Introduction
The heme-containing cytochrome P450 monooxygenases are critical components of many biosynthetic pathways: They catalyze regio- and stereospecific oxidations that are difficult or impossible to achieve in the laboratory, including hydroxylations, epoxidations, oxidative couplings and dealkylations that are required in the synthesis of high value-added products [1]. Recent work has shown that P450 enzymes can be adapted to produce abiological intermediates such as nitrenes and carbenes, further extending the usefulness of these enzymes in synthetic applications [2–4]. As such, the importance of the P450 superfamily to biotechnology is unarguable.
The variety of P450 substrate-product combinations is also remarkable: Substrates range in size from small hydrocarbons and terpenes to steroids, macrolide antibiotics, lipids and eicosanoids. Given the number of potential reaction sites even in small substrates, the combinatorial possibilities for CYP-catalyzed oxidations are vast.
An intriguing feature of the P450s is that, regardless of substrate, the same folding topology is maintained across the superfamily (Figure 1). The typical P450 (exclusive of membrane anchors) is ~400 amino acids in length. Clearly, the P450 fold was settled upon early in evolution as a stable platform for the activation of molecular oxygen. Such structural uniformity raises a number of interesting questions for the biotechnologist. Which sequence positions could be varied to evolve new functionality? Which must be conserved in order to maintain catalytic activity? Can one optimize an existing activity (e.g. throughput, efficiency) by rational engineering?
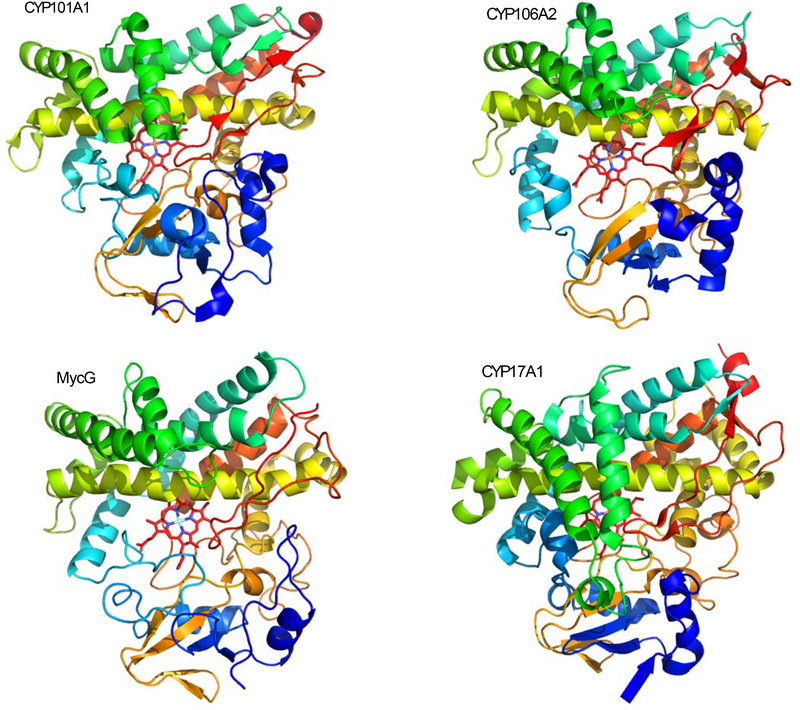
Figure 1.
Four cytochrome P450s currently under investigation using solution NMR methods. All structures are shown from approximately the same perspective, with rainbow coloring from N- (blue) to C- termini (red).
Top left: CYP101A1 (camphor hydroxylase from Pseudomonas putida), NMR structure 2L8M.
Top right: CYP106A1 (sterol 15-hydroxylase from Bacillus megaterium), X-ray structure 5XNT.
Lower left: MycG (mycinamycin IV hydroxylase-epoxidase from Micromonospora griseorubida), NMR structure 5UHU.
Lower right: CYP17A1 (steroid 17α-hydroxylase from Homo sapiens), X-ray structure 3RUK chain A.
