Abstract
Background
Several reports on the discovery of SARS-CoV-2 mutations and variations in Indonesia COVID-19 cases led to genomic dysregulation with the first pandemic cases in Wuhan, China. MicroRNA (miRNA) plays an important role in this genetic regulation and contributes to the enhancement of viral RNA binding through the host mRNA.
Objective
This research is aimed to detect miRNA targets of SARS-CoV-2 and examines their role in Indonesia cases against Wuhan cases.
Methods
SARS-CoV-2 sequences were obtained from GISAID (https://www.gisaid.org/), NCBI (https://ncbi.nlm.nih.gov), and National Genomics Data Center (https://bigd.big.ac.cn/gwh/) databases. MiRDB (https://github.com/gbnegrini/mirdb-custom-target-search) was used to annotate and predict target human mature miRNAs. For statistical analysis, we utilized a series chi-square test to obtain significant miRNA. DIANA-miRPath v3.0 (http://www.microrna.gr/miRPathv3) analyzed the Gene Ontology of mature miRNAs.
Result
The statistical results detected five significant miRNAs. Two miRNAs: hsa-miR-4778-5p and hsa-miR-4531 were consistently found in the majority of Wuhan samples, while they were only found in less than half of the Indonesia samples. The other three miRNA, hsa-miR-6844, hsa-miR-627-5p, and hsa-miR-3674, were discovered in most samples in both groups but with a significant difference ratio. Among these five significant miRNA targets, hsa-miR-6844 is the only miRNA that has an association with the ORF1ab gene of SARS-CoV-2.
Conclusion
The Gene Ontology analysis of five significant miRNA targets indicates a significant role in inflammation and the immune system. The specific detection of host miRNAs in this study shows that there are differences in the characteristics of SARS-CoV-2 between Indonesia and Wuhan.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13258-021-01119-7.
Keywords: miRNA, SARS-CoV-2, Indonesia, Wuhan, Genome data mining, Bioinformatics
Background
On December 12, 2019, the pandemic of coronavirus disease 2019 (COVID-19) was first reported in Wuhan by a novel coronavirus that has similarities with bat coronaviruses (SL-CoVZC45 and CoVZXC21) (Wu et al. 2020; Koyama et al. 2020). The novel RNA virus mutates rapidly and carries new variants spreading around the world (Mercatelli and Giorgi 2020). As of July 7, 2020, the majority of several clades of SARS-CoV-2 variants had been identified. Clade GR was most frequently observed worldwide. Meanwhile, clade L, the first observed clade of SARS-CoV-2, has the smallest proportion among all clades. The variants found in the Asian continent itself dominated by clade O (Hamed et al. 2020).
As of early October 2020, Indonesia has confirmed a total of 291,182 positive COVID-19 cases. The percentage of deaths in Indonesia is 3.72% (10,856 deaths), which is higher than the global death trend (2.98%) (WHO 2020). The surge of COVID-19 cases in Indonesia is expected to be related to mutations in the Spike Glycoprotein (S) gene that leads to the ability of the virus to bind to the ACE-2 (Angiotensin-Converting Enzyme 2) receptor in human hosts (Hoffmann et al. 2020). There are currently several in silico studies in Indonesia to understand the immunoinformatic aspect (Ansori et al. 2020b; Nidom et al. 2020), molecular docking (Parikesit and Nurdiansyah 2020a, b), genetic variant, and mutations (Ansori et al. 2020a, b; Nidom et al. 2020) of the SARS-CoV-2 genome from Indonesia COVID-19 cases. However, a microRNA (miRNA) SARS-CoV-2 study is rarely conducted in Indonesia. Therefore, we carried out an in silico approach for predicting and profiling miRNAs from Indonesia samples.
MicroRNA is a non-coding RNA that has a vital role in the gene expression of an organism. MicroRNA dysregulation can cause changes in expression patterns and gene mutations leading to disease severity (Winter et al. 2009; Krol et al. 2010; Mendell and Olson 2012; Finnegan and Pasquinelli 2013; Fernandez et al. 2017). Alterations of miRNA function also can change the genetic variation, which can be identified by detecting target miRNAs from miRNA sequences (Cammaerts et al. 2015; Martin-Guerrero et al. 2015; Fernandez et al. 2017). Additionally, pathogenicity and viral RNA replication are mediated by the direct binding of host miRNA to viral RNA. The binding of the miRNA-viral RNA process induces genetic dysregulation in the host. By mimicking cellular miRNAs host, viruses affect host regulatory pathways and directly change host transcriptomes (Skalsky and Cullen 2010; Trobaugh and Klimstra 2017).
