TO THE EDITOR:
Central nervous system (CNS) recurrence is a devastating outcome after initial therapy for diffuse large B-cell (DLBCL) or high-grade B-cell lymphoma (HGBL), yet predicting the risk of CNS recurrence and selecting patients for aggressive CNS-directed prophylaxis remains difficult. In the rituximab era, 2% to 4% of patients with DLBCL/HGBL experience a CNS recurrence.1-4 The CNS International Prognostic Index (CNS-IPI) can identify a higher-risk group (with incidence 10% to 12%), but half of events occur among patients with low/intermediate scores.1 Increased risk observed with the dual-expresser (MYC/BCL2) immunophenotype, double-hit HGBL with MYC and BCL2 and/or BCL6 rearrangements, as well as involvement of the bone marrow, testis, or breast, highlights the need to elucidate biology associated with the CNS invasion potential in DLBCL/HGBL.5-7
Recently, genomic analyses have expanded our understanding of the molecular heterogeneity of DLBCL, beyond the cell-of-origin (COO) classification based on gene expression profiling.8-11 Multiplatform analyses encompassing point mutations, structural variants, and copy-number alterations can classify tumors into biologically different clusters.9-12 So far, no subgroup defined by genetic profiling has been associated with secondary CNS invasion.4,9,13 We hypothesized that specific genetic signatures may be associated with CNS recurrence. Identifying them could improve prediction and inform patient selection for CNS-directed therapy.
From our institutional registry, we identified 26 patients with DLBCL/HGBL who experienced either isolated CNS (n = 13) or systemic (non-CNS) recurrence (n = 13) after rituximab-based immunochemotherapy (Table 1 and Figure 1A). These 2 groups did not significantly differ in clinical characteristics, including extranodal involvement, COO designation,14 dual-expresser status, or time from diagnosis (Figure 1C-D) or end of first-line therapy (P = .49). However, the CNS group contained 4 double-hit (MYC/BCL2) lymphomas.15 Two patients with CNS recurrence had high CNS-IPI (both double-hit lymphomas), but 85% had low/intermediate scores. None had CNS invasion detected at diagnosis.
Table 1.
Clinicopathologic characteristics of patients with CNS or systemic (non-CNS) recurrence
| All (n = 26) | CNS recurrence (n = 13) | Systemic recurrence (n = 13) | P | |
|---|---|---|---|---|
| Age, median (range), y | 60 (32-84) | 61 (43-71) | 57 (32-84) | .74 |
| Sex, n (%) | .69 | |||
| M | 16 (62) | 9 (69) | 7 (54) | |
| F | 10 (38) | 4 (31) | 6 (46) | |
| HIV+, n (%) | 2 (8) | 1 (8) | 1 (8) | .99 |
| Stage 3 or 4, n (%) | 21 (81) | 10 (77) | 11 (85) | .99 |
| LDH, median (IQR), U/L | 306 (172-537) | 375 (170-668) | 284 (198-315) | .49 |
| Extranodal involvement, n (%) | 19 (73) | 10 (77) | 9 (69) | .99 |
| Testis | 4 | 3 | 1 | |
| Bone marrow | 5 | 2 | 3 | |
| Kidney | 2 | 1 | 1 | |
| Bone | 3 | 1 | 2 | |
| Gastrointestinal | 3 | 1 | 2 | |
| Uterus | 1 | 1 | 0 | |
| Skin (leg-type DLBCL) | 0 | 1 | 0 | |
| Dual expresser, n (%)* | 10 (43) | 6 (60) | 4 (31) | .22 |
| Double-hit lymphoma, n (%) | 4 (15) | 4 (31) | 0 (0) | .10 |
| Complex karyotype, n (%) | 5 (19) | 3 (23) | 3 (23) | .99 |
| COO by Hans algorithm | .70 | |||
| GCB, n (%) | 14 (54) | 6 (46) | 8 (62) | |
| non-GCB, n (%) | 12 (46) | 7 (54) | 5 (38) | |
| CNS-IPI | .19 | |||
| Low, n (%) | 4 (15) | 3 (23) | 1 (8) | |
| Intermediate, n (%) | 20 (77) | 8 (62) | 12 (92) | |
| High, n (%) | 2 (8) | 2 (15) | 0 (0) | |
| First-line regimen, n (%) | .99 | |||
| R-CHOP | 16 (62) | 8 (62) | 8 (62) | |
| DA-EPOCH-R | 9 (35) | 5 (38) | 4 (31) | |
| Other | 1 (4) | 0 (0) | 1 (8) | |
| CNS prophylaxis, n (%) | 10 (38) | 7 (54) | 3 (23) | .23 |
| Intrathecal | 9 (35) | 6 (46) | 3 (23) | |
| Systemic methotrexate | 3 (12) | 3 (23) | 0 (0) | .22 |
Excluding 3 cases with missing data.
