Abstract
Tomato transformation is conventionally performed using Agrobacterium tumefaciens-infected cotyledons. Here, we propose a simple procedure for tomato transformation, by which A. tumefaciens cells were smeared onto floral buds of a tomato plant using a paintbrush. Sufficient numbers of fruits were obtained from them, although the smearing of an excess number of A. tumefaciens cells led to an adverse effect on the plant growth. Progeny plants were screened by growth on a kanamycin-containing selection medium plate. The nptII gene was detected in 10 plants among 1,599 progenies. These transformants were derived from fruits other than those obtained from the smeared buds. This suggested that A. tumefaciens cells moved to the buds located near the smeared buds and caused the transformation event. Our findings suggest that this procedure can be used for the introduction of a foreign gene into plant cells.
Keywords: Agrobacterium-mediated transformation, floral dip with smearing method, kanamycin resistance, mature tomato plant
Tomato transformation is generally performed using a method involving cotyledons of seedlings, in which cultured tissues obtained from the cotyledons are infected with Agrobacterium tumefaciens and regenerated on a selection medium (McCormick et al. 1986). This method often requires time and skilled labor to obtain a transformant line (Ling et al. 1998; van Roekel et al. 1993). Currently, transformation of Arabidopsis thaliana is performed using the floral dip method. For this method, A. tumefaciens cells harboring an appropriate gene are infected into an immature flower bud, by which the desired gene is introduced into the plant cells. Usually, transformant plants are obtained among plants from the resulting seeds (Clough and Bent 1998). In this study, we attempted to obtain a transformant tomato plant using a floral smearing method and established this method as a simple procedure for tomato transformation.
To determine the adequate number of A. tumefaciens cells for smearing on the floral buds, we investigated the transformation efficiency when a series of bacterial concentrations was used. For the leaf-disk method for tomato transformation, it has been reported that the concentration of A. tumefaciens cells may strongly affect the transformation efficiency, and that the highest efficiency was obtained when a cell suspension with OD600=0.2 was used (Pawar et al. 2013). In this study, we determined the influence of concentration of A. tumefaciens cells on the floral dip treatment.
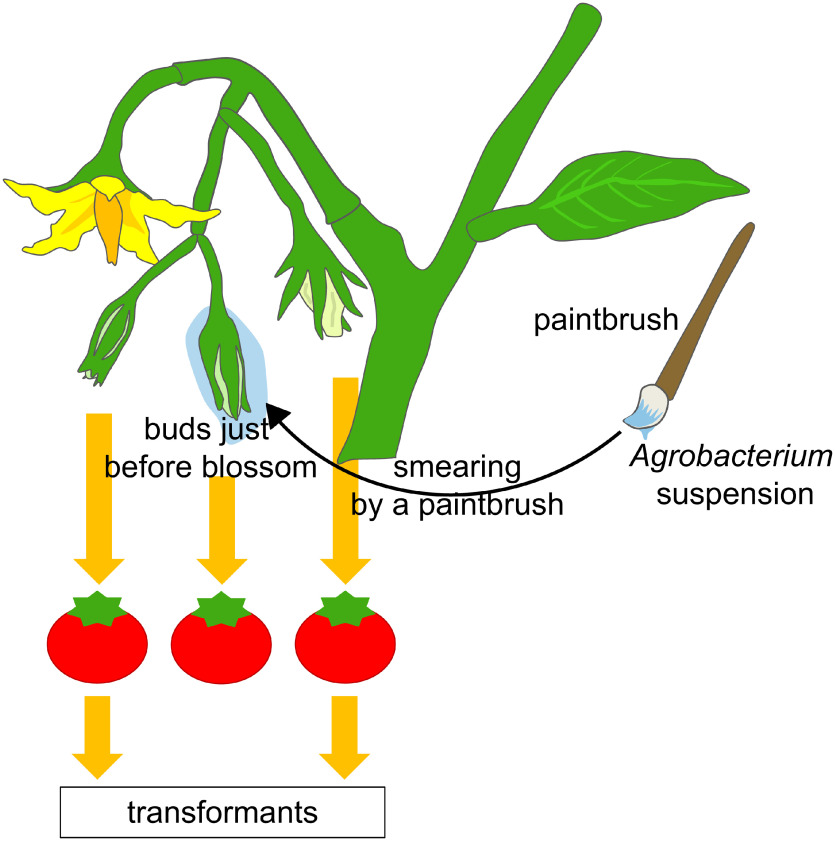
A. tumefaciens GV3101 strain harboring pBI121 vector was cultured in a liquid medium, and bacterial cells were harvested. They were suspended in the tomato infection medium (Murashige and Skoog Plant Salt Mixture (Sigma-Aldrich, St. Louis, MO, U.S.A.) 4.6 g l−1, MES 250 mg l−1, sucrose 50 g l−1, 1 N KOH 800 µl l−1, 6-Benzylaminopurine (BA) 1.0×10−2 mg l−1, Tween-20 200 µl l−1, acetosyringone 20 mg l−1) and used to prepare a series of A. tumefaciens suspension cocktails. Cell suspensions with a series of concentrations were adjusted to have OD600=0.2, OD600=0.4, OD600=0.6, and OD600=0.8, using the McFarland nephelometric method (Washington et al. 1972), with estimated concentrations of approximately 2.5×104, 5.0×104, 7.5×104, and 10.0×104 cells ml−1, respectively. Then, 1/100 amount of ‘Nenchaku-kun’ (Sumitomo Chemical