Relevant research using Asian SARS-CoV-2 isolates with various methods for predicting miRNA is usually investigated by conducting bioinformatics analysis. Sarma et al. (2020) compared China isolates to several countries to study miRNA interaction between humans and SARS-CoV-2. They applied VMir to identify several potentials pre-miRNAs and MatureBayes to identify and predict mature miRNAs. From their study, 22 significant potential miRNAs were predicted from five SARS-CoV-2 genome sequences. Another comparative study of the SARS CoV genome with SARS-CoV-2 from 24 countries conducted by Khan et al. (2020) who found 24 predicted miRNAs. They utilized miRNAFold software to predict pre-miRNAs from hairpin miRNAs, MFE (Minimum Free Energy) miRNA analysis software to find secondary structures, and MaturBayes to predict miRNAs. However, other studies directly predict without determining pre-miRNAs. That approach can be performed using web-based analyzing software, miRDB, to detect the targeted miRNAs. This software detects mature miRNAs that affect gene regulations directly with functional annotation (Fulzele et al. 2020; Haddad and Al-Zyoud 2020). Fulzele et al. (2020), observed the role of SARS-CoV-2 miRNA targets with human functional genes through KEGG pathway analysis and Gene Ontology analysis on DIANA-miRPath v.3.0 based on target prediction scores above 90 from miRDB. They compared isolates from several Asian countries to Wuhan isolates. The study shows that seven target miRNAs have significant scores from miRNA 558 SARS-CoV-2 isolates. Li et al. (2020) found that miR ‐ 16‐2‐3p, miR ‐ 6501‐5p, and miR ‐ 618 were significantly expressed on peripheral blood genome sequences from COVID-19 patients. This study of differential expression of miRNA is operated in miRdeep2 for filtering that is compared with the reference genome.
In this study, we used the SARS-CoV-2 samples found in Indonesia that were obtained from several public genome databases and compared them to the Wuhan samples to predict miRNA target using bioinformatics pipeline. This research aimed to find the target host miRNA of SARS-CoV-2 and the role of the pathogenesis of COVID-19 in Indonesia cases by applying bioinformatics analysis.
Methods
SARS-CoV-2 sequence data
All SARS-CoV-2 data were obtained from GISAID (https://www.gisaid.org/), NCBI (https://ncbi.nlm.nih.gov), and National Genomics Data Center (NGDC) which is a part of the China National Center for Bioinformation (https://bigd.big.ac.cn/gwh/). In total, we included 39 SARS-CoV-2 samples from Indonesia and 37 samples from Wuhan. The information of all samples from both groups can be found in Supplementary data Tables 1 and 2. All data were accessed on September 13, 2020.
Predicting miRNA target
MiRDB was used to annotate and predict target human mature miRNAs against SARS-CoV-2 sequences. MiRDB utilizes code from gbnegrini/mirdb-custom-target-search (https://github.com/gbnegrini/mirdb-custom-target-search) based on the version of the web-based software (http://mirdb.org/custom.html). We employed mirTarget2 algorithm based on Support Vector Machine to compute the miRNA target prediction score [30]. The threshold of the miRNA target prediction score used in our work is 80. Target miRNAs detected by miRDB are based on 38,589 mature miRNAs from the miRBase version 22.1 database (http://www.mirbase.org/). ChromosPros Version 2.1.9 software was used for quality control and trimming SARS-CoV-2 sequence data.
Statistical analysis
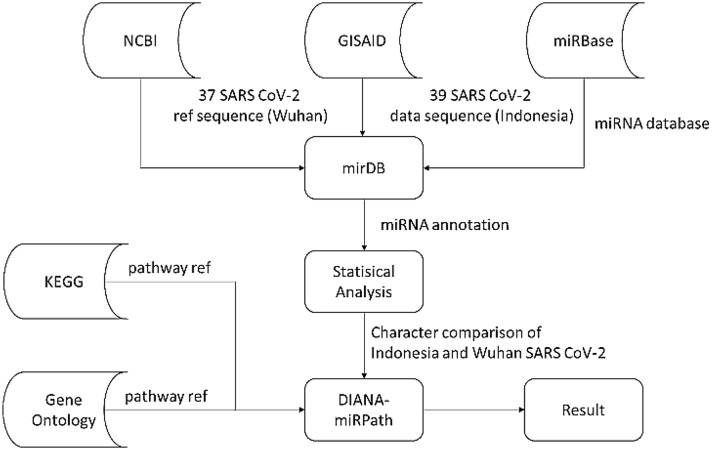
We performed a series chi-square test to obtain significant miRNA that can differentiate Indonesia and Wuhan isolates. This test captures the differences by seeing the proportion of miRNA presence rate in both groups. Only miRNAs with a p value < 0.05 were considered significant. The corresponding cross table for each miRNA in both groups was also generated to give a clear perspective on this comparative analysis. The whole research flow can be seen in (Fig. 1).
