Abstract
Epigenetic modifications, including DNA methylation, are involved in the regulatory mechanisms of gene expression in animals and plants. In this study, we investigated whether the action of 5-azacytidine (5-aza-Cd), which is a well-known DNA methylation inhibitor, in suspension-cultured tobacco cells is affected by treatment with nucleoside derivatives of 5-methylcytosine (5-mCs), namely 5-methylcytidine (5-mCd) and 5-methyl-2′-deoxycytidine (5-mdCd). In a tobacco cell line, 5-aza-Cd treatment reactivated an epigenetically silenced transgene containing the cauliflower mosaic virus 35S promoter fused to the β-glucuronidase coding region and the nopaline synthase polyadenylation signal. The reactivation was evident on the fifth day of treatment and was augmented during culture with application of 5-aza-Cd at every subcultivation. This treatment, provided only once in the initial culture, resulted in transient transgene reactivation, followed by attenuation of its activity. The reactivation induced by 5-aza-Cd was suppressed by concomitant treatment with either 5-mCd or 5-mdCd. These results suggest that the 5-mCs derivatives inhibit and/or reverse 5-aza-Cd-induced reactivation of a silent transgene in tobacco cells.
Keywords: DNA methylation, epigenetics, 5-azacytidine, 5-methylcytidine, gene silencing
Transgenes introduced into plant cells are occasionally inactivated. Influences of the surrounding DNA or chromatin structure at the insertion site of foreign genes into a plant chromosome have been implicated in the inactivation mechanism (Horsch et al. 1985). Early studies have demonstrated that this inactivation is associated with the cytosine methylation of DNA and that the inactivated, silent transgenes are often reactivated by treatment with nucleoside derivatives of 5-azacytosine (5-aza-Cs)—either 5-aza-Cd or 5-aza-2′-deoxycytidine (5-aza-dCd) (Amasino et al. 1984; Hepburn et al. 1983; Klaas et al. 1989; Weber et al. 1990). These agents are incorporated into DNA as 5-aza-Cs residues and inhibit DNA cytosine methyltransferase (Santi et al. 1983). Indeed, the reactivated transgenes subsequently undergo DNA demethylation (Amasino et al. 1984; Weber et al. 1990). In the mammalian cell system, 14C-labeled 5-aza-Cd was shown to have incorporated into both DNA (10 to 20% of total incorporated radioactivity) and RNA (80 to 90%) fractions (Li et al. 1970). A recent study demonstrated that 5-aza-Cd was converted into 5-aza-2′-deoxycytidine-5′-triphosphate (5-aza-dCTP) and 5-azacytidine-5′-triphosphate (5-aza-CTP) before incorporation into DNA and RNA, respectively, whereas 5-aza-dCd was converted only to 5-aza-dCTP (Rawson et al. 2016). To our knowledge, metabolism of neither 5-aza-Cd nor 5-aza-dCd has been investigated in plants.
Although there have been many studies on the application of 5-aza-Cs derivatives to animal and plant cells, only a few studies on 5-mCs derivatives, including 5-mCd and 5-mdCd, have been reported. Under normal conditions, treatment with these agents cannot alter the methylation state of DNA, because these are deaminated to thymidine and subsequently incorporated into DNA as thymine residues (Camiener 1967; Hotta and Stern 1961; Jekunen and Vilpo 1984; Jekunen et al. 1983; Sulimova et al. 1978). In human cells, three enzymatic reactions have been considered to function as barriers for preventing the misincorporation of 5-mCs derived from the degradation of methylated DNA in vivo as well as of 5-mCs applied exogenously: deamination at the deoxyribonucleoside and monophosphate levels and inability of diphosphate production (Vilpo and Vilpo 1995 for review). Regarding deamination at the deoxyribonucleoside level, it was demonstrated that human cytidine deaminase, which plays a role in salvaging pyrimidine nucleosides via deamination of cytidine and deoxycytidine to uridine and deoxyuridine, respectively, could also deaminate 5-mdCd to thymidine (Jekunen and Vilpo 1984; Zauri et al. 2015). Regarding deamination at the monophosphate level, it was shown that 5-methyl-2′-deoxycytidine-5′-monophosphate (5-mdCMP), whose intracellular synthesis from exogenous 5-mdCd has been proven, was very rapidly deaminated to thymidine monophosphate (Vilpo and Vilpo 1991). Additionally, Jost et al. (2002) reported that 5-mdCMP deaminase activity, in addition to 2′-deoxycytidine-5′-monophosphate (dCMP) deaminase activity, was present in human mature sperm cells. Regarding the inability of diphosphate production, it was shown that the cytidine monophosphate kinase CMPK1 could not phosphorylate 5-mdCMP due to its stringent specificity, serving as the main barrier to the formation of 5-methyl-2′-deoxycytidine-5′-triphosphate (5-mdCTP) and limiting its availability for DNA polymerases (Vilpo and Vilpo 1991, 1993; Zauri et al. 2015). Nevertheless, it was demonstrated that exogenous 5-mdCTP was incorporated into DNA in mammalian cells permeabilized by electroporation, leading to genomic methylation and gene silencing (Holliday and Ho 1991; Nyce 1991). These studies suggest that misincorporation of 5-mdCd into DNA occurs if the compound circumvents the metabolic barriers and is salvaged up to triphosphate formation.
