Abstract
Vibrio parahaemolyticus, the leading cause of seafood-associated gastroenteritis worldwide, has a strong ability to form biofilms on surfaces. Quorum sensing (QS) is a process widely used by bacteria to communicate with each other and control gene expression via the secretion and detection of autoinducers. OpaR is the master QS regulator of V. parahaemolyticus operating under high cell density (HCD). OpaR regulation of V. parahaemolyticus biofilm formation has been reported, but the regulatory mechanisms are still not fully understood. bis-(3′-5′)-cyclic di-GMP (c-di-GMP) is an omnipresent intracellular second messenger that regulates diverse behaviors of bacteria including activation of biofilm formation. In this work, we showed that OpaR repressed biofilm formation and decreased the intracellular concentration of c-di-GMP in V. parahaemolyticus RIMD2210633. The OpaR box-like sequences were detected within the regulatory DNA regions of scrA, scrG, VP0117, VPA0198, VPA1176, VP0699, and VP2979, encoding a group of GGDEF and/or EAL-type proteins. The results of qPCR, LacZ fusion, EMSA, and DNase I footprinting assays demonstrated that OpaR bound to the upstream DNA regions of scrA, VP0117, VPA0198, VPA1176, and VP0699 to repress their transcription, whereas it positively and directly regulated the transcription of scrG and VP2979. Thus, transcriptional regulation of these genes by OpaR led directly to changes in the intracellular concentration of c-di-GMP. The direct association between QS and c-di-GMP metabolism in V. parahaemolyticus RIMD2210633 would be conducive to precise control of gene transcription and bacterial behaviors such as biofilm formation.
Keywords: Vibrio parahaemolyticus, biofilm, quorum sensing, OpaR, c-di-GMP
Introduction
Vibrio parahaemolyticus, a Gram-negative, rod-shaped, halophilic bacterium, is responsible for the most common seafood-associated gastroenteritis that is often caused by the eating of raw or undercooked seafood (Broberg et al., 2011). V. parahaemolyticus has a strong ability to form biofilms on surfaces, which can enhance the survival rate of the bacterial cells in adverse growth conditions as well as the pathogenicity of the bacteria (Yildiz and Visick, 2009; Li et al., 2020). Biofilm formation of V. parahaemolyticus requires many specific structures and substances including flagella, type IV pili, capsular polysaccharide, exopolysaccharide, proteins, and extracellular DNA (Enos-Berlage et al., 2005; Shime-Hattori et al., 2006; Yildiz and Visick, 2009; Li et al., 2020). Some regulators such as OxyR (Chung et al., 2016), ToxR (Chen et al., 2018), QsvR (Enos-Berlage et al., 2005), and CpsR (Guvener and McCarter, 2003) and some regulatory processes such as quorum sensing (QS) (Yildiz and Visick, 2009; Ball et al., 2017; Lu et al., 2018) and bis-(3′-5′)-cyclic di-GMP (c-di-GMP) (Yildiz and Visick, 2009) are also required for the regulation of mature biofilm formation by V. parahaemolyticus.
Cyclic-di-GMP is a ubiquitous second messenger that controls diverse behaviors of bacteria including motility, virulence, and biofilm formation (Jenal et al., 2017). High concentrations of intracellular c-di-GMP promote the formation of biofilms but inhibit the motility of bacteria (Jenal et al., 2017). c-di-GMP synthesis is catalyzed by diguanylate cyclases (DCs) containing a GGDEF domain with GTP as the substrate, whereas its degradation is catalyzed by phosphodiesterases (PDEs) carrying either an HD-GYP or EAL domain (Jenal et al., 2017). V. parahaemolyticus harbors dozens of genes that encode proteins containing GGDEF and/or EAL (or HD-GYP) domains (Makino et al., 2003). The two proteins, ScrG and ScrC, contain both GGDEF and EAL domains, but they act only as PDEs to degrade c-di-GMP in wild-type V. parahaemolyticus strains (Boles and McCarter, 2002; Kim and McCarter, 2007; Ferreira et al., 2008; Trimble and McCarter, 2011). ScrC, which is encoded by the scrABC operon, acts as a DC to catalyze the synthesis of c-di-GMP in the absence of ScrA and ScrB, whereas it serves on a PDE to degrade c-di-GMP in the presence of ScrA and ScrB (Boles and McCarter, 2002; Ferreira et al., 2008; Trimble and McCarter, 2011). Overexpression of ScrG was sufficient to decrease biofilm formation and intracellular c-di-GMP, whereas removal of the EAL domain converts ScrG to an activator of biofilm formation and c-di-GMP synthesis (Kim and McCarter, 2007). Double gene mutant of scrG and scrC does not swarm but produces a more highly wrinkled colony than those of the single-gene mutants, suggesting that they have cumulative effects on biofilm formation (Kim and McCarter, 2007). ScrO, a newly described GGDEF-type protein, is encoded by the scrMNO operon (Kimbrough et al., 2020). Deleting scrO or the entire operon decreases biofilm formation and alters biofilm architecture and matrix production visualized on Congo red (Kimbrough et al., 2020). Three genes, scrJ and scrL which encode proteins with GGDEF domains, and lafV which encodes an EAL domain, were recently found to negatively influence swarming motility in V. parahaemolyticus (Kimbrough and McCarter, 2020).
Quorum sensing is a cell-to-cell communication process widely used by bacteria to control gene expression in response to the concentration change of signaling molecules called autoinducers (AIs) in surroundings (Ng and Bassler, 2009). V. parahaemolyticus produces three types of AIs: harveyi autoinducer 1 (HAI-1), autoinducer 2 (AI-2), and cholerae autoinducer 1 (CAI-1) (Zhang et al., 2012). At low cell density (LCD), low concentrations of AIs initiate a phosphorylation cascade, which ultimately leads to a high expression level of AphA and a low expression level of OpaR (Lu et al., 2018; Sun et al., 2012). Highly expressed AphA regulates many cellular pathways such as virulence gene expression, motility, and biofilm formation (Lu et al., 2018). By contrast, at high cell density (HCD), high levels of AIs lead to a low expression level of AphA and a high expression level of OpaR (Sun et al., 2012; Zhang et al., 2012). OpaR then regulates transcription of many genes including those associated with virulence, motility, and biofilm formation (Henke and Bassler, 2004; Gode-Potratz and McCarter, 2011; Zhou et al., 2013; Ball et al., 2017; Lu et al., 2019; Zhang et al., 2019). Thus, AphA and OpaR are the master QS regulators in V. parahaemolyticus that exert their major roles at LCD and HCD, respectively.
