Graphical abstract
Keywords: dynamics, nucleocapsid, RNA, SARS-CoV, cyclophilin-A
Abstract
The nucleocapsid protein is one of four structural proteins encoded by SARS-CoV-2 and plays a central role in packaging viral RNA and manipulating the host cell machinery, yet its dynamic behavior and promiscuity in nucleotide binding has made standard structural methods to address its atomic-resolution details difficult. To begin addressing the SARS-CoV-2 nucleocapsid protein interactions with both RNA and the host cell along with its dynamic behavior, we have specifically focused on the folded N-terminal domain (NTD) and its flanking regions using nuclear magnetic resonance solution studies. Studies performed here reveal a large repertoire of interactions, which includes a temperature-dependent self-association mediated by the disordered flanking regions that also serve as binding sites for host cell cyclophilin-A while nucleotide binding is largely mediated by the central NTD core. NMR studies that include relaxation experiments have revealed the complicated dynamic nature of this viral protein. Specifically, while much of the N-terminal core domain exhibits micro-millisecond motions, a central β-hairpin shows elevated inherent flexibility on the pico-nanosecond timescale and the serine/arginine-rich region of residues 176–209 undergoes multiple exchange phenomena. Collectively, these studies have begun to reveal the complexities of the nucleocapsid protein dynamics and its preferred interaction sites with its biological targets.
Introduction
Coronaviruses (CoVs) have been circulating in human populations for at least half a century leading to the current outbreak of SARS-CoV-2,1 which provides the impetus for fully characterizing their most abundantly expressed protein, the Nucleocapsid (N) protein.2 The N protein is one of the four structural proteins that also includes the envelope (E), the membrane (M), and the spike (S) proteins and its high immunogenicity could make the N protein a viable vaccine.3 For example, with 91% identity between SARS-CoV-1 and SARS-CoV-2, the N protein is more highly conserved than the S protein that is less than 80% identical between the two CoVs. The N protein plays the central role in packaging its genomic RNA and is implicated in multiple host protein interactions that increase replication and enhance infection.4, 5 For example, the most abundant host cell proline isomerase, cyclophilin-A (CypA), is targeted by N proteins from multiple CoVs to increase viral replication, although the explicit interaction sites and mechanism remain unknown.6, 7, 8, 9 In general, CoV N proteins comprise two well-folded core domains that include an N-terminal domain (NTD) and dimerize through their C-terminal domain (CTD), yet nearly half of their approximately 400 residues are predicted to be inherently disordered. Recent structural studies reveal that the SARS-CoV-2 N protein core domains are virtually identical to their SARS-CoV-1 N counterparts10, 11, 12 and small angle X-ray scattering (SAXS) indicates that these core domains may not interact and are therefore similar to beads on a string tethered by their disordered regions.13, 14 For the SARS-CoV-1 N protein, the inherently disordered regions enhance its binding affinity to a poly-U RNA 20-mer13 and we have previously shown that a central β-hairpin within the NTD of residues 90–108 that is implicated in targeting host proteins is inherently flexible.2, 15 A recent NMR study of the SARS-CoV-2 N protein NTD of residues 44–180 also revealed that the relatively conserved central β-hairpin interacts with small nucleotide fragments.10 This elegant NMR study utilized chemical shifts to model the NTD with small nucleotide fragments, which suggests that the SARS-CoV-2 N protein β-hairpin of residues 89–107 makes direct contact with nucleotides. However, the global dynamics and role of the disordered flanking regions in binding within the SARS-CoV-2 N protein remain largely unknown.
The SARS-CoV-2 N protein is 419 residues with its NTD core comprising residues 48–175 and its CTD core dimerization domain comprising residues 251–364 (Figure 1 (a)). The serine/arginine-rich (SR) region of residues 176–209 and remaining regions are largely disordered. Several recent studies have shown that the SARS-CoV-2 N protein forms condensates with RNA.16, 17, 18 These N protein/RNA condensates are mediated by N protein binding to RNA elements at the 5′ and 3′ ends of the genomic RNA followed by N protein binding to central regions within the genomic RNA.19 Interestingly, the SR region has been shown to be instrumental in facilitating condensate formation,16 suggesting that the disordered regions of the N protein comprise novel properties that may modulate self-association.
Figure 1.
Solution behavior of N protein constructs. (a) Schematic of the SARS-CoV-2 N protein with the folded core domains that include the NTD (blue) and CTD (green) shown along with the remaining disordered regions that include the SR region (gray). The folded X-ray crystal structures are also shown for the NTD, PDB accession 6M3M, and CTD, PDB accession 6WJI. (b) Analytical Superdex-75 (23.5 ml) analysis of N protein constructs comprising the NTD (blue), the dimerization domain (green), and the full-length N protein (red). Shown is the partition coefficient (kav) versus the log of the molecular weight with five standards used to construct the standard curve. (c) Estimated correlation times for NTD-containing constructs used here based on the R1 ρ /R1 ratios derived from residues 48–178 calculated using the method of Larsson et al.20 and molecular weights calculated from a standard curve previously determined in Kendrick et al.21 All analytical sizing experiments were conducted at 4 °C and all NMR relaxation experiments were conducted at 35 °C except for an additional data set collected for N 1–209 also collected at 20 °C.
To begin addressing the interactions and dynamic behavior of the SARS-CoV-2 N protein, here we have specifically focused on the SARS-CoV-2 N protein NTD and its flanking regions using solution NMR studies. Although the folded NTD exhibits no detectable interaction with the CTD, we have discovered that the NTD flanking regions induce self-association at lower temperatures while the SR region undergoes multiple dynamic exchange processes that leads to severe line-broadening at physiological temperature. Relaxation experiments confirm that the NTD central β-hairpin is inherently flexible and reveal slower timescale motions throughout the NTD core domain. NMR titration experiments with a series of small nucleotide fragments indicate that it is the NTD core domain along with the central β-hairpin that provide the initial interactions with these nucleotide fragments, while human CypA targets the disordered flanking regions. Interestingly, unlike some interactions that lead to the quenching of dynamics upon complex formation, relaxation experiments in the presence of a bound nucleotide indicate that the inherent dynamics of the NTD alone are largely retained. These studies have therefore identified dynamics on multiple timescales of the SARS-CoV-2 N protein, identified how the disordered regions are responsible for self-association, and have begun to elucidate the preferred sites of interactions with its biological targets.
Results
Engineering and solution behavior of SARS-CoV-2 N protein subregions
With the goal of characterizing the NTD and its interactions, we initially engineered a total of seven constructs. These included the full-length N protein as a reference along with three constructs comprising the NTD core alone or with various flanking regions (referred to as N 48–178, N 1–209, N 1–178) and three constructs comprising the CTD core alone along with various flanking regions (referred to as N 251–364, N 251–419, N 210–364), which collectively span the entirety of the N protein (Figure 1(a)). An additional construct was later generated to probe the role of the SR region in the context of self-association at low temperatures that comprises the NTD core and solely the SR region (N 48–209), which is further discussed within the following sections. All engineered SARS-CoV-2 N protein constructs were purified similarly with SDS-PAGE analysis illustrating their purities (Supplementary Figure S1(a)). The hydrodynamic behavior of all seven of these engineered constructs were first assessed using size-exclusion chromatography (Figure 1(b, c) and raw elution profiles shown in Supplementary Figure S1(b, c)). Constructs that comprised the flanking regions to the NTD and CTD migrate larger than their calculated molecular weights that is consistent with their inherent disorder but also roles in self-association further revealed by NMR studies for the NTD flanking regions at low temperatures described below. In contrast, the central core domains alone that comprised solely the NTD (residues 48–178) and CTD dimer (residues 251–364) migrate lower than their calculated molecular weights that are consistent with highly compact globular structures and a lack of self-association. Backbone assignments along with NMR relaxation experiments also detailed further below afforded the calculation of their respective correlation times for the three NTD-containing constructs and subsequent estimation of their apparent molecular weights (Figure 1(c), left). Specifically, the correlation times were extracted from the averaged R1ρ/R1 ratios for residues within the NTD (residues 48–178), as described by Larsson et al.20 These correlation times were then used to estimate their respective molecular weights calculated using a correlation curve previously published21 and were remarkably similar to their calculated molecular weights (Figure 1(c), right). Thus, although our initial studies below are largely focused on the NTD-containing constructs, we show here that these specific constructs used in this study comprising either of the folded core domains are amenable to recombinant purification.