DA-EPOCH-R, dose-adjusted etoposide, prednisone, vincristine, cyclophosphamide, doxorubicin, rituximab; F, female; GCB, germinal center B cell; IQR, interquartile range; LDH, lactate dehydrogenase; M, male; R-CHOP: rituximab, cyclophosphamide, doxorubicin, vincristine, prednisone.
Figure 1.
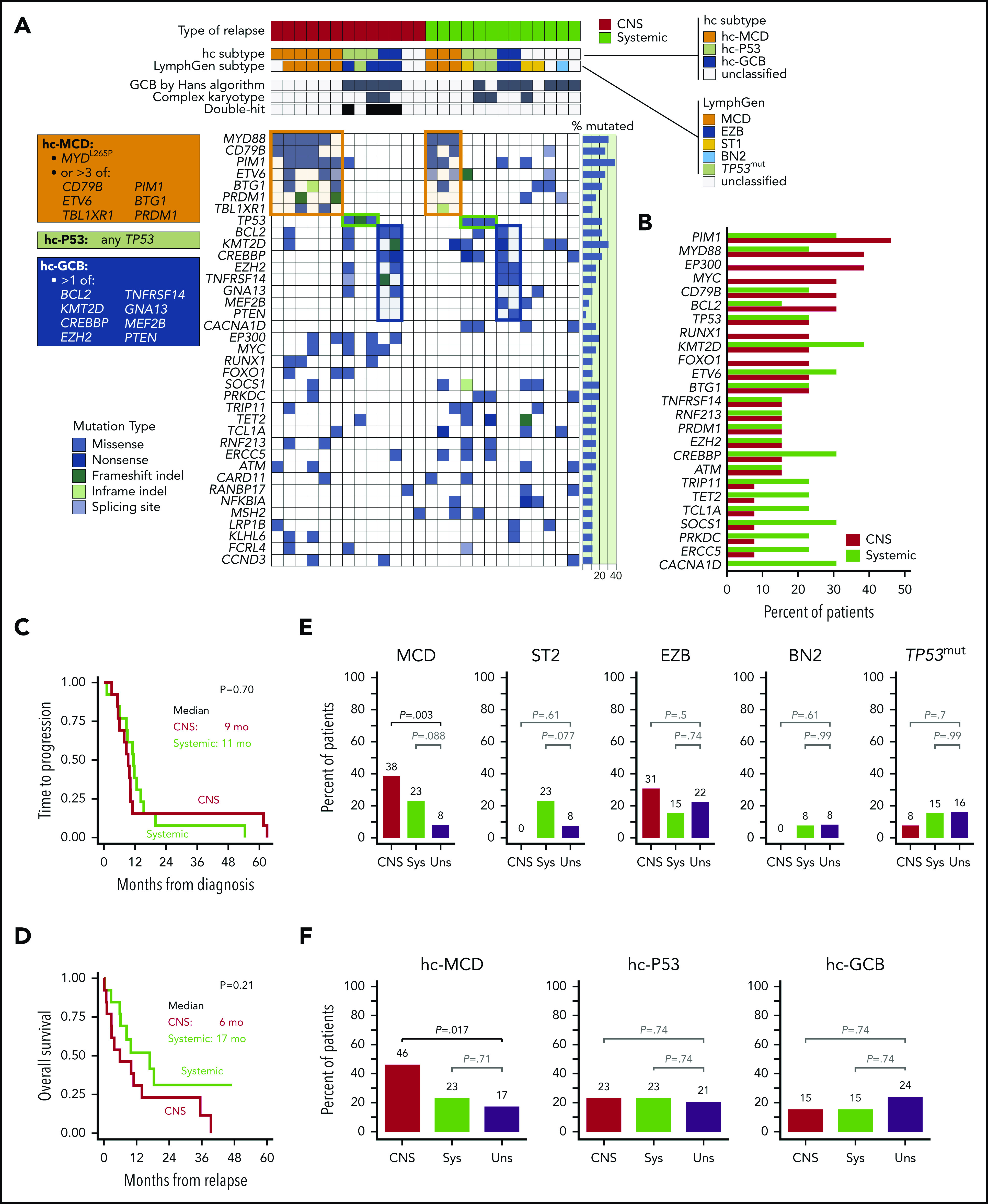
Clinicopathologic characteristics, mutations, and prevalence by lymphoma subtypes. (A) Clinicopathologic characteristics and mutations in patients with CNS or systemic (non-CNS) recurrence of DLBCL. Tumors were grouped into genomic subtypes according to the LymphGen classifier and hc. Boxes list definitions of the hc subgroups, and respective clusters are highlighted in the heatmap; only genes mutated in >10% patients are included. (B) Prevalence of mutated genes in groups with CNS or systemic (non-CNS) recurrence. Only genes mutated in >10% patients are included; no single gene was associated with CNS recurrence after adjustment for multiple testing. (C) Time from diagnosis to progression, stratified by type of recurrence. Median time to recurrence was 10 months (95% confidence interval, 8-11) and did not significantly differ between groups (log-rank P = .70). (D) Overall survival after recurrence. Median survival was shorter for patients with CNS recurrence (6 vs 17 months), but the difference was not statistically significant (log-rank P = .21). (E) Comparison of prevalence of specific LymphGen-determined DLBCL subtypes between patients with CNS or systemic-only (Sys) recurrence and unselected (Uns) DLBCL cases from 2 aggregated cohorts (N = 462)9,12,17; the MCD subtype was significantly more prevalent among patients with CNS recurrence (38% vs 8%, P = .003), whereas no CNS-relapsed tumor classified as ST2 or BN2 subtype. (F) Comparison of DLBCL subtypes defined by the hc between patients with CNS or systemic-only (Sys) recurrence and unselected DLBCL cases (Uns); the hc-MCD subtype was significantly more prevalent in tumors with CNS recurrence (46% vs 17%, P = .017), whereas other subtypes did not differ between groups.