Co., Tokyo, Japan), a spreading agent (a kind of pasted starch), was added to the cell suspensions. The resultant sticky A. tumefaciens suspension cocktails were smeared using a paintbrush on floral buds of Micro-Tom tomato, which was cultivated for 1 month after sowing. Each A. tumefaciens suspension cocktail was applied to three individual plants and smeared on five or six immature buds of each plant (Figure 1). After this treatment, tomato plants were put into a plastic container and decompressed for 10 min. Then, they were covered with a plastic wrapping film and incubated for 24 h in a growth chamber at 22°C under the long-day condition (16 h in light and 8 h in dark). After removal of the wrapping film, they were subsequently cultured in the growth cabinet under the same light condition.
Figure 1. Schematic representation for the procedure of Agrobacterium tumefaciens smearing onto tomato buds. A suspension of A. tumefaciens cells combined with a spreading agent was smeared using a paintbrush onto the buds just before they blossomed.
Sufficient numbers of fruits were obtained from the tomato plants treated with the A. tumefaciens suspensions with OD600=0.2, OD600=0.4, and OD600=0.6, but few fruits were generated from that with OD600=0.8 because of growth inhibition (Table 1). These findings suggested that an excess amount of A. tumefaciens resulted in an adverse effect on the plant growth.
Table 1. Number of fruits from the plants inoculated with Agrobacterium tumefaciens suspension.
| Experimental ID | Number of buds with treatment | Number of total fruits | Number of fruits from the buds with treatment | Number of fruits from the buds without treatment |
|---|---|---|---|---|
| OD600=0.2 #1 | 5 | 26 | 1 | 25 |
| OD600=0.2 #2 | 5 | 25 | 1 | 24 |
| OD600=0.2 #3 | 5 | 26 | 2 | 24 |
| OD600=0.4 #1 | 5 | 22 | 0 | 22 |
| OD600=0.4 #2 | 6 | 26 | 0 | 26 |
| OD600=0.4 #3 | 5 | 22 | 0 | 22 |
| OD600=0.6 #1 | 6 | 25 | 0 | 25 |
| OD600=0.6 #2 | 5 | 29 | 3 | 26 |
| OD600=0.6 #3 | 6 | 25 | 0 | 25 |
| OD600=0.8 #1 | 5 | 13 | 0 | 13 |
| OD600=0.8 #2 | 5 | 8 | 0 | 8 |
| OD600=0.8 #3 | 5 | 3 | 0 | 3 |
Inoculation of each A. tumefaciens cell suspension with a different concentration was performed on the buds of three individual plants. Experimental ID indicates the name of tomato plant corresponding to each experiment. Each experiment using a series of concentrations of A. tumefaciens cell suspension was performed in triplicate.
Seeds were harvested from fruits derived from the smeared buds and those located near these buds. The seeds of each plant were sown on a kanamycin-containing selection medium plate and germinated. Seedlings were cultivated for 4 weeks at 22°C under the long-day condition (16 h in light and 8 h in dark).
Usually, kanamycin-resistant tomato plants are selected by generation and elongation of a lateral root from the major root of the plants that are grown on a medium containing kanamycin. Although we attempted to select kanamycin-resistant plants in the same manner, it was rather difficult to detect the presence of kanamycin resistance because most of the plants produced a lateral root but only limited elongation occurred in 2 weeks of culturing. Therefore, we continued to culture them for at least 1 month and selected the well-grown plants (Table 1). In this way, we succeeded in the first screening and obtained a pool containing well-grown transformant plants.