Spectral comparisons reveal weak interactions of the NTD with its flanking regions
As NMR is ideal for monitoring even weak interactions, our recombinant expression and purification of isolated regions from the SARS-CoV-2 N protein allows for the interrogation of their potential interactions (Figure 2 ). Initial titration experiments were conducted to probe interactions between NTD-containing constructs and those comprising the CTD, which confirmed no detectable interaction. Specifically, no chemical shift perturbations (CSPs) were observed for labeled N 1–209 upon titration of unlabeled CTD constructs such as N 210–364 (Figure 2(a)). This is consistent with previous SAXS data that suggested the NTD and CTD do not interact.13, 14 We then took a systematic approach to interrogate interactions of the NTD with its flanking regions by both comparing and assigning the spectra of the three initial NTD constructs produced here (Figure 1). We note that the SR region of residues 176–209 is largely undetectable at the physiological temperatures of 35 °C used here that is due to chemical exchange further explored using temperature titrations within a following section below.
Figure 2.
Spectral comparisons of NTD-containing constructs reveal its flanking disordered regions induce global changes and that there is an absence of resonances from the SR region. (a) 15N-HSQC spectra of 15N-labeled N 1–209 alone (black) and in the presence of equimolar concentrations of unlabeled N 210–364 (green). (b) Left, comparison of 15N-HSQC spectra of N 48–178 (blue) and N 1–178 (red). Right, comparison of 15N-HSQC spectra of N 1–209 (black) and N 1–178 (red). (c) Left, amide CSPs are shown between of N 48–178 and N 1–178 with dotted line indicating the average (0.052 ppm) plus half the standard deviation (0.030 ppm), which is 0.082 ppm. Right, amide CSPs are shown between N 1–209 and N 1–178 with the dotted line indicating the average (0.038 ppm) plus half the standard deviation (0.030 ppm), which is 0.068 ppm. Contributions of half the standard deviation above the averages were used, as CSPs are relatively small under these conditions. (d) Left, amide spectral differences between N 48–178 and N 1–178 are mapped onto the NTD structure include A50, G69, N126, I130, 127–141, D144, R149, and N150 (red). Right, amide spectral differences between N 1–178 and N 1–209 are mapped onto the NTD structure include A35, L56, Q58, L64, I94, S105, L159, L161, and 172–174. All spectra were collected at 35 °C at 900 MHz.
Backbone assignments of all three NTD constructs reveal that both the N-terminal residues of 1–47 and SR region of residues 179–209 induce CSPs beyond the localized proximity to which they are attached (Figure 2(b, c)). For example, mapping CSPs induced by residues 1–47 indicate subtle changes to distal residues such as residues 137–144 (Figure 2(d), left). Thus, the N-terminal region of residues 1–47 likely associates directly with the NTD. To further address the interaction of residues 1–47, we recombinantly produced a peptide that corresponds to this disordered N-terminus (i.e., N 1–47) and compared its 15N-HSQC to that of N 1–209 (Supplementary Figure S2(a)). The distinct differences between resonances of this recombinant peptide and those in the context of its attached NTD further supports its interactions with the core NTD domain (Supplementary Figure S2(b)), which were also further probed below using NMR titrations. A comparison between N 1–178 and N 1–209 also revealed CSPs induced by the SR region that include the β-hairpin (Figure 2(d), right). Although most resonances from the SR region are absent at 35 °C and we have been unsuccessful in producing a recombinant peptide that corresponds to the full SR region, a study using a synthetically constructed peptide comprising part of the SR region residues 182–197 has recently been reported.16 Specifically, this region of the SR adopts a helical propensity in solution at low temperatures.16 This helical propensity was more recently confirmed by exploiting 13C direct detection to assign a construct of residues 1–248.22
The recombinant production of the N-terminal flanking region of N 1–47 allowed us to further confirm its interaction with the remaining NTD and do so in a temperature-dependent manner (Figure 3 ). Specifically, we generated another construct of N 48–209 in order to label this for titrations of N 1–47. We discovered here that the recombinant N 1–47 specifically interacts with the NTD core with much larger CSPs at 20 °C than at 35 °C (Figure 3(a, b)). These temperature-dependent CSPs are also consistent with data below that indicates both flanking regions to the NTD induce self-association at lower temperatures and therefore likely represent a significant contribution from intermolecular interactions. While this interaction is weak in nature, these titration data also provide proof of specificity. Specifically, the CSPs induced to N 48–209 by the addition of N 1–47 recapitulates the resonance frequencies observed within the full N 1–209, illustrating that the chemical environment is similar. While the CSPs induced are consistent with the differences described above between N 1–178 and N 48–178 (Figure 3(c), left), these perturbed residues form a surface here when the separate N 1–47 is titrated to N 48–209 (Figure 3(c), right). It is important to stress once again that based on these titrations and further data below that reveals the flanking regions induce self-association, it is likely that much of these interactions are intermolecular in the context of the fully folded N protein.
Figure 3.
The recombinant N-terminal flanking region interacts with the NTD core. (a) Titration experiments of 500 μM N 48–209 with unlabeled N 1–47 were conducted at both 20 °C and 35 °C at 900 MHz with 15N-HSQC spectra of two regions shown. (b) Amide CSPs of N 48–209 in the absence and presence of 2 mM N 1–47. 20 °C that are largely abrogated at 35 °C. Dotted line indicates the average (0.03 ppm for 20 °C and 0.002 ppm for 35 °C) plus half the standard deviation (0.024 ppm for 20 °C and 0.001 ppm for 35 °C). (c) Amide CSPs greater than one standard above the average at 20 °C are mapped onto the structure and include G69, Q70, N75, N77, T135, G137, A138, N140, T141, I157.
The disordered flanking regions of the NTD induce self-association
Initial studies of N 1–209 revealed that the dispersive quality observed within 15N-HSQC spectra is markedly diminished with lower temperatures, which could be due to either self-association or cold-denaturation. Specifically, resonances of the core region of residues 48–178 largely disappear at the lower temperatures. In order to distinguish between self-association and cold-denaturation, two strategies were taken that included subjecting additional constructs to temperature titrations and CA assignments of N 1–209 at lower temperature to quantify potential changes to secondary structure.
We first addressed whether the presence of the flanking regions is the underlying cause of the reduced spectral quality at lower temperatures. Both N 1–178 that comprises the N-terminal flanking region of residues 1–47 and N 48–209 that comprises the SR region exhibit a loss of resonances that correspond to the core folded region at lower temperatures (Figure 4 (a, b)). In contrast, N 48–178 that comprises the central core does not exhibit such drastic changes in its dispersion at low temperature with the vast majority of residues still observed (Figure 4(c)). Thus, despite the disordered nature of the flanking regions, the presence of either of these results in the loss of the central core. While this suggests that the flanking regions induce self-association at lower temperatures, we next sought to rule out potential changes to the structural integrity with temperature.
Figure 4.
Self-association of the N protein NTD constructs with temperature. 15N-HSQC spectra at 900 MHz as a function of temperature for (a) of N 1–178, (b) 48–209, and (c) 48–178. (d) CA deviations from their unstructured resonances at both 20 °C and 35 °C for N 1–209. Unstructured CA values were subtracted from measured CA values, as previously reported.23 Secondary structure from the corresponding X-ray crystal structure is shown as cartoon drawings on the top together with the specific residues (PDB accession 6M3M) along with CSI-predicted β-strands (black bars) calculated from all backbone chemical shift assignments of N 1–209.45 CSI-predicted β-strands include residues 51–59, 84–95, 107–112, 129–135, 158–162, and 171–173.