An obvious first step in answering these questions is to align P450 structures to identify variable residues in homologous positions. While strict conservation suggests that a given residue is important for maintaining function, variability does not necessarily identify positions involved in functional variation. An important lesson learned from directed evolution experiments on P450s is that residues remote from the active site can have a profound impact on enzyme function. It is difficult (if not impossible) to extract from a static crystal structure why a residue 20 Å away from substrate has an outsized impact on enzyme function [5, 6].
In order to begin to parse the contributions of particular positions to function, it is necessary to consider the role of protein dynamics in determining substrate selectivity and product specificity. Given the complex chemistry that must be directed, it is not surprising that the P450 enzymatic cycle is complex (Figure 2). For P450s involved in biosynthetic and catabolic pathways, P450s have evolved such that the following are true:
Substrate binding must be reasonably selective (although initial binding orientation need not, indeed is probably not, that giving rise to the final product).
The enzyme should not activate molecular oxygen in the absence of substrate, which would give rise to toxic reactive oxygen species.
Once activated, the reactive intermediate (Compound I, or Cpd-I, see Fig. 2) should not interact with any molecule except substrate, and then only at the position at which oxidation occurs. This ban obviously includes amino acid residues in the active site as well as the oxidation-sensitive heme porphyrin itself, as well as any solvent species that could be oxidized, such as water or halide ions.
Product release must be sufficiently fast so that product inhibition does not interfere with overall turnover, but slow enough so that hydrogen abstraction and oxygen rebound steps are complete for each reaction cycle.
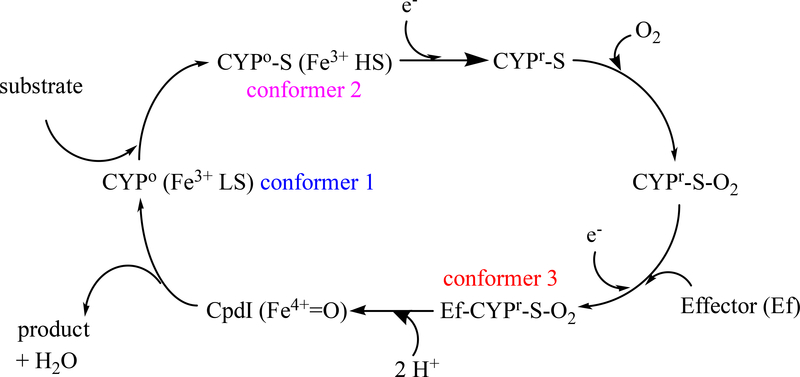
Figure 2.
Typical reaction cycle for cytochrome P450. Abbreviations: LS –low spin, HS –high spin, S –substrate, e− -electron, CpdI, activated oxygen species Compound I. Conformers identified by color are discrete conformations confirmed by NMR for CYP101A1.
It is logical to assume that the enzyme structure must undergo some form of conformational selection to ensure that each step listed above takes place from conformations that are separated by energetic barriers high enough to prevent rapid equilibration between steps, but low enough that the barriers can be crossed on a timescale required for efficient catalysis (Figure 2). Because nuclear magnetic resonance (NMR) provides access to dynamics on multiple time scales, it is an excellent tool for probing conformational changes in cytochromes P450 as a function of conditions (substrate or redox partner binding, oxidation and spin state changes). In combination with restrained molecular dynamics simulations, NMR data allows direct characterization of solution structural ensembles.
In this review, we will provide examples of how NMR has contributed to our understanding of the role of conformational changes and their time scales in controlling P450 function. We will limit discussion to solution NMR, but the reader should be aware that solid-state NMR is now widely used for examining membrane-bound P450s, particularly with lipid nanodiscs as vehicles for monomerizing membrane-bound P450s and examining their interactions with redox partners and effectors [7].