Although many plants soon stopped growing, some plants showed generation of more than three branched roots with green leaves, suggesting that they had survived on the kanamycin-containing medium. The proportions of these plants in those obtained from germinated seeds of plants treated with A. tumefaciens suspensions with OD600=0.2, OD600=0.4, and OD600=0.6 were 11.6%, 3.7%, and 7.2%, respectively. In total, we obtained 113 plants out of 1,599 progeny seeds from eight plants treated with the A. tumefaciens suspensions (Table 2).
Table 2. Number of seeds from fruits obtained from the plants smeared with Agrobacterium tumefaciens suspensions.
| Experimental ID | Number of total seeds harvested | Number of seeds germinated (A) | Number of grown plants | Number of plants in which the nptII gene was detected (B) | Ratio of (B)/(A) (%) |
|---|---|---|---|---|---|
| OD600=0.2 #1 | 253 | 210 | 26 | 2 | 0.95 |
| OD600=0.2 #2 | 219 | 189 | 18 | 0 | 0 |
| OD600=0.2 #3 | 39 | 30 | 6 | 0 | 0 |
| Subtotal | 511 | 429 | 50 | 2 | 0.47 |
| OD600=0.4 #1 | 201 | 164 | 5 | 0 | 0 |
| OD600=0.4 #2 | 298 | 225 | 3 | 0 | 0 |
| OD600=0.4 #3 | 196 | 155 | 12 | 1 | 0.65 |
| Subtotal | 695 | 544 | 20 | 1 | 0.18 |
| OD600=0.6 #1 | 49 | 36 | 0 | 0 | 0 |
| OD600=0.6 #2 | 336 | 307 | 23 | 0 | 0 |
| OD600=0.6 #3 | 275 | 256 | 20 | 7 | 2.73 |
| Subtotal | 660 | 599 | 43 | 7 | 1.17 |
| OD600=0.8 #1 | 18 | 12 | 0 | 0 | 0 |
| OD600=0.8 #2 | 20 | 15 | 0 | 0 | 0 |
| OD600=0.8 #3 | 0 | 0 | 0 | 0 | 0 |
| Subtotal | 38 | 27 | 0 | 0 | 0 |
| Total | 1,904 | 1,599 | 113 | 10 | 0.63 |
Experimental ID indicates the name of tomato plant corresponding to each experiment. Numbers of total seeds harvested from the individual tomato plants, numbers of seeds that germinated among them, numbers of grown plants selected on the kanamycin-containing medium, numbers of plants in which the nptII gene was detected, and the ratio of transformants in the germinated seeds are indicated. Subtotals show the total values in the experiments using the A. tumefaciens suspensions of the same concentration.
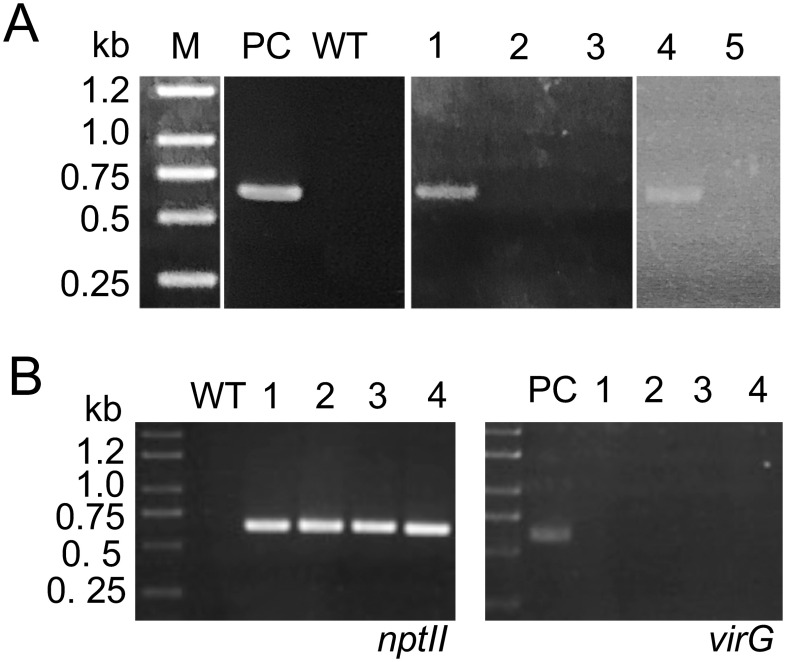
Next, we attempted to detect the kanamycin resistance gene nptII, derived from pBI121. Genomic DNA was prepared from leaves of the plants selected on the kanamycin-containing medium and used for analysis of the presence of nptII. Among the selected plants, we detected that some plants showed amplification of the specific DNA fragment corresponding to nptII, suggesting that they were the transformants containing nptII (Figure 2A). In addition, to detect the DNA fragment derived from the remaining Agrobacterium cells in the progeny plants, we were subjected to PCR-amplification of the DNA fragment corresponding to the virG gene in the Ti plasmid of A. tumefaciens using the Agrobacterium-specific primers. However, no corresponding fragment was detected in these plants (Figure 2B). These results suggest that they no longer contained Agrobacterium cells, and that these plants were transformed by the integration of the kanamycin resistance gene. We obtained two transformant plants derived from the fruits of a plant