Backbone chemical shifts have long been known to be sensitive measures of secondary structure23 and were therefore used here to determine whether there is any evidence of temperature-dependent unfolding for the NTD in addition to self-association. Specifically, CA resonances were made at 20 °C in order to compare with those at 35 °C for N 1–209. Even at 20 °C, many of the resonances for the core NTD are already severely line-broadened whereby the disordered flanking regions become more intense as further exploited in the next section. The secondary structure calculation using the Chemical Shift Index (CSI) at 35 °C is in agreement with the high-resolution structure of the NTD core10, 11 and these CA deviations from their random coil values at both 35 °C and 20 °C reveals no evidence of temperature-dependent unfolding (Figure 4(d)). Thus, these data suggest that the NTD self-associates in a temperature-dependent manner. There are, however, a subset of resonances that exhibit slight concentration-dependent resonance shifts at the higher temperature of 35 °C (Supplementary Figure S2(b)), thereby indicating that some weak self-association of the NTD persists even at this higher temperature. These concentration-dependent shifts are also consistent with the small CSPs induced by N 1–47 to N 48–209 at 35 °C relative to 20 °C, as well as with the specific CSPs themselves that indicates these interactions are the same (Figure 3). Self-association is therefore much weaker at the higher temperature, yet even weak self-association can affect relaxation rates, such as R2 relaxation rates, and is therefore considered below upon monitoring these. Interestingly, the SARS-CoV-1 N protein CTD dimer has been shown to self-associate into even higher order oligomers in a concentration-dependent manner as well24 and thus, our studies here could implicate further stabilizing intermolecular interactions within N proteins mediated by the NTD and its flanking regions.
The SR region exhibits exchange on multiple timescales
We next utilized temperature to explore the inherent properties of the SR region of residues 176–209 that comprises 14 serine and 7 arginine residues but was largely absent within spectra at 35 °C. Specifically, in contrast to multiple resonances that emerge within N 1–178 when compared to the central core of N 48–178, only the amides of A208 and R209 were observed when comparing N 1–178 to N 1–209 (see Figure 2(b) for spectral comparisons). Several possibilities could explain our inability to identify the majority of resonances from the SR region under these conditions (35 °C in 50 mM HEPES, pH 7.0, 150 mM NaCl), which includes fast exchange with solvent or an underlying dynamic exchange process within a timescale that leads to line broadening (on the micro-millisecond timescale, μs-ms). To rule out exchange with solvent, a comparison of N 1–178 and N 1–209 was also made at pH 5.5 to slow amide exchange (Supplementary Figure S2(c)), which still indicates an absence of new resonances that could be ascribed to the SR region. This suggests that an exchange with solvent is not the underlying cause of missing resonances from the SR region, which leads to the possibility that this region undergoes an inherent conformational exchange process that induces chemical exchange. Finally, we also tested whether the exchange of the SR region is an artifact of the truncated construct of N 1–209 by comparing spectra to the full-length N 1–419 (Supplementary Figure S2(d)). While several new resonances were present in the full-length N 1–419, these resonances arise from the C-terminal region as they are also present within N 251–419, suggesting that exchange of the SR region persists within the context of the full-length N protein. Thus, exchange within the SR region is not due to either amide exchange with solvent or due to probing only part of the N protein, but rather an intrinsic property of the SR region itself.
Because of the intrinsic properties of the disordered flanking regions in self-association described above, we were able to utilize temperature to “filter out” resonances from the folded core in order to primarily observe the disordered regions. Specifically, exchange can either be sped up with higher temperature to produce a single set of resonances averaged between sampled conformations (referred to as “fast exchange”) or slowed with lower temperature to observe resonances from a single sampled conformation (referred to as “slow exchange”). We first increased temperature, but this resulted in the appearance of only several extremely line broadened resonances within N 1–209 (Supplementary Figure S2(e)). Conversely, lowering the temperature to 0 °C where the resonances from the folded core are no longer observed revealed distinct differences between the disordered flanking regions within N 1–209 and N 1–178 whereby the SR region is finally visible (Figure 5 (a)). It is thus likely that the SR region is sampling multiple conformations within the μs-ms timescale that have significant chemical shift differences to induce the complete loss of intensities at higher temperatures and the lower temperature may shift and/or significantly shift sampled populations so that these resonances are observable. Interestingly, low temperatures were also necessary to probe the small synthetic peptide corresponding to part of the SR region,16 suggesting that exchange of the SR region even in the absence of interactions to the NTD core also occurs. By collecting HNCACB and HNcoCACB spectra at 5 °C on N 1–209, we were able to confirm that only resonances from the disordered flanking regions to the NTD that include the N-terminal residues and the SR region are visible at such low temperatures. Importantly, this allowed us to further corroborate the temperature dependence of exchange for the SR region that exhibits extreme line-broadening with higher temperatures as compared to resonances from the N-terminus (Figure 5(b), left). A quantifiable measure of this extreme line-broadening can also be illustrated by averaging the intensity ratios from those resonances that could be assigned within the SR region to resonances within the N-terminal region (Figure 5(b), right). Such a comparison of intensities between these two disordered flanking regions further illustrates how the SR region broadens within the range of 5 °C to 20 °C. Thus, despite the disordered nature of the SR region, this region undergoes an inherent exchange on the μs-ms timescale that leads to the loss of its resonances at physiological temperatures.
Figure 5.
The inherent exchange and temperature dependence of the SR region. (a) Comparison at 0 °C of 15N-HSQC spectra of N 1–209 and N 1–178 with a blow-up highlighting the differences collected at 900 MHz. (b) Temperature variation in 15N-HSQC spectra of N 1–209 at 5, 10, and 20 °C collected at 800 MHz and the associated changes to resonance intensities of the SR and N-terminal flanking regions. Along with an illustration of the 15N-HSQC spectra, the associated intensity ratios (SR/N-term ratio) are calculated from resonances within the SR region (S180, Q182, S183, G200, and S201) to those within the N-terminal region (I15, T16, S21, T24, A35). Peak heights were averaged between each group at each temperature with error bars calculated from the standard deviation. (c) Two-dimensional strips of N 1–209 at 5 °C extracted from an HNCACB (blue) and HNcoCACB (orange) at the indicated nitrogen planes with S198 and T199 resonances highlighted. Data were collected at 800 MHz. (d) 15N-HSQC spectra of N 1–209 at 20 °C alone (black) and in the presence of 250 μM (blue) and 500 μM (orange) CypA.
We also identified an additional even slower exchange process within the SR region mediated by proline isomerization and simultaneously identified the binding sites of host cell CypA. Specifically, these low temperature studies identified two new resonances that emerge within N 1–209 when compared to N 1–178 that comprise a serine-threonine pair, which were identified within HNCACB/HNcoCACB spectra at low temperature (Figure 5(c)). As there is only one such possibility within the SR region of S197-T198, the observation of two sets of resonances is immediately suggestive of proline isomerization caused by P199 isomerization. To further corroborate the existence of this cis conformation of P199, we utilized human CypA that enhances the rate of proline isomerization. Multiple viruses exploit human CypA to increase viral replication that includes CoVs thought to recruit CypA through their N proteins.7, 25, 26 Although CoV-mediated hijacking of host CypA is likely a complicated process, CypA inhibitors that target its active site reduce SARS-CoV-2 infection in vitro.9 However, this may require other unidentified host factors, such as those identified within hepatitis C.27 Here, we used CypA as a tool to further probe this proline isomerization and identify potential binding sites on the SARS-CoV-2 N protein. Specifically, we have shown that the cis conformation of CypA target sites line-broadens faster due to its higher affinity than the trans conformer.28 Thus, a similar effect on the N protein SR region was predicted upon addition of recombinant CypA. Indeed, the resonance corresponding to the minor conformation observed for T198 disappears upon addition of CypA and therefore further indicates that this resonance represents the amide of T198 in the cis conformation of P199 (Figure 5(d)). Interestingly, CSPs are also observed for both the dominant resonances of A12 and N41 within the N-terminal region that both precede proline residues, P13 and P42, respectively, indicating that there are multiple sites engaged by CypA within both flanking regions of the NTD core domain. Similar multi-site binding of host CypA to a structural protein within hepatitis C has also been reported.27 Although it is difficult to quantify affinities with such multi-site binding, their linear dependencies with added CypA indicate weak micromolar affinity interactions consistent with those observed for other viral/CypA interactions.29, 30 Thus, as proline isomerization itself is relatively slow with conformational sampling times in seconds,31 the identified proline isomerization of P199 here represents another dynamic exchange process exhibited by the SR region in addition to the μs-ms timescale dynamic described above. Moreover, the interactions of host CypA with the very same disordered flanking regions that we have discovered to modulate self-association may have wider implications for N protein-mediated condensation that is described further within the Discussion.