Fig. 1.
Research process diagram flow
miRNA gene ontology analysis
The miRNA targets were analyzed for their pathway using web-based software DIANA-miRPath v3.0 (http://www.microrna.gr/miRPathv3) based on biological process subcategory in Gene Ontology (Vlachos et al. 2015). DIANA-miRPath v3.0 applies DIANA-microT-CDS algorithm that predicts the CDS (Coding Sequence) mRNA and 3′ UTR regions with high accuracy (Paraskevopoulou et al. 2013). The whole research process is depicted in Table 3.
Table 3.
Gene Ontology Pathway of miRNA SARS-CoV-2 Indonesia vs Wuhan
| GO category | P value | Genes | miRNAs | miRNA List | Genes according to miRNAs |
|---|---|---|---|---|---|
| Cellular nitrogen compound metabolic process (GO:0034641) | < 1e-325 | 500 | 5 | hsa-miR-4778-5p | 225 |
| hsa-miR-4531 | 119 | ||||
| hsa-miR-6844 | 114 | ||||
| hsa-miR-627-5p | 31 | ||||
| hsa-miR-3674 | 83 | ||||
| Biosynthetic process (GO:0009058) | 3.33e-13 | 351 | 4 | hsa-miR-4778-5p | 176 |
| hsa-miR-4531 | 98 | ||||
| hsa-miR-6844 | 94 | ||||
| hsa-miR-627-5p | 28 | ||||
| Fc-epsilon receptor signaling pathway (GO:0038095) | 3.54e-10 | 34 | 3 | hsa-miR-4778-5p | 20 |
| hsa-miR-4531 | 9 | ||||
| hsa-miR-6844 | 8 | ||||
| Cellular protein modification process (GO:0006464) | 6.49e-10 | 220 | 3 | hsa-miR-4778-5p | 106 |
| hsa-miR-4531 | 61 | ||||
| hsa-miR-6844 | 68 |
Result
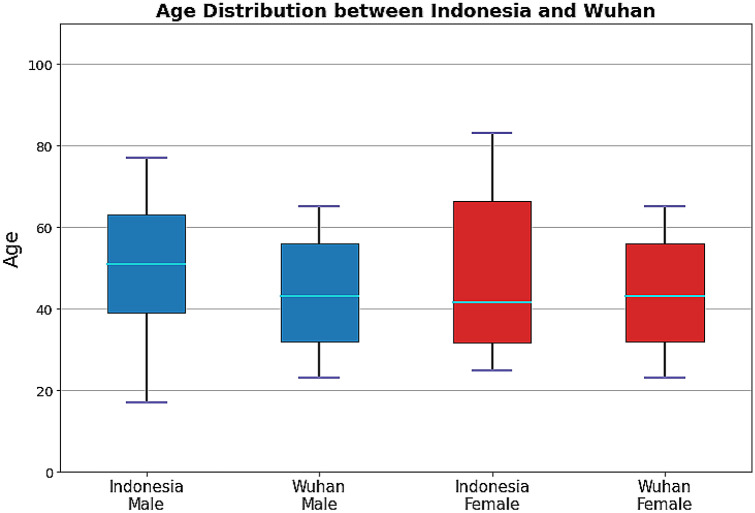
In total, there were 76 SARS-CoV-2 genome samples included in this study. The samples consist of 39 sequences from Indonesia and 37 sequences from Wuhan. Each sequence data is complemented with metadata, including age, gender, clade, and mutation data in the GISAID metadata to describe the SARS CoV-2 profile. The ages of patients infected with SARS CoV-2 in Indonesia ranged from 17-83 years, and the mean age of the patients was 50.79 years with a standard deviation of 17.88. Meanwhile, Wuhan patients who contracted COVID-19 ranged from 21 to 65 years old with an average age of 48.7 years and a standard deviation of 21.23. The gender profile of COVID-19 patients in Indonesia is 74.36% for men and 25.64% for women. Males and females composed 45.95 % and 54.05 % of the Wuhan population, respectively. In both countries, SARS CoV-2 infection was observed in people between the ages of 20 and 40 years old, as seen in the table of age distribution stratified by gender (Fig. 2). Based on this figure, in both countries, COVID-19 cases are often found in patients over 40 years of age in all populations. Based on the t-test analysis, the gender-related age distribution did not show any significant difference between the Indonesian and Wuhan populations.