In this study, we report on the reactivation of an epigenetically silenced transgene in tobacco cells treated with 5-aza-Cd and on the suppression of this reactivation following concomitant treatment with 5-aza-Cd and either 5-mCd or 5-mdCd.
Tobacco (Nicotiana tabacum ‘White Burley’) plants were aseptically grown at 25°C under a 17 h photoperiod on a semi-solid medium (pH, 5.7) containing inorganic salts and vitamins from Murashige and Skoog (MS) medium (Murashige and Skoog 1962), 3% sucrose, and 0.8% Bacto™ Agar (Difco Laboratories Inc., Detroit, MI, USA). Transgenic plants harboring a transgene containing the cauliflower mosaic virus 35S promoter fused to the β-glucuronidase (GUS) coding region and the nopaline synthase polyadenylation signal (NOS) (35S-GUS-NOS) were obtained by regenerating shoots from leaf discs following infection with Agrobacterium tumefaciens strain LBA4404 containing the plasmid vector pBI121 (Clontech Laboratories, Inc., Mountain View, CA, USA). Transgenic plants harboring the PKPI-GUS-NOS transgene were obtained by infecting with LBA4404 containing the 5′ promoter region approximately 1 kilo base pairs upstream of the potato Kunitz-type proteinase inhibitor (PKPI) gene (GenBank accession number X70376) inserted at the Xba I/Sma I site of pBI101 (Clontech Laboratories, Inc.) (Yamagishi 2015). The 5′ promoter region of PKPI was obtained using cassette ligation-mediated polymerase chain reaction (PCR) (Isegawa et al. 1992). Briefly, genomic DNA of potato (Solanum tuberosum ‘Irish Cobbler’) was digested with Xba I and ligated to a double-stranded DNA cassette possessing an Xba I site (TaKaRa Bio Inc., Shiga, Japan). The cassette-ligated restriction fragments were subjected to PCR using primers complementary to the cassette and PKPI cDNA (Yamagishi et al. 1991).
Calli from the leaf explants of transgenic and non-transgenic plants were propagated at 25°C under a 17 h photoperiod on a semi-solid medium (pH, 5.7) supplemented with 2 mg·l−1 2,4-dichlorophenoxyacetic acid (2,4-D) and 0.1 mg·l−1 6-benzyladenine (BA). To establish suspension culture, the calli were transferred to 100 ml Erlenmeyer flasks containing 25 ml of a liquid medium (pH, 5.7) containing inorganic salts and vitamins from MS medium, 3% sucrose, 2 mg·l−1 2,4-D, and 0.1 mg·l−1 BA. The cultures were incubated at 25°C in the dark, on a reciprocal shaker with agitation at 100 rpm. Subcultivation was conducted every 2 weeks.
5-aza-Cs, 5-aza-Cd, 5-aza-dCd, 5-mCs, 5-mCd, and 5-mdCd were purchased from Sigma-Aldrich Co. (Saint Louis, MO, USA). The chemical agents were dissolved in distilled water at 10 mM and filter-sterilized; 5-aza-Cs is insoluble in water and was dissolved in dimethyl sulfoxide. The chemical solutions were added to cell cultures at a given concentration on the 2nd day after subcultivation, unless otherwise stated.
