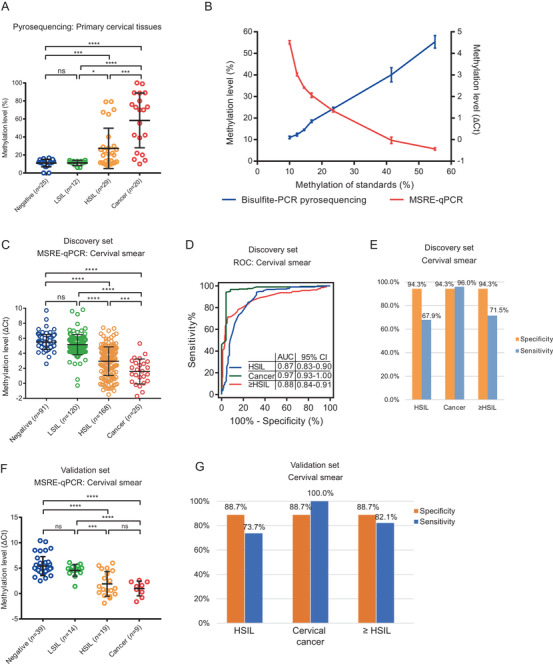
PCDHGB7 was specifically hypermethylated in cervical cancer and HSIL samples. (A) PCDHGB7 methylation level was detected by bisulfite‐PCR pyrosequencing in 86 primary cervical tissue samples. (B) The performance of bisulfite‐PCR (BS‐PCR) pyrosequencing and MSRE‐qPCR in detecting selected DNA methylation standard samples. The x‐axis indicates the DNA methylation level; seven standard samples were detected; the y‐axis in the left indicates the methylation level detected by bisulfite‐PCR pyrosequencing, the y‐axis in the right indicates the ΔCt detected by MSRE‐qPCR, and the ΔCt value reflects the DNA methylation. The repeats of pyrosequencing and MSRE‐qPCR were two and three for each grad, respectively. The mean ± SD values were plotted. (C) PCDHGB7 methylation level of 404 cervical smears in discovery set by MSRE‐qPCR. (D) The ROC curve in 404 cervical smears, and AUC values were illustrated. (E) The sensitivity and specificity of PCDHGB7 hypermethylation in HSIL, CC, and ≥HSIL group in cervical smears in discovery set. (F) PCDHGB7 methylation level of 81 cervical smears in validation set by MSRE‐qPCR. (G) The sensitivity and specificity of PCDHGB7 hypermethylation in HSIL, CC, and ≥HSIL group in cervical smears in validation set. In (A), (C), and (F), error bar represents upper quartile, lower quartile, and median. P values were calculated by the unpaired parametric test with GraphPad Prism 7.0. ns, not significant; *, P < 0.05; ***, P < 0.001; ****, P < 0.0001
Next, we comprehensively evaluated the performances of PCDHGB7 hypermethylation, hrHPV test, and TCT in CC screening (Table 1). For CC, the sensitivity of PCDHGB7 and hrHPV was similar (96% vs. 95.7%), while the specificity was improved dramatically (94.3% vs. 20.3%). It was also the case in HSIL. As for TCT, its specificity (51.2%) is much lower than that of PCDHGB7 in CC and HSIL samples. Furthermore, we evaluated the combined effect of PCDHGB7 hypermethylation, hrHPV test, and TCT. For screening clinical samples with ≥HSIL, if we define “positive” as both positive diagnosis for CC, PCDHGB7 combined with either hrHPV or TCT increased the specificity to 95.7% and 96.2%, which is higher than either of hrHPV (20.3%) or TCT (51.2%), or the combination of hrHPV and TCT (57.8%). However, the sensitivity of PCDHGB7 decreased due to these combinations. Similar results were found in three‐method combinations. These results demonstrated that hypermethylated PCDHGB7 by itself is an ideal alternative tool for CC screening, and there is no need for combining it with either hrPHV test or TCT. Additionally, the robustness of PCDHGB7 hypermethylation was also testified in the validation set, yielding 82.1% sensitivity and 88.7% specificity for ≥HSIL (Figure 2F); while the sensitivity could reach 100% with 88.7% specificity for identifying CC (Figure 2G).
TABLE 1.
