Summary
G-quadruplex structures are associated with various biological activities, while in vivo evidence is essential to confirm the formation of G-quadruplexes inside cells. Most conventional agents that recognize G-quadruplex, including antibodies and small-molecule G-quadruplex ligands, either stabilize the G-quadruplex or prevent G-quadruplex unfolding by helicase, thereby artificially increasing the G-quadruplex levels in cells. Unambiguous study of G-quadruplexes at natural cellular levels requires agents that do not enhance the stability of G-quadruplex. Herein, we report the first example of nonperturbative chemical nucleases that do not influence the stability of G-quadruplex telomeric DNA but can selectively cleave G-quadruplex DNA over duplex DNA. These chemical nucleases can be readily taken up by cells and promote selective cleavage of telomeric DNA with low levels of nonselective DNA cleavage of other regions of the genome. The cleavage of G-quadruplex telomeric DNA by nonperturbative chemical nucleases confirms the formation of G-quadruplex telomeric DNA in live cells.
Subject areas: Biochemistry, Molecular Biology
Graphical abstract

Highlights
-
•
Novel chemical nucleases exhibit no effect on G-quadruplex telomeric DNA stability
-
•
Selective nucleases cleave G-quadruplex DNA over duplex DNA
-
•
Cleavage of G-quadruplex telomeric DNA motifs confirms their existence in cells
Biochemistry; Molecular biology;
Introduction
Apart from the canonical double helix, DNA can adopt a variety of other secondary structures, including the G-quadruplex (G4) (Lane et al., 2008). The G4 motif is typically formed from guanine-rich sequences of DNA or RNA and may be functionally relevant in several biological processes (Hansel-Hertsch et al., 2016; Lipps and Rhodes, 2009; Varshney et al., 2020). Despite the abundance of in vitro chemical and biophysical studies of the G4 structure, in vivo evidence is essential to confirm the formation of G4s inside cells. There are few approaches for direct study of G4s in vivo (Lipps and Rhodes, 2009). While recent work has developed anti-G4 antibodies, such as BG4 that enable immunofluorescence imaging for G4 DNA and RNA in cells (Biffi et al., 2013, 2014), permeabilization agents are required to promote internalization into cells owing to the lack of membrane permeability by the antibody (Henderson et al., 2014). Ultimately, this undermines their potential for studying G4 in live cells or in vivo. Moreover, immunostaining with an antibody involves repeated blocking and washing steps to reduce nonselective interaction. The cellular level of G4 formation may be artificially increased if some G4 unwinding proteins, such as BLM, WRN, FANCJ, and Pif1, and their required cofactors, such as adenosine triphosphate, are removed during extensive washing steps (Huber et al., 2002; Wang et al., 2018; Wu and Spies, 2016; Wu et al., 2017). Moreover, anti-G4 antibodies exhibit very high binding affinity to G4 (the KD of BG4 to G4s ranges from ∼0.5 to 2 nM [Biffi et al., 2013]) and can decrease the unwinding rate of G4s by some helicases (Duan et al., 2015). In turn, this can potentially increase the levels of G4 by stabilizing the motif and inhibiting unwinding of the G4 during immunostaining. Indeed, not only the presence of G4s but also the actual levels of G4 lie at the heart of the G4 studies. In addition to anti-G4 antibodies, small-molecule G4 probes have also been developed to visualize G4 motifs (Di Antonio et al., 2020; Kumar et al., 2020; Liu et al., 2020b; Zhang et al., 2018). Polymerase stalling by G4 formation has also been used to develop a sequencing-based approach for detection of G4 formation, such as, G4-seq, rG4-seq, G4 ChIP-seq, and G4P-seq (Chambers et al., 2015; Hansel-Hertsch et al., 2018; Kwok et al., 2016; Marsico et al., 2019; Zheng et al., 2020). Herein, we report the use of novel chemical nucleases to investigate the occurrence of structured DNA G4s in cells as an alternative strategy to the use of conventional antibodies and small-molecule G4 probes. Two novel chemical nucleases that have no effect on the stability of G4 telomeric DNA were developed, and highly selective cleavage by these chemical nucleases revealed the cellular availability of G4s at their natural cellular levels.
Regulation of G4s with chemical tools may enable further investigation of the cellular activities of G4 structures, as well as open potential medicinal, diagnostic and biomedical applications (Neidle, 2017). Previous studies have reported a number of small-molecule G4 targeting ligands that can stabilize the G4 structure (Jamroskovic et al., 2020; Onizuka et al., 2019; Ruehl et al., 2019). For example, BioTASQ was used to pull down G4 RNA after cross-linking, and the captured G4 RNAs were identified through sequencing, known as G4RP-seq (Yang et al., 2018). In contrast to the well-reported G4-stabilizing ligands, chemical nucleases that can promote cleavage of the G4 motif represent a novel class of chemical tools to study G4 formation in cells. In fact, cleavage or downregulation of G4s by nucleases or small molecules is rarely reported (del Mundo et al., 2019; Nadai et al., 2018; O'Hagan et al., 2019a; O'Hagan et al., 2019b; Yu et al., 2019; Yu et al., 2015). Engineered protein-based nucleases have emerged as a powerful tool for the manipulation of DNA, such as CRISPR-cas9; however, these protein-based nucleases lack membrane permeability and target specific primary sequences of DNA rather than the secondary structure (Zhang et al., 2014). By contrast, the chemical nucleases described herein are secondary-structure-specific nucleases that can differentiate the G4 from the prevalent duplex DNA structure. Metal complexes have been used to investigate G4 structures and as molecular probes for cellular imaging (Liu et al., 2018, 2020a; Miron et al., 2021). A recent study has reported a Cu complex, NDI-Cu-DETA, for in vitro studies; however, NDI-Cu-DETA increases the stability of G4, while no in cellulo data with NDI-Cu-DETA was reported (Nadai et al., 2018). Our previous studies also have reported the design of selective G4 chemical nucleases for medicinal applications (Yu et al., 2015, 2019); however, there is still a need to develop nonperturbative chemical nucleases that have no effect on G4 stability to unambiguously study the natural level of G4s. To the best of our knowledge, herein, we report the first membrane-permeable metal complexes as nonperturbative chemical nucleases that exhibit no influence on the stability of G4 but cleave G4 DNA. This provides an alternative chemical tool to the use of conventional antibodies and G4-binding ligands for study of the cellular chemistry of this motif.
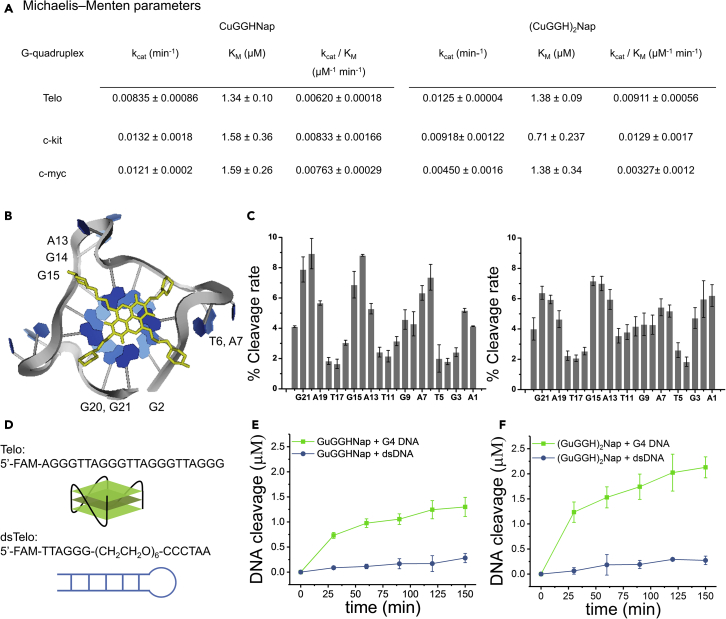
G4-targeting chemical nucleases were constructed by combining a permeable DNA-targeting domain with a DNA-cleavage motif. The low molecular weight and hydrophobicity of the targeting domain allows these chemical nucleases to cross cellular membranes in the absence of permeabilization agents, thereby addressing the major membrane permeability issue of approaches based on the use of conventional antibodies and protein-based nucleases. The DNA-cleaving motif is derived from the amino-terminal copper/nickel-binding motif (ATCUN) and can promote DNA cleavage under physiological conditions (Maiti et al., 2020; Yu and Cowan, 2018). By application of this strategy, we have developed a novel G4-binding naphthalene diimide derivative, Nap, that has no effect on the melting point of G4s and thereby constructed both a mononuclear Cu complex, CuGGHNap, and dinuclear Cu complex, (CuGGH)2Nap, respectively, by coupling Nap with one or two DNA-cleaving moieties, the Gly-Gly-His peptide (Figure 1A), while the latter can promote oxidative DNA break (Pinkham et al., 2018; Yu and Cowan, 2017b; Yu et al., 2020). These Cu complexes were expected to rapidly internalize into live cells and promote cellular cleavage of G4 DNA, confirming the in vivo presence of G4 formation and permitting downregulation of the G4 structure.
Figure 1.
Design of chemical nucleases and DNA binding affinity
(A) Chemical structures of the naphthalene diimide derivatives.
(B) FRET assays allow determination of both the binding affinity of naphthalene diimide derivatives to G-quadruplex DNA and cleavage activity of chemical nucleases in the presence of ascorbate and peroxide.