GUS enzyme activity was measured according to Jefferson (1987). The cultured cells were collected using a pipette, transferred to an Eppendorf tube, and washed with 1 ml distilled water three times. Cell extract was obtained by adding 300 µl extraction buffer and quartz sand, followed by grinding of cells using a plastic pestle. After centrifugation at 15,000 rpm (17,860×g) for 5 min, an aliquot of 30 µl from the supernatant was mixed with 45 µl assay buffer and incubated at 37°C for 60 min. The reaction was stopped by adding 1.425 ml stop buffer, and the concentration of a reaction product, 4-methylumbelliferone (4-MU), was measured using a spectrofluorimeter (F-3000, Hitachi, Ltd., Tokyo, Japan) using excitation at 365 nm and emission at 455 nm. GUS activity was obtained as picomoles of 4-MU produced per minute per milligram of protein and presented relative to the maximum activity in an experiment. Protein content was determined by the Bradford method (Bradford 1976). Data presented herein are representative of two to four independent experiments and are shown as mean and standard deviation of triplicate measurements in each experiment.
Statistical analysis was performed using Student’s t-test or Tukey’s multiple comparison test, and a p values of less than 0.05 was considered significant.
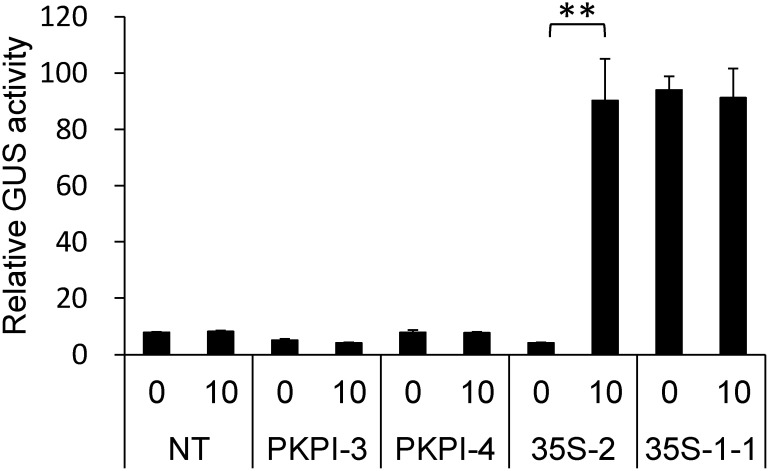
In a previous study, we had established suspension-cultured cell lines from transgenic tobacco plants harboring either PKPI-GUS-NOS or 35S-GUS-NOS transgene, whose promoter expression could be monitored by measuring GUS activity (Yamagishi 2015). The primary purpose was to investigate the promoter expression in response to a developmental, environmental, or hormonal factor, which has been shown to affect the accumulation of PKPI transcripts in potato tubers (Yamagishi et al. 1993, 1994a, b). In our experiments, we found that the transgenes in two PKPI-GUS-NOS cell lines, PKPI-3 and -4, and a 35S-GUS-NOS cell line, 35S-2, were inactivated, because GUS activity in these cell lines was much lower than that in the original plants and was equivalent to that in a non-transgenic cell line (Figure 1). To determine whether gene silencing due to DNA methylation had occurred, the cell lines were treated with 5-aza-Cd, which is a well-known DNA methylation inhibitor in vivo (Amasino et al. 1984; Klaas et al. 1989; Weber et al. 1990). GUS activity in 35S-2 cells treated with 5-aza-Cd for 5 days was reactivated to about 20-fold compared to that in untreated cells. However, 5-aza-Cd treatment of PKPI-3 and -4, two PKPI-GUS-NOS cell lines, and 35S-1-1, a cell line harboring active 35S-GUS-NOS transgene, showed no reactivation and inhibition of GUS activity, respectively (Figure 1).
Figure 1. Effects of 5-aza-Cd treatment on GUS activity in suspension-cultured tobacco cells. Non-transgenic (NT) and transgenic cells harboring inactive transgenes (PKPI-3, PKPI-4, and 35S-2) as well as 35S-1-1 cells harboring active transgene (35S-1-1) were treated with (10) or without (0) 10 µM 5-aza-Cd for 5 days. Statistical analysis was performed using Student’s t-test (** p<0.01).