Fig. 2.
Age distribution of SARS CoV-2 Metada from Indonesia and Wuhan samples stratified by gender
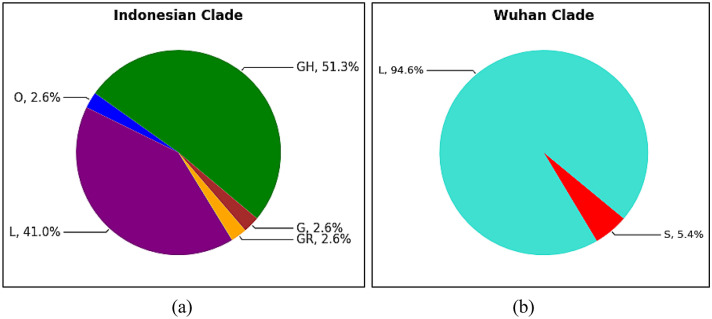
Based on GISAID metadata, the SARS CoV-2 clade profile between the Indonesian and Wuhan populations identified five and two distinct clades, respectively, as shown in Fig. 3. The clade GH and L dominated 51.3 % and 41 % of the clades found in the Indonesian population, respectively. Meanwhile, in Wuhan, clade L is the SARS CoV-2 group that controls almost all COVID-19 cases in this population. Clade L controls 94.6 % of the Wuhan population, with Clade S accounting for the remaining 5.4 percent. There is no significant difference between Clade L, which is mainly present in the Indonesian and Wuhan population. The mutation variants found from all the SARS CoV-2 clades were found in 72 variants in Indonesia and 24 in Wuhan. The Spike D614G and NSP12 P323L variants dominated SARS CoV-2 mutations in the Indonesian sample population, with both showing the same frequency of 30.56%. The NSP2 N92H and Spike N856K variants dominated the samples from Wuhan with frequencies of 25% and 12.5%.
Fig. 3.
Various SARS CoV-2 Clade in Indonesian (a) and Wuhan (b) population-Based on GISAID
The chi-square test outputs five significant miRNAs which differentiate Indonesia and Wuhan samples. Table 1 shows the list of these significant miRNA with the corresponding p value. The full contingency tables for these miRNAs can be seen in Table 2. hsa-miR-4778-5p (p value<0.001) and hsa-miR-4531 (p value=0.001) were two miRNAs with a clear contrast between these two groups. These two miRNAs were only predicted in less than half of genome samples from Indonesia, 48.72% and 25.64% respectively. On the other hand, these two miRNAs were predicted in the majority of Wuhan samples, with the former reach 100% of samples that contain this miRNA. The other three miRNAs, hsa-miR-6844 (p value=0.011), hsa-miR-627-5p (p value=0.023), and hsa-miR-3674 (p value=0.027), were consistently predicted in both sample groups despite of slight the differences in proportion. More than 90% of Indonesia samples were predicted to have these three miRNAs, whereas they are only detected in less than 85% of Wuhan samples. Furthermore, all samples from Indonesia were predicted to contain the hsa-miR-6844. Among these five significant miRNAs, only hsa-miR-6844 which associated with the ORF1ab gene.
Table 1.
Significant miRNA
| miRNA | P value |
|---|---|
| hsa-miR-4778-5p | < 0.001 |
| hsa-miR-4531 | 0.001 |
| hsa-miR-6844 | 0.011 |
| hsa-miR-627-5p | 0.023 |
| hsa-miR-3674 | 0.027 |
Table 2.