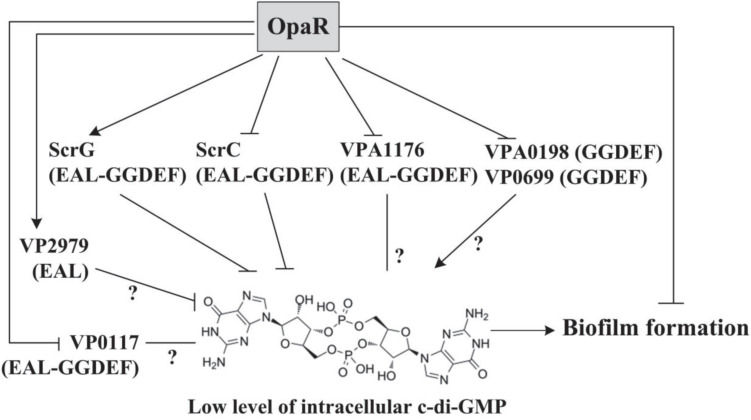
Vibrio parahaemolyticus QS regulation of biofilm formation via its master regulators AphA and OpaR has been reported (Enos-Berlage and McCarter, 2000; Guvener and McCarter, 2003; Enos-Berlage et al., 2005; Wang et al., 2013), but the regulation mechanisms are still not fully understood to date. In the present work, the results showed that OpaR repressed the biofilm formation by V. parahaemolyticus RIMD2210633 at least in part by reducing the intracellular c-di-GMP concentration (Figure 1). The data also showed that OpaR directly regulated the transcription of scrA, scrG, VP0117, VPA0198, VPA1176, VP0699, and VP2979, encoding a group of proteins with GGDEF and/or EAL domains (Figure 1). Thus, one of the mechanisms of OpaR-dependent biofilm formation in V. parahaemolyticus RIMD2210633 is that OpaR can alter the intracellular pool of c-di-GMP.
FIGURE 1.
Regulatory model. The arrow lines indicate positive regulation. The T-junctions indicate negative regulation. The question marks indicate that the roles of the protein need to be further investigated. The roles of ScrC and ScrG have been well studied in V. parahaemolyticus BB22.
Materials and Methods
Mutation of opaR and Complementation of the opaR Mutant
Vibrio parahaemolyticus RIMD2210633 was used as the wild-type (WT) strain in the current study (Makino et al., 2003). Non-polar opaR deletion mutant ΔopaR and the complementation of the opaR mutant (C-ΔopaR) were constructed in our previous studies (Zhang et al., 2012, 2019). Control strains were constructed by transforming the empty pBAD33 vector into WT and ΔopaR to counteract the effects of arabinose and chloramphenicol on bacterial growth and physiology (Sun et al., 2014). All the primers used are listed in Table 1.
TABLE 1.
Oligonucleotide primers used in this study.
| Target | Primers (forward/reverse, 5′–3′) |
| Protein expression | |
| opaR | AGCGGGATCCATGGACTCAATTGCAAAGAG/AGCGAAGCTTTTAGTGTTCGCGATTGTAG |
| qPCR | |
| scrA | CACACCACGAACACATTGC/TCAATAGCGTCACGGAATGC |
| scrG | AAGCCGTGGTGGAAGAAGG/GCGTGTTGAGTGCGTTGG |
| VP0117 | GACCACCTCAATAGTTATCTG/TAAGTAGGCTTGGACATCTC |
| VPA0198 | GCATCAGAATCAGCAAGAC/ATGCTTAGCTCCTCTTCTTC |
| VPA1176 | GCCATATTCCAAACTCGTTGTG/TGCGTAAGCCAAGTTGATGAG |
| VP0699 | CTGACACATCGTGATACTTC/TTGATGTTGCAGCTCTTG |
| VP2979 | GCAACTCTCAAGTCATCATC/CAACAACCGTCTTCTATGG |
| Primer extension | |
| scrA | /GACTTTAGTTCCACTTTTTTAGC |
| scrG | /ACTTAGTCAACAGTAAATCGTG |
| VP0117 | /GACAATCACACCGATAGCCAG |
| VPA1176 | /CACCCCATTCTGTCCAT |
| LacZ fusion | |
| scrA | GCGCGTCGACCATCAAGCCATTTTATGAAAC/GCGCGAATTCGTCGGCTGCGATTAGTCTG |
| scrG | GCGCGTCGACTAGCACGCTTGTGTTGGAC/GCGCGAATTCCAGGGAAATGAAGTAATCATGC |
| VP0117 | GCGCTCTAGACTCACACAACACTTTCTG/GCGCGAATTCAGACAATCACACCGATAG |
| VPA0198 | GCGCGTCGACCTCTGGTTCATTGTCTTG/GCGCGAATTCGTCTTGCTGATTCTGATG |
| VPA1176 | AAAGTCGACTCAGGTACGCTTGCTTCAC/GGGGAATTCCGTTGCTTGGTAGTGGTAATAG |
| VP0699 | GCGCGTCGACGGAGAATACCTAGCAGAG/GCGCGAATTCAGTATCACGATGTGTCAG |
| VP2979 | GCGCTCTAGATTTCTATCCGTTGGCTAC/GCGCGAATTCCTGACTTACATCGTGGAC |
| EMSA | |
| scrA | GAGCGTATATCCAAGTGGTTTG/GTCGGCTGCGATTAGTCTG |
| scrG | TAGCACGCTTGTGTTGGAC/CAGGGAAATGAAGTAATCATGC |
| VP0117 | CTCACACAACACTTTCTG/GCGCGAATTCAGACAATCACACCGATAG |
| VPA0198 | CTCTGGTTCATTGTCTTG/GTCTTGCTGATTCTGATG |
| VPA1176 | TCAGGTACGCTTGCTTCAC/CGTTGCTTGGTAGTGGTAATAG |
| VP0699 | GGAGAATACCTAGCAGAG/AGTATCACGATGTGTCAG |
| VP2979 | TTTCTATCCGTTGGCTAC/CTGACTTACATCGTGGAC |
| 16S rDNA | GACACGGTCCAGACTCCTAC/GGTGCTTCTTCTGTCGCTAAC |
| DNase I footprinting | |
| scrA | TCGCAGTTGAACAAATATGACG/CCATTAAGTGACTTTAGTTCC |
| scrG | TAAATACTATTAGGTAACGCG/CAGGGAAATGAAGTAATCATGC |
| VP0117 | CACACTAAAGGTCACAAGCAAG/AGACAATCACACCGATAG |
| VPA0198 | CTTACCTTGGCGTACCTCTTG/ACTGCTTTCTGGGTGAATAACG |
| VPA1176 | GCCATATTCCAAACTCGTTGTG/TGCGTAAGCCAAGTTGATGAG |
| VP0699 | CCACTCCTCTTACAAGAATGAG/AATATCGGATTCCAAGATTC |
| VP2979 | AGCAGTTCCTCGATTTACCTG/GCTTTTCTAGGCACAAAATGAC |
| 5′-RACE | |
| VPA0198 | GCTGATGGCGATGAATGAACACTG/AAAGCGAAAAAGAACAGTGC |
| GAACACTGCGTTTGCTGGCTTTGATG/ACTGCTTTCTGGGTGAATAACG | |
Growth Conditions
Vibrio parahaemolyticus strains were grown as described previously (Zhang et al., 2012; Lu et al., 2019). Briefly, bacterial cells were cultured in 2.5% (w/v) Bacto Heart Infusion (HI) broth (BD Biosciences, San Jose, CA, Untied States) at 37°C with shaking at 200 rpm for 12–14 h. The resultant culture was diluted 40-fold into the phosphate-buffered saline buffer (pH 7.2), and 150 μl of the diluted cells was spread onto an HI plate with a diameter of 5 cm. The bacterial cells were harvested after 6 h of incubation at 37°C to simulate the HCD condition at which OpaR was maximally produced (Lu et al., 2019). When necessary, the medium was supplemented with 100 μg/ml gentamicin, 5 μg/ml chloramphenicol, or 0.1% arabinose.