Substrate-induced spin state selection
In the absence of substrate, the P450 resting state is invariably a low-spin (S=1/2, λmax ~417 nm) ferric (Fe3+) form. The reduction potential of a low spin P450 is typically mismatched with that of physiological redox partners, preventing electron transfer in the absence of substrate. For bacterial P450s (as well as many biosynthetic eukaryotic CYPs), this redox partner is often a Fe2S2 ferredoxin, although NAD(P)H-dependent flavoproteins can also serve in this role [8]. Upon substrate binding, the heme shifts to a high-spin form (S=5/2, λmax ~390 nm) with a reduction potential appropriate for the first electron transfer [9]. While the substrate-induced shift from low to high spin is not always complete, there is evidence to suggest that at least some high spin needs to be present for the first electron transfer to occur under physiological conditions [10].
The spin state equilibrium has historically been ascribed to the presence/absence of an axial water ligand at the heme iron (the six-coordinate complex being low spin). However, recent evidence suggests that the origins of spin equilibrium are more complex. In CYP101A1 (cytochrome P450cam), binding of the native substrate (+)-camphor results in nearly complete shift to high spin, whereas non-native substrates (e.g., norcamphor, adamantanone) results in less complete shifts. NMR-detected titrations of CYP101A1 with these three substrates identified substrate-dependent perturbations remote from the active site that appear to track with the extent of spin state shifts, suggesting that the spin state equilibrium reflects a more general structural response by the enzyme, not merely the presence or absence of an axial heme ligand [11].
Another P450 that has been examined by NMR tends to support this more global view of substrate-dependent spin state selection. MycG, a bifunctional P450 that catalyzes the last two steps of mycinamicin II (M-II) biosynthesis, exhibits ~15% high spin formation upon binding of substrate mycinamicin IV (M-IV) [12]. Crystallographic structures of the MycG-(M-IV) complex show a high-occupancy water or hydroxide axial ligand, with M-IV substrate bound in an orientation not appropriate for the observed chemistry. NMR titration of MycG with M-IV indicates a different M-IV orientation than that observed crystallographically, with the site of oxidation closer to the heme iron, but still too distant to displace an axial ligand [12].
We have proposed that the spin state shift is a reflection of heme conformational equilibrium: Substrate binding that favors a “domed” heme conformation, moving the iron atom out of the plane of the equatorial pyrrole nitrogen ligands, would shift equilibrium shifts towards high-spin by reducing iron d-orbital splittings [10]. Given that there is considerable van der Waals contact between substrate and the heme (at least in CYP101A1 structures), the best-fit substrate would be that which most increases the fraction of “domed” heme. This doming could be enforced directly, by heme-substrate contacts, and indirectly, by packing interactions of substrate with residues that in turn pack against the heme porphyrin.
Initial binding, substrate re-orientation and product release: A role for effectors
The iron-oxo species (Fe4+=O) Cpd-I generated by cytochrome P450 is highly reactive [13]. Cpd-I abstracts a hydrogen atom from an unactivated C-H bond on the substrate, generating a radical that subsequently accepts the hydroxyl group from the Fe3+-OH (“oxygen rebound”), forming the alcohol product and returning the enzyme to its resting state. Cpd-I will react rapidly with water to form peroxides, and could potentially generate radical species from organic functionality in the enzyme active site. It is reasonable to assume that, in order to prevent the formation of electron-deficient oxo species and/or damaging the enzyme, the formation of Cpd-I should be accompanied by exclusion of water from and restricted access to the active site. Limited mobility of substrate or residues in contact with substrate would prevent their reaction with Cpd-I, preventing damage to the enzyme. On the other hand, such restricted access would likely inhibit substrate binding and product release, slowing substrate turnover considerably. Given the complicated reaction cycle, P450s are already relatively slow [14], so any process that further slows catalysis would clearly be disadvantageous. Indeed, P450-catalyzed steps in biosynthetic processes are often rate-determining for the entire pathway [15]. It would seem more efficient if initial substrate binding were to occur to an open “loose” enzyme conformer. Once the enzyme-substrate complex forms, it would be followed by a conformational change that reorients substrate appropriately for chemistry and isolates the active site from solvent. Such a conformational change could take place spontaneously, but would be more efficient if driven by the binding of an effector molecule. If the conformational change were slow on the time scale of the last steps of the reaction, re-opening of the active site would only occur after the reaction is complete, allowing product release and return to the enzyme resting state.