Performance of PCDHGB7 methylation detection, hrHPV test, and TCT in early cervical cancer screening
| Negative | LSIL | HSIL | Cervical cancer | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample type: cervical smear | Neg/All | Per | Neg/All | Per | Pos/All | Per | Sensitivity | Specificity | PPV | NPV | Pos/All | Per | Sensitivity | Specificity | PPV | NPV |
| hrHPV Test | 31/87 | 35.6% | 9/110 | 8.2% | 155/164 | 94.5% | 94.50% | 20.30% | 49.70% | 81.60% | 22/23 | 95.7% | 95.70% | 20.30% | 12.30% | 97.60% |
| TCT (> = ASCUS) | 68/89 | 76.4% | 36/114 | 31.6% | 122/163 | 74.8% | 74.80% | 51.20% | 55.20% | 71.70% | 17/23 | 73.9% | 73.90% | 51.20% | 14.70% | 94.50% |
| DNA methylation | 89/91 | 97.8% | 110/120 | 91.7% | 114/168 | 67.9% | 67.90% | 94.30% | 90.50% | 78.70% | 24/25 | 96.0% | 96.00% | 94.30% | 66.70% | 99.50% |
| hrHPV and TCT (> = ASCUS) (any one positive as positive) | 27/89 | 30.3% | 2/112 | 1.8% | 163/165 | 98.8% | 98.80% | 14.40% | 48.70% | 93.50% | 23/23 | 100.0% | 100.00% | 14.40% | 11.80% | 100.00% |
| hrHPV and TCT (> = ASCUS) (both two positives as positive) | 72/87 | 82.8% | 43/112 | 38.4% | 114/162 | 70.4% | 70.40% | 57.80% | 57.60% | 70.60% | 16/23 | 69.6% | 69.60% | 57.80% | 16.00% | 94.30% |
| DNA methylation and hrHPV (any one positive as positive) | 31/87 | 35.6% | 8/112 | 7.1% | 163/166 | 98.2% | 98.20% | 19.60% | 50.50% | 92.90% | 25/25 | 100.0% | 100.00% | 19.60% | 13.50% | 100.00% |
| DNA methylation and hrHPV (both two positives as positive) | 89/91 | 97.8% | 111/118 | 94.1% | 106/166 | 63.9% | 63.90% | 95.70% | 92.20% | 76.90% | 21/23 | 91.3% | 91.30% | 95.70% | 70.00% | 99.00% |
| DNA methylation and TCT (> = ASCUS) (any one positive as positive) | 67/89 | 75.3% | 34/115 | 29.6% | 153/167 | 91.6% | 91.60% | 49.50% | 59.80% | 87.80% | 25/25 | 100.0% | 100.00% | 49.50% | 19.50% | 100.00% |
| DNA methylation and TCT (> = ASCUS) (both two positives as positive) | 90/91 | 98.9% | 112/119 | 94.1% | 83/164 | 50.6% | 50.60% | 96.20% | 91.20% | 71.40% | 16/23 | 69.6% | 69.60% | 96.20% | 66.70% | 96.70% |
| Methylation and TCT (> = ASCUS) and hrHPV (any one positive as positive) | 27/89 | 30.3% | 2/114 | 1.8% | 165/167 | 98.8% | 98.80% | 14.30% | 48.70% | 93.50% | 25/25 | 100.0% | 100.00% | 14.30% | 12.60% | 100.00% |
| Methylation and TCT (> = ASCUS) and hrHPV (any two positive as positive) | 71/87 | 81.6% | 40/111 | 36.0% | 149/164 | 90.9% | 90.90% | 56.10% | 63.10% | 88.10% | 23/23 | 100.0% | 100.00% | 56.10% | 20.90% | 100.00% |
| Methylation and TCT (> = ASCUS) and hrHPV (all three positive as positive) | 90/91 | 98.9% | 6/119 | 5.0% | 77/164 | 47.0% | 47.00% | 45.70% | 40.30% | 52.50% | 15/23 | 65.2% | 65.20% | 45.70% | 11.60% | 92.30% |
ASCUS, atypical squamous cells of undetermined significance; HSIL, high‐grade squamous intraepithelial lesion; LSIL, low‐grade squamous intraepithelial lesion; NPV, negative predictive values; Per., percentage; Pos, positive; PPV, positive predictive values; TCT, ThinPrep cytology test.
Despite vaginal secretion being much easier to collect than cervical smears, its capacity in CC screening has long been ignored. In 273 vaginal secretions, we found the methylation level of PCDHGB7 represented by the lowering ΔCt of MSRE‐qPCR was significantly higher in ≥HSIL than in ≤LSIL (Figure 3A). When used for distinguishing patients with CC or HSIL, the AUC were 0.92 and 0.71, respectively (Figure 3B); with 90.4% specificity and 90.9% sensitivity for identifying CC (Figure 3C), these results demonstrated that vaginal secretion is an encouraging sample type for early CC screening by applying PCDHGB7 methylation detection.
FIGURE 3.
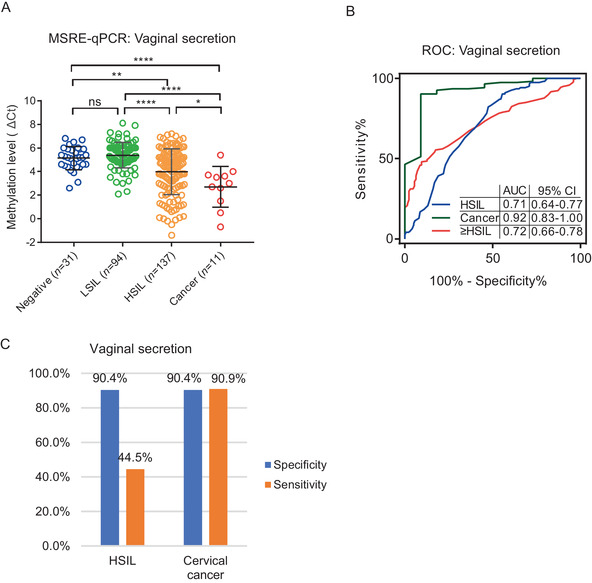
Application of hypermethylated PCDHGB7 detection for cervical cancer screening by vaginal secretions. (A, B) DNA methylation level (A), and ROC curve (B) in four stages of 273 vaginal secretions. Bars indicate the mean values. P values were calculated by the unpaired parametric test with GraphPad Prism 7.0. ns, not significance; **, P < 0.05; **, P < 0.01; ****, P < 0.0001. (C) The sensitivity and specificity of PCDHGB7 hypermethylation in HSIL and cervical cancer.
