Abstract
Licorice (Glycyrrhiza uralensis) is a medicinal plant that contains glycyrrhizin (GL), which has various pharmacological activities. Because licorice is a legume, it can establish a symbiotic relationship with nitrogen-fixing rhizobial bacteria. However, the effect of this symbiosis on GL production is unknown. Rhizobia were isolated from root nodules of Glycyrrhiza glabra, and a rhizobium that can form root nodules in G. uralensis was selected. Whole-genome analysis revealed a single circular chromosome of 6.7 Mbp. This rhizobium was classified as Mesorhizobium by phylogenetic analysis and was designated Mesorhizobium sp. J8. When G. uralensis plants grown from cuttings were inoculated with J8, root nodules formed. Shoot biomass and SPAD values of inoculated plants were significantly higher than those of uninoculated controls, and the GL content of the roots was 3.2 times that of controls. Because uninoculated plants from cuttings showed slight nodule formation, we grew plants from seeds in plant boxes filled with sterilized vermiculite, inoculated half of the seedlings with J8, and grew them with or without 100 µM KNO3. The SPAD values of inoculated plants were significantly higher than those of uninoculated plants. Furthermore, the expression level of the CYP88D6 gene, which is a marker of GL synthesis, was 2.5 times higher than in inoculated plants. These results indicate that rhizobial symbiosis promotes both biomass and GL production in G. uralensis.
Keywords: Glycyrrhiza uralensis, glycyrrhizin, nodulation, rhizobium, symbiosis
Introduction
Medicinal plants are the source of the active ingredients of many medicines and are a valuable resource. They have been used in disease treatment and prevention from ancient times. Among them, Glycyrrhiza uralensis Fisch. and Glycyrrhiza glabra L. are important sources of a crude drug that is used in Chinese herbal medicine (Ozaki and Shibano 2014). Glycyrrhiza uralensis is considered the ‘king’ of traditional Chinese medicine in China. G. uralensis is widely distributed in Central Asia, Mongolia, and China (Hayashi and Sudo 2009). In these regions, the native G. uralensis is usually collected for production of the crude drug, but overharvesting and desertification have caused major supply problems (Kusano et al. 2003). Glycyrrhiza uralensis is not common in Japan. Domestic cultivation could help to ensure a stable supply of G. uralensis.
Glycyrrhizin (GL, glycyrrhizic acid), which is the main component of the herbal medicine, is present in the roots and stolons of G. uralensis. GL belongs to a group of compounds called triterpenoid saponins, which have important pharmacological activities (Tang et al. 2015). For use as a medicine, G. uralensis must contain more than 2.0% GL on a crude dry-weight basis in the 17th edition of the Japanese Pharmacopoeia (JP; The Ministry of Health, Labor and Welfare, Japan, 2016).
Recently, efforts have been made to develop methods for domestic cultivation of G. uralensis that meets these standards. For example, Sato et al. (2004) tested the effects of adjusting the concentration of culture solution. Bioengineering approaches have also been tried, such as culture of a yeast transformed with the gene CYP88D6, which accumulated an intermediate of GL biosynthesis (Seki et al. 2008).
Because G. uralensis is a legume, it can form symbioses with rhizobial bacteria. The rhizobia induce the host plant to form root nodules, where they supply fixed atmospheric N2 to the plants in return for photosynthetic products. In this way, a mutually beneficial relationship is established between legumes and rhizobia (Mylona et al. 1995). N2 fixation by rhizobia plays a significant role in agricultural production and in geochemistry.
To date, no studies have investigated how rhizobial symbiosis affects GL production in G. uralensis. If rhizobial symbiosis promotes growth of G. uralensis and enhances the GL content of its roots, it may enhance the efficiency of agricultural GL production. In this study, we performed whole-genome analysis of rhizobium isolated from G. glabra nodules and examined how inoculation in G. uralensis affected host plant growth and GL production. This is the first report of the effect of inoculation of rhizobium on GL production in licorice.
Materials and methods
Isolation of Glycyrrhiza rhizobia
Glycyrrhiza glabra seedlings were transplanted into vinyl pots (diameter 15 cm, height 12 cm) filled with soil obtained from the field of Saga university, Japan. They were grown under long-day conditions (16 h light : 8 h dark, 25°C) for 6 months. Rhizobia isolated from nodules were grown in yeast mannitol liquid medium (Keele et al. 1969) and then suspended in sterilized distilled water (1.0×109 cells ml−1). Each plant was inoculated with 200 ml of suspension into G. uralensis and nodulation was recorded.
Extraction of genomic DNA from rhizobia
The isolated rhizobia were cultured in yeast extract–mannitol (YM) medium (Vincent 1970) with shaking at 28°C for 3 to 4 days. DNA was extracted using Wizard Genomic DNA Purification Kit (Promega, São Paulo, Brazil) according to the manufacturer’s instructions.
Sequencing and annotation
The genome sequence of J8 was determined on a PacBio RSII sequencer (performed by Macrogen Japan Corp., Kyoto, Japan). Reads were de novo assembled for circularization of the genome in HGAP4 software (SMRT Link v. 5.0.1) (Pacific Biosciences of California, Inc. California, USA). The draft sequence of J8 and the chromosomal sequence of Mesorhizobium japonicum MAFF303099 (Accession No.: BA000012, Kaneko et al. 2000; Keele et al. 1969; Saeki and Kouchi 2000) were compared using MUMmer (ver. 3.23) (Kurtz et al. 2004). Prokka v. 1.12 software (Illumina, Tokyo, Japan) designed for gene prediction in prokaryotes, was used to predict the locations of genes in the draft sequence of J8. Using the gene sequence of J8 (base sequence) as a query, a BLASTX search was performed for M. japonicum MAFF303099 genes (amino acid sequences). The genome was annotated with top hits (E-value ≤ 1E-5) and gene prediction data from Prokka.
Genome comparison and ortholog analysis
A circular genome was constructed on the basis of the database of Clusters of Orthologous Groups of proteins (COGs) of 6,325 predicted genes (Tatusov et al. 2001). The gene structure of J8 was similar to that of the predicted gene structure in the Ori region of the M. japonicum MAFF303099 genome sequence. The origin of replication was predicted by setting the position of the gene predicted to be closest to the Ori region (putative pyruvate, phosphate dikinase regulatory protein) as position 1. A phylogenetic tree was based on the 16S rRNA sequence. When a BLASTN search was performed using 5S rRNA, 16S rRNA and 23S rRNA gene sequences of M. japonicum MAFF303099 as a query, sequence similarity was detected in six places. Using the sequence of 16S rRNA, we searched the National Center for Biotechnology Information database (https://www.ncbi.nlm.nih.gov) for related species. In addition, we performed phylogenetic analysis of the combined amino acid sequences of nodA, nodB, and nodC of M. japonicum MAFF303099 and Mesorhizobium sp. M2A.F.Ca.ET.043.02.1.1 (Accsession No.: CP034445) using Unweighted Pari Group Method using arithmetic Average (UPGMA) method (Sneath and Sokal 1973).
Plant materials
Glycyrrhiza uralensis Fisch. (strain HI-1) and G. glabra L. (strain Hgl-1) were used. Voucher specimens (accession nos.: G. uralensis, GLY-URA-001; G. glabra, GLY-GLA-001) were deposited in the Herbarium of the Faculty of Pharmaceutical Sciences, Health Sciences University of Hokkaido, Japan (Kojoma et al. 1995, 2010). Soil was obtained from the field of Saga university, Japan. Cuttings were taken from the G. uralensis plants.