Most crystallographic structures of CYP101A1 in fact show a water-free active site with the substrate camphor poised precisely so that the 5-exo position (where hydroxylation occurs) is adjacent to the heme iron [16]. It was therefore surprising when NMR characterization of camphor binding to CYP101A1 showed the camphor to be bound in an orientation not seen in the crystal, but somewhat more removed from the heme iron, and with the camphor molecule in the wrong orientation for the observed chemistry. Upon titration with putidaredoxin (Pdx), the Fe2S2 ferredoxin that is the physiological reductant of CYP101A1, the camphor re-orients into the position seen crystallographically [17]. This rationalized the observation that, in the absence of Pdx, CYP101A1 does not turn over substrate, even if all of the other components and reducing agents are present [18]. Pdx is thus not only the reductant of CYP101A1, but an effector as well. Many P450s require the presence of an effector for turnover (often but not always a redox partner) [8], and we proposed that the role of the effector is twofold: Effector binding closes the active site prior to formation of Cpd-I, preventing peroxide formation and other undesirable side reactions, and re-orients bound substrate appropriately for the observed chemistry.
To determine precisely how effector binding accomplished these tasks, it was necessary to make sequential amide (15N-1H) assignments in the enzyme, providing a means of localizing Pdx-induced perturbations [19]. The Pdx binding site on CYP101A1 had been inferred from site-directed mutagenesis [20], and was later confirmed crystallographically [21, 22]. It is located on the proximal side of the heme, remote from the active site entrance on the distal side. It was surprising to find that the effects of Pdx binding on CYP101A1 was quite global, with effects being observed in a number of places on the distal side, including residues near the entrance to the active site [23]. Furthermore, the time scale of Pdx-induced perturbations were the same, regardless of where in the structure they occurred, indicating that a single event, with a rate constant ~700 s−1, was responsible. We proposed that a single Ile-Pro bond near the active site entrance undergoes a tran-cis isomerization upon Pdx binding, and that this isomerization is responsible for both active site closure and reorienting of substrate to that observed crystallographically (in which the Ile-Pro bond is cis) [24, 25]. It is worth noting that Val-Pro, Ile-Pro and Phe-Pro combinations are common at this position across the P450 superfamily, particularly for those enzymes that have relatively small substrates and catalyze selective regio- and stereochemical oxidations [1]. A recent crystallographic structure of the CYP101A1-Pdx covalent complex confirms that the Ile-Pro bond in question is capable of occupying both cis and trans conformations [26]. Pdx-induced perturbations in CYP101A1 have been tracked by paramagnetic relaxation, showing that non-productive encounter complexes form between the redox partners [27].
Effector- and substrate-induced NMR perturbations have also been tracked for the human P450 CYP17A1, a steroid hydroxylase that is the target for prostate cancer chemotherapies [28, 29]. The effector in this case is cytochrome b5, which has been found to modulate the activities of multiple cytochromes P450 [29]. Even with limited sequential resonance assignments available, the authors found evidence that the CYP17A1 structure responds to the binding of cytochrome b5 at many positions remote from the effector binding site, indicating a global structural response to effector binding.
Preliminary data on species-dependent variations of the interactions of vitamin D hydroxylase CYP24A1 with the effector and redox partner adrenodoxin have been reported [30].
Solution structural ensembles and substrate-induced conformational changes in cytochromes P450.