Contingency table of Significant miRNAs
| miRNA | Indonesia | Wuhan | ||
|---|---|---|---|---|
| Yes | No | Yes | No | |
| hsa-miR-4778-5p | 19 (48.72%) | 20 (51.28%) | 37(100%) | 0(0%) |
| hsa-miR-4531 | 10(25.64%) | 29(74.36%) | 24(64.86%) | 13(35.14%) |
| hsa-miR-6844 | 39(100%) | 0(0%) | 31(83.78%) | 6(16.22%) |
| hsa-miR-627-5p | 37(94.87%) | 2(5.13%) | 28(75.68%) | 9(24.32%) |
| hsa-miR-3674 | 38(97.44%) | 1(2.56%) | 30(81.08%) | 7(18.92%) |
In the subsequent analysis, the 5 significant miRNAs were then analyzed to obtain the related biological pathway using DIANA-miRPath v 3.0. Table 3 shows four pathways that were obtained based on the five significant miRNAs interest in Gene Ontology. Each of the associated biological pathways has a p-value less than 6.5e-10 and is associated with at least three significant miRNAs based on the distribution of probability densities from the hypergeometric test (Cao and Zhang 2014; Tomczak et al. 2018). The most significant biology pathway according to this analysis is Cellular Nitrogen Compound Metabolic Process (GO:0034641) (p value <1e-325). This biology pathway includes 500 related genes where almost half of these genes are associated with hsa-mir-4778-5p. This particular miRNA is also associated with the majority of genes in each other pathway.
Discussion
The proportion comparison of five significant miRNAs in our data, as listed in Table 1, clearly shows the difference of characteristics in the two sample groups. We observed that these five miRNAs are included in a list of 873 common miRNAs targeting the COVID-19 genome, which were found via in silico analysis by Fulzele et al. (2020) and Khan et al. (2020). They used COVID-19 isolates from various countries, including those from Wuhan and other Asian countries, excluding Indonesia. However, we found none of these five miRNAs was significant in their results. Therefore, more thorough investigations on these locally unique miRNAs are needed. Aside from COVID-19, our significant miRNAs have been reported by previous studies on other diseases. Most of them have associations with several cancers (Cummins et al. 2006; Vaz et al. 2010; Jima et al. 2010; Persson et al. 2011).
Gene Ontology analysis shows four significant pathways related to the role of hsa-miR-4778-5p, hsa-miR-4531, and hsa-miR-6844 in the biological process (Table 3). Cellular Protein Modification Process (GO: 0006464) indicates that our significant miRNAs play roles in PTM. The reduction of phosphorylation, as one of the PTM mechanisms, is associated with NSP12 mutation (Sun et al. 2020), which was found in the majority of our Indonesian samples. NSP12 is well-known to be associated with ORF1AB (Yoshimoto 2020), whereas one of the significant miRNAs in our finding, hsa-miR-6844, is also related to ORF1AB. Therefore, NSP12 mutation may have an association with this miRNA. Thus, our result shows a consistent finding with the previous study. The other two biological processes, Cellular Nitrogen Compound Metabolic Process (GO: 0034641) and Biosynthetic Process (GO: 0009058) have a role in the inflammatory process during SARS-CoV-2 infection (Valko et al. 2007; Arisan et al. 2020; Bouhaddou et al. 2020; Hu et al. 2020b; Abu-Farha et al. 2020). Lastly, Fc-epsilon Receptor Signaling Pathway (GO: 0038095) plays an important role in the immunological reaction. A preliminary proteomics study (Kothapalli et al. 2020) with clinical samples related to lung injury caused by COVID-19 showed the downregulation of several proteins in the Fc-epsilon Receptor Signaling Pathway. Further research is needed to understand our predicted miRNA target in the inflammatory process and immune system in COVID-19 based on this gene ontology analysis.
The results of our miRNA target prediction analysis indicate some differences between SARS-CoV-2 found in Indonesia and Wuhan. The differences among the same virus are commonly caused by either mutation (subtle genetics changes) or recombination (major genetics changes) (Wimmer and Goldbach 1992). However, in this preliminary study, we merely focus on the mutations that may cause the differences in SARS-CoV-2 characteristics from these two origins. Supplementary data Tables 3 and 4 show mutations found in each sample from both origins. In our Indonesian samples, the top three most common mutations are Spike D614G, NSP12 P323L, and N3 Q57H. These three mutations are not identical with the three most common mutations in Wuhan samples, namely NSP2 N92H, Spike N856K, and NS8 L84S. The effect of Spike D614G mutation on the infectivity of SARS-CoV-2, as the most common mutation, is well reported in several recent studies (Plante et al. 2020; Mercatelli and Giorgi 2020; Korber et al. 2020; Mohammad et al. 2020; Yurkovetskiy et al. 2020; Franco-Muñoz et al. 2020; Jackson et al. 2020). This mutation is highly associated with the infectious rate on human lung cells, and colon cells (Yurkovetskiy et al. 2020). Additionally, the combination of Spike D614G and NSP12 P323L in all detected SARS-CoV-2 variants indicates the importance of these two mutations in terms of transmission and pathogenicity rate (Hartley et al. 2020; Kannan et al. 2020; Mutlu et al. 2020). A study from Vietnam cases, which also found D614G and P323L as the most common mutations, suggests that there is a high probability of the European origin of viruses in their samples as these two mutations are highly dominant in European samples (Nguyen et al. 2020). Unlike the previous two mutations, only a few studies discuss the N3 Q57H mutation (Wang et al. 2020; Soratto et al. 2020; Hassan et al. 2020). This mutation was found in 70% of the United States and Singapore samples (Wang et al. 2020).