We performed next-generation sequencing (NGS) using a clinically validated 592-gene assay (custom Agilent SureSelect XT assay; Illumina NextSeq platform; Caris Life Sciences, Irving, TX; supplemental Table 1, available on the Blood Web site) on formalin-fixed, paraffin-embedded specimens (77% from the initial diagnosis). All pathogenic or unclassified variants were included for analysis, while synonymous mutations were excluded according to published guidelines (supplemental Table 2).16 We input the data into the novel LymphGen classifier, which categorized DLBCL into subgroups defined as MCD (characterized by MYD88L265P, CD79B mutations, and immune evasion), ST2 (featuring SGK2 and TET2 mutations), EZB (characterized by epigenetic dysregulation), and BN2 (with NOTCH2 activation and frequent BCL6 rearrangements; supplemental Figure 1).12 Since the A53 (aneuploid/TP53-mutated) subtype could not be predicted without copy-number alteration data, we designated LymphGen-unclassifiable tumors with TP53 mutations as TP53mut. In addition, we constructed a simplified hierarchical classifier (hc) based on commonly mutated genes that are included in widely available, clinically validated NGS panels (augmented by BCL6 rearrangement from routine fluorescence in situ hybridization). This classifier distinguished 3 subtypes frequent in relapsed DLBCL (Figure 1A): (1) hc-MCD (tumors with MYD88L265P or >3 mutations in CD79B, PIM1, ETV6, BTG1, PRDM1, or TBL1XR1), (2) hc-P53 (all other tumors with TP53 mutations), and (3) hc-GCB (all remaining tumors with >1 mutation in BCL2, CREBBP, EZH2, KMT2D, TNFRSF14, GNA13, MEF2B, or PTEN). We compared prevalence of these subtypes within our groups with CNS or systemic (non-CNS) relapse vs 2 aggregated reference cohorts of unselected DLBCL from Dana-Farber Cancer Institute (N = 135) and British Columbia Cancer Agency (N = 327).9,12,17
Among our 26 cases of relapsed DLBCL/HBCL, PIM1 was the most frequently mutated gene (38%), followed by MYD88 and KMT2D (31%). No single gene was significantly associated with CNS-specific recurrence (Figure 1B). When compared with the reference cohorts, DLBCL with CNS recurrence had a significantly higher prevalence of the LymphGen MCD subtype (38% vs 8%, P = .003 on Fisher’s exact test; Figure 1E). The EZB subtype was also common (31%) but fully overlapped with the double-hit phenotype. No subset other than MCD significantly differed from the reference cohorts, although notably, none of the tumors with CNS recurrence were classified as ST2.