As useful as chemical shift and dynamic information may be for localizing conformational changes in P450s, they are not structural data per se. For structural information, we make use of residual dipolar couplings (RDCs) as restraints in molecular dynamics simulations to generate average structures of P450s as a function of conditions (e.g., effector and substrate binding) [23, 31–34]. RDCs are the result of partial orientation of solute molecules in the presence of magnetically or mechanically aligned additives in solution NMR samples [35]. Common aligning agents include nematic liquid crystals, bacteriophage or stretched polyacrylamide gels. As a result of such fractional alignment, the normally field-independent scalar couplings between atoms are modulated, with the magnitude and sign of these modulations (RDCs) dependent upon the orientation of the bonds to which they are assigned relative to an alignment tensor. For a single-domain protein, RDC-derived structures show the average orientations of secondary structural features in the molecular frame of reference. Because these are solution structures, they are free of the constraints imposed by the crystal lattice (which can be significant).
RDCs have been used to compare the conformational changes that occur upon substrate binding in two unrelated P450s, CYP101A1 and MycG [31–34, 36]. CYP101A1 binds a small hydrophobic substrate (camphor) while MycG binds a large macrolactone antibiotic precursor (M-IV, vide supra). Not surprisingly, the active sites of the two enzymes are quite different: while camphor is completely enclosed in the CYP101A1 active site, much of the MycG molecule extends past the active site entrance into solvent. What was surprising was the consistency of the conformational changes that take place upon substrate binding in both enzymes [31]. In both CYP101A1 and MycG, the same secondary structural features reorient to accommodate the substrate. Furthermore, secondary structures move as units, so that the largest NMR spectral changes are observed at hinge regions that connect secondary structural features, rather than within the features themselves. Another noteworthy observation is that the features that are not perturbed upon substrate binding are the same in both enzymes: These include the “β-meander” that contains the axial heme thiolate ligand and helices associated with the meander. Furthermore, while overall sequence homology between the two enzymes is low (<24% identity), those regions that are unperturbed by substrate binding show much greater homology (63% identity, 78% similarity, no gaps). This suggests to us that these regions represent the core of the P450 molecule that must be conserved in order for the enzyme to be functional [19].
A preliminary report from the Estrada lab describes the effect of bimodal ligand binding on CYP121, a target for anti-tuberculosis drugs [37]. As spectra are of high quality, it is likely that CYP121 will prove to be a fruitful target for solution NMR investigations in the course of development of new drugs targeting this endemic disease.
Mining NMR-derived P450 structures as a guide to enzyme engineering.
An early report on directed evolution in CYP101A1 by the Arnold group noted that a single mutation remote from the active site was necessary to obtain the desired modified chemistry [38]. Later, our NMR characterization of substrate binding in that enzyme identified a series of hydrogen bonds that connected the mutated site to residues in the active site directly involved in substrate orientation [11]. Subsequent NMR-directed mutations in CYP101A1 confirmed that it is possible to identify functionally important positions in the P450 fold based on NMR observations [32].
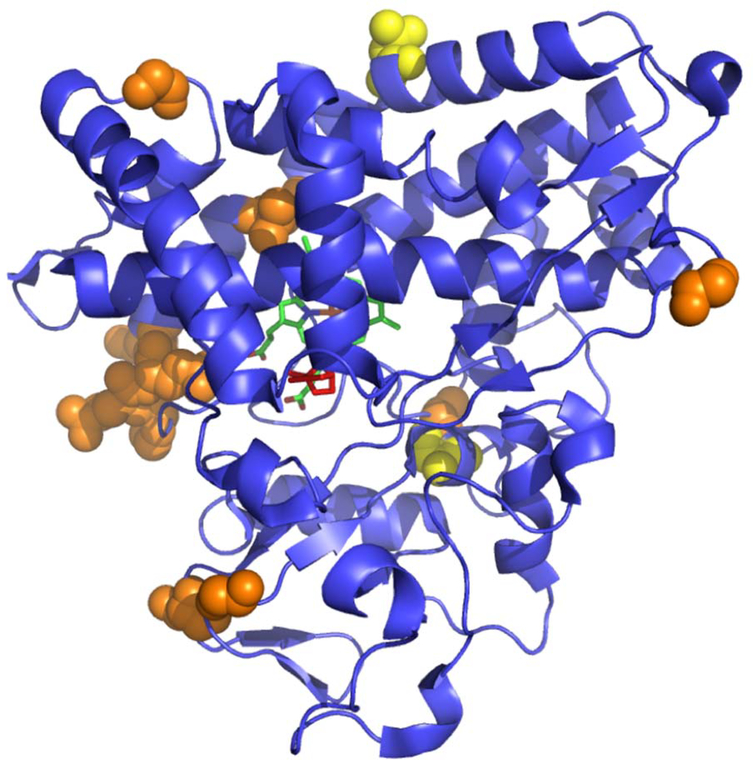
Of course, it is a long way between knowing which positions are functionally important and using that information to re-engineer an enzyme to a new function or improve upon an existing one. We are currently envisioning a hybrid NMR and computational approach to this problem. Using RDC-restrained molecular dynamics (MD) simulations, we have identified mechanical coupling pathways in the CYP101A1 structure that are sensitive both to the presence and orientation of substrate in the active site of the enzyme [39]. Long-range normal modes were identified that are active in presence of substrate but quiescent in the absence thereof. The mechanical couplings responsible for these modes pass through the active site (and substrate), linking points on opposing surfaces of the enzyme (Figure 3).