Colony Morphology
The colony morphology assay was performed as previously described (Fang et al., 2013). Briefly, V. parahaemolyticus strains were grown overnight in HI broth, and 2 μl of each resultant culture was spotted onto the HI agar. The plates were incubated at 37°C for at least 2 days.
Crystal Violet Staining
The crystal violet (CV) staining was performed as previously described (Fang et al., 2013). Briefly, overnight cell cultures were diluted 50-fold into 5 ml of fresh HI broth and cultured at 37°C to OD600 = 1.2–1.4. The resulting cultures were 50-fold diluted respectively into 2 ml of fresh Difco marine (M) broth 2216 (BD Biosciences, Untied States) containing 0.1% arabinose and 5 μg/ml chloramphenicol in glass tubes and then grown at 30°C with shaking at 150 rpm for 48 h. Media with planktonic cells were collected for measurement of OD600 values. The surface-attached cells were stained with 0.1% CV. The bound CV was then dissolved with 20% ethanol, and the OD570 values were determined. Relative biofilm formation was calculated with the formula: 100 × OD570/OD600.
Quantification of c-di-GMP
The bacterial cells were harvested after 6 h of incubation at 37°C to measure the intracellular concentration of c-di-GMP, which was performed exactly as previously described (Gao et al., 2020).
Quantitative PCR
The quantitative PCR (qPCR) assay was performed as previously described (Gao et al., 2011). Briefly, total RNAs were extracted from the WT and ΔopaR strains, respectively. The cDNAs were generated using 1–2 μg of total RNAs and 3 μg of random hexamer primers. The relative mRNA levels of each target gene were determined based on the standard curve for 16S rRNA expression for each RNA preparation. The experiment was performed at least three independent times.
LacZ Fusion and β-Galactosidase Assay
For the lacZ fusion assay (Sun et al., 2014; Zhang et al., 2017), the regulatory DNA region of each gene was cloned into the pHRP309 plasmid harboring a promoterless lacZ gene and a gentamicin resistance gene (Parales and Harwood, 1993). After verification by DNA sequencing, each recombinant plasmid was transferred to WT and ΔopaR, respectively. The resulting transformants were cultured and lysed to measure β-galactosidase activity in the supernatants using a β-Galactosidase Enzyme Assay System (Promega, Untied States). The two-plasmid reporter assay in Escherichia coli was exactly performed as previously described (Qiu et al., 2020). Briefly, E. coli 100 λpir (Epicentre) bearing the indicated opaR expression plasmid pBAD33-opaR or the empty pBAD33 plasmid and one of the recombinant lacZ plasmids was cultured overnight in Luria–Bertani (LB) broth at 37°C. The overnight cultures were diluted 1:100 into 5 ml of fresh LB broth containing 0.2% arabinose, and incubated at 37°C to the mid-log phase (OD600 = 1.2).
Preparation of 6× His-Tagged OpaR
The coding region of opaR was cloned into the pET28a plasmid (Novagen, Madison, WI, Untied States) and then transferred into E. coli BL21λDE3 to express His-OpaR (Kleber-Janke and Becker, 2000). Purification of overexpressed His-tagged OpaR (His-OpaR) was performed as previously described (Zhang et al., 2012). The dialyzed His-OpaR was concentrated to a final concentration of 0.3 to 0.6 mg/ml. The purity of the resulting protein was analyzed by SDS-12% PAGE.
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay (EMSA) was performed exactly as previously described (Zhang et al., 2012). The EMSA result was detected by autoradiography using the Fuji Medical X-ray film (Fuji Photo Film Co., Tokyo, Japan).
DNase I Footprinting
The DNase I footprinting assay was performed exactly as previously described (Zhang et al., 2012, 2017). The results were detected by autoradiography using the Fuji Medical X-ray film (Fuji Photo Film Co., Tokyo, Japan).
Primer Extension Assay
The primer extension assay was performed exactly as previously described (Zhang et al., 2012, 2017). The results were detected by autoradiography using the Fuji Medical X-ray film (Fuji Photo Film Co., Tokyo, Japan).
Experimental Replicates and Statistical Methods
Colony morphology, primer extension, EMSA, and DNase I footprinting assays were the result of two independent experiments with similar (or the same) results. The c-di-GMP quantification, CV staining, LacZ fusion, and qPCR results were of at least three independent bacterial cultures, with values expressed as the means ± standard deviation (SD). Paired Student’s t-tests were used to calculate statistical significance with P < 0.01 considered significant.
Results
OpaR Represses Biofilm Formation of V. parahaemolyticus
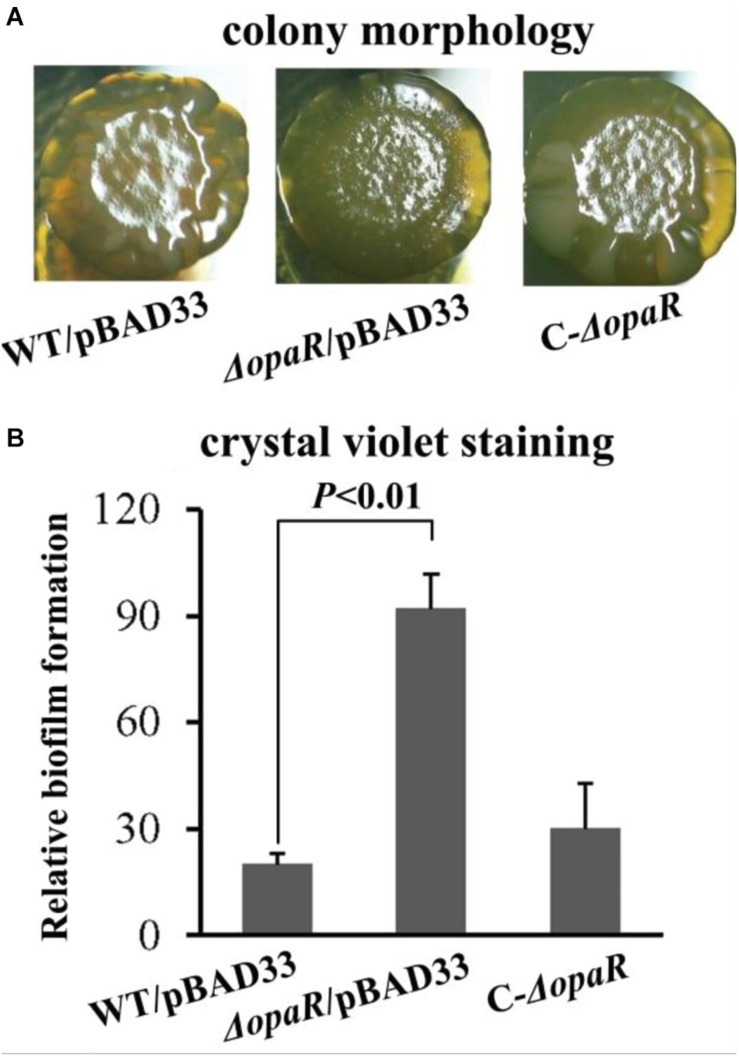
Reports have demonstrated that OpaR controls the expression of numerous genes including those involved in biofilm formation in V. parahaemolyticus BB22 (Gode-Potratz and McCarter, 2011; Gode-Potratz et al., 2011; Kernell Burke et al., 2015). However, a study revealed that there are a number of differences in genome sequences between BB22 and RIMD2210633 (Jensen et al., 2013), which may result in different regulatory patterns of OpaR on its target genes between the two strains. In this study, we characterized the biofilm formation regulated by OpaR in V. parahaemolyticus RIMD2210633 by comparing biofilm morphology and quantity in WT and ΔopaR. As shown in Figure 2A, ΔopaR/pBAD33 produced a wrinkled colony, whereas WT/pBAD33 and the complementary opaR mutant (C-ΔopaR) formed much smoother colonies than ΔopaR/pBAD33. The in vitro surface-attached biofilms of V. parahaemolyticus strains were subsequently assessed using the CV staining. As shown in Figure 2B, ΔopaR/pBAD33 produced a significant increase in normalized CV staining relative to WT/pBAD33 and C-ΔopaR, while C-ΔopaR gave a restored CV staining. These results suggested that OpaR repressed the biofilm formation by V. parahaemolyticus RIMD2210633.