The simplified hierarchical classifier designated 84% of LymphGen MCD tumors as hc-MCD, and this subtype was identified in 46% of CNS recurrences (vs 17% in the reference cohorts; P = .017; Figure 1F). Furthermore, 55% of cases with low-to-intermediate CNS-IPI who subsequently experienced a CNS recurrence were classified as hc-MCD. In contrast, hc-MCD was not significantly overrepresented among tumors with systemic (non-CNS) recurrence (23%; P = .09). The hc-P53 and hc-GCB subtypes encompassed the double-hit cases in the CNS group, but their overall prevalence did not significantly differ from unselected DLBCL (P = .74). Overall, 85% of DLBCL/HGBL tumors with CNS recurrence could be assigned a specific hc subtype (vs 62% in reference datasets; P = .10).
We further validated our findings in 2 external datasets from a study by Reddy et al10 and GOYA, the largest phase 3 trial in DLBCL to date (supplemental Figures 2 and 3).18 Similar to our data, no single gene in these studies was significantly associated with CNS recurrence (vs other relapses). However, in the Reddy et al study, both the hc-MCD (29% vs 15%, P = .023) and LymphGen MCD (15% vs 4%; P = .009) subtypes were uniquely more prevalent in CNS recurrence. In GOYA, as in our data, 53% of patients experiencing CNS recurrence had low/intermediate CNS-IPI, and the dual-expresser status was not prognostic (P = .32), although combined CNS-IPI and COO improved the prediction of CNS recurrence.4 NGS, performed on 12 tumors with CNS relapse, found that CDKN2A deletion was the only alteration significantly associated with CNS recurrence (67%). When we applied the hc classifier to GOYA cases, 42% of cases with CNS relapse classified as hc-MCD; no cases had TP53 mutations or double-hit HGBL.
Our analysis of DLBCL/HGBL tumors with CNS recurrence compliments prior observations showing high prevalence of the MCD subtype in primary CNS lymphoma and in testicular, breast, or cutaneous leg-type DLBCL—extranodal sites historically associated with CNS invasion.7,12 The MCD subtype constituted nearly half of our cases with CNS recurrence, while other cases included mainly TP53-mutated or double-hit HGBL. CDKN2A deletion, a potentially targetable alteration,19,20 was significantly associated with CNS recurrence in both GOYA (P = .002) and Reddy datasets (P < .001), but was unfortunately not determined in our study. It might further improve CNS risk prediction.
To our knowledge, this analysis is the first to raise the hypothesis that a meaningful genetic prediction of CNS recurrence could be inferred from a commercially available NGS assay applicable in routine clinical practice. This prediction could neither be derived from any single-gene mutation, nor is it at present practical to obtain from comprehensive multiplatform analyses like the one used to build the LymphGen classifier. We found that most tumors with CNS recurrence are well defined molecularly and fall into 2 categories. The most frequent hc-MCD subtype (corresponding to DFCI cluster 5,9 the MCD LymphGen subtype,12 or cluster B by the LYSA classifier21) is easy to identify and molecularly resembles primary CNS lymphoma.13 Its association with some extranodal locations suggests that molecular underpinning rather than anatomic location may drive the risk of CNS invasion.7,22 The second subgroup encompasses high-grade tumors characterized by double-hit biology or TP53 mutations, which frequently exhibit HGBL signature on gene expression profiling.17,23 Further application of the molecular definition of DLBCL at risk for CNS recurrence will need validation in a prospective clinical trial. Identifying high-risk subsets using commercially available NGS panels at diagnosis may offer an opportunity to more precisely select patients for aggressive CNS-directed prophylaxis using high-dose methotrexate or intensive intrathecal therapy.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgment
Support for this study was provided by Caris Life Sciences.
Footnotes
Sequencing and clinical information have been submitted to COSMIC (COSP identifier: COSP48173).