treated with A. tumefaciens suspension with OD600=0.2 (named as OD600=0.2 #1), one plant derived from a plant treated with the suspension with OD600=0.4 (OD600=0.4 #3), and seven plants derived from a plant treated with the suspension with OD600=0.6 (OD600=0.6 #3). This finding also indicated that our procedure could be used for the introduction of a foreign gene into plant cells. When the A. tumefaciens suspension with OD600=0.6 was smeared on the buds, the highest transformation ratio was achieved. In this case, the ratio of transformants in the germinated seeds was 2.73% (Table 2).
Figure 2. Detection of the transgene. (A) Detection of the nptII gene in the transformants. PC, pBI121 DNA; WT, genomic DNA of the wild-type plant; 1 to 5, genomic DNA prepared from representative progenies. 1 to 3 are derived from the progenies of OD600=0.2 #1, and 4 and 5 are derived from those of OD600=0.6 #3. A specific DNA fragment corresponding to nptII was polymerase chain reaction (PCR)-amplified from the genomic DNA using a set of primers, 5′-AATATCACGGGTAGCCAACG-3′ and 5′-GCTTGGGTGGAGAGGCTATT-3′, which was based on the plasmid pBI121 (acc. number: AF485783). (B) Evaluation of the transformants. The existence of nptII (left panel) and virG (right panel) genes was examined in the transformants. Numbers indicate the progenies derived from the buds of OD600=0.6 #3. Each number corresponds to the same individual plant. A specific fragment corresponding to virG (acc. number: X04965) was PCR-amplified using a set of primers, 5′-TCGATGACGACGTCGCTATG-3′ and 5′-CGCAGCCTCAAAATGAGAAC-3′, which was based on the Ti plasmid. PC, Agrobacterium tumefaciens DNA; WT, genomic DNA of the wild-type plant; 1 to 4, genomic DNA prepared from representative transformants.
We analyzed transformation frequency for the seeds of fruits derived from buds inoculated with A. tumefaciens cell suspension, and those located near these buds. The ratio of transformants obtained from the buds smeared with A. tumefaciens suspension was compared with that for the buds located near them. Some of the smeared buds were falling before ripening, and only a small number of fruits was generated from them. No transformant was obtained among the seeds derived from the fruits derived from these buds. All transformants containing the kanamycin resistance gene were derived from the buds other than the smeared buds (Table 3). This finding suggested that A. tumefaciens moved to the buds located near the smeared buds and caused the transformation event.
Table 3. Number of transformants derived from buds of treated plants.
| Experimental ID | Seeds from smeared buds | Seeds from other buds | ||
|---|---|---|---|---|
| Number of seeds | Number of transformants | Number of seeds | Number of transformants | |
| OD600=0.2 #1 | 5 | 0 | 248 | 2 |
| OD600=0.4 #3 | 0 | 0 | 196 | 1 |
| OD600=0.6 #3 | 0 | 0 | 275 | 7 |
Experimental ID indicates the name of tomato plant that produced transformants containing the nptII gene. Numbers of seeds and transformants in the progenies generated by the buds with/without Agrobacterium tumefaciens treatment are shown.