Preparation of rhizobia
J8 grown in yeast mannitol liquid medium was harvested and then suspended in sterilized distilled water (1.0×109 cells ml−1). Each plant was inoculated with 200 ml of suspension. For the closed system experiments (inoculation test using plant boxes filled with sterilized vermiculite), J8 was diluted in Broughton and Dilworth B&D medium (Broughton and Dilworth 1971) which is N-free and inoculated at 1.0×107 cells ml−1.
Cuttings
The soil used was a mixture of equal parts by volume of vermiculite, river sand and sod soil (Green Sangyo). Cuttings were prepared by cutting shoots from the parent plants which were genetically identical at three leaves from the top. Basal ends of shoots were cut horizontally and immediately immersed in water at 25°C for 30 min. After immersion, cuttings were inserted vertically into wet soil in vinyl pots (diameter 9 cm, height 8 cm). Water was sprayed on the leaves of the cuttings every day. Cuttings were supplied with tap water at their base for about a week after cutting and then with liquid B&D medium containing 1,000 µM KNO3. Plants were grown for 4 weeks before being used for nodulation tests. In prior testing, no nodule formed when plants were grown in B&D medium. Therefore, KNO3 was added (Egamberdieva et al. 2016; Zheng et al. 2006).
Nodulation test using plants grown from cuttings
J8 was diluted with liquid B&D medium containing 1,000 µM KNO3 to 1.0×109 cells ml−1 and 200 ml of it was added to the soil. As a control, liquid B&D medium containing 1,000 µM KNO3 was given. The surface of the soil was covered to prevent contamination. Plants were grown for 90 days at 25°C. During the test, liquid B&D medium containing 1,000 µM KNO3 was applied to the roots about once a week.
Nodulation test using plant boxes
Seeds of G. uralensis were soaked in distilled water for 3 days. Seed coats were removed with tweezers. A plant box (6 cm width×6 cm length×10 cm depth) was filled with 250 ml of vermiculite, and 150 ml of liquid B&D medium with (100 µM KNO3) or without (0 µM KNO3) N was added. Six seeds were sown per box. A box was covered with other plant box as a lid to prevent the formation of nodules on uninoculated plants. Plants were grown at 25°C for 60 days after inoculation.
Measurement of SPAD value
The SPAD value was measured with a SPAD chlorophyll meter (SPAD-502Plus, Minolta Camera Co. Osaka, Japan). The average value of three leaves was used for each plant.
Acetylene reduction assay (ARA) for nitrogenase activity
One plant was placed in a 34-ml test tube, which was then covered with a rubber serum cap and degassed for 30 s. Acetylene that was diluted five times was injected into the tube, which was then incubated in a 25°C chamber. After 1 h of incubation, the amount of ethylene was determined by using a Shimadzu Gas Chromatograph (GC-14).
qRT-PCR
After growth measurements were taken, plant roots were quickly frozen in liquid N2 and stored at −80°C. Total RNA was prepared with a RNeasy Plant Mini Kit (Qiagen, Basel, Switzerland). DNase I treatment was performed with RT-Grade DNase (Wako, Osaka, Japan). A one-step SYBR PrimeScript RT-PCR kit (TaKaRa, Shiga, Japan) was used. Gene-specific primer sequences were as follows: 18S ribosomal RNA (Accession No.: X02623) (5′-CTC AAC ACG GGG AAA CTT AC-3′ and 5′-AGA CAA ATC GCT CCA CCA AC-3′) and GuCYP88D6 (Accession No.: AB433179) (5′-CGG AAC ATA GGC TTT TTC GAC-3′ and 5′-TAC ATT GCT AGC GCC TTG TG-3′). The specific primers for GuCYP88D6 were designed in Primer 3 software. The 18S ribosomal RNA gene was used as an internal control (Shabani et al. 2010).
Measurement of glycyrrhizin content