(C). Dissociation constants (μM) evaluating DNA binding affinity for the naphthalene diimide derivatives.
Results
Chemical nucleases targeting G4 and nuclease activity
Fine-tuning of naphthalene diimide derivatives has resulted in a candidate, Nap, that exhibits no effect on the stability of G4 telomeric DNA, c-kit, and c-myc promoters (Table 1). The synthesis of the naphthalene diimide derivatives is described in the Method details of supplemental information and includes Nap, the control analog lacking the DNA cleavage motif, and two chemical nucleases with one or two DNA-cleaving motifs derived from Gly-Gly-His peptide (CuGGHNap and (CuGGH)2Nap, respectively). All naphthalene diimide derivatives typically display an emission centered at 662 nm (λex = 620 nm). The excitation spectrum of naphthalene diimide derivatives overlaps with the emission from fluorescein (Figure S2), which enables potential Förster resonance energy transfer (FRET) from fluorescein to the naphthalene diimide moiety. To confirm the G4 binding affinity of these naphthalene diimide derivatives toward G4 DNA, a telomeric oligonucleotide was labeled with a 5′ fluorescein label (Telo: fluorescein-d(AGGGTTAGGGTTAGGGTTAGGG)), while fluorescein and oligonucleotide were separated by a flexible A1 residue to prevent steric hindrance and allow the detection of ligand stacking in the G-tetrad. Given that these naphthalene diimide derivatives have a significantly lower quantum yield (<100 fold) than fluorescein (data not shown), a decrease of emission intensity of fluorescein was observed when naphthalene diimide derivatives were titrated into G4 DNA as a result of the FRET response.
Table 1.
Influence of naphthalene diimide derivatives on the melting points for G-quadruplex DNAs
| Melting points (°C) | Telo | c-kit | c-myc |
|---|---|---|---|
| DNA only | 67.5 ± 0.3 | 64.7 ± 1.2 | 86.8 ± 1.0 |
| Nap | 68.6 ± 2.6 | 66.0 ± 0.5 | 82.1 ± 1.0 |
| CuGGHNap | 64.6 ± 0.9 | 87.5 ± 0.3 | 71.7 ± 2.4 |
| (CuGGH)2Nap | 65.9 ± 2.0 | 75.3 ± 0.2 | 70.4 ± 1.0 |
The Cu complexes, CuGGHNap and (CuGGH)2Nap, as well as the analogue lacking CuGGH moiety (Nap), display a similar binding affinity to telomeric G4 DNA with measured KDs in the range of 0.1–0.3 μM (Figures 1C and S3). In addition to telomeric DNA, both c-myc and c-kit promoter DNA were also used to represent the proposed G4s believed to exist within the cell to test the selectivity of these compounds for various types of G4s. Moderate binding affinity is also demonstrated for the c-myc (KDs of 0.27–0.54 μM) and c-kit (KDs of 0.19–0.97 μM) G4 DNA, but both of these G4 species have binding affinities almost 1.3 to 3 times lower than the Telo DNA (Figures 1C and S4). In additional, DNA binding affinity to calf thymus DNA (CT-DNA) was also determined by use of a competition assay with Telo G4 DNA. (CuGGH)2Nap, with two CuGGH moieties, exhibits a dominant preference for Telo G4 DNA over duplex DNA with a KD of ∼64 μM and a specificity factor of ∼432, while the specificity factors for Nap and CuGGHNap are only ∼7 and ∼36, respectively (Figure 1C).
Overall, introduction of the CuGGH moiety barely influences the DNA binding affinity of Nap toward G4 DNA, but dramatically undermines the undesired interaction with duplex CT-DNA and improves the DNA binding selectivity. In contrast to the high binding affinity (low nanomolar range) of the anti-G4 antibody toward G4s, which can impede the unwinding of G4 by helicases (Duan et al., 2015), these chemical nucleases only exhibit moderate binding affinity and minimize potential artificial effects on G4 formation. No enhancement of G4 formation by naphthalene diimides at 37°C was observed (Figure S5). This laid the foundation for the possible cellular application of these naphthalene diimide derivatives because they do not enhance G4 levels in cells but only recognize the G4 naturally formed in cells. In addition, the melting point for Telo DNA was measured by circular dichroism (CD) at 290 nm (Table 1 and Figure S5). Within error, all the naphthalene diimide derivatives exhibit a negligible influence (<5°C) on the melting point of Telo DNA, indicating no stabilizing effect of these compounds on G4 formation by the telomeric DNA sequence. In the case of c-kit and c-myc promoter, the melting points varied significantly when the CuGGHNap and (CuGGH)2Nap compounds were added to solution indicating a significant difference in the mode of binding for c-kit and c-myc promoter versus telomeric G4es. Given that the solution structures of c-kit and c-myc motifs are very different from the telomeric G4, structural differences in the positioning of catalyst, sites targeted for cleavage, and intrinsic rates of nuclease activity, as well as binding contacts, would be expected. In addition to variations in reactivity, differences in stability for the bound structure and melting temperatures would be anticipated. While the G4 is predominant in CD melting experiments, and the naphthalene diimide motif exhibits a preference for G4 DNA binding at 37°C, both CuGGHNap and (CuGGH)2Nap may also bind to unfolded DNA at higher temperatures during melting, thereby decreasing the Tm of c-myc. In contrast to Nap, the GGH moiety of CuGGHNap and (CuGGH)2Nap may result in different binding affinity to the unfolded telo, c-kit, and c-myc, and so, CuGGHNap and (CuGGH)2Nap may demonstrate a different effect on the Tm of G4 from Nap.
DNA cleavage reactivity of CuGGHNap and (CuGGH)2Nap requires the presence of the biologically relevant coreagents, ascorbate, and H2O2 (data not shown). The catalytic domain promotes the formation of metal-associated reactive oxygen species (ROS) from copper redox chemistry (Alexander et al., 2017, 2018; Yu and Cowan, 2017a) that subsequently attack the deoxyribose ring and induce DNA breaks (Yu and Cowan, 2018; Yu et al., 2015). The fluorescein-labelled G4 DNA oligonucleotide was used as a substrate to investigate the activity of the G4-targeting chemical nucleases against G4 telomeric DNA and the G4 c-myc and c-kit promoters. In the presence of ascorbate and peroxide, a time-dependent change of 520-nm emission intensity was observed after the cleavage of G4 DNA by CuGGHNap or (CuGGH)2Nap, indicating either a potential change of microenvironment or dissociation of naphthalene derivatives from the reaction products after cleavage (Ross et al., 2015). The emission intensities of both starting material before cleavage, and the product after complete cleavage, were used to quantitate the concentration of in situ cleavage product formed during the reaction and calculation of the initial reaction velocity.
Various concentrations of substrate were used to investigate the Michaelis-Menten kinetics of these Cu complexes. The Michaelis-Menten parameters for (CuGGH)2Nap with two Cu centers resulted in improved enzyme efficiency relative to CuGGHNap with one Cu center (Figures 2A and S4). The KMs of both Cu complexes with all G4 substrates were similar, consistent with the results that both (CuGGH)2Nap and CuGGHNap exhibit identical binding affinity to substrate. Nevertheless, the Cu complex with two Cu centers exhibits a larger kcat, by ∼50%, relative to CuGGHNap in the case of the Telo DNA. Therefore, the improved enzyme efficiency (kcat/KM) of (CuGGH)2Nap mainly arises from the faster cleavage rate of the substrate at the catalytic step promoted by the two Cu centers when it is bound to its specified telomeric G4 DNA target.
Figure 2.
Reaction site selectivity for cleavage of G-quadruplex telomeric DNA and substrate selectivity
(A) Michaelis–Menten parameters for G-quadruplex DNA cleavage.
(B) Major cleavage sites are highlighted in a reported G4-Napthalene complex (PDB: 3UYH).
(C) Time-dependent DNA cleavage by CuGGHNap (left) and (CuGGH)2Nap (right) was analyzed by use of denaturing PAGE to determine the relative initial cleavage rates for reactions at each position.
(D) The design of substrates includes a G-quadruplex telomeric DNA and a hairpin telomeric sequence representing dsDNA.
(E and F) Substrate selectivity for the chemical nucleases was revealed by comparing the patterns of time-dependent DNA cleavage of G-quadruplex telomeric DNA and the duplex telomeric DNA control by CuGGHNap (E) and (CuGGH)2Nap (F).
Based on the binding data summarized in Figure 1C and the changes in melting temperatures for the oncogenes demonstrated in Table 1, it would be expected that cleavage of oncogene quadruplexes would be less predictable, relative to Telo DNA. This is especially relevant to the case of the c-myc oncogene where destabilizing effects were observed when the Nap compound, and the ATCUN Nap derivatives in particular, was added to c-myc. Interestingly, the addition of a second copper-binding site had the opposite effect on the kcat values for the oncogenes, relative to the case of Telo DNA, indicating that the binding of the CuGGHNap and (CuGGH)2Nap to the oncogene is markedly different in comparison with that of Telo DNA.