Authorship
Contribution: T.A.O., H.K., N.R.P., and A.J.O. designed the research; J.W. collected clinical information; J.V. and A.S. provided support collecting pathology; T.A.O., P.M.D., H.K., and A.J.O. interpreted the data; T.A.O., J.W., A.J.O., D.O.T., P.M.D., H.K., and N.R.P. wrote the manuscript; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interest.
Correspondence: Thomas A. Ollila, Alpert Medical School of Brown University, Rhode Island Hospital, 593 Eddy St, Providence, RI 02903; e-mail: thomas_ollila@brown.edu.
REFERENCES
- 1.Schmitz N, Zeynalova S, Nickelsen M, et al. CNS International Prognostic Index: a risk model for CNS relapse in patients with diffuse large B-cell lymphoma treated with R-CHOP. J Clin Oncol. 2016;34(26):3150-3156. [DOI] [PubMed] [Google Scholar]
- 2.Gleeson M, Counsell N, Cunningham D, et al. Central nervous system relapse of diffuse large B-cell lymphoma in the rituximab era: results of the UK NCRI R-CHOP-14 versus 21 trial. Ann Oncol. 2017;28(10):2511-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett NL, Wilson WH, Jung SH, et al. Dose-adjusted EPOCH-R compared with R-CHOP as frontline therapy for diffuse large B-cell lymphoma: clinical outcomes of the phase III Intergroup Trial Alliance/CALGB 50303. J Clin Oncol. 2019;37(21):1790-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klanova M, Sehn LH, Bence-Bruckler I, et al. Integration of cell of origin into the clinical CNS International Prognostic Index improves CNS relapse prediction in DLBCL. Blood. 2019;133(9):919-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savage KJ, Slack GW, Mottok A, et al. Impact of dual expression of MYC and BCL2 by immunohistochemistry on the risk of CNS relapse in DLBCL. Blood. 2016;127(18):2182-2188. [DOI] [PubMed] [Google Scholar]
- 6.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ollila TA, Olszewski AJ. Extranodal diffuse large B cell lymphoma: molecular features, prognosis, and risk of central nervous system recurrence. Curr Treat Options Oncol. 2018;19(8):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503-511. [DOI] [PubMed] [Google Scholar]
- 9.Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes [published correction appears in Nat Med. 2018;24:1292]. Nat Med. 2018;24(5):679-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy A, Zhang J, Davis NS, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171(2):481-494.e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright GW, Huang DW, Phelan JD, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 2020;37(4):551-568.e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapuy B, Roemer MG, Stewart C, et al. Targetable genetic features of primary testicular and primary central nervous system lymphomas. Blood. 2016;127(7):869-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275-282. [DOI] [PubMed] [Google Scholar]
- 15.Decker DP, Egan PC, Zayac AS, Treaba DO, Olszewski AJ. Treatment strategies and risk of central nervous system recurrence in high-grade B-cell and Burkitt lymphoma. Leuk Lymphoma. 2020;61(1):198-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ennishi D, Jiang A, Boyle M, et al. Double-hit gene expression signature defines a distinct subgroup of germinal center B-cell-like diffuse large B-cell lymphoma. J Clin Oncol. 2019;37(3):190-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jardin F, Jais JP, Molina TJ, et al. Diffuse large B-cell lymphomas with CDKN2A deletion have a distinct gene expression signature and a poor prognosis under R-CHOP treatment: a GELA study. Blood. 2010;116(7):1092-1104. [DOI] [PubMed] [Google Scholar]
- 20.Karube K, Enjuanes A, Dlouhy I, et al. Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets. Leukemia. 2018;32(3):675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubois S, Tesson B, Mareschal S, et al. ; Lymphoma Study Association (LYSA) investigators . Refining diffuse large B-cell lymphoma subgroups using integrated analysis of molecular profiles. EBioMedicine. 2019;48:58-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardette E, Maraval A, Brunet-Possenti F, et al. Central nervous system involvement of primary cutaneous diffuse large B-cell lymphoma, leg type: 13 cases. J Eur Acad Dermatol Venereol. 2017;31(11):e498-e501. [DOI] [PubMed] [Google Scholar]
- 23.Sha C, Barrans S, Cucco F, et al. Molecular high-grade B-cell lymphoma: defining a poor-risk group that requires different approaches to therapy. J Clin Oncol. 2019;37(3):202-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