In addition to the mutation factor, we also included clade analysis to compare the characteristics of SARS-CoV-2 from Indonesia and Wuhan. Clade can be associated with the phylogenetics analysis which categories certain variants of virus based on the lineage. SARS-CoV-2 clade affects the diversity of miRNA profiles in a region including Indonesian and Wuhan samples in the current study. Among SARS-CoV-2 Indonesia samples, the majority of them were grouped into clade GH (50%) and L (40%), with the rest of them, are categorized to clade G, O, and GR. On the other hand, only one sample is considered as clade S, while the rest of the samples from Wuhan are considered as clade L which is dominantly represented in Asian samples (Mercatelli and Giorgi 2020). Interestingly, clade GH, as the most common group in Indonesia samples, has been largely detected in the Americas (Hu et al. 2020a; Mercatelli and Giorgi 2020). This clade is derived from the mutations D614G and Q57H as commonly found in Indonesia samples. Currently, clade GH alongside GR is the most commonly observed clades in all SARS-CoV-2 sequenced samples globally (Mercatelli and Giorgi 2020). From the temporal analysis, it is believed that the peak of GH clade counts was in May 2020 (Alm et al. 2020).
Conclusion
In the present study, we compared the predicted miRNAs from two groups of SARS-CoV-2 samples from Indonesia and Wuhan (China). This comparative analysis detected various miRNA targets with different proportions in both groups. However, our statistical analysis predicted only five significant miRNAs that were associated with SARS-CoV-2 infection. Further analysis on the biology pathway prediction indicates four pathways that are related to the majority of significant miRNAs. The differences in miRNAs proportion in both groups also lead to the disparity of other SARS-CoV-2 characteristics such as clade and mutation. This in silico research can be used as the initial foundation for further research and our significant miRNA target can be used as a potential biomarker or treatment for COVID-19 cases in Indonesia. In future studies, we will expand our samples and compare them with several ASEAN countries to validate our significant miRNA target. We will also conduct in vitro and in vivo research for the study expression and role of significant target miRNAs in the severity of COVID-19.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 Additional file 1 of Information data with the mutation from China (Wuhan) and Indonesia SARS-CoV-2 which were accessed from the public database. (DOCX 25 KB)
Abbreviations
- pre-miRNA
Precursor-microRNA
- miRDB
MicroRNA target prediction database
- DIANA-miRPath
DNA intelligent analysis–micro Pathway
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GO
Gene ontology
- NCBI
National Center for Biotechnology Information
- GISAID
Global initiative on sharing all influenza data
- UTR regions
Untranslated region
- NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- MAP2K7
Mitogen-activated protein kinase kinase 7
- SOS1
SOS Ras/Rac Guanine Nucleotide Exchange Factor 1
- IgE
Immunoglobulin E
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abu-Farha M, Thanaraj TA, Qaddoumi MG, et al. The role of lipid metabolism in COVID-19 virus infection and as a drug target. Int J Mol Sci. 2020;21(10):3544. doi: 10.3390/ijms21103544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm E, Broberg EK, Connor T, et al. Geographical and temporal distribution of SARS-CoV-2 clades in the WHO European Region, January to June 2020. Eurosurveillance. 2020;25(32):2001410. doi: 10.2807/1560-7917.ES.2020.25.32.2001410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansori A, Kharisma V, Sabilil MS, et al. Genetic Variant of SARS-CoV-2 isolates in indonesia: spike glycoprotein gene. J Pure Appl Microbiol. 2020;14:971–978. doi: 10.22207/JPAM.14.SPL1.35. [DOI] [Google Scholar]
- Ansori ANM, Kusala MKJ, Normalina I, et al. Immunoinformatic investigation of three structural protein genes in Indonesian SARS-CoV-2 isolates. Syst Rev Pharm. 2020;11:422–434. [Google Scholar]