Given that CuGGHNap and (CuGGH)2Nap can destabilize c-myc and stabilize c-kit, we focused our study on G4 telomeric DNA to avoid artifacts from G4 formation levels in cells. To further study the selective pattern of DNA cleavage, denaturing polyacrylamide gel electrophoresis was used to identify the cleavage sites in G4 DNA for each Cu complex. Following the reported crystal structure of a G4-Naphthalene complex (Micco et al., 2013), major cleavage sites were observed at A1, G2, T6, A7, A13-G15, G20, and G21 (Figures 2B and 2C), where two Cu centers of (CuGGH)2Nap are most likely positioned. One Cu center primarily affects A13-G15. The second Cu center can attack either the residues forming G-tetrad (G2, G20, G21) or residues in a loop (T6, A7), reflecting the flexibility of the propyl linker that allows this Cu center to attack two distinct groups of residues. The specific DNA cleavage pattern of these nucleases is associated with their specific DNA-binding site. Interestingly, the cleavage sites observed for CuGGHNap are similar to those identified for (CuGGH)2Nap, indicating two distinct binding patterns for CuGGHNap, namely, the single Cu center of CuGGHNap can residue in two possible positions where two Cu centers of (CuGGH)2Nap are placed. Minor occurrences of nonselective DNA cleavage at other sites may result from secondary binding/cleavage and diffusion of ROS. In addition, a self-complementary duplex telomeric DNA model was also tested as a substrate under similar DNA cleavage conditions to study the selectivity of DNA cleavage by Cu complexes. A substrate with the same length as G4 Telo was used at the beginning; however, this substrate exhibits an extremely high stability and was not denatured by 6 M urea and 25% formamide, therefore, a shorter self-complementary duplex telomeric DNA model (dsTelo: 5′-fluorescein-d(TTAGGG)-(CH2CH2O)6-d(CCCTAA)) was tested (Figure 2D). To avoid formation of intermolecular G4 from multiple strands of dsTelo, 100 mM LiCl instead of KCl was used as counter ions to destabilize G4 formation, where LiCl should have a negligible impact on the stability of the DNA duplex. Both CuGGHNap and (CuGGH)2Nap exhibit more robust DNA cleavage of G4 telomeric DNA, relative to duplex telomeric DNA (Figures 2E and 2F). Moreover, the DNA-cleavage activity of (CuGGH)2Nap, with two CuGGH motifs, was more selective than observed for CuGGHNap with one Cu center, consistent with the higher DNA-binding selectivity of (CuGGH)2Nap.
Cleavage of G4 telomeric DNA in cells promoted by chemical nucleases
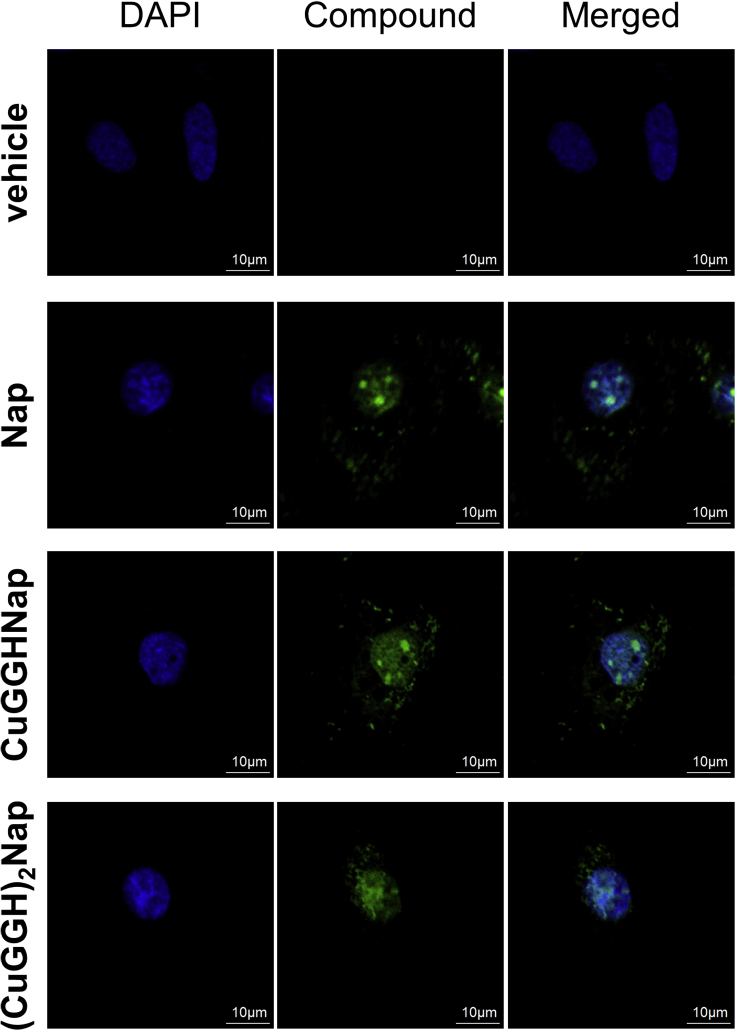
Although these naphthalene diimide derivatives can recognize G4 selectively, unfortunately, they only exhibit a slight increase of emission intensity (∼2–3 times) when bound to G4 (data no shown). While the potential low signal-to-noise ratio may hinder the use of the naphthalene diimide derivatives for direct staining and detection of G4s in vivo, nevertheless the intrinsic fluorescence of the naphthalene diimide core (λem = 633 nm) was still useful for determination of the general cellular localization of each chemical nuclease (CuGGHNap and (CuGGH)2Nap). In the absence of any permeabilization agents, all naphthalene diimide derivatives are rapidly internalized into HuH7 cells. With the cellular nucleus stained by DAPI, the overlap of both DAPI and naphthalene diimide fluorescence signals indicate the presence of Cu complexes in the nucleus. Both CuGGHNap and (CuGGH)2Nap exhibit significant membrane permeability and accumulate in the nucleus after 12-h incubation in HuH7 cells by virtue of their low molecular weight and hydrophobicity (Figure 3). Because of the low levels of enhancement of the emission intensity when the Nap derivatives are bound to G4 (data not shown, ∼2- 3 fold), the foci revealed in the confocal microscope should reflect the overall localization of these compounds, disregarding the DNA-binding status. In addition, a few minor cytoplasmic foci may also indicate cytoplasmic RNA G4 recognition by the naphthalene diimide derivatives in the cytoplasm. Given that CuGGHNap and (CuGGH)2Nap primarily accumulate in the nucleus, in this work we focus on the potential nuclease effects of CuGGHNap and (CuGGH)2Nap on G4 DNA instead of G4 RNA; however, there are obvious applications of these chemical nucleases for future studies of cytoplasmic G4 RNA.
Figure 3.
Colocalization of naphthalene diimide derivatives and the cellular nucleus
HuH7 cells were incubated with a 2-μM concentration of the indicated compounds for 12 h, and DAPI was used to stain the cellular nucleus.
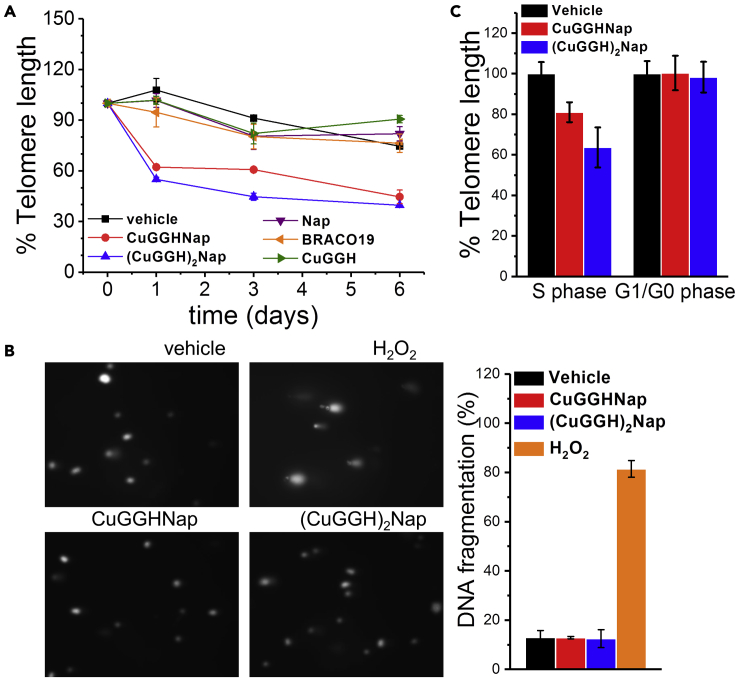
The viability of HuH7 human hepatoma cells under various concentrations of chemical nucleases was measured by use of the MTT assay (Figure S11), yielding moderate cytotoxicity (IC50's of ∼12–35 μM). To minimize artificial effects from cell death and nonselective DNA cleavage at high concentration of chemical nucleases, cells were typically incubated at a nontoxic concentration (2 μM). To investigate the impact on telomeric DNA after incubation with these naphthalene diimide derivatives, a reported quantitative PCR (q-PCR) assay was used to monitor the change of telomere length of the mammalian cells (O'Callaghan and Fenech, 2011). Rapid reduction of telomere length was observed in HuH7 (∼40–50%) after 1-day incubation with CuGGHNap or (CuGGH)2Nap (Figure 4A).
Figure 4.
DNA fragmentation and telomere reduction in HuH7 cells after treatment with the indicated compounds
(A) The relative telomere length of HuH7 was measured by use of an q-PCR assay after incubation with 2 μM of the indicated compounds. Telomere length at day 0 is normalized to 100%.
(B) Representative images from an alkaline Comet assay revealing the level of DNA fragmentation in HuH7 cells following incubation with 2 μM CuGGHNap or (CuGGH)2Nap for 1 day. A positive control was obtained by incubating cells with 50 μM H2O2 for 30 min. Histograms are the average results from >100 comets.