The inherent dynamics of the NTD include millisecond motions within its folded core and a disordered β-hairpin
R2, R1 ρ, and R1 relaxation rates collected at 35 °C on the three initially produced NTD containing constructs (N 1–209, N 1–178, and N 48–178) reveal the inherent dynamic nature of this region (Figure 6 (a–c)). Overall, both R2 and R1 ρ relaxation rates increase while R1 relaxation decreases with the addition of each flanking region relative to the NTD core (residues 48–178). Such overall rate changes are expected with their slowed tumbling due to the presence of the disordered flanking regions and their weak self-association, which are reflective in their calculated correlation times (Figure 1(c)). Relaxation rates were also collected at 20 °C for N 1–209 to confirm the expected increase in R2 and R1 ρ relaxation rates with a concomitant decrease in R1 relaxation rates due to slower tumbling (Supplementary Figure 3(a–c)), which is also reflective in the calculated correlation time (Figure 1(c)). The disordered nature of both the N-terminal residues 1–47 and the β-hairpin of residues 89–107 is readily apparent by their diminished R2 and R1 ρ relaxation rates along with their increased R1 relaxation rates, which indicate elevated ps-ns timescale motions relative to the rest of the protein. Thus, residues 89–107 are largely disordered in solution but have a propensity to form a β-hairpin, as indicated by their CSI (in Figure 4(b)) and stabilized within the X-ray crystal structures of SARS-CoV-1 and SARS-CoV-2 N protein NTDs.11, 32 These results were expected based on our previous R1 relaxation data for the SARS-CoV-1 N protein NTD core domain15 and the fact that all but one residue is conserved within this β-hairpin between SARS-CoV-1 N and SARS-CoV-2 N proteins (V96 → I94).
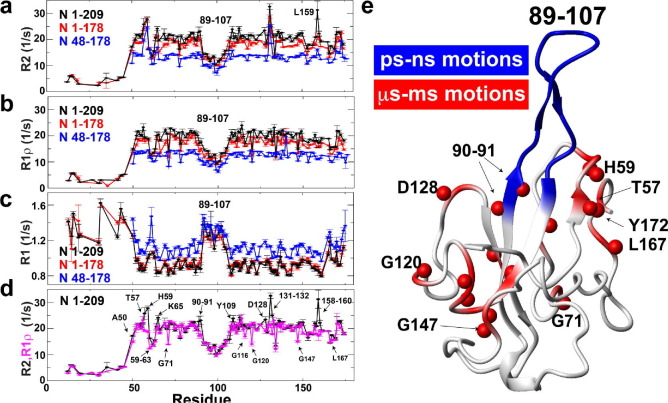
Figure 6.
Relaxation rates monitored for N protein N-terminal constructs identify dynamics on multiple timescales. Relaxation rates are shown for all three NTD constructs that include N 1–209 (black), 1–178 (red), and 48–178 (blue). (a) R2 relaxation rates, (b) R1 ρ relaxation rates, and (c) R1 relaxation rates. (d) Overlay of R2 (black) and R1 ρ (magenta) relaxation rates for N 1–209. (e) Mapping of dynamic regions include the dynamic β-hairpin of residues 89–107 highlighted (blue on ribbon) and residues exhibiting exchange of more than 2 s−1, taken as the difference between R2 and R1 ρ (red on ribbon and amides as red spheres). Only exchanging residues that do not exhibit a concentration dependence are shown, as in Supplementary Figure 1. These include T57, 59–60, K65, G71, 89–90, Y111, L113, T115, G116, G120, D128, V133, G147, L167, and F172. All data were collected at 900 MHz at 35 0C with 500 μM protein.
Next we sought to identify slower motions that may exist within the NTD. Chemical exchange due to μs-ms timescale motions monitored via R2 relaxation can be partially quenched during R1 ρ relaxation due to the high refocusing frequencies utilized within the latter’s relaxation delay. Here, a comparative analysis between R2 and R1 ρ relaxation rates clearly identifies contributions from such exchange (Figure 6(d)). However, caution must be taken due to potential contributions from self-association that we described above persists even at 35 °C. Thus, in addition to collecting relaxation data at 500 μM N 1–209, relaxation experiments were also collected at 250 μM protein to delineate exchange contributions due to self-association versus intramolecular motions (Supplementary Figure S3(d, e)). The majority of residues exhibiting differences between R2 and R1 ρ relaxation rates were similar at both concentrations with an identical average difference of 1.2 s−1, which is in agreement with the reduced self-association at the higher temperatures described above. Concentration-dependent changes between R2 and R1 ρ relaxation rates were observed for two linear sequences of residues 131–132 and 158–160, which could be ascribed to self-association. Thus, mapping residues that exhibit a quenching of their R2 relaxation rates by 2 s−1 or more at both concentrations illustrates the presence of slower motions throughout the NTD core that exist along with the higher-amplitude faster dynamics of the β-hairpin (Figure 6(e)).
Small nucleotide fragments interact with similar regions of the NTD
We performed NMR titrations with multiple nucleotide fragments in order to identify the preferred sites of interactions on the NTD and its flanking regions. As CoV N proteins comprise RNA chaperone activity that is defined by their broad ability to bind various sequences,33 we initiated studies using two RNA fragments that included a 30-mer stem loop derived from a bacterial ribosome (referred to as H25 30-mer) and a GU-rich 12-mer (referred to as GU 12-mer) used as a model RNA for a previous study.34 However, while initiating such titrations with these RNA fragments, preferred SARS-CoV-2 N protein occupancies within its genomic RNA was discovered.19 These include several single-stranded stretches adjacent to larger stem loops, such as nucleotides 127–148 within the 5′-UTR of the genomic RNA. Thus, we also performed additional titrations of fragments derived from this region that included a central 5-mer of nucleotides 138–142 (referred to as AAUAA), a 7-mer RNA with deoxy terminal nucleotides derived from nucleotides 137–143 (referred to as dUAAUAAdC, where “d” refers to the deoxy form), and finally a 7-mer DNA based on this site (referred to as TAATAAC). The impetus for probing hybrid RNA with deoxy termini and DNA was that despite the rigorous purification of our N protein constructs and the use of RNAse inhibitors, these ligands could be amenable to longer relaxation experiments performed at the 35 °C and described within the next section. Furthermore, N proteins are known to bind host DNA that may play different roles during the viral life-cycle that includes DNA competition for cell-cycle arrest.35 While structural models of the NTD core domain were recently calculated with a 7-mer (CUAAACG) and a 10-mer (UCUCUAAACG) derived from a stem-loop upstream from our sites within the SARS-CoV-2 genome,10 our findings suggest that the flanking regions are largely not utilized for such small nucleotide fragments.