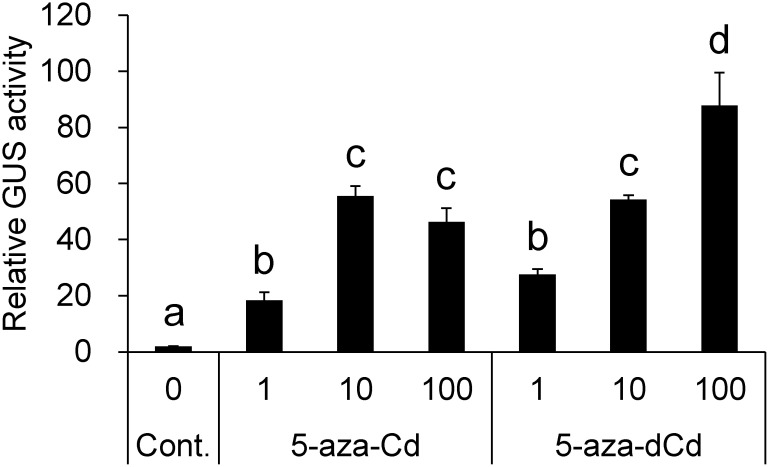
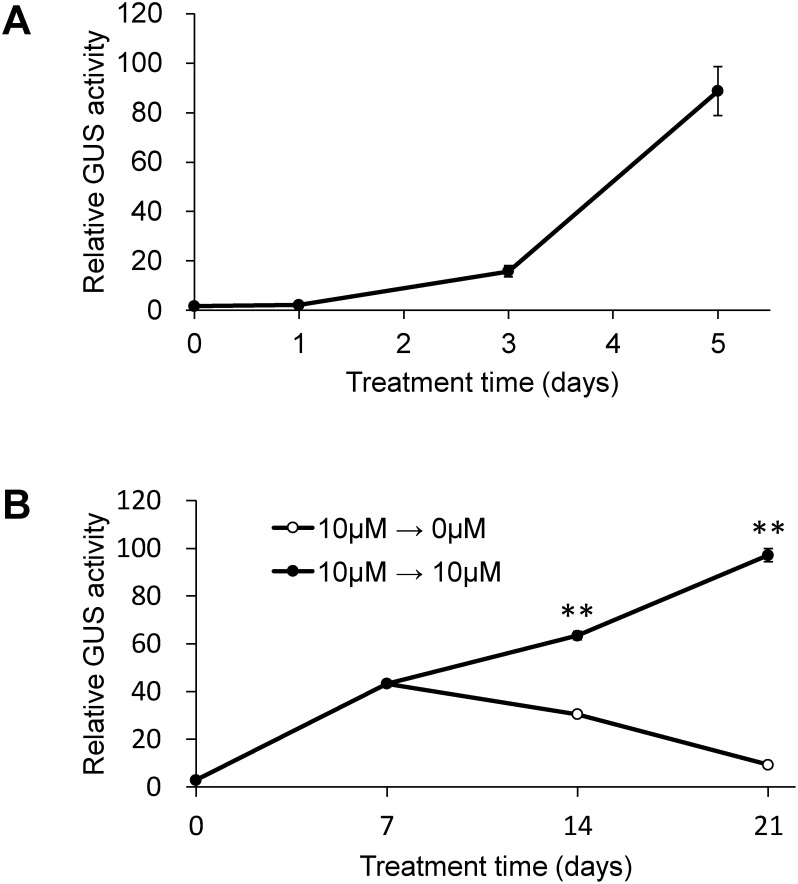
In dose response assays, 5-aza-Cd treatment at 10 and 100 µM induced almost the same level of reactivation. Moreover, 5-aza-dCd treatment at 10 µM induced reactivation equivalent to 5-aza-Cd treatment, while 5-aza-dCd treatment at 100 µM induced 1.6-fold higher reactivation (Figure 2). No reactivation was detected after 5-aza-Cs treatment (data not shown). We selected 5-aza-Cd for subsequent assays, since it has been used more commonly in studies of plants than 5-aza-dCd and was therefore expected to provide common understanding of the results obtained. Time course after 5-aza-Cd treatment showed that reactivation was detectable on day 3 and became evident on day 5 (Figure 3A). Next, we examined whether the reactivated state in 35S-2 cells was maintained during subcultivation (Figure 3B). Application of 5-aza-Cd only once in the initial culture resulted in transient reactivation, which peaked on day 7, followed by the attenuation of activity. Application of 5-aza-Cd at every subcultivation resulted in continuous increase in GUS activity, which was 33-fold on day 21 compared to that on day 0.