The GL content of G. uralensis roots were measured as described by Kojoma et al. (2010). The HPLC system used consisted of a high-performance liquid chromatography (Chromaster 5310, Hitachi, Tokyo, Japan) and an InsertSustain C18, 5 µm HPLC column (GL Sciences Inc. Tokyo, Japan). Acetonitrile and formic acid were used as the solvents. The column oven was set to 40°C, the column flow rate was set to 0.6 ml min−1, and detection was performed at 240 to 260 nm.
Results
Phylogeny of the rhizobium strain isolated from G. glabra
At the beginning of this study, we didn’t have G. uralensis as material. So, we used G. glabra for isolation of rhizobia which establish symbiotic relationships with Glycyrrhiza species. After that, we inoculated G. glabra with isolated strains again, but no strain could form nodules in G. glabra at 4 weeks after inoculation. Therefore, we inoculated G. uralensis with these strains because we had already obtained G. uralensis at that time. Based on the sequence analysis of 16S rRNA gene, 14 strains were identified. Ability of nodulation and nitrogen fixation for these strains were shown in Table 1. Seven strains formed nodules on G. glabra and J8 showed the highest ARA value.
Table 1. List of rhizobial strains isolated from root nodules of Glycyrrhiza glabra growing in the soil obtained from the field of Saga university.
| Strains | Closest species and strain (accession No.), max identity | Nodulation on G.uralensis | ARA |
|---|---|---|---|
| J1 | Pseudomonas veronii strain CIP 104663 (NR_028706.1) 98% | − | − |
| J2 | Pseudomonas rhodesiae strain CIP 104664 (NR_024911.1) 97% | + | n.d. |
| J3 | Pseudomonas proteolytica strain CMS 64 (NR_025588.1) 96% | − | − |
| J4 | Cedecea lapagei strain DSM 4587 (NR_126318.1) 99% | − | − |
| J5 | Enterobacter ludwigii strain EN-119 (NR_042349.1) 99% | − | − |
| J6 | Enterobacter cloacae ssp. dissolvens strain LMG 2683 (NR_044978.1) 97% | − | − |
| E1 (J7) | Bradyrhizobium viridifuturi strain SEMIA 690 (NR_145860.1) 97% | + | n.d. |
| J8 | Mesorhizobium amorphae strain ACCC 19665 (NR_024879.1) 96% | + | + |
| J9 | Rhizobium oryzae strain Alt 505 (NR_044393.1) 94% | + | n.d. |
| J10 | Rhizobium oryzicola strain ZYY136 (NR_137225.1) 95% | + | n.d. |
| J11 | Paenibacillus taiwanensis strain BCRC 17411 (NR_044007.1) 94% | − | − |
| J12 | Bacillus velezensis strain FZB42 (NR_075005.2) 98% | + | + |
| J13 | Bacillus cereus strain ATCC 14579 (NR_074540.1) 99% | + | + |
| J14 | Bacillus toyonensis strain BCT-7112 (NR_121761.1) 97% | − | − |
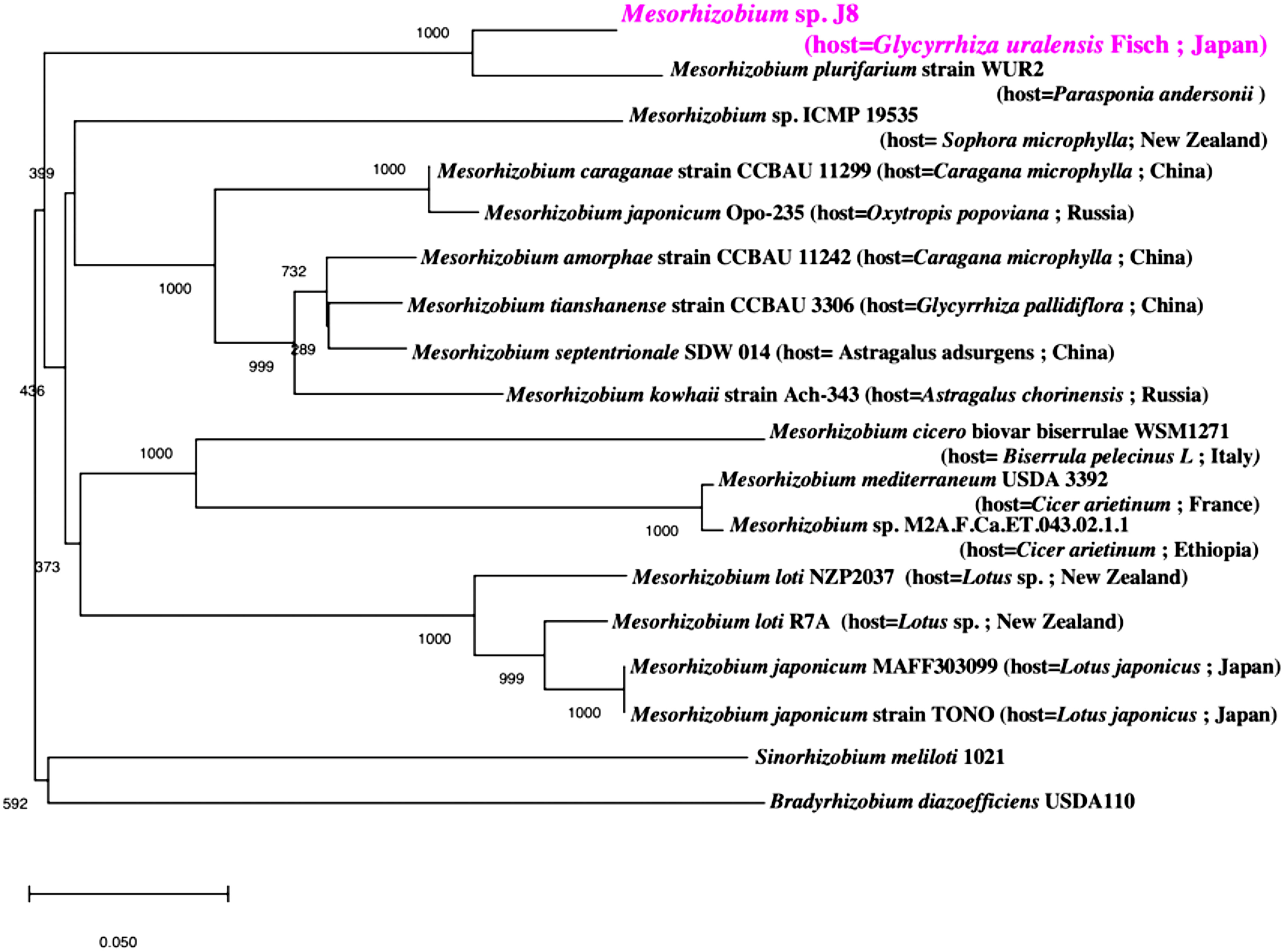
Phylogenetic analysis of 16S rRNA sequences classified J8 as Mesorhizobium, and the most closely related species is Mesorhizobium sp. M2A.F.Ca.ET.043.02.1.1, isolated from root nodules of Cicer arietinum (Figure 1). Phylogenetic analysis of the combined amino acid sequences of nodA, nodB, and nodC from J8 and other Mesorhizobium strains showed that the species most closely related to J8 is Mesorhizobium plurifarium strain WUR2 (Figure 2). These two strains formed a clade distinct from that formed by Mesorhizobium tianshanense strain CCBAU 3306, Mesorhizobium kowhaii Ach-343, and Mesorhizobium japonicum Opo-235, three strains that have been confirmed to establish symbiosis with G. uralensis (Chen et al. 2008; Safronova et al. 2019).
Figure 1. Phylogenetic relationship of Mesorhizobium sp. J8 and related bacteria based on 16S rRNA gene sequences. The tree was built using the unweighted pair group method with arithmetic mean.
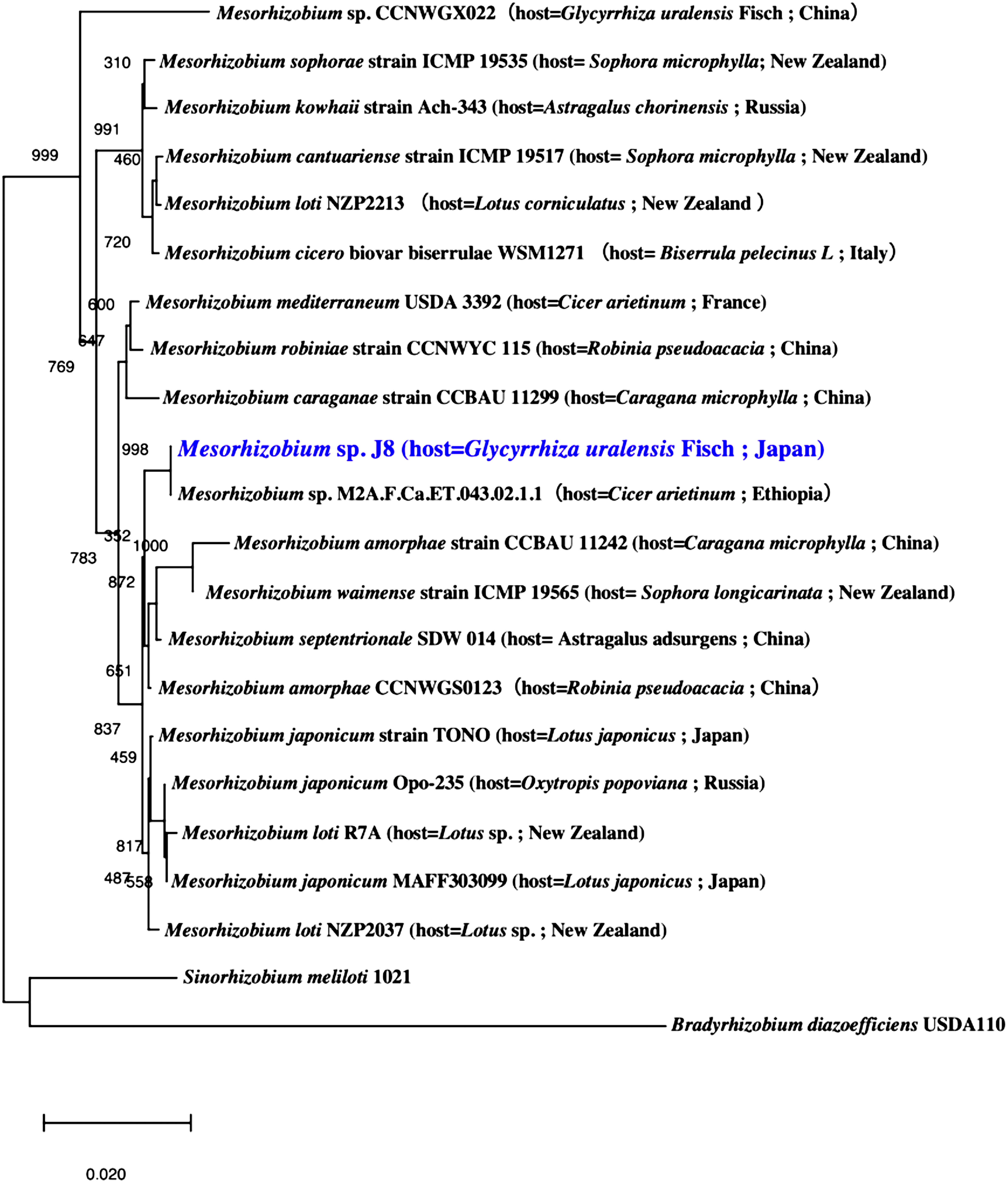
Figure 2. Phylogenetic relationship of Mesorhizobium sp. J8 and related bacteria based on the amino acid sequences of nodA, nodB, and nodC. The tree was built using the unweighted pair group method with arithmetic mean.