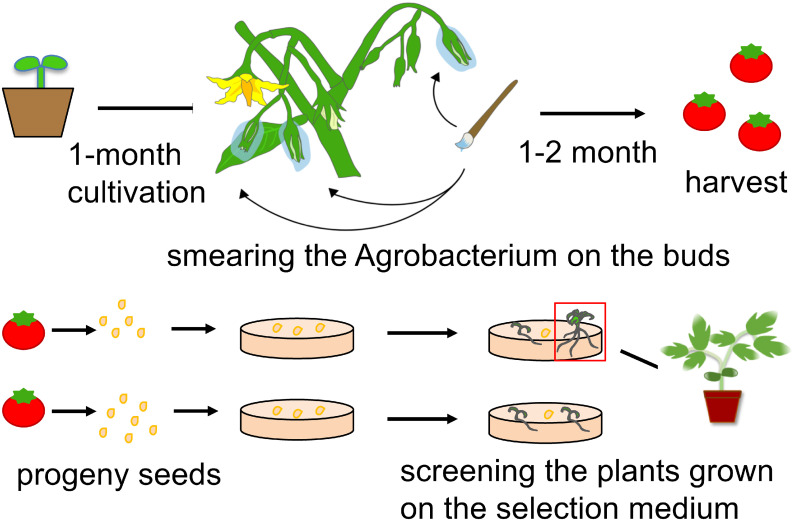
Transformation of tomato using the floral dip method based on a technique different from the usual procedure has been reported, with a transformation efficiency of 0.25–0.5% for floral dips or floral injections into the buds (Sharada et al. 2017). A procedure for floral dip transformation of Brassica rapa has been proposed by Hu et al. (2019). They suggested that efficient transformation occurs when a plant at an immature stage is used. Here, we proposed a simple procedure for tomato transformation by smearing A. tumefaciens suspension onto floral buds (Figure 3). The conventional method for tomato transformation has been established (McCormick et al. 1986). Our method exhibited a similar efficiency to the conventional one, and it would be convenient to achieve the tomato transformation because it requires less skilled labor under the clean experimental condition.
Figure 3. Schematic representation of the proposed procedure of tomato transformation. Agrobacterium tumefaciens smearing was performed on the buds of 1-month-old tomato plants after germination. Seeds were harvested from the fruits generated on the treated plants. They were sown on the selection medium containing kanamycin. Among the plants that grew well, transformants were selected.
Although some buds were falling before ripening and no transformant was obtained from the fruits derived from the smeared buds, we found that the generation of transformants occurred from the seeds derived from buds located near the buds smeared with A. tumefaciens cells. Our findings indicated that A. tumefaciens moved to other buds on the plant with the smeared buds and led to a transformation event in embryos located in the other buds. Therefore, these findings indicate that this procedure results in a sufficient number of transformants when well-grown plants with multiple flowers are used. In addition, this procedure may be applied to other tomato cultivars and contribute to their molecular breeding.
Acknowledgments
Micro-Tom tomato was provided by the Tomato Genetics Resource Center at the University of California, Davis. We thank M. Kihira, H. Onodera, M. Ohnuma, R. Suzuki, T. Imamura, A. Ikeda, Y. Kouzuki, A. Otsuka, and R. Shimada for their valuable suggestions.
Abbreviations
- nptII
the gene for kanamycin resistance
- OD600
optical density at 600 nm
References
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Hu D, Bent AF, Hou X, Li Y (2019) Agrobacterium-mediated vacuum infiltration and floral dip transformation of rapid-cycling Brassica rapa. BMC Plant Biol 19: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H-Q, Kriseleit D, Ganal MW (1998) Effect of ticarcillin potassium clavulanate on callus growth and shoot regeneration in Agrobacterium-mediated transformation of tomato (Lycopersicon esculentum Mill.). Plant Cell Rep 17: 843–847 [DOI] [PubMed] [Google Scholar]
- McCormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, Fraley R (1986) Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep 15: 81–84 [DOI] [PubMed] [Google Scholar]
- Pawar BD, Jadhav AS, Kale AA, Chimote VP, Pawar SV (2013) Effect of explants, bacterial cell density and overgrowth-control antibiotics on transformation efficiency in tomato (Solanum lycopersicum L.). J Appl Hortic 15: 95–99 [Google Scholar]
- Sharada MS, Kumari A, Pandey AK, Sharma S, Sharma P, Sreelakshmi Y, Sharma R (2017) Generation of genetically stable transformants by Agrobacterium using tomato floral buds. Plant Cell Tissue Organ Cult 129: 299–312 [Google Scholar]
- van Roekel JS, Damm B, Melchers LS, Hoekema A (1993) Factors influencing transformation frequency of tomato (Lycopersicon esculentum). Plant Cell Rep 12: 644–647 [DOI] [PubMed] [Google Scholar]
- Washington JA II, Warren E, Karlson AG (1972) Stability of barium sulfate turbidity standards. Appl Microbiol 24: 1013. [DOI] [PMC free article] [PubMed] [Google Scholar]