(C). The relative telomere length of synchronized HuH7 cells. S-phase cells obtained from the double-thymidine block: after retreat of thymidine, 2 μM compound and 100 ng/mL nocodazole were added to the synchronized cells for 10 h (control: 100 ng/mL nocodazole only); G1/G0-phase cells obtained from serum starvation: cells were incubated with 2 μM compound in serum-free medium. Telomere length of the vehicle control of synchronized HuH7 cells is normalized to 100%, respectively.
Previous studies have demonstrated inhibition of telomerase activity with a direct inhibitor or G4 ligand; however, these telomerase inhibitors cannot reduce telomere length promptly. Rather, a prolonged treatment is required (Damm et al., 2001; Shammas et al., 2004). For instance, BIBR1532 induces ∼60% erosion of telomere only after 140 population doublings of NCI-H460 cancer cells (Damm et al., 2001), and telomestatin shortens the telomere of MMS cancer cells by ∼18% only after a 14-day incubation period (Shammas et al., 2004). These data reported in previous studies are consistent with a slow telomere reduction rate in normal cells, corresponding to only ∼ 50 to 200 nt shortening per cell division, where telomerase activity is absent (Huffman et al., 2000). In other words, the rapid telomere attrition regulated by CuGGHNap and (CuGGH)2Nap do not result from telomerase inhibition but from direct DNA cleavage. Moreover, Nap, the control compound lacking the DNA cleavage moiety, and BRACO19, a reported G4 ligand that may suppress telomerase activity through stabilization of G4 telomeric DNA, induce negligible telomere reduction even after 6 days, confirming that inhibition of telomerase can barely shorten telomere length in the short term (Figure 4A).
Given that there is no increase of the melting point of G4 telomeric DNA in the presence of these chemical nucleases, these catalysts cannot artificially increase the level of G4 formation in cells, but only target those G4 structures naturally formed from their biological activities in live cells, which can address the potential problems identified with anti-G4 antibodies. Despite the prevailing duplex structure of telomeric DNA, the transient formation of G4 telomeric DNA can be recognized and cleaved by these G4-targeting Cu complexes, promoting a rapid reduction in telomere length. These results confirm the formation of G4 telomeric DNA in live cells, providing in vivo evidence for G4 formation complementary to those visualized by use of anti-G4 antibody in the previous studies. Control experiments show that CuGGH, the peptide lacking a DNA-targeting moiety, does not reduce telomere length, confirming that the substrate selectivity of CuGGHNap and (CuGGH)2Nap is indispensable for telomere shortening. In addition, an Annexin V/PI assay was used to study the possibility of cell death induced by these Cu complexes. Under the same conditions as the q-PCR experiments, essentially no increase (<5%) in cell death was observed to be induced by either CuGGHNap or (CuGGH)2Nap at these nontoxic concentrations, either by apoptosis or necrosis (Figure S9).
Because the endogenous nucleases upregulated during programmed cell death can also lead to an increase of DNA fragmentation level, our results exclude the possibility that DNA cleavage of telomeric DNA was induced by apoptosis-associated nuclease activity. In other words, the rapid reduction observed in telomere length should arise from DNA cleavage activity of the Cu complexes rather than apoptosis-associated nuclease activity. Moreover, overall DNA fragmentation in cells was also evaluated by use of the comet assay under alkaline conditions (Figure 4B), where both single-stranded and double-stranded breaks of DNA could be revealed. A positive control with 50 μM H2O2 can induce ∼70–80% DNA fragmentation of nuclear DNA through random oxidative cleavage of DNA. Nevertheless, the observation of low CuGGHNap and (CuGGH)2Nap promoted DNA fragmentation levels (∼10%) in HuH7 cells indicates few nonspecific DNA breaks induced by these G4-targeting Cu complexes. On the other hand, these results are consistent with the aforementioned Annexin V/PI experiments that showed no increase in the rate of cell death at nontoxic concentrations of the Cu complexes, given that DNA fragmentation represents one of the distinct features of apoptosis/necrosis. Therefore, these results also rule out the possibility that the observed telomere reduction by CuGGHNap and (CuGGH)2Nap results from random nonselective DNA cleavage or cell death and confirm the DNA cleavage selectivity of these nucleases in live cells. Namely, these chemical nucleases preferentially cleave G4 DNA over regular duplex DNA.
To investigate telomere reduction under different cellular events, we measured the telomere reduction rate of cells in different phases of the cell cycle. Cells were arrested at the G1/G0 phase through serum starvation, while S phase cells were obtained through double thymidine blocks. After release from the thymidine block, cells undergoing S-phase transition were incubated with G4-targeting Cu complexes. Cell cycle analysis confirms the cellular synchronization and reveals that it took 1–2 h for cells to recover from the thymidine block and another 5–6 h to proceed to the S phase (data not shown). After entering the S phase, another 3-h incubation was performed to allow all cells to accomplish S-phase cycling completely because incomplete DNA synthesis during the S phase may cause false-positive results for telomere reduction. Therefore, S-phase cells were incubated with Cu complexes for a total of 10 h. In addition, to avoid further telomere reduction in the subsequent cell cycle, nocodazole was also added to trap cells in the G2 phase. G4-targeting Cu complexes CuGGHNap and (CuGGH)2Nap demonstrate a more robust telomere reduction in S-phase cells (∼20–40%), relative to G1/G0-phase cells (Figure 4C), consistent with enhanced G4 telomeric DNA formation in S-phase cells (Biffi et al., 2013).
Normal cells have a limited ability to proliferate (Hayflick limit) because cellular senescence can arise when telomere length for normal cells is shortened to a critical level. However, cancer cells should not be arrested into cellular senescence because of the activity of highly expressed telomerase. Senescence-associated β-galactosidase is a biomarker that is upregulated during senescence and can be detected by histochemical staining of cells using its substrate, X-gal. In fact, cellular senescence in HuH7 cells was significantly promoted after incubation with CuGGHNap and (CuGGH)2Nap (Figure S10). These results confirm the downstream cellular events that are associated with rapid telomere reduction by chemical nucleases.
Discussion
In this work, we have established a strategy for the design of permeable chemical nuclease mimics that target the G4 motif in cells. Following this strategy, we have developed mononuclear and dinuclear Cu complexes as chemical nucleases that selectively promote DNA cleavage of G4es, including telomeric DNA, c-kit, and c-myc promoters. G4-targeting Cu complexes, as well as an analogue lacking the CuGGH motif, exhibit similar DNA binding affinity to each of the three G4 DNAs, disregarding the sequence and type of G4s. These results indicate π-π stacking between the naphthalene diimide core and G-tetrad to be the primary interaction between these G4 ligands and DNA, while the neutral-charged CuGGH motif contributes little to DNA binding. In fact, the Cu complexes with CuGGH motif only exhibit a slight increase (∼2–3 fold) of binding affinity to G4 telomeric DNA and c-kit promoter, relative to the analogue lacking the CuGGH motif.
Interestingly, the chemical nucleases, CuGGHNap and (CuGGH)2Nap, display different impacts on the stability of the three G4es tested, namely, stabilizing c-kit, destabilizing c-myc, and no effect on telomeric DNA. These three G4s exhibit different folding topology and contain different loop sequence among the G-tetrads (Ambrus et al., 2005; Phan et al., 2007), and so, these results most likely reflect different binding modes for the naphthalene diimide derivatives to these G4s. This is most pronounced in the interactions or steric constraints between loops of specific G4s and the CuGGH motif(s) of the naphthalene diimide derivatives because the analogue lacking the CuGGH motif, Nap, exhibited no significant change in melting point for each of the three G4s. The CuGGH motif may either interact with the loops of the G4s or introduce steric hindrance against the loops of the G4s owing to its bulky size. For example, CuGGHNap with one CuGGH motif increases Tm of c-kit by ∼24°C, reflecting potential favorable interaction of CuGGH and a loop of c-kit that promotes G4 formation, while the second additional CuGGH motif of (CuGGH)2Nap may repel another loop of c-kit, thereby, resulting in a net change of Tm by only ∼10°C. Similarly, the lowering of Tm of c-myc by CuGGHNap may result from the steric effect of the CuGGH motif, while the second additional CuGGH motif of (CuGGH)2Nap may have no interaction or repulsion with c-myc and lead to no further destabilization of c-myc G4.
Because of the unfavorable destabilization effects, the introduction of a second CuGGH motif undermines the kcat of chemical nucleases for the cleavage of c-myc and c-kit, relative to the mononuclear Cu complex CuGGHNap, while the kcat of (CuGGH)2Nap is 50% faster than CuGGHNap for the cleavage of G4 telomeric DNA substrates. Moreover, the incorporation of a CuGGH moiety to the naphthalene core can diminish the nonselective binding affinity of Cu complexes to duplex CT-DNA. The bulky CuGGH moiety appears to introduce steric constraints that disrupt nonselective binding to duplex DNA, while the replacement of a protonated NH2 group by the neutral CuGGH motif can also decrease the overall positive charge of the G4 ligand and weaken nonselective electrostatic interaction with duplex DNA. Overall, the CuGGH motif contributes a slight improvement to G4 binding affinity but appears to significantly enhance binding selectivity between G4 and duplex DNA and modulate the stability of different G4s.