FIGURE 2.
OpaR represses biofilm formation by V. parahaemolyticus RIMD2210633. Relative biofilms of V. parahaemolyticus RIMD2210633 were measured by rugose colony morphology (A) and intensity of crystal violet staining (B). Pictures presented were representative of three independent experiments with three replicates each.
Deletion of opaR Increases the Intracellular c-di-GMP Level
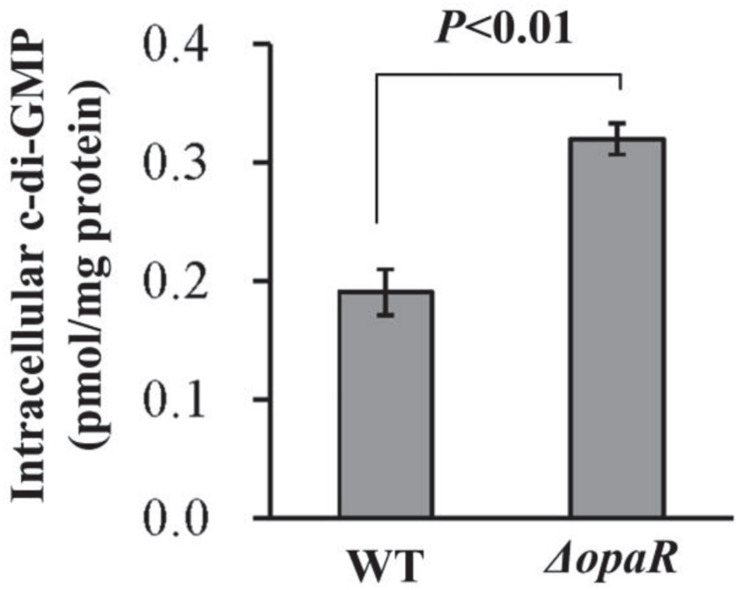
In Vibrio, increased c-di-GMP level results in decreased swarming motility and enhanced biofilm formation (Yildiz and Visick, 2009). Thus, increased ability of biofilm formation by ΔopaR promoted us to detect whether OpaR regulated c-di-GMP production in V. parahaemolyticus RIMD2210633. As shown in Figure 3, a significantly enhanced intracellular c-di-GMP level was observed for ΔopaR relative to that for WT, suggesting that OpaR inhibited the production of c-di-GMP in V. parahaemolyticus RIMD2210633.
FIGURE 3.
Intracellular c-di-GMP concentrations in V. parahaemolyticus RIMD2210633. Bacterial cells were harvested after 6 h of incubation at 37°C on HI plates. The data are expressed as the mean ± SD of at least three independent experiments.
Predicted OpaR Box-Like Sequences Within the Regulatory Regions of Target Genes
The V. parahaemolyticus RIMD2210633 genome harbors at least 50 genes that encode proteins containing GGDEF and/or EAL (or HD-GYP) domains, and their expression or not may influence the intracellular c-di-GMP levels and biofilm formation (Makino et al., 2003). The 500-bp upstream DNA sequences of these genes were downloaded from the RIMD2210633 genome with the online “retrieve-seq” program (van Helden, 2003). The DNA-binding box OpaR was then applied to predict the presence of OpaR box-like sequences within these DNA regions using the matrix scan tool (van Helden, 2003; Zhang et al., 2012). The method generated weight scores for each DNA sequence. A higher score represented a higher probability for direct binding of OpaR. When a score of 6 was used as the cutoff value, the OpaR box-like sequences were identified in the regulatory regions of scrA, scrG, VP0117, VPA0198, VPA1176, VP0699, and VP2979 (Table 2), and thus, these genes were selected as target genes for subsequent gene regulation studies.
TABLE 2.
Predicted OpaR box-like sequences within target promoters.
| Operon | First gene | OpaR box-like sequence |
||||
| Position& | Start | End | Sequence | Score | ||
| scrABC | scrA | D | −260 | −241 | TATAGATAAAACTATTATTA | 11.23 |
| scrG | D | −254 | −235 | TAATGAGTATTCAGTCAATT | 10.31 | |
| VP0117 | D | −55 | −36 | TATTAACAGAAATGTCAGTA | 12.13 | |
| VPA0198 | D | −63 | −44 | TAATAAGAATTTTAACAATA | 10.6 | |
| VPA1175-VPA1176 | VPA1176 | D | −175 | −156 | TAATGACAATTCAATCATTT | 10.76 |
| VP0699 | D | −66 | −47 | TATAAAGTTTTTTGTTATTA | 8.1 | |
| VP2979 | D | −232 | −213 | TATGAATAAAAATGTCATTA | 9.5 | |
&“D”indicates the direct sequence. Minus numbers denote the nucleotide positions upstream of the translation start sites of the indicated genes.