All five nucleotide fragments were titrated into 100 μM 15N-labeled N 1–209, which revealed that similar regions within the NTD core domain are involved in engaging small nucleotide fragments (Figure 7 ). For example, despite the very different nucleotide sequences of these five fragments, CSPs were induced to many of the same residues that included R149 and S105 (Figure 7(a–e), top two panels). For several nucleotide fragments, a subset of resonances immediately disappeared upon the first titration point, such as the amide of S105 shown here, and these do not reemerge with higher concentrations of added nucleotide fragments. Such severe line broadening could arise from conformational dynamics but also from binding to multiple sites within nucleotide fragments. Evidence for multiple binding sites is clear for the larger fragments. For example, the H25 30-mer stem loop is quickly saturated at sub stoichiometric concentrations that suggests multiple binding sites (Figure 7(a)). Furthermore, the H25 30-mer stem loop induces the loss of resonances within its 15N-HSQC compared to the other titrations (Supplementary Figure S4(a–e)), which is consistent with multiple sites that would lead to the formation of a larger complex. Similar aggregation has also been observed for nucleotide binding to the CTD of SARS-CoV-1 N.24 Direct evidence of multiple binding sites is also directly observed for the GU 12-mer that requires at least a 2-site binding isotherm to fit its CSPs (Figure 7(b)). Interestingly, while the shorter nucleotide fragments bind weaker (Figure 7(c–e)), the dUAAUAAdC also induces similar severe line broadening (Figure 7(d)), which may reflect a dynamic exchange within the complex compared to its more malleable DNA counterpart (Figure 7(e)). Binding isotherms also indicate that the larger nucleotide fragments used here bind tighter than smaller fragments and that the more malleable DNA binds tighter than RNA (Table 1 ). The fact that the linear GU 12-mer sequence is engaged tighter than the H25 30-mer stem loop is consistent with the recently identified preference in N protein occupancies for linear regions within the viral genome.19 Finally, we also performed the reverse titration of the H25 30-mer step loop upon N 1–209 binding (Supplementary Figure S4(f)). As this stem loop has recently been assigned,34 these titrations confirm the expected line-broadening due to binding N 1–209 in that the RNA imino protons from both sides of the stem loop exhibit such broadening. Thus, studies here suggest that the N protein NTD exhibits no clear preference for short nucleotide fragments tested here and future screens may benefit from the use of larger genomic RNA fragments.
Figure 7.
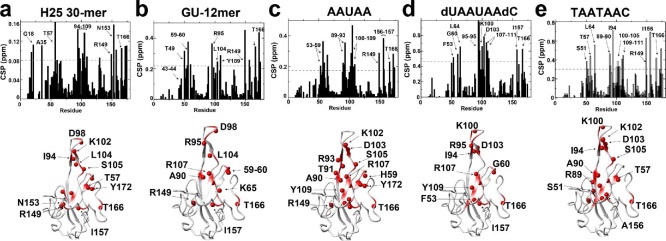
Nucleotide fragment titrations and their associated binding isotherms for N 1–209. Each sample comprised 100 μM 15N-labeled N 1–209 at 35 °C at 900 MHz in the presence of indicated concentrations of the nucleotide fragments with the 15N-HSQC of two regions shown and their respective binding isotherms. Data were collected for the following titrations with calculated binding affinities reported in Table 1: (a) 30-mer H25 stem loop. (b) GU 12-mer. c) AAUAA. (d) dUAAUAAdC. (e) TAATAAC.
Table 1.
Titration analysis from binding isotherms of individual nucleotide fragments.
| Nucleotide fragment | Kd (μM) |
|---|---|
| H25 30-mera | 19.9 ± 4.7 |
| GU 12-merb | 6.0 ± 2.0 >1000 |
| AAUAA | 189 ± 22 |
| dUAAUAAdC | 138 ± 12 |
| TAATAAC | 110 ± 8 |
Concentration points above 20 μM were unusable due to severe line broadening shown in Supplementary Figure 4.
2-site binding isotherm was required to fit the data.
A comparison of the overall CSPs induced by the five nucleotide fragments further highlights that similar regions within the NTD are affected by nucleotide binding and that the NTD flanking regions are minimally involved in binding to at least these smaller fragments (Figure 8 (a–e), top). For example, residues 50–60 (the first β-strand), 86–109 (the β-hairpin), and residues within 149–174 exhibit CSPs across all 5 titrations. These regions are adjacent to each other when mapped onto the structure of the NTD core (Figure 8(a–e), bottom). In general, the disordered flanking region of residues 1–47 only exhibit significant CSPs to the longest fragment used (Figure 8(a)). Furthermore, no new resonances were observed during titrations that would correspond to the SR region, which suggests that this region still undergoes an inherent chemical exchange that leads to severe line broadening even in the presence of these bound nucleotide fragments. This continued dynamic exchange of the SR region is also consistent with our relaxation experiments below that indicate a minimal change to the inherent dynamics of the remaining N 1–209 as well. Thus, the centrally folded NTD core is the primary site of small nucleotide fragment interactions but may become more important for larger nucleotide regions.
Figure 8.
CSPs induced for N 1–209 by nucleotide fragments. (a) 30-mer H25 stem loop, the dotted line represents the average (0.04 ppm) plus one standard deviation (0.04 ppm), which is 0.08 ppm. (b) 12mer, the dotted line represents the average (0.10 ppm) plus one standard deviation (0.21 ppm), which is 0.21 ppm. (c) AAUAA, the dotted line represents the average (0.09 ppm) plus one standard deviation (0.09 ppm), which is 0.18 ppm. (d) dUAAUAAdC, the dotted line represents the average (0.21 ppm) plus one standard deviation (0.24 ppm), which is 0.45 ppm. (e) TAATAAC, the dotted line represents the average (0.15 ppm) plus one standard deviation (0.15 ppm), which is 0.30 ppm. Amide CSPs larger than one standard deviation above the average are mapped onto the structure (red spheres).
The inherent dynamics of the NTD are only partially altered in the presence of a nucleotide fragment
The conformational dynamics of macromolecules are often altered upon complex formation. For example, we have recently shown that disordered loops within the active sites of an oxidoreductase family are quenched upon coenzyme engagement.36, 37 For N 1–209, extended incubations with RNA fragments indicated that these fragments are susceptible to RNase cleavage over the several days and high temperature necessary for data collection that persisted even in the presence of RNase inhibitors. This RNA degradation was surprising considering the multi-column purification that led to a single band on an SDS-PAGE denaturing gel (Figure 1(b)). While these N protein constructs could potentially be further purified by other methods, such as reverse-phase chromatography, our titration data described above confirms that DNA binds similarly to the RNA and therefore could be utilized to obtain dynamic insight within an N protein complex. Thus, relaxation data were acquired for N 1–209 in complex with the DNA fragment, TAATAAC.
We collected standard R2, R1 ρ, and R1 relaxation data on a 500 μM N 1–209 sample in the presence of 2.5 mM TAATAAC in order to compare the dynamics within this complex to that of the free N 1–209. Considering the relatively weak binding affinity of these nucleotide fragments (Table 1), caution should be taken to not over-interpret R2 and R1 ρ ρ relaxation rates that may have contributions from exchange between the bound and free forms. For example, under these conditions, approximately 5% of the protein would not be bound, which could contribute to these relaxation rates and preclude a strict R2/R1 ρ analysis as conducted for the free protein (Figure 6(d)). Nonetheless, it is apparent that the trends for both R2 and R1 ρ were very similar to that of the free protein (Figure 9 (a)). For R1 that is only sensitive to the ps-ns timescale, relaxation is strictly a weighted average between bound and free forms and were therefore directly compared to the free form (Figure 9(b)). Once again, these rates indicate that the dynamics in the bound state are largely similar to that of the free protein with only subtle changes largely within the β-hairpin that include residues 92–96 and K102. This slight reduction in the amplitudes of R1 relaxation rates indicates a reduction in ps-ns motions, which are evidence of a partial quenching of motions. Interestingly, the opposite is true for residues 1–47 that exhibit an overall increase in R1 relaxation rates relative to the remaining NTD core region that suggest an increase in ps-ns timescale motions within the complex (Figure 9(b)). This may be explained by the weak interaction of residues 1–47 with the NTD core (Figures 2 and 3), which are likely occluded by nucleotide binding leading to an increase in the flexibility of this region.