- Arisan ED, Dart A, Grant GH, et al. The Prediction of miRNAs in SARS-CoV-2 Genomes: hsa-miR databases identify 7 Key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses. 2020;12:614. doi: 10.3390/v12060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhaddou M, Memon D, Meyer B, et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammaerts S, Strazisar M, De Rijk P, Del Favero J. Genetic variants in microRNA genes: impact on microRNA expression, function, and disease. Front Genet. 2015;6:186. doi: 10.3389/fgene.2015.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Zhang S. A Bayesian extension of the hypergeometric test for functional enrichment analysis. Biometrics. 2014;70:84–94. doi: 10.1111/biom.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez N, Cordiner RA, Young RS, et al. Genetic variation and RNA structure regulate microRNA biogenesis. Nat Commun. 2017;8:15114. doi: 10.1038/ncomms15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Muñoz C, Álvarez-Díaz DA, Laiton-Donato K, et al. Substitutions in Spike and Nucleocapsid proteins of SARS-CoV-2 circulating in South America. Infect Genet Evol. 2020;85:104557. doi: 10.1016/j.meegid.2020.104557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulzele S, Sahay B, Yusufu I, et al. COVID-19 virulence in aged patients might be impacted by the host cellular microRNAs abundance/profile. Aging Dis. 2020;11:509. doi: 10.14336/AD.2020.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad H, Al-Zyoud W. miRNA target prediction might explain the reduced transmission of SARS-CoV-2 in Jordan, Middle East. Non-Coding RNA Res. 2020;5:135–143. doi: 10.1016/j.ncrna.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamed S, Elkhatib W, Khairallah A, Noreddin A. Global dynamics of SARS-CoV-2 clades and their relation to COVID-19 epidemiology. Sci Rep. 2020;11:8435. doi: 10.1038/s41598-021-87713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Tillett RL, AuCoin DP, et al. Genomic surveillance of Nevada patients revealed prevalence of unique SARS-CoV-2 variants bearing mutations in the RdRp gene. medRxiv. 2020 doi: 10.1101/2020.08.21.20178863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan SS, Choudhury PP, Roy B, Jana SS. Missense mutations in SARS-CoV2 genomes from Indian patients. Genomics. 2020;112:4622–4627. doi: 10.1016/j.ygeno.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2020;19(3):141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Cai X, Song X, et al. Possible SARS-coronavirus 2 inhibitor revealed by simulated molecular docking to viral main protease and host toll-like receptor. Future Virol. 2020;15(6):359–368. doi: 10.2217/fvl-2020-0099. [DOI] [Google Scholar]
- Jackson CB, Zhang L, Farzan M, Choe H. Functional importance of the D614G mutation in the SARS-CoV-2 spike protein. Biochem Biophys Res Commun. 2020;538:108–115. doi: 10.1016/j.bbrc.2020.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jima DD, Zhang J, Jacobs C, et al. Deep sequencing of the small RNA transcriptome of normal and malignant human B cells identifies hundreds of novel microRNAs. Blood. 2010;116:e118–e127. doi: 10.1182/blood-2010-05-285403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan SR, Spratt AN, Quinn TP, et al. Infectivity of SARS-CoV-2: there is something more than D614G? J Neuroimmune Pharmacol. 2020;15(4):574–577. doi: 10.1007/s11481-020-09954-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MAAK, Sany MRU, Islam MS, et al. Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2 and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front Genet. 2020;11:765. doi: 10.3389/fgene.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothapalli KSD, Park HG, Brenna JT. Polyunsaturated fatty acid biosynthesis pathway and genetics implications for interindividual variability in prothrombotic, inflammatory conditions such as COVID-19. Prostaglandins Leukot Essent Fat Acids. 2020;162:102183. doi: 10.1016/j.plefa.2020.102183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Platt D, Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ. 2020;98:495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Li C, Hu X, Li L, Li J. Differential microRNA expression in the peripheral blood from human patients with COVID-19. J Clin Lab Anal. 2020;34:e23590. doi: 10.1002/jcla.23590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Guerrero I, Gutierrez-Camino A, Lopez-Lopez E, et al. Genetic variants in miRNA processing genes and pre-miRNAs are associated with the risk of chronic lymphocytic leukemia. PLoS ONE. 2015;10:e0118905. doi: 10.1371/journal.pone.0118905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli D, Giorgi FM. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad A, Alshawaf E, Marafie SK, et al. Higher binding affinity of Furin to SARS-CoV-2 spike (S) protein D614G could be associated with higher SARS-CoV-2 infectivity. Int J Infect Dis. 2020;103:611–616. doi: 10.1016/j.ijid.2020.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu O, Ugurel OM, Sariyer E, et al. Targeting SARS-CoV-2 Nsp12/Nsp8 interaction interface with approved and investigational drugs: an in silico structure-based approach. J Biomol Struct Dyn. 2020;16:1–13. doi: 10.1080/07391102.2020.1819882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TT, Pham TN, Van TD, et al. Genetic diversity of SARS-CoV-2 and clinical, epidemiological characteristics of COVID-19 patients in Hanoi, Vietnam. PLoS ONE. 2020;15:e0242537. doi: 10.1371/journal.pone.0242537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nidom RV, Indrasari S, Normalina I, et al. Investigation of the D614G mutation and antibody-dependent enhancement sequences in Indonesian SARS-CoV-2 isolates and comparison to southeast Asian isolates. Syst Rev Pharm. 2020;11:203–213. [Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5. 0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikesit AA, Nurdiansyah R. The Predicted Structure for the Anti-Sense siRNA of the RNA Polymerase Enzyme (RdRp) gene of the SARS-CoV-2. Ber Biol. 2020;19:97–108. [Google Scholar]
- Parikesit AA, Nurdiansyah R. Drug repurposing option for COVID-19 with structural bioinformatics of chemical interactions approach. Cermin Dunia Kedokt. 2020;47:222–226. [Google Scholar]
- Persson H, Kvist A, Rego N, et al. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res. 2011;71:78–86. doi: 10.1158/0008-5472.CAN-10-1869. [DOI] [PubMed] [Google Scholar]
- Plante JA, Liu Y, Liu J, et al. Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility. bioRxiv. 2020 doi: 10.1101/2020.09.01.278689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma A, Phukan H, Halder N, Madanan MG (2020) An in-silico approach to study the possible interactions of miRNA between human and SARS-CoV2. Comput Biol Chem 88:107352. 10.1016/j.compbiolchem.2020.107352 [DOI] [PMC free article] [PubMed]
- Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soratto TAT, Darban H, Bjerkner A, et al. Four SARS-CoV-2 genome sequences from late april in Stockholm, Sweden, reveal a rare mutation in the spike protein. Microbiol Resour Announc. 2020;9(35):e00934–e1020. doi: 10.1128/mra.00934-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Ren K, Zhang X, et al. Mass spectrometry analysis of newly emerging Coronavirus HCoV-19 spike protein and human ACE2 reveals camouflaging glycans and unique post-translational modifications. Engineering. 2020 doi: 10.1016/j.eng.2020.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczak A, Mortensen JM, Winnenburg R, et al. Interpretation of biological experiments changes with evolution of the Gene Ontology and its annotations. Sci Rep. 2018;8:5115. doi: 10.1038/s41598-018-23395-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobaugh DW, Klimstra WB. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol Med. 2017;23:80–93. doi: 10.1016/j.molmed.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valko M, Leibfritz D, Moncol J, et al. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Vaz C, Ahmad HM, Sharma P, et al. Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genom. 2010;11:288. doi: 10.1186/1471-2164-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-miRPath v3. 0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Chen J, Gao K, et al. Characterizing SARS-CoV-2 mutations in the United States. Res Sq. 2020 doi: 10.21203/rs.3.rs-49671/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2020) Coronavirus disease 2019 (COVID-19). https://www.who.int/indonesia/news/novel-coronavirus/situation-reports. Accessed 22 Oct 2020
- Wimmer E, Goldbach R. Viral genetics. Curr Opin Genet Dev. 1992;2:59–60. doi: 10.1016/S0959-437X(05)80322-3. [DOI] [Google Scholar]
- Winter J, Jung S, Keller S, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto FK. The proteins of severe acute respiratory syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L, Wang X, Pascal KE, et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 Additional file 1 of Information data with the mutation from China (Wuhan) and Indonesia SARS-CoV-2 which were accessed from the public database. (DOCX 25 KB)