General description of genome structure of Mesorhizobium sp. J8 isolated from G. glabra
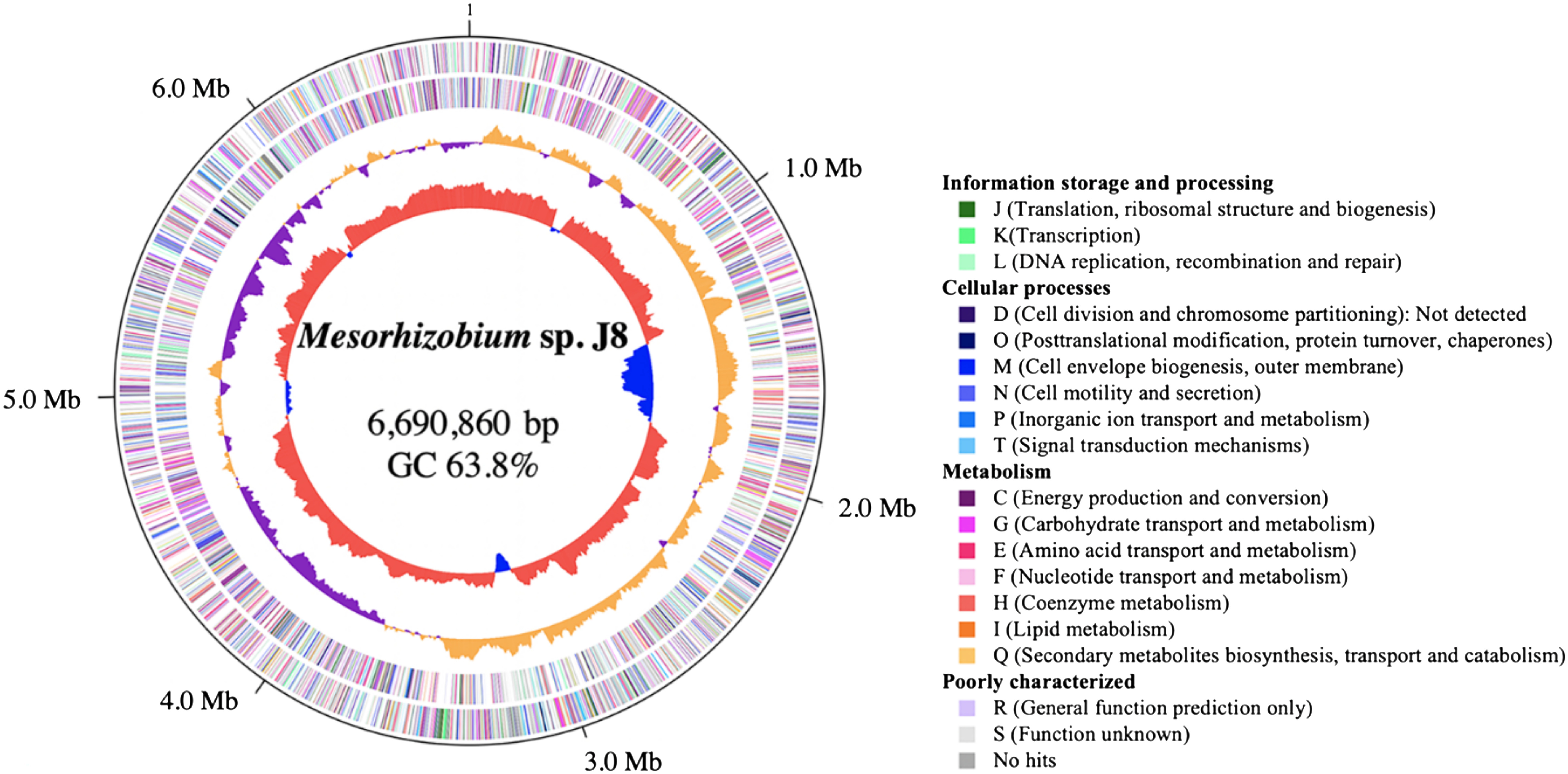
Genome sequencing of Mesorhizobium sp. J8 revealed a single circular chromosome of 6,690,860 base pairs, with an average GC content of 63.8% (Figure 3). A total of 6,325 protein coding genes and 52 tRNA genes were predicted on the genome of J8. The putative origin of replication was predicted on the basis of the position of a GC skew shift (position: 0 Mb) and coincided with the occurrence of a gene cluster typically associated with origins of replication in circular chromosomes of M. japonicum MAFF303099.
Figure 3. The genome structure of Mesorhizobium sp. J8. A circular genome map was created using in silico Molecular Cloning (Ohyama et al. 2006). The outmost circle and the second show the positions of the putative protein-encoding genes in the clockwise and counterclockwise directions, respectively. Genes whose functions could be deduced by sequence similarity to genes of known function are depicted in green, blue and warm colors, and those whose function could not be deduced are in light colors by COG assignment. The innermost and second-innermost circles show the GC content and the GC skew, respectively. The GC content circle shows the deviation from average GC content of the entire sequence (red, higher than average GC content; blue, lower than average content). The GC skew circle shows the deviation from average skew of the entire sequence (orange, higher than average GC skew; purple, lower than average content). The labels outside the outmost circle represent genome positions (in Mb).