Figure 9.

Relaxation rates of N 1–209 in complex with TAATAAC. (a) R2 relaxation rates (red) and R1 ρ relaxations (green). (b) R1 relaxation rates for both N 1–209 alone (black) and in complex with TAATAAC (red). All data were collected at 900 MHz at 35 0C with 500 μM protein and 2.5 mM TAATAAC.
Discussion
Here, we have used NMR to experimentally explore the dynamics and interactions of the SARS-CoV-2 N protein N-terminal region with other regions of the N protein itself as well as with its interacting partners that include RNA and host cell CypA. While initial studies confirmed that the NTD does not associate with the CTD, as was expected based on recent SAXS data,14 we discovered that the disordered flanking regions are responsible for self-association. This was supported by comparative analyses of NTD-containing constructs along with titration experiments of independently purified regions. For example, titration of the N-terminal flanking region of N 1–47 with the remaining N 48–209 identified a surface within the NTD core that likely mediates such self-association (Figure 10 ). Our studies also provide insight to both the complicated array of internal dynamics of the N protein and its interactions described herein.
Figure 10.

The primary interaction sites of the NTD and its flanking region identified here. Shown are all the amides that incur CSPs upon titration of any of the five nucleotide fragments used within this study (red and amides as balls) with a cartoon representation of the proline residues targeted by host cell CypA. Sites identified as important for self-association are delineated with arrows with the surface of the central NTD core domain shown to bind the recombinant N-terminal disordered peptide of N 1–47 identified via titration studies also shown (red surface representation). The disordered flexible regions that induce self-association are the same sites where CypA binds.
The N protein NTD and its flanking regions exhibit an array of inherent dynamics on multiple timescales. For example, relaxation rates collected on multiple NTD constructs reveal μs-ms motions of much of the NTD core domain and elevated ps-ns motions within the β-hairpin of residues 89–107 that illustrates its inherent flexibility. This can be compared to the β-hairpin within the SARS-CoV-1 N protein NTD that we have also shown exhibits similar flexibility.15 Such an inherent dynamic within the N protein NTD may be especially important when considering that the primary function is to broadly bind multiple RNA sequences within the viral genome, which is also consistent with the chaperone role that N proteins play in RNA folding.33, 38 In addition to the dynamics of the NTD core domain, multiple exchange phenomena were identified within the functionally important SR region of residues 176–209. These motions include a temperature-dependent fast exchange (μs-ms) leading to the disappearance of SR resonances at physiological temperatures and a slower inherent exchange mediated by P199 isomerization. Recent studies have begun to elucidate the clinical importance of the SR region, which is a “hot spot” for evolutionary changes, N protein-mediated condensation with RNA, and is a primary target site of phosphorylation that inhibits condensation.16, 39 Thus, the dynamic nature of this SR region on multiple timescales may play a role in its unique properties that drive condensation and ultimately its interactions.
We utilized NMR to begin probing the interactions of the N protein NTD and its flanking regions with its biological targets (Figure 10), which may provide critical insight into these complicated interactions that may underlie viral infection. First, titrations of small nucleotide fragments indicate that the NTD central core is the primary region responsible for their binding, which are consistent with CSPs observed in a recent NMR study that utilized solely the NTD core region.10 Although only small CSPs were observed for the disordered flanking regions indicating that these regions are not the primary binding site for small nucleotide fragments, the larger CSPs observed for the N-terminal flanking region with the largest RNA fragment may suggest secondary roles in RNA binding. While flexibility within the NTD core may be important for nucleotide interactions, our findings here also reveal that the inherent dynamics of the NTD are not significantly perturbed by its binding to a small nucleotide fragment. For example, complex formation with the small DNA fragment induces only a slight reduction in the amplitude of R1 relaxation rates within the NTD β-hairpin indicating a slight reduction in mobility. Conversely, the R1 amplitudes increase within the N-terminal flanking region indicating higher mobility within this DNA complex that is consistent with the nucleotide fragment occluding the weaker N-terminus interactions with the NTD core. It is therefore possible that motions would be more dramatically altered with larger genomic RNA fragments that comprise the full-length N protein. Second, potentially the most interesting finding here is that host cell CypA weakly engages both disordered flanking regions that are the same regions that we have shown to be responsible for self-association. This may begin to explain an indirect role of host cell CypA in viral replication. Namely, host CypA enhances viral replication for multiple CoVs that include SARS-CoV-2 through an unknown mechanism.6, 9 However, recent studies have now shown the SARS-CoV-2 N protein mediates condensate formation with enrichment of the replication machinery.16 Thus, the fact that we have discovered here that the disordered flanking regions mediate self-association and are also the binding sites for host CypA may begin to reveal the complicated way in which CypA indirectly modulates viral replication through modulating N protein condensates. Such NMR studies now provide hypotheses for testing condensate formation in the presence of CypA using both in vitro and cellular studies.
It remains an ongoing effort to understand how CoV N proteins engage both RNA and possibly DNA, but also how the N protein orchestrates genomic RNA packaging. Viral packaging is remarkably selective in that genomic RNA must be delineated from a crowded cellular environment that also comprises sub-genomic viral RNA and host RNA. The final steps of RNA packaging appear to be dependent on a stem loop within the genome known as a packaging signal (PS),40, 41 yet recent cross-linking studies reveal that the N protein preferentially targets several other regions to initiate this process.19 The promiscuity of N protein NTD binding to small nucleotide fragments shown here suggests that genomic interactions are more complicated, which likely includes the CTD or N protein oligomerization mediated by the genomic RNA itself. The inherent self-association properties of the NTD mediated by the disordered regions and their interactions with host CypA observed here illustrate the complicated nature of these interactions. Furthermore, our findings that illustrate how dynamics persist within the N 1–209/DNA complex may also indicate that this complex remains dynamic in order to further engage other sequences that results in more intimate RNA interactions. It will be interesting to determine whether larger fragments may be necessary to confer selectivity and whether the inherent disordered regions and the SR regions become ordered within larger complexes. For example, biochemical methods may be used with larger regions of the 5′-UTR to determine whether adjacent stem loops may play a role in guiding the N protein to specific interaction sites or whether there are RNA fragments that bind more tightly than those used here. Although larger fragments may not be amenable to standard NMR studies, the identification of a preferred binding interaction for the entirety of the N protein could result in a homogeneous sample amenable to other methods that include cryo electron microscopy (cryoEM). Moreover, if smaller regions are found to simultaneously engage the entire N protein, specifically labeling methods may be used in order to quantify NMR relaxation rates of the N protein within these larger complexes to determine whether motions are quenched. Finally, considering the high immunogenicity of the N protein combined with its relatively conserved sequence compared to the S protein, the N protein has been proposed as a viable vaccine.3 The confirmation here of the inherent flexibility of the β-hairpin within the NTD and the fact that it is broadly involved in nucleotide binding may suggest that epitopes derived from this region could make excellent vaccine candidates.