Figure 2. Dose response of GUS activity in 35S-2 cells treated with the nucleoside derivatives of 5-aza-Cs. The cells were treated with either 5-aza-Cd or 5-aza-dCd at a concentration of 1 µM (1), 10 µM (10), or 100 µM (100) for 5 days. Statistical analysis was performed using Tukey’s multiple comparison test. Different letters above the bars indicate a significant difference at p<0.05.
Figure 3. Time course of GUS activity in 35S-2 cells treated with 5-aza-Cd. The cells were cultured in a medium containing 10 µM 5-aza-Cd for 0, 1, 3, or 5 days (A). The cells were cultured in a medium containing 10 µM 5-aza-Cd for 7 days and then subcultivated in a medium with (●) or without (○) 5-aza-Cd every 7 days (B). Statistical analysis was performed using Student’s t-test (** p<0.01).
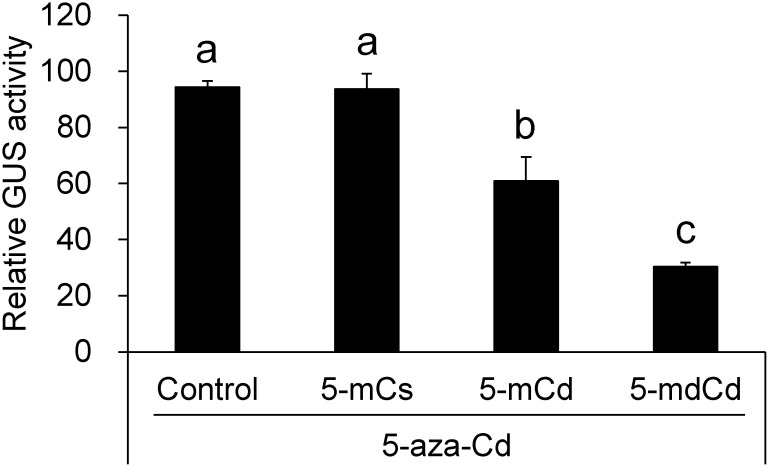
Assuming that the 5-aza-Cd-induced reactivation was a consequence of DNA demethylation, we investigated the effects of 5-mCs, its ribonucleoside (5-mCd), and its deoxyribonucleoside (5-mdCd) on the reactivated transgene in 35S-2 cells. Concomitant treatment with 5-aza-Cd and either 5-mCd or 5-mdCd suppressed transgene reactivation by 36% and 68%, respectively, while 5-mCs showed no suppression (Figure 4). Both nucleosides also inhibited intrinsic GUS activity in 35S-1-1 cells with the active transgene, albeit slightly [2% (p>0.05) and 19% (p>0.05), respectively (data not shown)].
Figure 4. Effects of 5-mCs and its nucleoside derivatives on GUS activity induced by 5-aza-Cd. The cells were treated solely with 10 µM 5-aza-Cd (control) and concomitantly with 10 µM 5-aza-Cd and either 10 µM each of 5-mCs, 5-mCd, or 5-mdCd for 5 days. Statistical analysis was performed using Tukey’s multiple comparison test. Different letters above the bars indicate a significant difference at p<0.05.
This study demonstrated that the silent 35S-GUS-NOS transgene in 35S-2 cells can be reactivated by treatment with DNA methylation inhibitors, such as 5-aza-Cd and 5-aza-dCd. Further investigation revealed that reactivation by the former can be suppressed by the 5-mCs derivatives 5-mCd and 5-mdCd. PKPI-GUS-NOS transgenes in PKPI-3 and -4 cells were never reactivated, suggesting a cause other than genomic methylation, such as the loss or mutation of the transgene. Thus, 35S-2 cells exhibit a useful trait for exploring the mechanisms of epigenetic DNA methylation.