OpaR Regulates the Transcription of c-di-GMP Metabolism-Related Genes
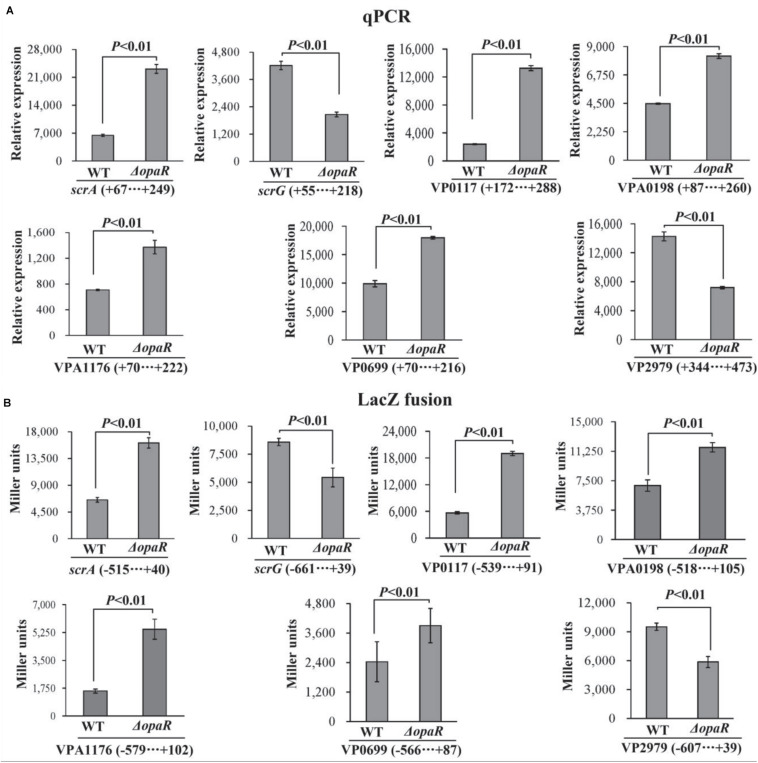
The bacterial cells were harvested after 6 h of incubation at 37°C on HI plates, at which the expression level of OpaR was the highest (Lu et al., 2019). The results of qPCR showed that the mRNA levels of scrA, VP0117, VPA0198, VPA1176, and VP0699 significantly increased, whereas those of scrG and VP2979 significantly decreased, in ΔopaR relative to those in WT (Figure 4A). To further investigate whether OpaR has regulatory actions on the promoter activity of each target gene, the upstream DNA region of each target gene was cloned into the pHRP309 vector containing a promoterless lacZ gene. The recombinant plasmid for each target gene was then transferred into ΔopaR and WT to test β-galactosidase activity in the supernatants. As shown in Figure 4B, the promoter activities of scrA, VP0117, VPA0198, VPA1176, and VP0699 were all much higher in ΔopaR than those in WT, whereas those of scrG and VP2979 were significantly lower in ΔopaR relative to those in WT. These results suggested that V. parahaemolyticus RIMD2210633 OpaR acted as a negative regulator of scrA, VP0117, VPA0198, VPA1176, and VP0699, but it acted as a positive regulator of scrG and VP2979.
FIGURE 4.
OpaR regulates the transcription of c-di-GMP metabolism-related genes in V. parahaemolyticus RIMD2210633. The negative and positive numbers indicate the nucleotide positions upstream and downstream of indicated genes, respectively. Bacterial cells were harvested after 6 h of incubation at 37°C on HI plates, and total RNAs were extracted using TRIzol Reagent. (A) qPCR assay was used to determine the relative mRNA levels for each target gene in ΔopaR and WT using a standard curve of 16S rRNA expression. (B) LacZ fusion. The regulatory DNA region of each target gene was cloned into the pHRP309 plasmid and then transferred into ΔopaR and WT to determine the promoter activity in cellular extracts.
OpaR Regulates the Expression of Target Genes in a Heterologous Host
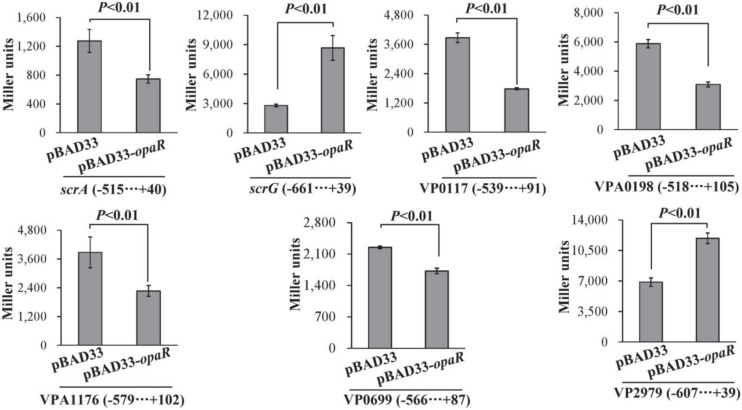
To detect whether OpaR can regulate the expression of target genes in a heterologous host, we expressed OpaR from the pBAD33 arabinose-inducible promoter in the E. coli 100 λpir strain, which also contained the recombinant pHRP309 plasmid of one of the target genes (Qiu et al., 2020). As shown in Figure 5, the expression of opaR from pBAD33-opaR led to a significantly lower promoter activity for each of scrA, VP0117, VPA0198, VPA1176, and VP0699 but a significantly higher promoter activity for each of scrG and VP2979, when compared to the E. coli bearing the empty pBAD33. These results suggested that, in E. coli, OpaR bound to the promoter DNA regions of scrA, VP0117, VPA0198, VPA1176, and VP0699 to repress their expression, whereas it bound to the promoter DNA regions of scrG and VP2979 to activate their transcription.
FIGURE 5.
Two-plasmid reporter assay. E. coli strains containing the empty pBAD33 plasmid or the indicated opaR expression plasmid pBAD33-opaR and one of the recombinant lacZ plasmids were grown in LB broth containing 0.2% arabinose to mid-log phase (OD600 = 1.2), at which point aliquots were collected and assayed for lacZ expression using the β-galactosidase assay. The negative and positive numbers indicate the nucleotide positions upstream and downstream of the indicated genes, respectively.