Comparison with other Mesorhizobium strains
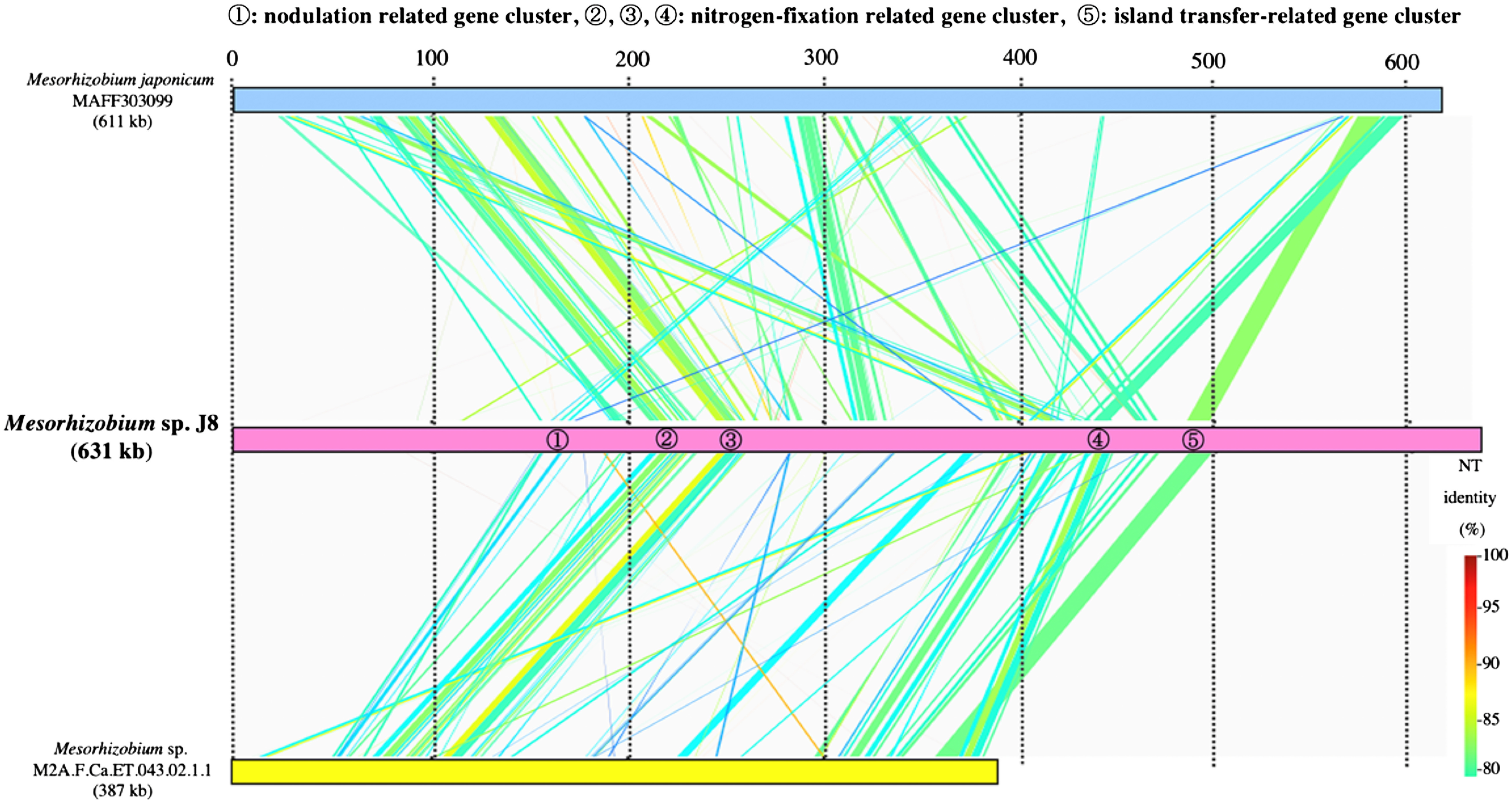
To identify the structural features of J8 genome, we performed comparative analysis with M. japonicum MAFF303099. Based on a dot plot similarity analysis of overall the J8 genome with M. japonicum MAFF303099, it was considered that these two strains share collinear similarity throughout the chromosome with multiple interruptions by the putative genomic islands including symbiosis islands of both genomes (Figure 4). According to the syntenic relation with MAFF303099 and GC content in the J8 genome, the symbiotic island of J8 was estimated to be between 1.3 and 1.9 Mb (Figure 4). Although three tRNA genes including Phe-tRNA gene, on which symbiotic island of MAFF303099 is integrated, were predicted in this region of the J8 genome, the duplication of 3′ terminal portion of the gene was not detected for none of these tRNA genes indicating that symbiotic island of J8 could have been targeted the genome sequence other than tRNA gene. By comparing the similarity of estimated symbiotic island region of the J8 genome against symbiotic islands of M. japonicus MAFF303099 and Mesorhizobium sp. M2A.F.Ca.ET.043.02.1.1, it was revealed that the estimated symbiotic island of the J8 carries a nodulation related gene cluster, nitrogen-fixation related gene clusters and an island transfer-related gene cluster, and these gene clusters are moderately conserved (around 85% identity) among the three Mesorhizobium strains with different host plant species (Figure 5).
Figure 4. Comparison of chromosomal sequences of Mesorhizobium japonicum MAFF303099 generated using MUMmer (ver. 3.23). The positions on chromosomes of Mesorhizobium japonicum MAFF303099 and Mesorhizobium sp. J8 are indicated on the x-axis and y-axis. The dot color indicates the percentage similarity, as indicated in the key.
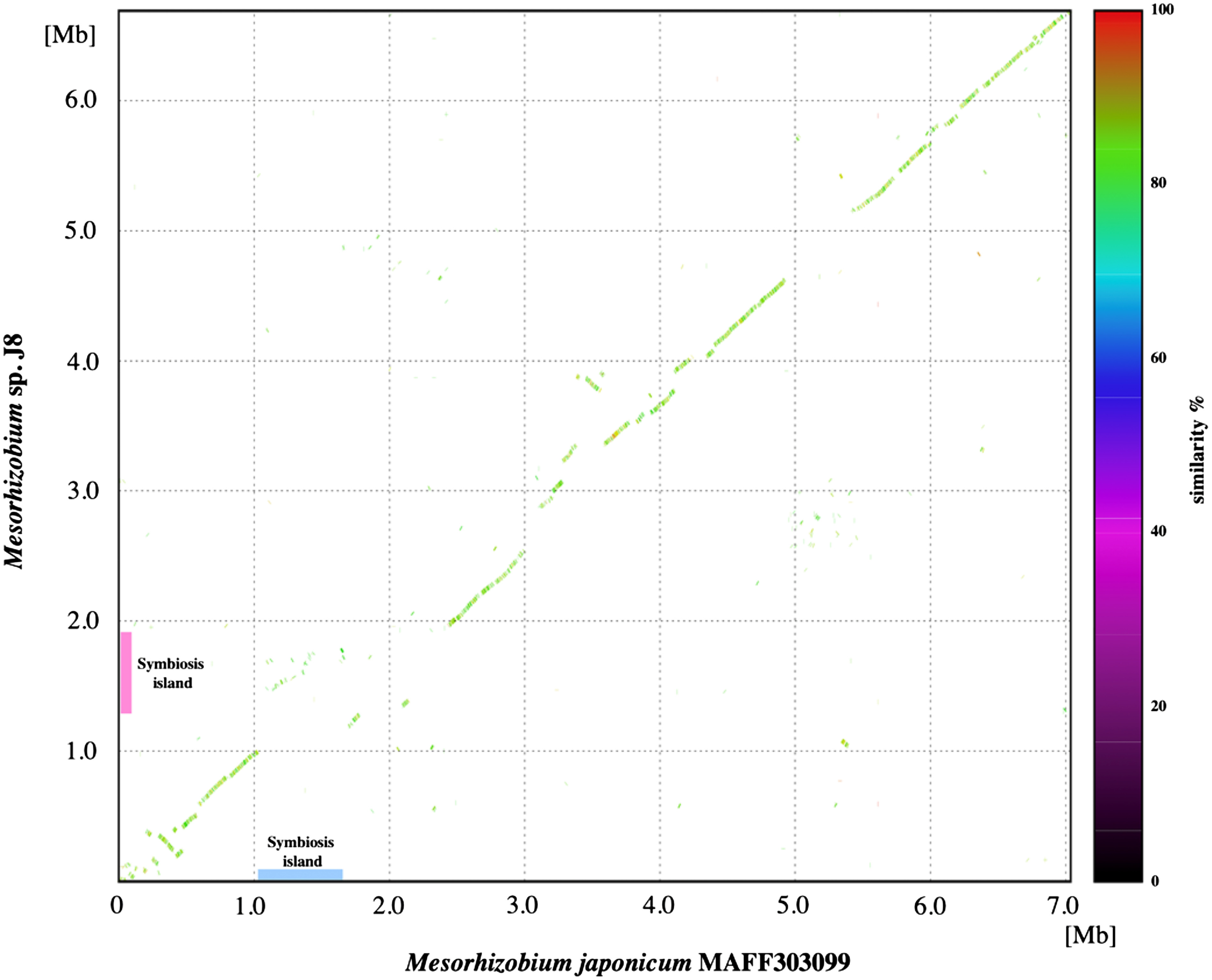
Figure 5. Comparison of nodulation gene clusters in Mesorhizobium sp. J8 and related bacteria using GenomeMatcher (Ohtsubo et al. 2008). Colored strips represent the conserved gene regions between the compared strains, and the color indicates the percentage similarity, as detailed in the key.