Collectively, hypermethylated PCDHGB7 is identified as a novel UCOM marker and an ideal biomarker for distinguishing HSIL from LSIL. The introduction of PCDHGB7 makes vaginal secretions feasible for CC screening, which will allow testing to be more easily applied and adopted.
CONFLICT OF INTEREST
Wenqiang Yu and Shihua Dong report having a pending patent application. The other authors disclosed no potential conflicts of interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Samples were collected from Xijing Hospital of Air Force Military Medical University, Jinshan Hospital of Fudan University, and International Peace Maternity and Child Health Hospital. Written informed consent was provided to all patients before sample collection. Institutional Review Board approval for research on human subjects was obtained from hospitals.
AUTHOR CONTRIBUTIONS
D. S. H., Y. W. Q., and L. Q. designed and initiated the project. D. S. H. and Y. W. Q. supervised the project. D. S. H., X. P., L. Q., C. L. M., D. X. L., M. Z. R., Z. B. L., Y. W. Q., and S. L. generated the data, acquired and managed patients, and provided facilities. D. S. H., X. P., and M. Z. R. performed analysis and interpretation of data. X. P., D. S. H. and Y. W. Q. wrote the manuscript. X. P. and D. S. H. drew the graphical abstract. All the authors read and approved the final manuscript.
DATA AVAILABILITY STATEMENT
The DNA methylation data are available from UCSC Xena browser (https://xenabrowser.net/), and the expression data are downloaded from TCGA Hub (https://tcga.xenahubs.net). CTCF ChIP‐Seq data were downloaded from ENCODE database.
Supporting information
Supporting information
TableS1‐S2
ACKNOWLEDGEMENTS
We thank Yue Yu for editorial help and comments on the manuscript. We thank Jiangjing Yuan at International Peace Maternity and Child Health Hospital for assistance in collecting clinical samples. This work was supported by the National Key R&D Program of China (Grant No. 2018YFC1005004), the Science and Technology Innovation Action Plan of Shanghai (Grant No. 17411950900), the National Natural Science Foundation of China (Grant No. 31671308, 31872814, 81172477, 81272295, 81402135, 81701398), Major Special Projects of Basic Research of Shanghai Science and Technology Commission (Grant No. 18JC1411101, 18JC1411104), Science and Technology Commission of Shanghai Municipality (Grant No. 12ZR1402200, 17441907400, 18411963600), the Ministry of Education of the People's Republic of China (Grant No. 2009CB825600), and the Innovation Group Project of Shanghai Municipal Health Commission (Grant No. 2019CXJQ03), Shanghai Municipal Key Clinical Specialty (Grant No. shslczdzk06302), and Shanghai Jiao Tong University Medicine‐Engineering Fund (Grant No. YG2017MS41).
Shihua Dong, Qi Lu, Peng Xu, and Limei Chen contributed equally to this work.
Contributor Information
Qi Lu, Email: hathorl@163.com.
Long Sui, Email: suilong@fudan.edu.cn.
Yudong Wang, Email: owangydong@126.com.
Wenqiang Yu, Email: wenqiangyu@fudan.edu.cn.
REFERENCES
- 1. Koch A, Joosten SC, Feng Z, et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018;15:459‐466. [DOI] [PubMed] [Google Scholar]
- 2. Dong S, Li W, Wang L, et al. Histone‐related genes are hypermethylated in lung cancer and hypermethylated HIST1H4F could serve as a pan‐cancer biomarker. Cancer Res. 2019;79:6101‐6112. [DOI] [PubMed] [Google Scholar]
- 3. Junghans D, Haas IG, Kemler R. Mammalian cadherins and protocadherins: about cell death, synapses and processing. Curr Opin Cell Biol. 2005;17:446‐452. [DOI] [PubMed] [Google Scholar]
- 4. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: gLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 5. Cuzick J, Clavel C, Petry KU, et al. Overview of the European and North American studies on HPV testing in primary cervical cancer screening. Int J Cancer. 2006;119:1095‐1101. [DOI] [PubMed] [Google Scholar]
- 6. Cuzick J, Arbyn M, Sankaranarayanan R, et al. Overview of human papillomavirus‐based and other novel options for cervical cancer screening in developed and developing countries. Vaccine;26(Suppl 10):2008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
TableS1‐S2
Data Availability Statement
The DNA methylation data are available from UCSC Xena browser (https://xenabrowser.net/), and the expression data are downloaded from TCGA Hub (https://tcga.xenahubs.net). CTCF ChIP‐Seq data were downloaded from ENCODE database.