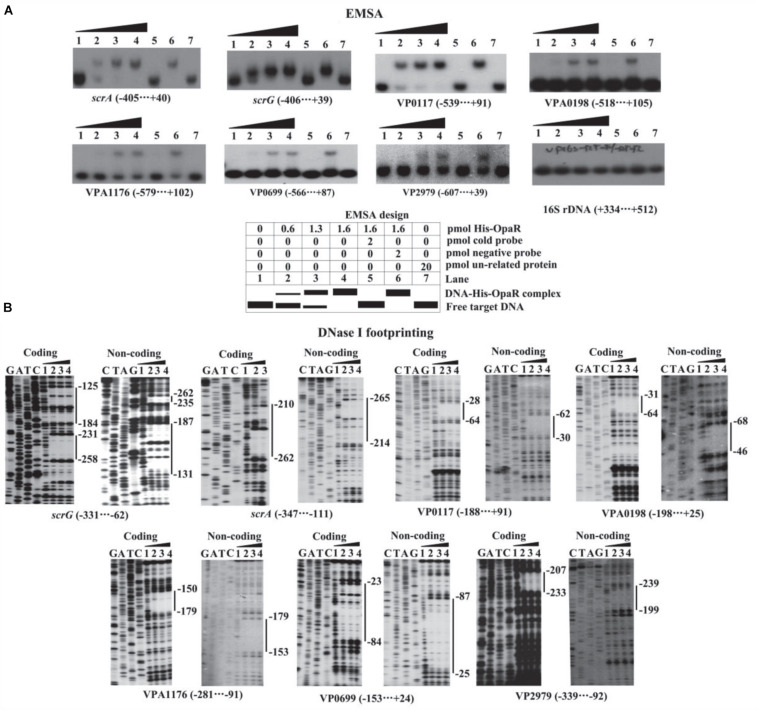
His-OpaR Bound to the Regulatory DNA Regions of Target Genes
The EMSA and DNase I footprinting assays were applied to detect the binding of His-OpaR to the regulatory DNA regions of scrA, scrG, VP0117, VPA0198, VPA1176, VP0699, and VP2979. The results of EMSA showed that His-OpaR bound to the regulatory DNA fragment of each target gene in a dose-dependent manner but was unable to bind the DNA fragment of 16S rDNA used as the negative control (Figure 6A). As further demonstrated by DNase I footprinting (Figure 6B), His-OpaR protected a single region for each upstream DNA region of scrA, VP0117, VPA0198, VPA1176, VP0699, and VP2979, located from 265 to 210 bp, from 64 to 28 bp, from 68 to 31 bp, from 179 to 150 bp, from 87 to 23 bp, and from 239 to 199 bp upstream of the translation start site of the each corresponding gene, respectively. His-OpaR protected two different DNA regions within the regulatory region of scrG located from 262 to 231 and from 187 to 125 upstream of the coding region. These results demonstrated that OpaR directly regulated the transcription of scrA, scrG, VP0117, VPA0198, VPA1176, VP0699, and VP2979 in V. parahaemolyticus RIMD2210633.
FIGURE 6.
Binding of His-OpaR to the target promoters. The negative and positive numbers indicated the nucleotide positions upstream and downstream of the indicated genes, respectively. (A) EMSA. The regulatory DNA fragment of each target gene was incubated with increasing amounts of His-OpaR and then subjected to 6% (w/v) polyacrylamide gel electrophoresis. Schematic representation of the EMSA design was shown below. (B) DNase I footprinting assay. Labeled coding or noncoding DNA probes were incubated with increasing amounts of His-OpaR (Lanes 1, 2, 3, and 4 containing 0, 6, 9, and 12 pmol, respectively) and then subjected to DNase I footprinting assay. Lanes G, A, T, and C represent the Sanger sequencing reactions. The protected regions are indicated by the vertical bars on the right side of the image.
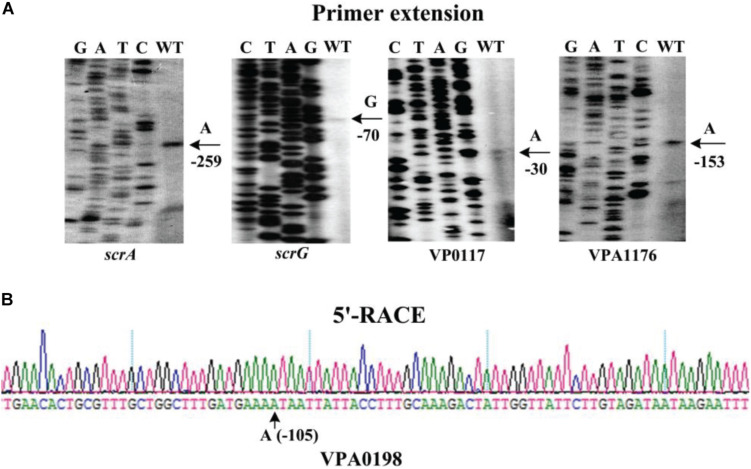
Identification of the Transcription Start Sites for the Target Genes
The transcription start sites of each target gene were detected by primer extension assay (Figure 7A). The assay detected a single transcription start site for scrA, scrG, VP0117, and VPA1176 located at 259, 70, 30, and 68 bp upstream of the corresponding gene, respectively. The putative −10 and −35 elements of each of the transcription start site matched with the consensus prokaryotic sequence (Figure 8). However, the primer extension assay did not detect any transcription start sites for VPA0198, VP0699, and VP2979. Thus, 5′-RACE was further employed to map the transcription start sites for the three genes (Gao et al., 2020). As shown in Figure 7B, the assay only detected one transcription start site for VPA0198 located 105 bp upstream of the translation start site, and its −10 and −35 elements were a good match with the consensus prokaryotic sequence (Figure 8). The assay also did not map any transcription start sites for VP0699 and VP2979. However, a transcription start site located at 37 and 64 bp upstream of the translation start site of VP0699 and VP2979 was predicted separately by using the online tool SoftBerry1 (Figure 8). The putative −10 and −35 elements of each start was also a good match with the consensus (Figure 8). Thus, they were considered as the most likely transcription start sites for VP0699 and VP2979.
FIGURE 7.
Transcription start sites for target genes. Negative numbers represent nucleotide positions upstream of the translation start site for each target gene. The transcription start sites are marked with arrows and positions. Lanes G, A, T, and C represent the Sanger sequencing reactions. (A) Primer extension. An oligonucleotide primer complementary to the mRNA of each target gene was designed. The primer extension products were analyzed with an 8 M urea-6% acrylamide sequencing gel. (B) 5′ RACE. The 5′-end of transcripts of target genes were analyzed using the FirstChoice®RLM-RACE (Invitrogen, United States) according to the manufacturer’s instructions.
FIGURE 8.
Structural organization of targeted promoters. The DNA sequences were derived from V. parahaemolyticus RIMD2210633. Transcription/translation start sites were marked with bent arrows. Shine–Dalgarno (SD) and –10/–35 elements were enclosed in boxes. The OpaR-box like sequences was marked in red. The OpaR sites were underlined with solid lines.
Discussion
OpaR was first described to regulate the variation of opaque (OP)–translucent (TR) colony in V. parahaemolyticus BB22, which is associated with the production of capsular polysaccharide (McCarter, 1998; Enos-Berlage and McCarter, 2000; Chen et al., 2010). Disruption of opaR on the chromosome of an OP strain produced a TR strain, whereas overexpression of opaR in a TR strain increased production and converted the strain to an OP type (McCarter, 1998). Approximately 15% of total genes of V. parahaemolyticus BB22 were regulated by OpaR, including those involved in capsular polysaccharide synthesis, c-di-GMP metabolism, and type IV pili assemblage (Gode-Potratz and McCarter, 2011; Kernell Burke et al., 2015). Both OP and TR strains formed stable but distinguishable structured biofilms; the OP strain displayed a rapid adherence and then a rapid decline, reaching a plateau at the early stage of the kinetics of biofilm formation, which was higher than that of the TR plateau value (Enos-Berlage et al., 2005). The gene cluster VPA1403–VPA1412 is responsible for the production of exopolysaccharide, a major component of V. parahaemolyticus biofilm (Yildiz and Visick, 2009; Chen et al., 2010). OpaR positively regulated the transcription of cpsA (VPA1403) and increased the cellular c-di-GMP level (Guvener and McCarter, 2003; Gode-Potratz and McCarter, 2011). Thus, the biofilms formed by V. parahaemolyticus BB22 are under the positive control of OpaR.