Effect of inoculation of Mesorhizobium sp. J8 on the growth and symbiosis parameters of G. uralensis
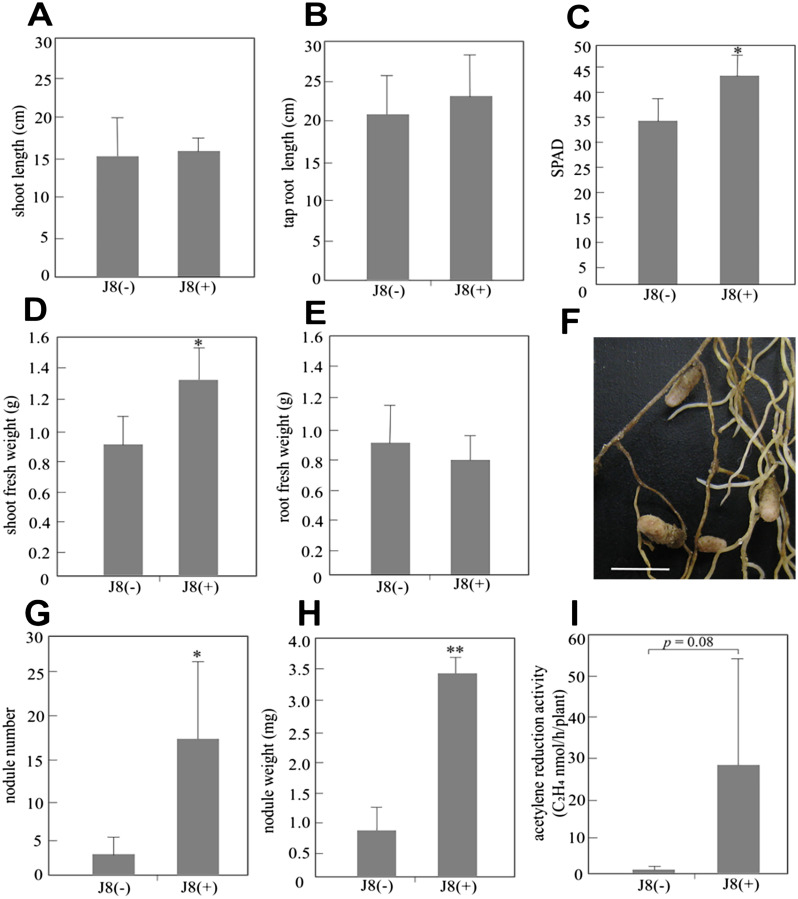
Comparison of the growth and symbiosis parameters of inoculated (J8(+)) and uninoculated (J8(−)) G. uralensis plants grown from cuttings giving liquid B&D medium containing 1,000 µM KNO3 at 90 days after inoculation showed no significant differences in shoot length, tap root length, or root fresh weight (Figure 6A, B, E), but shoot fresh weight was significantly larger in J8(+) plants than in J8(−) plants (Figure 6D). The SPAD value, which indicates the chlorophyll content of leaves, of J8(+) plants was significantly higher than that of J8(−) plants (Figure 6C). Generally, as the N content of the plant increases, the SPAD value also increases. Therefore, J8(+) plants likely contained more N than the control. Although a few J8(−) plants formed small nodules (Figure 6G), both nodule number and nodule weight were significantly higher in J8(+) plants (Figure 6G, H). The ARA value per J8(+) plant was slightly higher than that of J8(−) plants (p=0.08) (Figure 6I). These results indicate that enhanced N fixation in J8(+) plants induced increased shoot growth and SPAD values.
Figure 6. Comparison of the growth and symbiosis parameters of inoculated (J8(+), n=6) and uninoculated (J8(−), n=5) G. uralensis plants grown from cuttings at 90 days after inoculation. The level of shoot length (A), tap root length (B), shoot fresh weight (D) and root fresh weight (E) were measured. (C) The leaf SPAD value. Three leaves of each plant were measured and average values were calculated. (F) Phenotype of nodules of Glycyrrhiza uralensis plants with (J8+) Mesorhizobium sp. J8 inoculation (scale bar=5 mm). The nodule number (G), nodule weight (H) and acetylene reduction activity (I) of uninoculated plants and inoculated plants were analyzed. The error bars represent the standard deviation (SD). Significant differences from the uninoculated plants are shown (Student’s t-test: * indicates p<0.05 and ** indicates p<0.01).
Effect of rhizobial inoculation on expression of the CYP88D6 gene in G. uralensis
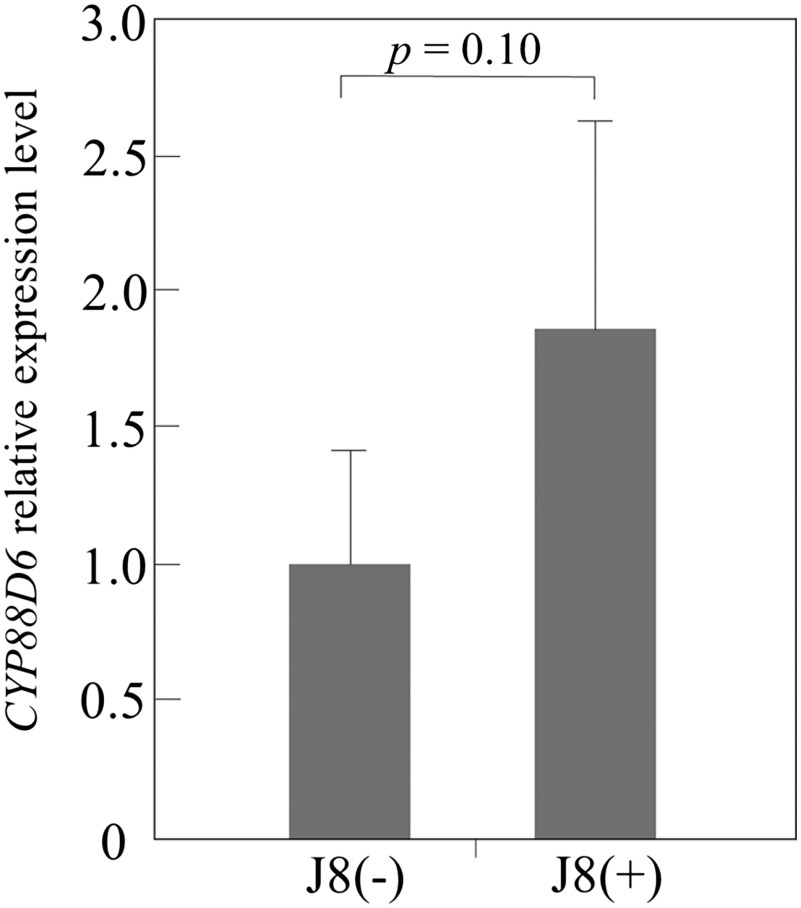
We investigated whether inoculation with J8 affects the expression level of the marker gene for GL synthesis in the roots of G. uralensis plants prepared from cuttings. CYP88D6 encodes β-amyrin 11-oxidase, which acts in the glycyrrhizin synthetic pathway, and its expression level is correlated with GL content (Seki et al. 2008).
CYP88D6 expression was measured by qRT-PCR in the roots 90 days after inoculation. CYP88D6 expression of J8(+) plants was 1.8 times that of the control (p=0.1) (Figure 7). This result indicates that GL production was enhanced in inoculated plants.
Figure 7. Expression level of CYP88D6 gene in root tissue of J8 uninoculated plants (J8(−), n=5) and inoculated plants (J8(+), n=6) prepared from cuttings, by qRT-PCR. 18S ribosomal RNA gene was used as an internal control. The expression data represent the mean±SD of 6 (J8(−)) and 5 (J8(+)) independent plants, each having three technical replicates.