Figure 3.
Endpoints of normal modes active in CYP101A1 in the presence (orange) and absence (yellow) of bound substrate (+)-camphor. Heme is shown in green, camphor in red. Note that the substrate activated modes essentially bisect the molecule.
Particularly interesting is the sensitivity that these normal modes exhibit with respect to substrate orientation. In a series of MD runs, we imposed a dihedral restraint on substrate orientation: If the restraint keeps the substrate in the orientation most populated in the absence of that restraint, normal modes are damped relative to the unrestrained simulations, but follow the same mechanical coupling pathways. However, reorientation of the substrate by even a relatively small amount (10o) results in dramatic changes in those pathways. New, shorter modes appear with different endpoints than in the unrestrained calculations. Our interpretation of this phenomenon is that the CYP101A1 structural dynamics have evolved not only to select for a particular substrate, but to ensure the correct substrate orientation upon binding.
An intriguing example of the use of combined NMR and neutron diffraction data to examine collective motions in the thermostable CYP117 has been published from the Jain group [40].
While normal mode vibrational frequencies have not yet been calculated from the current simulations, it seems reasonable to assume that the longer the distance involved in the mode (wavelength), the lower the frequency of that mode, and the lower the energy of the associated vibration. It is true that substrate binding typically stabilizes a P450 (and many/most other enzymes!). As such, lowering the energies of vibrational modes provides a potential mechanism for this stabilization, and hints at a new approach to enzyme engineering. We are currently attempting to implement an NMR-directed computational/mutational approach to the problem of enzyme design. If best-fit substrates will activate the longest (i.e., “native-like”) normal modes, it should be possible to predict from MD simulations which mutations are required for the enzyme to accept a new substrate.
Outlook: The future of NMR for cytochromes P450.
Despite the fact that solution NMR of biomolecules is a relatively mature field, direct application to P450s is still relatively rare. Early published examples were almost exclusively centered on the interaction of substrate with the heme iron, making use of paramagnetic relaxation restraints to orient the substrate within the active site [41]. Two factors contribute to the reluctance of many researchers to apply NMR in their research. The first is simply spectral complexity: The large number of resonances to be assigned is daunting, and although it has now been accomplished for number of P450s, it is not a task to be undertaken lightly. Samples are typically perdeuterated and uniformly 15N and 13C labeled, requiring efficient expression in minimal and/or defined media, which can be expensive. Multiple selectively labeled samples are required for sorting resonances by amino acid type, decreasing ambiguity in the assignment process. While these are surmountable problems for soluble monomeric P450s, eukaryotic P450s are usually membrane-bound in vivo and often multimerize in solution, giving rise to severely broadened and uninformative spectra. Still, recent strides in using membrane mimetics such as nanodiscs [42] and improved techniques for examining membrane-bound biomolecules by NMR [43] make it likely that the use of solution NMR methods will continue to help unravel the mysteries of the P450 superfamily.