The data presented here demonstrated that mutation of opaR increases the biofilm formation of V. parahaemolyticus RIMD2210633 (Figure 2) and the intracellular c-di-GMP concentration (Figure 3). These results are contrary to the previous observations in V. parahaemolyticus BB22 (Guvener and McCarter, 2003; Gode-Potratz and McCarter, 2011). We believe that the discrepancies between the presented data and the previous results resulted from the bacterial genetic background. This is because the BB22 genome contains a number of unique genes that the RIMD2210633 genome does not contain, and vice versa (Jensen et al., 2013). Especially, a single missense mutation in luxO of V. parahaemolyticus BB22 created a quorum-regulatory protein with an altered, constitutively active function, resulting in reduced OpaR activity but a functional QS system, whereas mutations of luxO in other Vibrio (such as Vibrio cholerae) were thought to lock the system in the pathogenic state (Vance et al., 2003; Gode-Potratz and McCarter, 2011). Actually, a similar phenomenon has been also seen in V. cholerae that HapR, an OpaR homolog, acts as a repressor of exopolysaccharide production, c-di-GMP synthesis, and biofilm formation (Hammer and Bassler, 2003; Waters et al., 2008).
Vibrio parahaemolyticus RIMD2210633 harbors more than 50 genes encoding proteins involved in controlling the intracellular c-di-GMP concentration (Makino et al., 2003). The presented work selected seven genes that might be directly regulated by OpaR as the target genes to investigate the regulatory actions of OpaR on them. The data showed that OpaR bound to the promoter–proximal DNA regions of scrA, VP0117, VPA0198, VPA1176, and VP0699 to repress their transcription, whereas it positively and directly regulated the transcription of scrG and VP2979. Thus, OpaR transcriptional regulation of seven genes encoding GGDEF and/or EAL-type proteins indeed led directly to changes in the intracellular concentration of c-di-GMP. The QS cascade and c-di-GMP molecule are two important signaling relays controlling gene expression in V. parahaemolyticus. The direct association between the master QS regulator OpaR and c-di-GMP metabolism in V. parahaemolyticus RIMD2210633 would be conducive to precise control of gene transcription and bacterial behaviors. However, with the exception of scrA and scrG, functions of the other genes in c-di-GMP metabolism are completely unknown and should be clarified in future studies.
We reconstructed the OpaR-dependent promoter organization of scrA, scrG, VP0117, VPA0198, VPA1176, VP0699, and VP2979 by collecting data on the transcription start site, core promoter −10 and −35 elements, translation start site, OpaR-binding sites, and Shine–Dalgarno sequences (Figure 8). The OpaR site for each of the regulatory DNA regions of scrA, VP0117, VPA1176, and VP0699 overlaps with the −35 and/or −10 elements and/or the transcription start site, and thus, OpaR is thought to repress their transcription by directly interfering with RNA polymerase (RNAP) action. The OpaR-binding site in the promoter–proximal DNA region of VPA0198 locates downstream of the transcriptional start, and thus, the binding of OpaR may block the elongation of RNAP to repress the transcription of VPA0198. The OpaR sites for scrG and VP2979 are located upstream of the −35 promoter element, and thus, OpaR positively regulated scrG and VP2979 transcription should require class I transcriptional stimulation that depends on the RNAP subunit C-terminal domain to function (Ishihama, 2000).
Taken together, this work reports that the master QS regulator OpaR acts as a repressor of the biofilm formation by V. parahaemolyticus RIMD2210633. OpaR regulates the transcription of seven genes encoding a group of GGDEF and/or EAL-type proteins and decreases the intracellular concentration of c-di-GMP. The regulation by OpaR to these genes leads to a reduction in cellular c-di-GMP concentration and then a decrease in biofilm formation.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YZ, DZ, and RL designed, organized and supervised the experiments, interpreted the results, and edited the manuscript. YZ, YQ, HG, JS, XL, MZ, XX, WY, BN, LH, and ZY performed the laboratory experiments. YZ drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by grants from the China Postdoctoral Science Foundation (Grant No. 2020M681513), the National Natural Science Foundation of China (Grant No. 82072239), and the Jiangsu Planned Projects for Postdoctoral Research Funds (Grant No. 2020Z102).
References
- Ball A. S., Chaparian R. R., van Kessel J. C. (2017). Quorum sensing gene regulation by LuxR/HapR master regulators in vibrios. J. Bacteriol. 199:e00105-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles B. R., McCarter L. L. (2002). Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 184 5946–5954. 10.1128/jb.184.21.5946-5954.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberg C. A., Calder T. J., Orth K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13 992–1001. 10.1016/j.micinf.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Qiu Y., Tang H., Hu L. F., Yang W. H., Zhu X. J., et al. (2018). ToxR is required for biofilm formation and motility of Vibrio parahaemolyticus. Biomed. Environ. Sci. 31 848–850. [DOI] [PubMed] [Google Scholar]
- Chen Y., Dai J., Morris J. G., Jr., Johnson J. A. (2010). Genetic analysis of the capsule polysaccharide (K antigen) and exopolysaccharide genes in pandemic Vibrio parahaemolyticus O3:K6. BMC Microbiol. 10:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. H., Fen S. Y., Yu S. C., Wong H. C. (2016). Influence of oxyR on growth, biofilm formation, and mobility of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 82 788–796. 10.1128/aem.02818-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enos-Berlage J. L., Guvener Z. T., Keenan C. E., McCarter L. L. (2005). Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol. Microbiol. 55 1160–1182. 10.1111/j.1365-2958.2004.04453.x [DOI] [PubMed] [Google Scholar]
- Enos-Berlage J. L., McCarter L. L. (2000). Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J. Bacteriol. 182 5513–5520. 10.1128/jb.182.19.5513-5520.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang N., Gao H., Wang L., Qu S., Zhang Y. Q., Yang R. F., et al. (2013). Optimized methods for biofilm analysis in Yersinia pestis. Biomed. Environ. Sci. 26 408–411. [DOI] [PubMed] [Google Scholar]
- Ferreira R. B., Antunes L. C., Greenberg E. P., McCarter L. L. (2008). Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J. Bacteriol. 190 851–860. 10.1128/jb.01462-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Ma L., Qin Q., Qiu Y., Zhang J., Li J., et al. (2020). Fur represses Vibrio cholerae biofilm formation via direct regulation of vieSAB, cdgD, vpsU, and vpsA-K transcription. Front. Microbiol. 11: 587159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Zhang Y., Yang L., Liu X., Guo Z., Tan Y., et al. (2011). Regulatory effects of cAMP receptor protein (CRP) on porin genes and its own gene in Yersinia pestis. BMC Microbiol. 11:40. 10.1186/1471-2180-11-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gode-Potratz C. J., Kustusch R. J., Breheny P. J., Weiss D. S., McCarter L. L. (2011). Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol. Microbiol. 79 240–263. 10.1111/j.1365-2958.2010.07445.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gode-Potratz C. J., McCarter L. L. (2011). Quorum sensing and silencing in Vibrio parahaemolyticus. J. Bacteriol. 193 4224–4237. 10.1128/jb.00432-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvener Z. T., McCarter L. L. (2003). Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus. J. Bacteriol. 185 5431–5441. 10.1128/jb.185.18.5431-5441.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer B. K., Bassler B. L. (2003). Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50 101–104. 10.1046/j.1365-2958.2003.03688.x [DOI] [PubMed] [Google Scholar]