Although the chemical nucleases may destabilize c-myc and stabilize c-kit, they show no effect on the stability of G4 telomeric DNA, laying the essential foundation for studying the natural levels of G4 telomeric DNA in host cells. That is, there is no artificial increase or decrease of G4 telomeric DNA formation when these chemical nucleases are applied to cells, which addresses the artifact problem of anti-G4 antibodies. Accordingly, we focused our studies on using chemical nucleases for the cleavage of G4 telomeric DNA instead of c-myc and c-kit.
The selective DNA binding pattern of the G4-targeting motif spatially restrains the position of the Cu center, relative to the G4 DNA. In other words, only those residues (A1, G2, T6, A7, A13-G15, G21, and G22) that lie in close proximity to the Cu center are susceptible to the ROS formed by the Cu center(s). In fact, the selectivities of DNA cleavage sites by the Cu complexes confirm this binding pattern and indicate the positioning of the Cu center(s). G4-targeting Cu complexes promote a more robust cleavage of G4 telomeric DNA, relative to the duplex structure of telomeric DNA. Substrate selectivity should majorly arise from a combination of binding selectivity and the appropriate placement of the Cu-bound ROS. As noted earlier, the requirement for an “active” placement and orientation of the Cu-bound ROS, relative to scissile bonds, to promote efficient cleavage chemistry will serve as a second “filter” for selective substrate cleavage (Yu and Cowan, 2017a). Overall, the observed substrate selectivity should be controlled by this double-filter mechanism.
G4-targeting Cu complexes, CuGGHNap and (CuGGH)2Nap, can be rapidly internalized into cells by virtue of their low molecular weight and hydrophobicity, which addresses the uptake problem of protein-based engineered nucleases and antibodies. The localization of these chemical nucleases to the cellular nucleus indicates that they can target G4 DNA or RNA in cells. The chemical nucleases, CuGGHNap and (CuGGH)2Nap, can promote rapid telomere reduction of HuH7 cells after a 1- to 6-day incubation period at nontoxic concentrations. Cellular senescence is a permanent arrest from the cell replication cycle, a downstream event of telomere shortening that is typically associated with aging in normal tissues. Both chemical nucleases, CuGGHNap and (CuGGH)2Nap, can induce cellular senescence in hepatoma HuH7 cells through telomere reduction, consistent with cellular pathways for normal cells during aging.
In contrast to the rapid telomere reduction promoted by chemical nucleases, CuGGHNap and (CuGGH)2Nap, neither the targeting Nap motifs lacking the cleavage motif nor the catalytic CuGGH lacking the G4-targeting motif can induce any significant telomere reduction in the same incubation time. Both the G4-targeting motif and the DNA cleavage motif are prerequisites for the observed reduction in telomere length, implicating direct cleavage of G4 telomeric DNA to be promoted by these Cu complexes. In addition, the low DNA fragmentation level in cells revealed by the comet assay, after incubation with Cu complexes, also reflects the cellular cleavage selectivity of these chemical nucleases and is consistent with their substrate selectivity in vitro. Previous studies have shown that inhibition of telomerase cannot reduce telomere length rapidly; therefore, the robust telomere reduction promoted by CuGGHNap and (CuGGH)2Nap should arise from DNA cleavage of telomeric DNA in cells rather than potential telomerase inhibition by the complexes. Moreover, neither CuGGHNap nor (CuGGH)2Nap induces apoptosis/necrosis or random DNA damage at nontoxic concentrations, as demonstrated by the combination of Annexin V/PI and comet assays, and the possibility that cleavage of telomeric DNA arises from apoptosis-associated nuclease activity or nonselective DNA cleavage can be excluded. Accordingly, we propose that the G4-targeting chemical nucleases described herein can recognize the transient formation of G4 telomeric DNA in cells and induce irreversible breaks to telomeric DNA and that the successive cleavage of telomeric DNA promoted by these Cu complexes leads to rapid telomere reduction. Given that no increase in melting point was observed for G4 DNA in the presence of CuGGHNap or (CuGGH)2Nap, these chemical nucleases cannot artificially increase G4 levels in cells; namely, the observed cleavage of G4 telomeric DNA may reflect the natural levels of G4 in cells, which can potentially address those problems associated with anti-G4 antibodies. Overall, the observed rapid telomere reduction by G4-targeting nucleases confirms that telomeric DNA can form G4 motifs in cells.
While the G4 can spontaneously form from ssDNA in vitro, the formation of G4 is dynamic and transient (Maizels, 2006). Previous studies have shown enhanced G4 formation during the S phase of the cell cycle by use of a G4-recognizing antibody (Biffi et al., 2013). The increase of G4 telomeric DNA formation during the S phase may result from the unwinding of duplex telomeric DNA during DNA replication because ssDNA can form a G4 structure more readily than dsDNA (Kreig et al., 2015). In fact, both CuGGHNap and (CuGGH)2Nap induce more robust telomere reduction in S-phase cells, relative to G1/G0-phase cells, reflecting the increased prevalence of the G4 structure in the S phase of the cell cycle. These results demonstrate that the nuclease activity of these Cu complexes is associated with different levels of G4 formation during different cellular events, such as the different phases of the cell cycle.
In conclusion, we have developed membrane-permeable Cu complexes that target the G4 structure as secondary-structure-specific nucleases. These serve as a component of a chemical toolbox that is complementary to the application of conventional G4-targeting antibodies and small-molecule G4 probes but overcome the permeability problems of antibodies and the potential for being artifactual increases in the cellular prevalence of the G4 motif by G4-stabilizing agents. Moreover, the Cu complexes that we have designed are the first-reported chemical nucleases that do not impact the stability of G4 telomeric DNA and can be used to study G4 formation of telomeric DNA at their natural cellular levels. In contrast to the slow reduction in telomere length caused by telomerase inhibitors, these G4-targeting Cu complexes can promote direct cleavage of G4 telomeric DNA, resulting in a rapid and robust telomere reduction in cells that eventually induce cellular senescence. The cleavage of telomeric G4 DNA by these chemical nucleases confirms the in vivo evidence for the formation of G4 telomeric DNA that was previously revealed through an anti-G4 antibody and G4 probe. The activity of chemical nucleases can be used to reveal the formation of G4 structures in different cellular events and potentially regulate their functional roles by direct cleavage. While in this work, we focus on cellular cleavage of G4 telomeric DNA, further tuning of the activity and selectivity of these G4 chemical nucleases should extend their application to other G4 DNA or RNA motifs of interest.
Limitations of the study
This study has some limitations that will require future research to address. While the chemical nucleases described in this work exhibit no effect on the stability of G4 telomeric DNA, they do either increase or decrease the melting points of c-kit and c-myc G4, respectively. Aside from the G4 DNA investigated in this study, other guanine-rich DNA or RNA sequences may also potentially form a G4 motif. A more comprehensive evaluation of all possible G4s is beyond the scope of a single study. Accordingly, the effect of these chemical nucleases in modulating the G4 landscape in cells, especially other G4s of DNA or RNA distinct from telomeric DNA, is not clear. In addition, the potential cross-interaction between anti-G4 antibodies and small-molecular G4 ligands and other G4-binding agents were not studied. However, the unique features of the chemical nucleases described herein provides guidance for development of chemical nucleases that either selectively target other G4s or universal nucleases that exhibit no effect on the stability of a broad spectrum of G4 motifs.
STAR★methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| BRACO19 | Sigma Aldrich | SML0560 |
| GGH | Bachem | 4006117 |
| Nap | This paper | N/A |
| CuGGHNap | This paper | N/A |
| (CuGGH)2Nap | This paper | N/A |
| Critical commercial assays | ||
| Dead Cell Apoptosis Kit with Annexin V Alexa Fluor™ 488 & Propidium Iodide (PI) | ThermoFisher Scientific | V13241 |
| Experimental models: Cell lines | ||
| Huh7 | Japanese Collection of Research Bioresources | JCRB0403 |
| Oligonucleotides | ||
| fluorescein-AGGGTTAGGGTTAGGGTTAGGG | This paper | N/A |
| fluorescein-AGGGAGGGCGCTGGGAGGAGGG | This paper | N/A |
| fluorescein-AGGGTGGGTAGGGTGGG | This paper | N/A |
| fluorescein-d(TTAGGG)-(CH2CH2O)6-d(CCCTAA) | This paper | N/A |
| teloF primer: TTGTTTGGGTTTGGGTTTGGGTTTGGGTT TGGGTT | O'Callaghan and Fenech, 2011 | N/A |
| teloR primer: GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT | O'Callaghan and Fenech, 2011 | N/A |
| 36B4F primer: GCAAGTGGGAAGGTGTAATCC | O'Callaghan and Fenech, 2011 | N/A |
| 36B4R primer: CCCATTCTATCATCAACGGGTACAA | O'Callaghan and Fenech, 2011 | N/A |
| 36B4STD: AAGTGGGAAGGTGTAATCC GTCTCCACAGACAAGGCCAGGACT CGTTTGTACCCGTTGATGATAGAATGGG | O'Callaghan and Fenech, 2011 | N/A |
| teloSTD: ((TTAGG)14) | O'Callaghan and Fenech, 2011 | N/A |
| Software and algorithms | ||
| FlowJo | FlowJo, LLC | https://www.flowjo.com/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| CASPlab | CASPlab | https://casplab.com/ |
| OriginPro | OriginLab Corporation | https://www.originlab.com/ |
Resource availability
Lead contact
J. A. Cowan, Department of Chemistry and Biochemistry, Ohio State University, Columbus, OH 43210, USA. E-mail: cowan.2@osu.edu. Tel: +1 614 292 2703
Materials availability
Synthesis of chemical nucleases is described in method details and are available from the corresponding author on request.