Materials and methods
Protein expression and purification
Seven N protein constructs were engineered that include the full-length N protein of residues 1–419, N 48–178, N 1–178, N 1–209, N 48–209, N 251–364, N 251–419, and N 210–364, which were designed with a C-terminal 6xhis-tag in pET29 and purchased from Twist Bioscience (San Francisco, CA). Unlabeled proteins were grown in luria broth (LB) while labeled proteins were grown in M9 minimal media (6 g/l Na2HPO4, 3 g/l KH2PO4, 0.5 g/l NaCl, 1 g/l NH4Cl, 2 g/l glucose, 2 ml of 1 M MgSO4 100 ml of 1 M NaCl CaCl2, 10 mg/l thiamine). 15N-labeled proteins were grown in M9 media supplemented with 15N-labeled NH4Cl and 13C,15N-labeled proteins were grown in M9 media with both 15N-labeled NH4Cl and 13C-labeled glucose. A typical growth for both unlabeled and labeled protein comprised 4 L of LB begun from a single colony supplemented with kanamycin. For unlabeled protein, media was induced with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at an A 600 of 0.8 for 3 hours at 37 °C. For labeled protein, growths were subjected to a media switch whereby at an A 600 of 0.5 the cells were centrifuged for 15 min at 3500 g and cell pellets allowed to further grow in M9 media for an hour before inducing with IPTG. Pellets were lysed via sonication in Ni buffer (50 mM Na2HPO4, pH 7.0, 500 mM NaCl, 10 mM imidazole), centrifuged, and supernatants applied to a 20 mL Ni affinity column (Sigma). Eluted proteins were dialyzed against SP ion exchange buffer (50 mM Tris, pH 7.0, 150 mM NaCl, 1 mM EDTA) and applied to a 40 mL SP fast-flow column (Cytiva). Eluted protein was concentrated to 3–4 mL and applied to a Superdex-75 column (Cytiva, 120 mL total bed volume) in final NMR buffer (50 mM HEPES, pH 7.0, 150 mM NaCl). Eluted proteins were concentrated and frozen until further use. Analytical sizing utilized an analytical Superdex-75 column (23.5 mL total bed volume) with 200 μL of 50 μM protein.
For the N 1–47 peptide, we engineered an N-terminal 6xhis-tag followed by the small Ubiquitin-like Modifier (SUMO), and then finally the sequence for N 1–47. The peptide was recombinantly expressed and applied to Ni affinity identically as to that described above. Elutions were dialyzed back into Ni Buffer, cleaved with 100 μg of SUMO Protease produced in-house (also known as Ulp1p, UniProt accession A0A0L8VFW2), and re-applied to the Ni affinity column to strip the 6xhis-tagged SUMO. The flow-through fractions from this second Ni affinity were concentrated to 3–4 mL and applied to a Superose-12 column (Cytiva, 120 mL total bed volume) in NMR buffer. Eluted protein was concentrated and frozen until further use.
Cyclophilin-A was expressed and purified as we have previously described.42 However, the final purification step of size exclusion chromatograph using a Superdex-75 column was performed within the same NMR buffer as all N protein constructs.
All final samples for NMR studies below comprised 330 μL total volume and were supplemented with 10% D2O.
Nucleotide reagents
All RNA fragments were purchased from Horizon Discovery (Boulder, CO) and DNA from IDT (Coralville, IA). These included GGUGGGAGCCUCUUUUAUGGGGUGACUGCC (referred to as H25 30-mer), GUGUGUGCCUGU (referred to as GU 12-mer), AAUAA (referred to as AAUAA), dUAAUAAdC (referred to as dUAAUAAdC), and the DNA of TAATAAC (referred to as TAATAAC). Each sample titrated with RNA also included RNase inhibitors (Thermo Fisher and NEB).
Nuclear magnetic resonance spectroscopy
For backbone assignment, standard HNCACB and CBCA(co)NH spectra for N 48–178, N 1–178, and N 1–209 were all collected on 500 μM 13C,15N-labeled samples at 35 °C at 800 MHz equipped with a cryo-probe at the National High Magnetic Field Laboratory, Tallahassee, FL. Assignments for the full N 1–209 at these conditions have been deposited into the Biological Magnetic Resonance Data Bank (BMRB ID 50835). Additionally, assignments were also collected for 13C,15N-labeled N 1–209 at 20 °C. For both titration and relaxation data, all spectra were collected on the Rocky Mountain Varian 900 equipped with a cryo-probe at the University of Colorado Anschutz Medical School, Aurora, CO. Standard Biopack sequences were used for all relaxation data. For both R2 and R1 ρ relaxation rates on the free proteins, mixing times of 0.01, 0.03, 0.05, 0.09, 0.11, 0.13, and 0.15 s were used and for R1 relaxation rates, mixing times of 0.01, 0.1, 0.3, 0.5, 0.9, 1.1, 1.3, 1.5 s were used. These mixing times were also used for the complex of 0.5 mM N 1–209 with 2.5 mM TAATAAC. However, as relaxation was much faster for both R2 and R1 ρ relaxation rates for the folded NTD region within the complex, data were re-acquired with mixing times of 0.01, 0.03, 0.05, and 0.07 s and the number of scans were doubled from 8 to 16 scans to obtain better signal-to-noise. Thus, reported R2 and R1 ρ ρ relaxation rates here for the complex with DNA utilized these later data for the central folded NTD that had lower uncertainties but the longer relaxation times for the flanking regions. All spectra were processed using NMRPipe43 and all data were analyzed using CCPNmr software.44 Finally, all binding affinities were derived from the indicated titration data and it using GraphPad Prism version 4.0 using the indicated binding isotherms of single or double sites (GraphPad Software Inc., San Diego, CA).
Acknowledgments
Acknowledgements
EZE was supported by NSF application number 1807326, NIH application number R56 CA230069, and NIH application number, R21AI146295. BV was supported by NIH Grant R01GM130694-01A1 and NSF Grant 1917254. Use of the Rocky Mountain NMR facility at Anschutz campus of the University of Colorado is supported by University of Colorado Cancer Center Support Grant P30 CA046934. The National High Magnetic Field Laboratory is supported by National Science Foundation through NSF/DMR-1644779 and the State of Florida.
Author contributions
E.Z.E., A.I., A.D., and K.C.H. helped design all plasmids. J.R., E.L., and E.Z.E. expressed and purified all proteins. A.B., B.V., and E.Z.E. assigned NMR spectra. A.B. and E.L. collected NMR data. P.N. and B.V. helped design and provided nucleotide fragments.
Data availability
Data supporting the findings of this manuscript are available from the corresponding authors upon reasonable request. NMR assignments of N 1-209 have been deposited in the BMRB (BMRB ID 50835).
Conflict of Interest Statement
The authors declare no conflicts of interest.