Klaas et al. (1989) demonstrated that a transgenic tobacco cell line treated with either 5-aza-Cd or 5-aza-dCd showed genomic demethylation and reactivation of a transcriptionally silent, hypermethylated T-DNA gene. Weber et al. (1990) demonstrated that protoplasts derived from a transgenic tobacco plant with a silent 35S-GUS-NOS transgene showed a strong reactivation of the transgene after treatment with 10–20 µM 5-aza-Cd for 4–5 days. Bochardt et al. (1992) showed that the treatment of a transgenic tobacco cell line with 5 µM 5-aza-Cd for everyday for either 5 or 10 consecutive days produced both short-term (up to 8 weeks) and long-term (9–13 weeks) effects on the reactivation of silent 35S-GUS-NOS transgene. These effects involved an early decrease in DNA methylation level, followed by reactivation of the GUS gene in a small fraction of cells and subsequent dramatic increase in the fraction of GUS-expressing cells. Furthermore, transient decrease in the fraction of reactivated cells was noted during the 4th week in the period of short-term effects. The reactivation events observed in our study are consistent with previous reports and can be consequently regarded as the short-term effect, since no sign of dramatic increase in activity due to the long-term effect was detected regardless of the treatment method (Figure 3B). We found that single 5-aza-Cd treatment reactivated the transgene to the level on day 0 following transient reactivation, whereas multiple 5-aza-Cd treatments continuously increased the activity. These results suggest that the reactivated state under the short-term effect is reversible in the absence of the agent, probably due to the susceptibility of the transgene to cellular processes of foreign gene silencing, such as DNA remethylation.
To our knowledge, the metabolism of neither 5-aza-Cd or 5-aza-dCd has been investigated in plants. However, involvement of the pyrimidine salvage pathway has been presumed based on studies of mammalian cells (Li et al. 1970; Rawson et al. 2016). In potato tubers, cytidine is exclusively salvaged to cytidine-5′-monophosphate (CMP) by uridine/cytidine kinase and non-specific nucleoside phosphotransferase, and a small amount of cytidine is deaminated to uridine by cytidine deaminase. The de novo synthesis of deoxyribonucleotide via ribonucleotide reductase and the deoxycytidine salvage pathway also operate in potato. The triphosphate form of deoxycytidine, a substrate for DNA polymerase, is produced by sequential phosphorylation. Non-specific nucleoside phosphotransferase produces monophosphate, which is then converted into diphosphate by nucleoside monophosphate kinase and subsequently to triphosphate by nucleoside diphosphate kinase (Katahira and Ashihara 2002). If the exogenous 5-aza-Cd is salvaged through the same metabolic pathway as cytidine, a period of 5 days preceding the evident reactivation (Figure 3A) would be required for metabolizing 5-aza-Cd up to 5-aza-dCTP and subsequently incorporating it into DNA during DNA replication and cell division, causing demethylation of the transgene locus. Demethylation of both DNA strands would be necessary for the reactivation, since hemimethylation of the 35S-GUS-NOS gene leads to complete inhibition of transient gene expression (Weber and Graessmann 1989). During this 5-day period, 2–3 cell doublings occur, which is consistent with the theory that demethylation of both strands requires a cell to pass through two cell cycles without DNA methylation (Bochardt et al. 1992; Klaas et al. 1989).
5-aza-Cs could not reactivate the transgene even after the prerequisite 5-day period (data not shown). Moreover, it could not induce the expression of a silent T-DNA gene in a suspension culture of tobacco cells (Klaas et al. 1989). In potato tubers, cytosine was not metabolized and cytosine-metabolizing enzymes, including cytosine deaminase, could not be detected (Katahira and Ashihara 2002). Tobacco plants lack cytosine deaminase activity (Stougaard 1993). Thus, 5-aza-Cs is probably excluded from the pyrimidine salvage pathway and therefore from demethylation machinery for reactivating the silent transgenes. For the same reason, 5-mCs is unlikely to be salvaged either and therefore would fail to suppress the reactivation (Figure 4).
Although the precise nature of the events underlying the suppression of 5-aza-Cd-induced reactivation of silent transgenes by the nucleoside derivatives of 5-mCs remains unclear, we propose a certain idea in the present study; deoxyribonucleoside form is more effective in suppression than the ribonucleoside form, suggesting the presence of a salvage pathway toward DNA rather than RNA is closely related to the suppression mechanism.