Acknowledgements:
TCP acknowledges partial support from NIH grant R01-GM……….
References
- 1.Pochapsky TC et al. (2010) Conformational plasticity and structure/function relationships in cytochromes P450. Antioxidants & Redox Signaling 13 (8), 1273–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen K and Arnold FH (2020) Engineering new catalytic activities in enzymes. Nature Catalysis 3 (3), 203–213. [Google Scholar]
- 3.Arnold FH (2018) Directed Evolution: Bringing New Chemistry to Life. Angewandte Chemie International Edition 57 (16), 4143–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farwell CC et al. (2015) Enantioselective Enzyme-Catalyzed Aziridination Enabled by Active-Site Evolution of a Cytochrome P450. Acs Central Science 1 (2), 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold FH (2015) The nature of chemical innovation: new enzymes by evolution. Quarterly Reviews of Biophysics 48 (4), 404–410. [DOI] [PubMed] [Google Scholar]
- 6.Jung ST et al. (2011) Cytochrome P450: taming a wild type enzyme. Current Opinion in Biotechnology 22, 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnaba C and Ramamoorthy A (2018) Picturing the Membrane-assisted Choreography of Cytochrome P450 with Lipid Nanodiscs. ChemPhysChem 19 (20), 2603–2613. [DOI] [PubMed] [Google Scholar]
- 8.Li S et al. (2020) Redox Partners: Function Modulators of Bacterial P450 Enzymes. Trends in Microbiology 28 (6), 445–454. [DOI] [PubMed] [Google Scholar]
- 9.Sligar SG (1976) Coupling of spin, substrate and redox equilibria in cytochrome P450. Biochemistry 15, 5399–5406. [DOI] [PubMed] [Google Scholar]
- 10.Pochapsky TC et al. (2017) NADH reduction of nitroaromatics as a probe for residual ferric form high-spin in a cytochrome P450. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1866, 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang M et al. (2011) Spring-loading the active site of cytochrome P450cam. Metallomics 3 (4), 339–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li SY et al. (2012) Substrate recognition by the multifunctional cytochrome P450 MycG in mycinamicin hydroxylation and epoxidation reactions. Journal of Biological Chemistry 287 (45). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rittle J and Green MT (2010) Cytochrome P450 Compound I: Capture, Characterization, and C-H Bond Activation Kinetics. Science 330, 933–937. [DOI] [PubMed] [Google Scholar]
- 14.Korzekwa K (2014) Enzyme Kinetics of Oxidative Metabolism: Cytochromes P450, Humana Press. [DOI] [PubMed] [Google Scholar]
- 15.Paddon CJ et al. (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496 (7446), 528–532. [DOI] [PubMed] [Google Scholar]
- 16.Poulos TL et al. (1987) High-resolution crystal structure of cytochrome P450cam. J Mol Biol 195 (3), 687–700. [DOI] [PubMed] [Google Scholar]
- 17.Wei JY et al. (2005) Detection of a high-barrier conformational change in the active site of cytochrome P450(cam) upon binding of putidaredoxin. Journal Of The American Chemical Society 127 (19), 6974–6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipscomb JD et al. (1976) Autooxidation and hydroxylation reactions of oxygenated cytochrome P450cam. Journal of Biological Chemistry 251 (4), 1116–1124. [PubMed] [Google Scholar]
- 19.Pochapsky TC and Pochapsky SS (2019) What your crystal structure will not tell you about enzyme function. Accounts of Chemical Research 52 (5), 1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura K et al. (1994) Significant contribution of arginine 112 and Its positive charge of Pseudomonas putida cytochrome P450(Cam) in the electron transport from putidaredoxin. Biochimica Et Biophysica Acta-Protein Structure and Molecular Enzymology 1207 (1), 40–48. [DOI] [PubMed] [Google Scholar]
- 21.Tripathi S et al. (2013) Structural basis for effector control and redox partner recognition in cytochrome P450. Science 340 (6137), 1227–1230. [DOI] [PubMed] [Google Scholar]