- Henke J. M., Bassler B. L. (2004). Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 186 3794–3805. 10.1128/jb.186.12.3794-3805.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A. (2000). Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54 499–518. [DOI] [PubMed] [Google Scholar]
- Jenal U., Reinders A., Lori C. (2017). Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 15 271–284. 10.1038/nrmicro.2016.190 [DOI] [PubMed] [Google Scholar]
- Jensen R. V., Depasquale S. M., Harbolick E. A., Hong T., Kernell A. L., Kruchko D. H., et al. (2013). Complete genome sequence of prepandemic Vibrio parahaemolyticus BB22OP. Genome Announc. 1:e00002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernell Burke A., Guthrie L. T., Modise T., Cormier G., Jensen R. V., McCarter L. L., et al. (2015). OpaR controls a network of downstream transcription factors in Vibrio parahaemolyticus BB22OP. PLoS One 10:e0121863. 10.1371/journal.pone.0121863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. K., McCarter L. L. (2007). ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J. Bacteriol. 189 4094–4107. 10.1128/jb.01510-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbrough J. H., Cribbs J. T., McCarter L. L. (2020). Homologous c-di-GMP-binding scr transcription factors orchestrate biofilm development in Vibrio parahaemolyticus. J. Bacteriol. 202:e00723-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbrough J. H., McCarter L. L. (2020). Identification of three new GGDEF and EAL domain-containing proteins participating in the Scr surface colonization regulatory network in Vibrio parahaemolyticus. J. Bacteriol. 203:e00409-20. 10.1128/JB.00409-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleber-Janke T., Becker W. M. (2000). Use of modified BL21(DE3) Escherichia coli cells for high-level expression of recombinant peanut allergens affected by poor codon usage. Protein Expr. Purif. 19 419–424. 10.1006/prep.2000.1265 [DOI] [PubMed] [Google Scholar]
- Li W., Wang J. J., Qian H., Tan L., Zhang Z., Liu H., et al. (2020). Insights into the role of extracellular DNA and extracellular proteins in biofilm formation of Vibrio parahaemolyticus. Front. Microbiol. 11:813. 10.3389/fmicb.2020.00813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Osei-Adjei G., Huang X., Zhang Y. (2018). Role and regulation of the orphan AphA protein of quorum sensing in pathogenic Vibrios. Future Microbiol. 13 383–391. 10.2217/fmb-2017-0165 [DOI] [PubMed] [Google Scholar]
- Lu R., Tang H., Qiu Y., Yang W., Yang H., Zhou D., et al. (2019). Quorum sensing regulates the transcription of lateral flagellar genes in Vibrio parahaemolyticus. Future Microbiol. 14 1043–1053. 10.2217/fmb-2019-0048 [DOI] [PubMed] [Google Scholar]
- Makino K., Oshima K., Kurokawa K., Yokoyama K., Uda T., Tagomori K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361 743–749. 10.1016/s0140-6736(03)12659-1 [DOI] [PubMed] [Google Scholar]
- McCarter L. L. (1998). OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J. Bacteriol. 180 3166–3173. 10.1128/jb.180.12.3166-3173.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W. L., Bassler B. L. (2009). Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43 197–222. 10.1146/annurev-genet-102108-134304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales R. E., Harwood C. S. (1993). Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for gram- bacteria. Gene 133 23–30. 10.1016/0378-1119(93)90220-w [DOI] [PubMed] [Google Scholar]
- Qiu Y., Hu L., Yang W., Yin Z., Zhou D., Yang H., et al. (2020). The type VI secretion system 2 of Vibrio parahaemolyticus is regulated by QsvR. Microb. Pathog. 149:104579. 10.1016/j.micpath.2020.104579 [DOI] [PubMed] [Google Scholar]
- Shime-Hattori A., Iida T., Arita M., Park K. S., Kodama T., Honda T. (2006). Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiol. Lett. 264 89–97. 10.1111/j.1574-6968.2006.00438.x [DOI] [PubMed] [Google Scholar]
- Sun F., Zhang Y., Qiu Y., Yang H., Yang W., Yin Z., et al. (2014). H-NS is a repressor of major virulence gene loci in Vibrio parahaemolyticus. Front. Microbiol. 5:675. 10.3389/fmicb.2014.00675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Zhang Y., Wang L., Yan X., Tan Y., Guo Z., et al. (2012). Molecular characterization of direct target genes and cis-acting consensus recognized by quorum-sensing regulator AphA in Vibrio parahaemolyticus. PLoS One 7:e44210. 10.1371/journal.pone.0044210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble M. J., McCarter L. L. (2011). Bis-(3’-5’)-cyclic dimeric GMP-linked quorum sensing controls swarming in Vibrio parahaemolyticus. Proc. Natl. Acad. Sci. U.S.A. 108 18079–18084. 10.1073/pnas.1113790108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J. (2003). Regulatory sequence analysis tools. Nucleic Acids Res. 31 3593–3596. 10.1093/nar/gkg567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance R. E., Zhu J., Mekalanos J. J. (2003). A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun. 71 2571–2576. 10.1128/iai.71.5.2571-2576.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Ling Y., Jiang H., Qiu Y., Qiu J., Chen H., et al. (2013). AphA is required for biofilm formation, motility, and virulence in pandemic Vibrio parahaemolyticus. Int. J. Food Microbiol. 160 245–251. 10.1016/j.ijfoodmicro.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Waters C. M., Lu W., Rabinowitz J. D., Bassler B. L. (2008). Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190 2527–2536. 10.1128/jb.01756-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz F. H., Visick K. L. (2009). Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17 109–118. 10.1016/j.tim.2008.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gao H., Osei-Adjei G., Yang W., Yang H., Yin Z., et al. (2017). Transcriptional regulation of the type VI secretion system 1 genes by quorum sensing and ToxR in Vibrio parahaemolyticus. Front. Microbiol. 8:2005. 10.3389/fmicb.2017.02005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Hu L., Qiu Y., Osei-Adjei G., Tang H., Zhang Y., et al. (2019). QsvR integrates into quorum sensing circuit to control Vibrio parahaemolyticus virulence. Environ. Microbiol. 21 1054–1067. 10.1111/1462-2920.14524 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Qiu Y., Tan Y., Guo Z., Yang R., Zhou D. (2012). Transcriptional regulation of opaR, qrr2-4 and aphA by the master quorum-sensing regulator OpaR in Vibrio parahaemolyticus. PLoS One 7:e34622. 10.1371/journal.pone.0034622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Yan X., Qu F., Wang L., Zhang Y., Hou J., et al. (2013). Quorum sensing modulates transcription of cpsQ-mfpABC and mfpABC in Vibrio parahaemolyticus. Int. J. Food Microbiol. 166 458–463. 10.1016/j.ijfoodmicro.2013.07.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.