In this study, we reported that 5-aza-Cd-induced reactivation of an epigenetically silenced transgene was suppressed by 5-mCs derivatives. Further investigation is warranted to elucidate the nature of events, including the determination of metabolic fates of the 5-mCs derivatives and methylation frequency of the transgene under treatment with 5-mCs derivatives and/or 5-aza-Cd. Nevertheless, this study provides an approach to the estimation of epigenetics-associated compounds. Specifically, an assay combining an agent that reversibly alters the epigenetic status (i.e., 5-aza-Cd) with a cell line whose reporter gene expression is sensitive to that agent (i.e., 35S-2 cells) can provide novel insights into the actions of the tested compounds (i.e., nucleoside derivatives of 5-mCs) in epigenetics.
Abbreviations
- 5-aza-Cd
5-azacytidine
- 5-aza-Cs
5-azacytosine
- 5-aza-dCd
5-aza-2′-deoxycytidine
- 5-aza-dCTP
5-aza-2′-deoxycytidine-5′-triphosphate
- 5-mCd
5-methylcytidine
- 5-mCs
5-methylcytosine
- 5-mdCd
5-methyl-2′-deoxycytidine
- 5-mdCMP
5-methyl-2′-deoxycytidine-5′-monophosphate
- 5-mdCTP
5-methyl-2′-deoxycytidine-5′-triphosphate
- PKPI
potato Kunitz-type proteinase inhibitor
References
- Amasino RM, Powell ALT, Gordon MP (1984) Changes in T-DNA methylation and expression are associated with phenotypic variation and plant regeneration in a crown gall tumor line. Mol Gen Genet 197: 437–446 [DOI] [PubMed] [Google Scholar]
- Bochardt A, Hodal L, Palmgren G, Mattsson O, Okkels FT (1992) DNA methylation is involved in maintenance of an unusual expression pattern of an introduced gene. Plant Physiol 99: 409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principles of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Camiener GW (1967) Studies of the enzymatic deamination of cytosine arabinoside—III: Substrate requirements and inhibitors of the deaminase of human liver. Biochem Pharmacol 16: 1691–1702 [DOI] [PubMed] [Google Scholar]
- Hepburn AG, Clarke LE, Pearson L, White J (1983) The role of cytosine methylation in the control of nopaline synthase gene expression in a plant tumor. J Mol Appl Genet 2: 315–329 [PubMed] [Google Scholar]
- Holliday R, Ho T (1991) Gene silencing in mammalian cells by uptake of 5-methyl deoxycytidine-5′-triphosphate. Somat Cell Mol Genet 17: 537–542 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Wallroth M, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Hotta Y, Stern H (1961) Deamination of deoxycytidine and 5-methyldeoxycytidine in developing anthers of Lilium longiflorum (var. Croft). J Biophys Biochem Cytol 9: 279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isegawa Y, Sheng J, Sokawa Y, Yamanishi K, Nakagomi O, Ueda S (1992) Selective amplification of cDNA sequence from total RNA by cassette-ligation mediated polymerase chain reaction (PCR): Application to sequencing 6.5 kb genome segment of hantavirus strain B-1. Mol Cell Probes 6: 467–475 [DOI] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol Biol Report 5: 387–405 [Google Scholar]
- Jekunen A, Puukka M, Vilpo J (1983) Exclusion of exogenous 5-methyl-2′-deoxycytidine from DNA in human leukemic cells: A study with [2-14C]- and [methyl-14C]5-methyl-2′-deoxycytidine. Biochem Pharmacol 32: 1165–1168 [DOI] [PubMed] [Google Scholar]
- Jekunen A, Vilpo JA (1984) 5-Methyl-2′-deoxycytidine: Metabolism and effects on cell lethality studied with human leukemic cells in vitro. Mol Pharmacol 25: 431–435 [PubMed] [Google Scholar]
- Jost JP, Thiry S, Siegmann M (2002) 5-Methyldeoxycytidine monophosphate deaminase and 5-methylcytidyl-DNA deaminase activities are present in human mature sperm cells. FEBS Lett 519: 128–134 [DOI] [PubMed] [Google Scholar]