Data and code availability
The data used for generation of figures in this work are included in this manuscript; Additional data sets that are not included have not been deposited in a public repository but are available from the corresponding author on request.
Experimental model and subject details
Cell culture
Human cancer cell lines HuH-7 were cultured in DMEM (Dulbecco’s modified Eagle's medium) supplemented with 10% fetal bovine serum in an atmosphere containing 5% CO2 at 37°C.
Method details
Materials and instruments
All reagents were commercially available. 1H- and 13C-NMR spectra were recorded at 298 K using a Bruker DPX-400 spectrometer and standard pulse sequences. Electrospray mass spectra (ESI-MS) were measured by use of a Bruker MicroTOF, and the predicted isotope distribution patterns were calculated using the prediction program provided by the manufacturer. GGH peptide was purchased from Bachem, protected by di-tert-butyl-dicarbonate. All oligonucleotides, both with and without 5’ fluorescein, were purchased from Integrated DNA Technologies, Telo: 5’-d(AGGGTTAGGGTTAGGG), dsTelo: 5’-d(TTAGGG)-(CH2CH2O)6-d(CCCTAA), c-kit promoter: 5’-d(AGGGAGGGCGCTGGGAGGAGGG), c-myc promoter 5’-d(AGGGTGGGTAGGGTGGG).
Synthesis of Nap
The precursor, 4,9-dibromo-2,7-bis(3-morpholinopropyl)benzo[lmn][3,8]phenanthroline-1,3,6,8-(2H,7H)-tetraone, was synthesized using a reported method (Micco et al., 2013). A total of 100 mg of this compound was added slowly to 0.218 g 1,3-propylenediamine in 10 mL DMF, yielding a reddish solution. After stirring at RT for 15 hr in the dark, solvent was removed by rotavap. Crude product was dissolved in 1 M HCl and reverse-phase semi-preparative HPLC carried out by use of an HP 1100 Series HPLC apparatus (linear elution gradient: 95% H2O 5% CH3CN 0.1% TFA to 70% H2O 30% CH3CN 0.1% TFA in 25 min, 5 mL/min) with detection at 550 nm, and separation achieved with a C18 Vydac column (5 μM, 250 mm length, 10 mm I.D.). Yield: 63%. 1H NMR (400 MHz, D2O) δ 7.53 (s, 2H), 4.05 (d, J = 13.9 Hz, 8H), 3.76 (t, J = 12.1 Hz, 4H), 3.49 (d, J = 11.8 Hz, 8H), 3.25 (t, J = 7.6 Hz, 4H), 3.14 (t, J = 7.6 Hz, 8H), 2.09 (t, J = 7.1 Hz, 8H). 13C NMR (100 MHz, D2O) δ 165.33, 163.29, 148.39, 124.45, 120.15, 117.38, 101.07, 63.77, 54.82, 51.76, 39.69, 37.49, 37.28, 26.70, 22.06. HRMS (ESI) calcd for [C34H49O8N6]+ (M + H)+: m/z 665.3775, found 665.3679.
Synthesis of GGHNap and (GGH)2Nap
To a solution of 34.0 mg Boc-GGH peptide dissolved in 1 mL DMF, 12.8 mg N,N′-diisopropylcarbodiimide (DIC) and 11.7 mg N-hydroxysuccinimide (NHS) were added. The above solution was stirred in the dark for 30 min, a solution of 15.0 mg 1 dissolved in 1 mL DMF was slowly added, and the resulting solution was stirred in the dark at room temperature for 24 hr. Reverse-phase semi-preparative HPLC was carried out by use of an HP 1100 Series HPLC apparatus (isocratic: 12% CH3CN with 0.1% TFA in water, 5 mL/min) with detection at 550 nm, and separation achieved with a C18 Vydac column (5 μM, 250 mm length, 10 mm I.D.). GGHNap, yield: 31%. 1H NMR (400 MHz, D2O) δ 8.56 (s, 1H), 7.60 (d, J = 4.6 Hz, 2H), 7.25 (s, 1H), 4.64 (t, J = 4.8 Hz, 1H), 4.11 (t, J = 6.4 Hz, 2H), 4.06 (dd, J = 13.0, 2.1 Hz, 4H), 3.92 (s, 4H), 3.81 (s, 4H), 3.75 (t, J = 12.3 Hz, 4H), 3.49 (d, J = 12.5 Hz, 2H), 3.43–3.30 (m, 6H), 3.29–3.07 (m, 12H), 2.09 (t, J = 7.4 Hz, 4H), 1.90 (m, 4H). HRMS (ESI) calcd for [C44H61N13O9]+ (M + H)+: m/z 915.4715, found 915.459. (GGH)2Nap, yield: 22%. 1H NMR (400 MHz, D2O) δ 8.49 (d, J = 1.3 Hz, 2H), 7.46 (d, J = 6.0 Hz, 2H), 7.18 (s, 2H), 4.61 (dd, J = 8.5, 5.6 Hz, 2H), 3.99 (m, 8H), 3.86 (s, 4H), 3.75 (s, 4H), 3.69 (t, J = 12.5 Hz, 4H), 3.42 (d, J = 12.4 Hz, 4H), 3.30 (m, 8H), 3.23–3.01 (m, 12H), 2.02 (quint, J = 7.6 Hz, 4H), 1.84 (t, J = 6.2 Hz, 4H). HRMS (ESI) calcd for [C54H75N18O12]+ (M + H)+: m/z 1167.5812, found 1167.5964.
Synthesis of CuGGHNap and (CuGGH)2Nap
Independent UV titrations at 240 nm, or fluorescence titrations (λex = 620 nm), were performed with both Cu(II) and Ni(II) to quantify the concentration of metal-free ligand GGHNap and (GGH)2Nap. An equimolar and 2 equimolar amount of CuCl2 was added to ligand GGHNap and (GGH)2Nap, respectively, in 10 mM Tris-HCl (pH = 7.4) to yield solutions of CuGGHNap and (CuGGH)2Nap complexes.
FRET assay
G4 ligands were titrated into a solution of 1 μM fluorescein-labelled oligonucleotides, Telo (fluorescein-d(AGGGTTAGGGTTAGGGTTAGGG)), c-kit promoter (fluorescein-d(AGGGAGGG CGCTGGGAGGAGGG)) or c-myc promoter (fluorescein-d(AGGGTGGGTAGGGTGGG)) in 10 mM Tris-HCl and 100 mM KCl (pH 7.4). The change of emission intensity at 520 nm (λex = 495 nm) was monitored for the measurement of KD for G-quadruplex DNA. The KD for calf thymus DNA (CT-DNA) was calculated by use of a competition assay whereby G4 ligands were titrated to a solution containing 1 μM Telo and 100 μM CT-DNA in 10 mM Tris-HCl and 100 mM KCl (pH 7.4).
DNA cleavage was performed in the presence of 1 mM ascorbate, 1 mM H2O2, and 0.5 μM copper complex in 10 mM Tris-HCl and 100 mM KCl (pH 7.4) at 37°C, and the change of emission intensity at 520 nm (λex = 495 nm) was monitored.
Thermal melts
Thermal melting curves were collected on a JASCO J-815 Circular Dichroism Spectrometer in a 1 mm pathlength quartz cuvette. Measurements were collected from 320 to 200 nm with a scan rate of 200 nm/min with increasing temperature from 25°C to 95°C at a ramp rate of 1°C/min. Measurements of CD signal intensity were taken every 2°C. Samples contained 10 μM of Telo, c-kit, or c-myc DNA in the absence or presence of 10 μM naphthalene diimide derivatives in 10 mM tris-HCl (pH 7.4), 100 mM KCl. DNA samples were annealed by heating to 92°C for 1 min and then allowed to slowly reach room temperature before thermal melt measurements were taken.
Denaturing PAGE
Solutions containing 6 μM (strand concentration) of Telo, 4 μM Cu complexes, 1 mM ascorbate and 1 mM H2O2 in 100 mM Tris-HCl and 100 mM KCl (pH 7.4) were incubated at 37°C for 0, 30, 60, 90, 120, 150 min. Reactions were quenched by loading buffer containing 10% DMSO and 1 mM EDTA. A 15% denaturing polyacrylamide gel containing 6 M urea was used for gel electrophoresis, and the resulting gel was imaged by use of a Typhoon Trio Imager (GE). The integration of bands was performed by use of the software provided by the manufacturer.
To maintain the same concentration of total nucleotides as Telo, 11 μM (strand concentration) of dsTelo DNA was incubated with 4 μM Cu complexes, 1 mM ascorbate and 1 mM H2O2. A 10 mM Tris-HCl buffer containing 100 mM LiCl (pH 7.4) was utilized to avoid the formation of intermolecular G-quadruplex without influencing duplex DNA. Reactions were incubated and quenched under the same conditions as Telo, followed by gel electrophoresis in a 25% denaturing polyacrylamide gel containing 7 M urea and 15% formamide.