- 22.Hiruma Y et al. (2013) The structure of the cytochrome P450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography. Journal of Molecular Biology 425 (22), 4353–4365. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W et al. (2008) Solution NMR Structure of Putidaredoxin-Cytochrome P450cam Complex via a Combined Residual Dipolar Coupling-Spin Labeling Approach Suggests a Role for Trp106 of Putidaredoxin in Complex Formation. Journal of Molecular Biology 384 (2), 349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.OuYang B et al. (2008) A functional proline switch in cytochrome P450(cam). Structure 16 (6), 916–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asciutto EK et al. (2009) Structural and dynamic implications of an effector-induced backbone amide cis–trans Isomerization in cytochrome P450cam. Journal of Molecular Biology 388, 801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Follmer AH et al. (2019) Ligand and Redox Partner Binding Generates a New Conformational State in Cytochrome P450cam (CYP101A1). Journal of the American Chemical Society 141 (6), 2678–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrałojć W et al. (2017) Identification of productive and futile encounters in an electron transfer protein complex. Proceedings of the National Academy of Sciences of the United States of America 114 (10), E1840–E1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Estrada DF et al. (2014) Human Cytochrome P450 17A1 Conformational Selection. Journal of Biological Chemistry 289 (20), 14310–14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bart AG and Scott EE (2017) Structural and functional effects of cytochrome b5 interactions with human cytochrome P450 enzymes. J Biol Chem 292 (51), 20818–20833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estrada DF (2018) The cytochrome P450 24A1 interaction with adrenodoxin relies on multiple recognition sites that vary among species. Journal of Biological Chemistry 293 (11), 4167–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tietz DR et al. (2017) Substrate recognition by two different P450s: Evidence for conserved roles in a common fold. Scientific Reports 7, 13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colthart AM et al. (2016) Detection of substrate-dependent conformational changes in the P450 fold by nuclear magnetic resonance. Scientific Reports 6, 22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asciutto EK et al. (2011) Experimentally restrained molecular dynamics simulations for characterizing the open states of cytochrome P450(cam). Biochemistry 50 (10), 1664–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asciutto EK et al. (2012) Solution structural ensembles of substrate-free cytochrome P450cam. Biochemistry 51, 3383–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prestegard JH et al. (2004) Residual dipolar couplings in structure determination of biomolecules. Chemical Reviews 104 (8), 3519–3540. [DOI] [PubMed] [Google Scholar]
- 36.Tietz DR et al. (2017) Solution conformations and dynamics of substrate-bound cytochrome P450 MycG. Biochemistry 56, 2701–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Estrada DF et al. (2020) Solution NMR Studies of CYP121 of Mycobacterium tuberculosis; Evidence that Ligand Binding is Bi-modal. The FASEB Journal 34 (S1), 1–1. [Google Scholar]
- 38.Joo H et al. (1999) Laboratory evolution of peroxide-mediated cytochrome P450 hydroxylation. Nature 399 (6737), 670–673. [DOI] [PubMed] [Google Scholar]
- 39.Asciutto EK and Pochapsky TC (2018) Some surprising implications of NMR-directed simulations of substrate recognition and binding by cytochrome P450cam (CYP101A1). Journal of Molecular Biology 430 (9), 1295–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J et al. (2018) Entropic contribution to enhanced thermal stability in the thermostable P450 CYP119. Proceedings of the National Academy of Sciences 115, 201807473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Modi S et al. (1996) The catalytic mechanism of cytochrome P450 BM3 involves a 6 angstrom movement of the bound substrate on reduction. Nature Structural Biology 3 (5), 414–417. [DOI] [PubMed] [Google Scholar]
- 42.Mazhab-Jafari MT et al. (2015) Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proceedings of the National Academy of Sciences 112 (21), 6625–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puthenveetil R and Vinogradova O (2019) Solution NMR: A powerful tool for structural and functional studies of membrane proteins in reconstituted environments. Journal of Biological Chemistry 294 (44), 15914–15931. [DOI] [PMC free article] [PubMed] [Google Scholar]