- Katahira R, Ashihara H (2002) Profiles of pyrimidine biosynthesis, salvage and degradation in disks of potato (Solanum tuberosum L.) tubers. Planta 215: 821–828 [DOI] [PubMed] [Google Scholar]
- Klaas M, John MC, Crowell DN, Amasino RM (1989) Rapid induction of genomic demethylation and T-DNA gene expression in plant cells by 5-azacytosine derivatives. Plant Mol Biol 12: 413–423 [DOI] [PubMed] [Google Scholar]
- Li LH, Olin EJ, Buskirk HH, Reineke LM (1970) Cytotoxicity and mode of action of 5-azacytidine on L1210 leukemia. Cancer Res 30: 2760–2769 [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nyce J (1991) Gene silencing in mammalian cells by direct incorporation of electroporated 5-methyl-2′-deoxycytidine 5′-triphosphate. Somat Cell Mol Genet 17: 543–550 [DOI] [PubMed] [Google Scholar]
- Rawson JMO, Daly MB, Xie J, Clouser CL, Landman SR, Reilly CS, Bonnac L, Kim B, Patterson SE, Mansky LM (2016) 5-Azacytidine enhances the mutagenesis of HIV-1 by reduction to 5-aza-2′-deoxycytidine. Antimicrob Agents Chemother 60: 2318–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi DV, Garrett CE, Barr PJ (1983) On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell 33: 9–10 [DOI] [PubMed] [Google Scholar]
- Stougaard J (1993) Substrate-dependent negative selection in plants using a bacterial cytosine deaminase gene. Plant J 3: 755–761 [Google Scholar]
- Sulimova GE, Vaniushin BF, Khoíka L, Fridrikh A, Bulgakov R, Cherny B (1978) On the impossibility of the incorporation of 5-methylcytosine and its nucleosides into higher plant DNA. Biokhimiia 43: 240–245 (in Russian) [PubMed] [Google Scholar]
- Vilpo JA, Vilpo LM (1991) Biochemical mechanisms by which reutilization of DNA 5-methylcytosine is prevented in human cells. Mutat Res 256: 29–35 [DOI] [PubMed] [Google Scholar]
- Vilpo JA, Vilpo LM (1993) Nucleoside monophosphate kinase may be the key enzyme preventing salvage of DNA 5-methylcytosine. Mutat Res 286: 217–220 [DOI] [PubMed] [Google Scholar]
- Vilpo JA, Vilpo LM (1995) Prevention of DNA 5-methylcytosine reutilization in human cells. Somat Cell Mol Genet 21: 285–288 [DOI] [PubMed] [Google Scholar]
- Weber H, Graessmann A (1989) Biological activity of hemimethylated and single-stranded DNA after direct gene transfer into tobacco protoplasts. FEBS Lett 253: 163–166 [Google Scholar]
- Weber H, Ziechmann C, Graessmann A (1990) In vitro DNA methylation inhibits gene expression in transgenic tobacco. EMBO J 9: 4409–4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi K (2015) Analysis of the promoter activity of a potato Kunitz-type proteinase inhibitor gene in transgenic tobacco. Bull Koen Gakuen Women’s Jr College 13: 41–48 (in Japanese) [Google Scholar]
- Yamagishi K, Mitsumori C, Kikuta Y (1991) Nucleotide sequence of a cDNA encoding the putative trypsin inhibitor in potato tuber. Plant Mol Biol 17: 287–288 [DOI] [PubMed] [Google Scholar]
- Yamagishi K, Mitsumori C, Takahashi K, Fujino K, Koda Y, Kikuta Y (1993) Jasmonic acid-inducible gene expression of a Kunitz-type proteinase inhibitor in potato tuber disks. Plant Mol Biol 21: 539–541 [DOI] [PubMed] [Google Scholar]
- Yamagishi K, Nagatani K, Fukase T, Mitsumori C, Isejima EM, Kikuta Y (1994a) Gene expression of a Kunitz-type proteinase inhibitor and patatin in response to storage temperature of potato tubers. Jpn J Crop Sci 63: 368–369 [Google Scholar]
- Yamagishi K, Nagatani K, Fukase T, Mitsumori C, Kikuta Y (1994b) Molecular characterization of a Kunitz-type proteinase inhibitor expressing specifically in potato tubers. J Fac Agric Hokkaido Univ 66: 1–12 [Google Scholar]
- Zauri M, Berridge G, Thézénas ML, Pugh KM, Goldin R, Kessler BM, Kriaucionis S (2015) CDA directs metabolism of epigenetic nucleosides revealing a therapeutic window in cancer. Nature 524: 114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]