Effect of inoculation on GL production in G. uralensis plants grown from cuttings
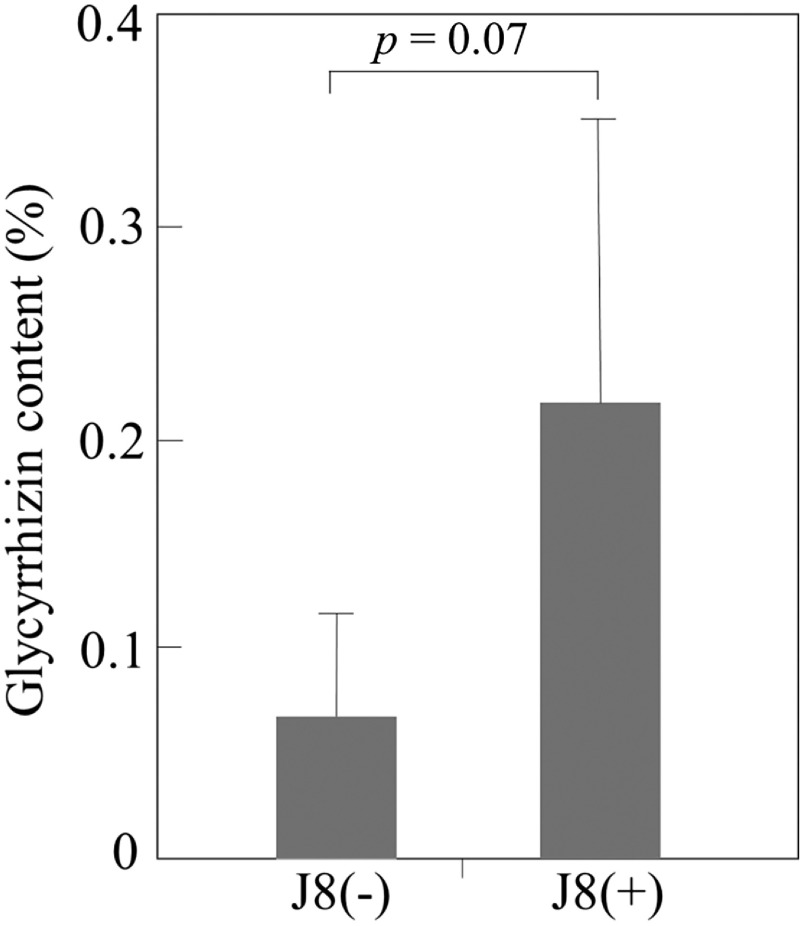
The GL content per unit root weight was higher in J8(+) plants than in J8(−) plants (Figure 8). Although nodules were formed on the roots of J8(−) plants (Figure 6G), nodule number and weight and N fixation activity were extremely restricted. Therefore, the effects of nodulation on GL production in uninoculated plants can be ignored. These results indicate that inoculation of adequate rhizobium onto G. uralensis induced enhanced GL production.
Figure 8. Effects of J8 inoculation on GL content per unit weight in Glycyrrhiza uralensis roots 90 days after inoculation: GL content in uninoculated (J8(−), n=5) vs. inoculated (J8(+), n=6) plants. Error bars represent standard deviation (SD). The significance of differences between treatments was determined by Student’s t-test (p=0.07).
Effect of inoculation of Mesorhizobium sp. J8 on the growth and symbiotic parameters of G. uralensis plants grown from seeds
Since nodules formed in uninoculated plants, we conducted nodulation tests in a closed system. As shown in Table 2, shoot fresh weight with or without J8 inoculation tended to be higher in the treatments supplied with N (p=0.08). Moreover, no G. uralensis plants formed nodules under any N condition tested. In plants grown in soil containing 0 and 100 µM KNO3, although no significant differences were observed in shoot length, tap root length, shoot fresh weight, or root fresh weight, SPAD value of J8(+) plants was significantly higher than that of control plants. However, no significant difference was observed in nodule number or nodule weight between J8(+) plants grown under different N concentrations. ARA values tended to decrease as N concentration increased (Table 2).
Table 2. Comparison with the effect of Mesorhizobium sp. J8 inoculation on growth and nodulation in Glycyrrhiza uralensis plant prepared from seeds grown in soil with or without N.
| Trait | 0 µM KNO3 | 100 µM KNO3 | ||
|---|---|---|---|---|
| J8(−) | J8(+) | J8(−) | J8(+) | |
| Shoot length (cm) | 9.7±1.8 | 10.2±2.0 | 11.3±2.3 | 12.2±3.6 |
| Tap root length (cm) | 12.6±4.1 | 10.7±2.6 | 14.1±7.4 | 13.1±5.0 |
| Shoot fresh weight (mg) | 76.9±23.8 | 76.1±27.4 | 89.6±31.9 | 126.3±74.7 |
| Root fresh weight (mg) | 65.8±32.4 | 49.4±25.9 | 85.2±46.2 | 90.0±81.1 |
| SPAD | 21.3±4.1 | 25.6±6.7* | 22.3±4.3 | 30.3±5.0** |
| Nodule number | n.d. | 10.1±4.9 | n.d. | 11.1±8.6 |
| Nodule weight (mg) | n.d. | 11.7±15.7 | n.d. | 10.2±8.2 |
| ARA (C2H4 nmol/h/plant) | n.d. | 19.6±14.3 | n.d. | 14.7±15.1 |
Values are means±SD of three replicate experiments (n=6 in each experiment). *and** represent significant difference at p<0.05 and p<0.01 between J8(−) and J8(+) in each nitrogen concentration.
Effect of inoculation with Mesorhizobium sp. J8 on the expression of CYP88D6 in G. uralensis plants grown from seeds
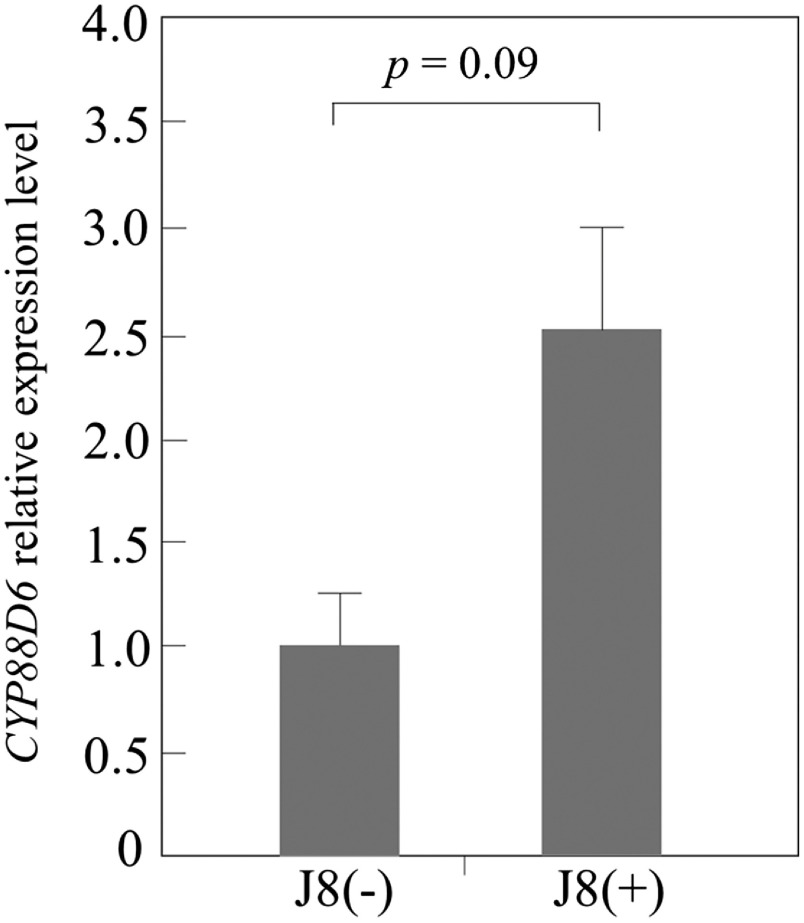
It appears that nodulation and growth of inoculated plants supplemented with 100 µM KNO3 were improved in closed system conditions (Table 2). Although we tried to compare GL content between inoculated and uninoculated plants, it was so low that it was difficult to measure accurately. Therefore, we measured the expression level of CYP88D6 by qRT-PCR. As shown in Figure 9, that in J8(+) plants was 2.5 times that in the control. This result supports the proposal that inoculation with J8 enhanced GL production (Figure 9).