Cell synchronization
S-phase cells and G1/G0-phase cells were obtained through the double-thymidine block and serum starvation, respectively. Propidium iodide was used to assess the cell cycle, using a BD LSR II Flow Cytometer System to confirm cell synchronization. Additionally, for S-phase HuH7 cells, after the retreat of thymidine, cells were incubated with 2 μM compound and 100 ng/mL nocodazole for 10 hr (control: 100 ng/mL nocodazole). Additionally, cell cycle analysis was performed every hour until 10 hr had passed. It was observed that under our experimental conditions HuH7 cells recover from thymidine block treatment in the first 1–2 hr, subsequently undergo S-phase in the following 5–6 hr and are eventually trapped in the G2/M-phase due to nocodazole inhibition (data not shown). For G1/G0-phase HuH7 cells, cells were incubated with 2 μM compound in serum-free medium.
Telomere length measured by real-time PCR
A total of 1 × 105 cells was incubated with the test articles. After the indicated incubation period, the medium was withdrawn, and the cells rinsed three times with PBS. Genomic DNA was extracted, and the telomere length of each sample was subsequently measured by real-time PCR using a reported method (O'Callaghan and Fenech, 2011). Telomere length measured by this q-PCR assay was reported to be consistent with that measured by South Blotting (O'Callaghan and Fenech, 2011). Briefly, a pair of primers teloF (TTGTTTGGGTTTGGGTTTGGGTTTGGGTT TGGGTT) and teloR (GGCTTGCCTTACCCTTACC CTTACCCTTACCCTTACCCT) were used to measure the amount of telomeric DNA in the sample, while another pair of primers 36B4F (GCAAGTGGGAAGGTGTAATCC) and 36B4R (CCCATTCTATCATCAACGGGTACAA) were used to measure the amount of a human single-copy gene 36B4 in the same sample but in a separate well. A 10 μL solution containing 80 ng of genomic DNA extracted from cells and 2 μM of each primer (either for telomeric DNA or 36B4) in water was mixed with 10 μL Fast SYBR Green Master Mix (purchased from Life technologies), yielding totally 20 μL reaction solution in each well of a 96-well PCR plate. Standard wells contain known amount of either teloSTD ((TTAGG)14) or 36B4STD(AAGTGGGAAGGTGTAATCC GTCTCCACAGACAAGGCCAGGACT CGTTTGTACCCGTTGATGATAGAATGGG), 80 ng pUC19 plasmid DNA and the same quantity of primers and SYBR Green Master Mix. Real-time PCR was performed by the Applied Biosystems 7900HT Fast Real-Time PCR System using the below cycling conditions (optimized by manufacturer): 2 min at 95°C, then 20 s at 95°C, followed by 40 cycles of 95°C for 15 s, 60°C for 1 min, then followed by a dissociation (or melt) curve. The amount of telomeric DNA or 36B4 in each sample was calculated from the standard plot of telomeric DNA or 36B4, respectively. To correct the difference between samples, 36B4 was used as an internal reference to normalize the amount of telomeric DNA in each sample.
Alkaline comet assay
Cells were incubated with metal complexes at the indicated concentration for 24 hr. A positive control was obtained by incubating cells in 50 μM H2O2 for 30 min. After cells were harvested through trypsinization, 1.5 × 103 cells were embedded in 0.75% low-melting point agarose on Comet slides (Trevigen). Cells were lysed in an alkaline solution containing 0.3 M NaOH (pH > 13) in the dark for 15 hr at 4°C. Electrophoresis was performed for 10 min at 22 V in alkaline-EDTA buffer (pH > 12). After electrophoresis, the cells were rinsed by H2O, followed by staining with 10 μg/mL propidium iodide for 20 min. Fluorescence imaging was performed by use of a Zeiss Axioskop microscope. CASPlab software was used to score a total of 100 cells for each sample.
Colocalization studied by confocal microscope
Cells were cultured on glass slips and incubated with the indicated compounds for 12 hr. Subsequently, the cells were rinsed with PBS 3 times, followed by fixation with 3% formaldehyde. A solution of 100 ng/mL DAPI in PBS was used to stain the cellular nucleus. Cells were again rinsed 3 times with PBS and visualized by use of an Olympus Confocal Microscopy FV1000. The intrinsic emission of naphthalene diimide derivatives was monitored following excitation by a 633 nm laser line.
FITC-annexin V/PI assay
Approximately 2.5 × 104 cells were seeded into a 6-well plate and allowed to attach for 24 hr. After incubating the cells with the test articles for the indicated time, the cells were then harvested by trypsinization. Following resuspension in 10 mM HEPES (pH 7.4), 2.5 mM CaCl2 and 140 mM NaCl, the cells were counted. Approximately 1 × 105 cells were subsequently stained by FITC-Annexin V and PI in 10 mM HEPES (pH 7.4), 2.5 mM CaCl2 and 140 mM NaCl. FACS (Fluorescence-Activated Cell Sorting) analysis was performed by use of a BD LSR II Flow Cytometer System.
Senescence-associated β-galactosidase assay
Approximately 2.5 × 104 HuH7 cells were seeded onto a glass slip and allowed to attach for 24 hr before the addition of the test article. After the indicated incubation period, the medium was withdrawn, and cells were rinsed with PBS, followed by fixation with 3% formaldehyde in PBS. Following removal of the formaldehyde solution, the cells were rinsed with PBS and stained by use of an X-gal solution (1 mg/mL, pH 6) for 16 h at 37°C as described in the reported method. Cells were visualized by use of a Zeiss Axioskop microscopy by phase-contrast imaging.
MTT assay
A total of 3,000 cells were seeded into each well of a 96-well plate, followed by 24 h incubation that allowed the cells to attach. After 72 h incubation with each of the test compounds, 80 μg of MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was added to each well and subsequently incubated at 37°C for 4 h. After the medium was carefully withdrawn, a 200 μL volume of DMSO was added to each well and the absorbance at 560 nm was measured by use of a SpectraMax M5 Multi-Mode Microplate Reader.
Quantification and statistical analysis
All experiments were performed in triplicate. All data are expressed as the mean ± standard deviation.
Acknowledgments
This work was supported by grants from the National Science Foundation [CHE-1800239]. Z.Y. was supported by the Pelotonia Fellowship Program.
Author contributions
Z.Y. and A.L.H. performed the experiments. Z.Y., A.L.H., and J.A.C wrote the manuscript and interpreted the data.
Declaration of interests
The authors declare no competing financial interests.
Published: June 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102661.
Supplemental information
References
- Alexander J.L., Thompson Z., Cowan J.A. Antimicrobial metallopeptides. ACS Chem. Biol. 2018;13:844–853. doi: 10.1021/acschembio.7b00989. [DOI] [PubMed] [Google Scholar]
- Alexander J.L., Yu Z., Cowan J.A. Amino terminal copper and nickel binding motif derivatives of ovispirin-3 display increased antimicrobial activity via lipid oxidation. J. Med. Chem. 2017;60:10047–10055. doi: 10.1021/acs.jmedchem.7b01117. [DOI] [PubMed] [Google Scholar]
- Ambrus A., Chen D., Dai J.X., Jones R.A., Yang D.Z. Solution structure of the biologically relevant g-quadruplex element in the human c-MYC promoter. implications for g-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- Biffi G., Di Antonio M., Tannahill D., Balasubramanian S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014;6:75–80. doi: 10.1038/nchem.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- Damm K., Hemmann U., Garin-Chesa P., Hauel N., Kauffmann I., Priepke H., Niestroj C., Daiber C., Enenkel B., Guilliard B. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 2001;20:6958–6968. doi: 10.1093/emboj/20.24.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Mundo I.M.A., Vasquez K.M., Wang G.L. Modulation of DNA structure formation using small molecules. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:118539. doi: 10.1016/j.bbamcr.2019.118539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Antonio M., Ponjavic A., Radzevicius A., Ranasinghe R.T., Catalano M., Zhang X., Shen J., Needham L.M., Lee S.F., Klenerman D. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat. Chem. 2020;12:832–837. doi: 10.1038/s41557-020-0506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X.L., Liu N.N., Yang Y.T., Li H.H., Li M., Dou S.X., Xi X.G. G-quadruplexes significantly stimulate Pif1 helicase-catalyzed duplex DNA unwinding. J. Biol. Chem. 2015;290:7722–7735. doi: 10.1074/jbc.M114.628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016;48:1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- Hansel-Hertsch R., Spiegel J., Marsico G., Tannahill D., Balasubramanian S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2018;13:551–564. doi: 10.1038/nprot.2017.150. [DOI] [PubMed] [Google Scholar]
- Henderson A., Wu Y.L., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M., Sen D., Lansdorp P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M.D., Lee D.C., Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman K.E., Levene S.D., Tesmer V.M., Shay J.W., Wright W.E. Telomere shortening is proportional to the size of the G-rich telomeric 3 '-overhang. J. Biol. Chem. 2000;275:19719–19722. doi: 10.1074/jbc.M002843200. [DOI] [PubMed] [Google Scholar]