Edited by Prof. M.F. Summers
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2021.167108.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nature Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00468-6. 1740–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Surjit M., Lal S.K. The SARS-CoV nucleocapsid protein: A protein with multifarious activities. Infect. Genet. Evol. 2008;8:397–405. doi: 10.1016/j.meegid.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutta N.K., Mazumdar K., Gordy J.T. The nucleocapsid protein of SARS–CoV-2: A target for vaccine development. J. Virol. 2020;94:e000647. doi: 10.1128/JVI.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Surjit M., Liu B.P., Chow V.T.K., Lal S.K. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Y., Li W., Gao T., Cui Y., Jin Y.W., Li P., et al. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J. Virol. 2017;91 doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carbajo-Lozoya J., Ma-Lauer Y., Malesevic M., Theuerkorn M., Kahlert V., Prell E., et al. Human coronavirus NL63 replication is cyclophilin A-dependent and inhibited by non-immunosuppressive cyclosporine A-derivatives including Alisporivir. Virus Res. 2014;184:44–53. doi: 10.1016/j.virusres.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Brunn A., Ciesek S., von Brunn B., Carbajo-Lozoya J. Genetic deficiency and polymorphisms of cyclophilin A reveal its essential role for Human Coronavirus 229E replication. Curr. Opin. Virol. 2015;14:56–61. doi: 10.1016/j.coviro.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pawlotsky J.M. COVID-19 Pandemic: Time to Revive the Cyclophilin Inhibitor Alisporivir. Clin. Infect. Dis. 2020;71:2191–2194. doi: 10.1093/cid/ciaa587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Softic L., Brillet R., Berry F., Ahnou N., Nevers Q., Morin-Dewaele M., et al. Inhibition of SARS-CoV-2 Infection by the Cyclophilin Inhibitor Alisporivir (Debio 025) Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00876-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinesh D.C., Chalupska D., Silhan J., Koutna E., Nencka R., Veverka V., et al. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. PLoS Pathog. 2020;16:e1009100. doi: 10.1371/journal.ppat.1009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang S.S., Yang M., Hong Z.S., Zhang L.P., Huang Z.X., Chen X.X., et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou R., Zeng R., von Brunn A., Lei J. Structural characterization of the C-terminal domain of SARS-CoV-2 nucleocapsid. Mol. Biomed. 2020;202:1. doi: 10.1186/s43556-020-00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang C.K., Hsu Y.L., Chang Y.H., Chao F.A., Wu M.C., Huang Y.S., et al. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: Implications for ribonucleocapsid protein packaging. J. Virol. 2009;83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng W.H., Liu G.F., Ma H., Zhao D., Yang Y.R., Liu M.Y., et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527:618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarkson M.W., Lei M., Eisenmesser E.Z., Labeikovsky W., Redfield A., Kern D. Mesodynamics in the SARS nucleocapsid measured by NMR field cycling. J. Biomol. NMR. 2009;45:217–225. doi: 10.1007/s10858-009-9347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savastano A., de Opakua A.I., Rankovic M., Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nature Commun. 2020:11. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perdikari T.M., Murthy A.C., Ryan V.H., Watters S., Naik M.T., Fawzi N. L., SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. Embo J. 2021 doi: 10.15252/embj.2020106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cascarina S.M., Ross E.D. A proposed role for the SARS-CoV-2 nucleocapsid protein in the formation and regulation of biomolecular condensates. Faseb J. 2020;34:9832–9842. doi: 10.1096/fj.202001351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iserman C., Roden C.A., Boerneke M.A., Sealfon R.S.G., McLaughlin G.A., Jungreis I., et al. Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. Mol. Cell. 2020;80:1078–1091. doi: 10.1016/j.molcel.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsson G., Martinez G., Schleucher J., Wijmenga S.S. Detection of nan-second internal motion and determination of the overall tumbling times independent of the time scale of internal motion in proteins from NMR relaxation data. J. Biomol. NMR. 2003;27:291–312. doi: 10.1023/a:1025836018993. [DOI] [PubMed] [Google Scholar]
- 21.Kendrick A.A., Holliday M., Isern N.G., Zhang F., Camilloni C., Huynh C., et al. The dynamics of interleukin-8 and its interaction with human CXC receptor I peptide. Protein Sci. 2014;23:464–480. doi: 10.1002/pro.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schiavina M., Pontoriero L., Uversky V.N., Felli I.C., Pierattelli R. The highly flexible disordered regions of the SARS-CoV-2 nucleocapsid N protein. Biomol. NMR Assign. 2021 doi: 10.1007/s12104-021-10009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wishart D.S., Sykes B.D. Chemical-shifts as a tool for structure determination. Nuclear Magn. Reson. Pt C. 1994:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 24.Luo H.B., Chen J., Chen K.X., Shen X., Jiang H.L. Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: Self-association analysis and nucleic acid binding characterization. Biochemistry. 2006;45:11827–11835. doi: 10.1021/bi0609319. [DOI] [PubMed] [Google Scholar]
- 25.Ishii N., Watashi K., Hishiki T., Goto K., Inoue D., Hijikata M., et al. Diverse effects of cyclosporine on hepatitis C virus strain replication. J. Virol. 2006;80:4510–4520. doi: 10.1128/JVI.80.9.4510-4520.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Wilde A.H., Zevenhoven-Dobbe J.C., van der Meer Y., Thiel V., Narayanan K., Makino S., et al. Cyclosporin A inhibits the replication of diverse coronaviruses. J. Gen. Virol. 2011;92:2542–2548. doi: 10.1099/vir.0.034983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grise H., Frausto S., Logan T., Tang H. A conserved tandem cyclophilin-binding site in hepatitis C virus nonstructural protein 5A regulates alisporivir susceptibility. J. Virol. 2012;86:4811–4822. doi: 10.1128/JVI.06641-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlegel J., Redzic J.S., Porter C., Yurchenko V., Bukrinsky M., Armstrong G.S., et al. Solution characterization of the extracellular region of CD147 and its interaction with its enzyme ligand cyclophilin-A. J. Mol. Biol. 2009;391:518–535. doi: 10.1016/j.jmb.2009.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosco D.A., Eisenmesser E.Z., Pochapsky S., Sundquist W.I., Kern D. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5247–5252. doi: 10.1073/pnas.082100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanoulle X., Badillo A., Wieruszeski J.M., Verdegem D., Landrieu I., Bartenschlager R., et al. Hepatitis C virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. J. Biol. Chem. 2009;284:13589–13601. doi: 10.1074/jbc.M809244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grathwohl C., Wuthrich K. NMR-studies of the rates of proline cis-trans isomerization in oligopeptides. Biopolymers. 1981;20:2623–2633. [Google Scholar]
- 32.Jayaram H., Fan H., Bowman B.R., Ooi A., Jaaram J., Collisson E.W., et al. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: Implications for nucleocapsid formation. J. Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuniga S., Sola I., Moreno J.L., Sabella P., Plana-Duran J., Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357:215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nichols P.J., Henen M.A., Born A., Strotz D., Guntert P., Vogeli B. High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints. Commun. Biol. 2018:1. doi: 10.1038/s42003-018-0067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perrin-Cocon L., Diaz O., Jacquemin C., Barthel V., Ogire E., Ramiere C., et al. The current landscape of coronavirus-host protein-protein interactions. J. Translat. Med. 2020;18 doi: 10.1186/s12967-020-02480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paukovich N., Xue M.J., Elder J.R., Redzic J.S., Blue A., Pike H., et al. Biliverdin reductase B dynamics are coupled to coenzyme binding. J. Mol. Biol. 2018;430:3234–3250. doi: 10.1016/j.jmb.2018.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duff M., Redzic J., Ryan L., Paukovich N., Zhao R., Nix J., et al. Structure, dynamics, and function of the evolutionarily changing biliverdin. J. Biochem. 2020:1756–2651. doi: 10.1093/jb/mvaa039. 101093/jb/mvaa039. [DOI] [PubMed] [Google Scholar]
- 38.Zuniga S., Cruz J.L.G., Sola I., Mateos-Gomez P.A., Palacio L., Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahman M.S., Islam M.R., Ul A.A., Islam I., Hoque M.N., Akter S., et al. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J. Med. Virol. 2020 doi: 10.1002/jmv.26626. [DOI] [PubMed] [Google Scholar]
- 40.Woo J., Lee E.Y., Lee M., Kim T., Cho Y.E. An in vivo cell-based assay for investigating the specific interaction between the SARS-CoV N-protein and its viral RNA packaging sequence. Biochem. Biophys. Res. Commun. 2019;520:499–506. doi: 10.1016/j.bbrc.2019.09.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masters P.S. Coronavirus genomic RNA packaging. Virology. 2019;537:198–207. doi: 10.1016/j.virol.2019.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlegel J., Armstrong G.S., Redzic J.S., Zhang F.L., Eisenmesser E.Z. Characterizing and controlling the inherent dynamics of cyclophilin-A. Protein Sci. 2009;18:811–824. doi: 10.1002/pro.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMR pipe – A multidimensional spectral processing system based on Unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas P., et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins-Struct. Funct. Bioinf. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 45.Hafsa N.E., Wishart D.S. CSI 2.0: A significantly improved version of the Chemical Shift Index. J. Biomol. NMR. 2014;60:131–146. doi: 10.1007/s10858-014-9863-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this manuscript are available from the corresponding authors upon reasonable request. NMR assignments of N 1-209 have been deposited in the BMRB (BMRB ID 50835).