- Jamroskovic J., Doimo M., Chand K., Obi I., Kumar R., Brannstrom K., Hedenstrom M., Das R.N., Akhunzianov A., Deiana M. Quinazoline ligands induce cancer cell death through selective STAT3 inhibition and G-quadruplex stabilization. J. Am. Chem. Soc. 2020;142:2876–2888. doi: 10.1021/jacs.9b11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreig A., Calvert J., Sanoica J., Cullum E., Tipanna R., Myong S. G-quadruplex formation in double strand DNA probed by NMM and CV fluorescence. Nucleic Acids Res. 2015;43:7961–7970. doi: 10.1093/nar/gkv749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Chand K., Bhowmik S., Das R.N., Bhattacharjee S., Hedenstrom M., Chorell E. Subtle structural alterations in G-quadruplex DNA regulate site specificity of fluorescence light-up probes. Nucleic Acids Res. 2020;48:1108–1119. doi: 10.1093/nar/gkz1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok C.K., Marsico G., Sahakyan A.B., Chambers V.S., Balasubramanian S. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods. 2016;13:841. doi: 10.1038/nmeth.3965. [DOI] [PubMed] [Google Scholar]
- Lane A.N., Chaires J.B., Gray R.D., Trent J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps H.J., Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Liu L.Y., Fang H., Chen Q., Chan M.H., Ng M., Wang K.N., Liu W., Tian Z., Diao J., Mao Z.W. Multiple-color platinum complex with super-large Stokes shift for super-resolution imaging of autolysosome escape. Angew. Chem. Int. Ed. Engl. 2020;59:19229–19236. doi: 10.1002/anie.202007878. [DOI] [PubMed] [Google Scholar]
- Liu L.Y., Liu W.T., Wang K.N., Zhu B.C., Xia X.Y., Ji L.N., Mao Z.W. Quantitative detection of G-quadruplex DNA in live cells based on photon counts and complex structure discrimination. Angew. Chem. Int. Ed. 2020;59:9719–9726. doi: 10.1002/anie.202002422. [DOI] [PubMed] [Google Scholar]
- Liu W., Zhong Y.F., Liu L.Y., Shen C.T., Zeng W., Wang F., Yang D., Mao Z.W. Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding. Nat. Commun. 2018;9:3496. doi: 10.1038/s41467-018-05810-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti B.K., Govil N., Kundu T., Moura J.J.G. Designed metal-ATCUN derivatives: redox- and non-redox-based applications relevant for chemistry, biology, and medicine. iScience. 2020;23:101792. doi: 10.1016/j.isci.2020.101792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- Marsico G., Chambers V.S., Sahakyan A.B., McCauley P., Boutell J.M., Di Antonio M., Balasubramanian S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019;47:3862–3874. doi: 10.1093/nar/gkz179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micco M., Collie G.W., Dale A.G., Ohnmacht S.A., Pazitna I., Gunaratnam M., Reszka A.P., Neidle S. Structure-based design and evaluation of naphthalene diimide G-quadruplex ligands as telomere targeting agents in pancreatic cancer cells. J. Med. Chem. 2013;56:2959–2974. doi: 10.1021/jm301899y. [DOI] [PubMed] [Google Scholar]
- Miron C.E., van Staalduinen L., Rangaswamy A.M., Chen M., Liang Y., Jia Z., Mergny J.L., Petitjean A. Going platinum to the tune of a remarkable guanine quadruplex binder: solution- and solid-state investigations. Angew. Chem. Int. Ed. Engl. 2021;60:2500–2507. doi: 10.1002/anie.202012520. [DOI] [PubMed] [Google Scholar]
- Nadai M., Doria F., Scalabrin M., Pirota V., Grande V., Bergamaschi G., Amendola V., Winnerdy F.R., Phan A.T., Richter S.N. A catalytic and selective scissoring molecular tool for quadruplex nucleic acids. J. Am. Chem. Soc. 2018;140:14528–14532. doi: 10.1021/jacs.8b05337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017;1:0041. [Google Scholar]
- O'Callaghan N.J., Fenech M. A quantitative PCR method for measuring absolute telomere length. Biol. Proced. Online. 2011;13:3. doi: 10.1186/1480-9222-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan M.P., Haldar S., Duchi M., Oliver T.A.A., Mulholland A.J., Morales J.C., Galan M.C. A photoresponsive stiff-stilbene ligand fuels the reversible unfolding of G-quadruplex DNA. Angew. Chem. Int. Ed. 2019;58:4334–4338. doi: 10.1002/anie.201900740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan M.P., Morales J.C., Galan M.C. Binding and beyond: what else can G-quadruplex ligands do? Eur. J. Org. Chem. 2019;2019:4995–5017. [Google Scholar]
- Onizuka K., Hazemi M.E., Sato N., Tsuji G., Ishikawa S., Ozawa M., Tanno K., Yamada K., Nagatsugi F. Reactive OFF-ON type alkylating agents for higher-ordered structures of nucleic acids. Nucleic Acids Res. 2019;47:6578–6589. doi: 10.1093/nar/gkz512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A.T., Kuryavyi V., Burge S., Neidle S., Patel D.J. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkham A.M., Yu Z., Cowan J.A. Attenuation of west nile virus NS2B/NS3 protease by amino terminal copper and nickel binding (ATCUN) peptides. J. Med. Chem. 2018;61:980–988. doi: 10.1021/acs.jmedchem.7b01409. [DOI] [PubMed] [Google Scholar]
- Ross M.J., Bradford S.S., Cowan J.A. Catalytic metallodrugs based on the LaR2C peptide target HCV SLIV IRES RNA. Dalton T. 2015;44:20972–20982. doi: 10.1039/c5dt02837j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruehl C.L., Lim A.H.M., Kench T., Mann D.J., Vilar R. An octahedral cobalt(III) complex with axial NH3 ligands that templates and selectively stabilises G-quadruplex DNA. Chem. Eur. J. 2019;25:9691–9700. doi: 10.1002/chem.201901569. [DOI] [PubMed] [Google Scholar]
- Shammas M.A., Reis R.J.S., Li C., Koley H., Hurley L.H., Anderson K.C., Munshi N.C. Telomerase inhibition and cell growth arrest after telomestatin treatment in multiple myeloma. Clin. Cancer Res. 2004;10:770–776. doi: 10.1158/1078-0432.ccr-0793-03. [DOI] [PubMed] [Google Scholar]
- Varshney D., Spiegel J., Zyner K., Tannahill D., Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020;21:459–474. doi: 10.1038/s41580-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang Q.M., Wang Y.R., Xi X.G., Hou X.M. DNA-unwinding activity of Saccharomyces cerevisiae Pif1 is modulated by thermal stability, folding conformation, and loop lengths of G-quadruplex DNA. J. Biol. Chem. 2018;293:18504–18513. doi: 10.1074/jbc.RA118.005071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.G., Spies M. G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 2016;44:8742–8753. doi: 10.1093/nar/gkw574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.Q., Hou X.M., Zhang B., Fosse P., Rene B., Mauffret O., Li M., Dou S.X., Xi X.G. Single-molecule studies reveal reciprocating of WRN helicase core along ssDNA during DNA unwinding. Sci. Rep. 2017;7:43954. doi: 10.1038/srep43954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.Y., Lejault P., Chevrier S., Boidot R., Robertson A.G., Wong J.M.Y., Monchaud D. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat. Commun. 2018;9:4730. doi: 10.1038/s41467-018-07224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Cowan J.A. Catalytic metallodrugs: substrate-selective metal catalysts as therapeutics. Chem. Eur. J. 2017;23:14113–14127. doi: 10.1002/chem.201701714. [DOI] [PubMed] [Google Scholar]
- Yu Z., Cowan J.A. Design of artificial glycosidases: metallopeptides that remove H antigen from human erythrocytes. Angew. Chem. Int. Ed. 2017;56:2763–2766. doi: 10.1002/anie.201612079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Cowan J.A. Metal complexes promoting catalytic cleavage of nucleic acids-biochemical tools and therapeutics. Curr. Opin. Chem. Biol. 2018;43:37–42. doi: 10.1016/j.cbpa.2017.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Fenk K.D., Huang D., Sen S., Cowan J.A. Rapid telomere reduction in cancer cells induced by G-quadruplex-targeting copper complexes. J. Med. Chem. 2019;62:5040–5048. doi: 10.1021/acs.jmedchem.9b00215. [DOI] [PubMed] [Google Scholar]
- Yu Z., Han M.L., Cowan J.A. Toward the design of a catalytic metallodrug: selective cleavage of G-quadruplex telomeric DNA by an anticancer copper-acridine-ATCUN complex. Angew. Chem. Int. Edit. 2015;54:1901–1905. doi: 10.1002/anie.201410434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Thompson Z., Behnke S.L., Fenk K.D., Huang D., Shafaat H.S., Cowan J.A. Metalloglycosidase mimics: oxidative cleavage of saccharides promoted by multinuclear copper complexes under physiological conditions. Inorg. Chem. 2020;59:11218–11222. doi: 10.1021/acs.inorgchem.0c01193. [DOI] [PubMed] [Google Scholar]
- Zhang F., Wen Y., Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum. Mol. Genet. 2014;23:R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- Zhang S.G., Sun H.X., Wang L.X., Liu Y., Chen H.B., Li Q., Guan A.J., Liu M.R., Tang Y.L. Real-time monitoring of DNA G-quadruplexes in living cells with a small-molecule fluorescent probe. Nucleic Acids Res. 2018;46:7522–7532. doi: 10.1093/nar/gky665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K.W., Zhang J.Y., He Y.D., Gong J.Y., Wen C.J., Chen J.N., Hao Y.H., Zhao Y., Tan Z. Detection of genomic G-quadruplexes in living cells using a small artificial protein. Nucleic Acids Res. 2020;48:11706–11720. doi: 10.1093/nar/gkaa841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used for generation of figures in this work are included in this manuscript; Additional data sets that are not included have not been deposited in a public repository but are available from the corresponding author on request.