Figure 9. Expression level of CYP88D6 gene in root tissue of J8 uninoculated plants (J8(−)) and inoculated plants (J8(+)) prepared from seeds grown in soil containing 100 µM KNO3, by qRT-PCR. 18S ribosomal RNA gene was used as an internal control. The expression data represent the mean±SD of 5 independent plants.
Discussion
We isolated a strain of Mesorhizobium from G. glabra that can establish symbiosis with G. uralensis. We conducted a genome analysis of the rhizobium and examined its effect on growth and GL production of G. uralensis. Several papers have reported on symbiotic endobacteria isolated from root nodules of G. uralensis. For example, Mesorhizobium sp. strain CCNWGX022 was isolated from arid and semi-arid regions of China (Wei et al. 2007). Mesorhizobium kowhai strain Ach-343 isolated from Astragalus chorinensis Bunge and M. japonicum strain Opo-235 isolated from Oxytropis popoviana Peschkova in Buryatia in the Lake Baikal region of Russia were able to establish symbiosis with G. uralensis (Safronova et al. 2019).
Although there are several reports on the isolation of bacteria from nodules of G. uralensis as mentioned, only some studies tested the ability to form root nodules. For example, a total of 159 strains of endophytic bacteria were isolated from surface-sterilized root nodules of wild Glycyrrhiza spp. growing at 40 sites in central and northwestern China. Not all bacteria formed nodules, and some strains were suspected to play a role in assisting symbiotic interactions or promoting plant growth (Li et al. 2012).
The genome size of Mesorhizobium sp. J8 is 6.69 Mbp, comparable to that of M. japonicum MAFF303099, at 7.04 Mb. Although MAFF303099 has two plasmids, named pMLa (0.35 Mb) and pMLb (0.21 Mb) (Kaneko et al. 2000), J8 does not contain any plasmids. Rhizobia can be classified into two types depending on whether the symbiosis region, in which the genes essential for symbiotic N2 fixation are concentrated, is a symbiosis island or a symbiosis plasmid. J8 has a symbiotic island, as M. japonicum MAFF303099 and Bradyrhizobium diazoefficiens USDA 110 have (Kaneko et al. 2000, 2002). Moreover, the size of the symbiotic island in J8 is closer to that of M. japonicum MAFF303099 than to that of Mesorhizobium sp. M2A.F.Ca.ET.043.02.1.1, isolated from C. arietinum, and the arrangement of the nodulation related gene cluster, nitrogen-fixation related gene clusters and an island transfer-related gene cluster in J8 is more similar to that of the latter strain than to that of MAFF303099 (Figure 5). Although the gene that is related to enhanced GL production in J8 is not yet identified, screening for such genes could be performed using gene disruption libraries or such as STM (signature-tagged mutagenesis) library of MAFF303099 (Shimoda et al. 2008).
In the nodulation test using plants grown from cuttings, growth was measured 90 days after inoculation with J8. The positive effect of J8 inoculation could be detected in the amount of shoot biomass (Figure 6D). Inoculation also increased N2 fixation (p=0.08) (Figure 6I), which was reflected in significantly higher SPAD values (p=0.02) (Figure 6C). However, small nodules were formed even on uninoculated plants grown from cuttings (Figure 6G). We repeated these experiments several times, but each time uninoculated plants showed slight root nodule formation. Therefore, we conducted nodulation tests using plant boxes to eliminate contamination (closed system). Initially, we grew plants for 90 days in the boxes. However, because the boxes appeared to impose a size constraint that impaired plant growth, we subsequently collected data at 60 days after inoculation. As expected, J8(−) plants did not form nodules in the closed system, allowing us to clearly determine the effects of nodule formation on plant growth. Shoot biomass was slightly higher in J8(+) plants grown in soil containing N (100 µM KNO3, p=0.07), and no difference was observed between J8(+) and J8(−) plants grown in soil without N (Table 2). On the other hand, root biomass production was tended to inhibited in J8(+) plants grown in soil without N (p=0.1). This suggests that inoculation with J8 slightly inhibits root biomass production at least in the early stages of growth in soil without N. However, in soil containing 100 µM KNO3, inoculation with J8 did not inhibit root biomass production. Therefore, it appears that in a closed system, providing 100 µM KNO3 promotes growth of G. uralensis when inoculated with J8. No significant difference was detected in the number of root nodules formed at any N concentration. Furthermore, there was no significant difference in nodule weight in soil containing 0 or 100 µM KNO3. N2 fixation activity was detected at each concentration, but was higher in soil without N. The SPAD value was significantly increased by inoculation with J8 in soil containing 0 or 100 µM KNO3 (Table 2).
The expression level of CYP88D6, which is correlated with GL content, in the roots of J8(+) G. uralensis grown from seeds grown in soil containing 100 µM KNO3 was 2.5 times that in the controls (Figure 9). Also, J8(+) cuttings grown for 90 days tended to have a higher GL content than the control (p=0.07), as well as increased expression of CYP88D6 (p=0.1) (Figures 7, 8). These results indicate that inoculated plants had enhanced GL production. The GL content per unit root weight in J8(+) plants was about 3.2 times that in the control, and that per plant was also about 2.8 times as high in J8(+) plants (Figure 8).
Various methods to induce rapid root biomass production have been tried in Glycyrrhiza cultivation. For example, Itoh et al. (2018) cultivated G. uralensis in tubes for complete harvest of stolons. Sato et al. (2004) examined the relation between the concentration of nutrients applied and GL content by culturing G. glabra in a hydroponic system. In general, these methods do not stress the plants. However, the production of secondary metabolites such as terpenoids tends to be activated when plants are under stress (Isah 2019). The rhizobium inoculation method used here ensures synthesis of secondary metabolites such as GL by applying the biological stress of rhizobium inoculation while promoting plant growth.
Here, we inoculated G. uralensis with rhizobia. It would also be worthwhile to evaluate the effects of inoculation with mycorrhizal fungi, which infect plant roots and supply phosphorus and water. There are several reports the effects of inoculation with mycorrhizal fungi on Glycyrrhiza spp. (Chen et al. 2017; Xie et al. 2018). For example, Orujei et al. (2013) used two arbuscular mycorrhizal species (Glomus mosseae and G. intraradices) to evaluate the effects of inoculation on G. glabra growth and secondary metabolite production. The GL content of G. glabra increased 3 to 9 times compared with the control at 3 and 6 months after inoculation. Since many leguminous plants are simultaneously colonized by both rhizobia and mycorrhizal fungi (Sakamoto et al. 2013, 2019), it would be worthwhile to evaluate the effect of co-inoculation on GL production. Future work should investigate the long-term effects of inoculation with J8 on G. uralensis, because here it was grown for at most 90 days.
Acknowledgments
This work was supported by Grants-in-Aid for Challenging Exploratory Research (No. 19K22314).
Abbreviations
- ARA
acetylene reduction assay
- GL
glycyrrhizin
- SPAD
soil plant analysis development
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
Accession numbers
Nucleotide sequence data reported in this study are available in the DDBJ/EMBL/GenBank database under the following accession number: AP024109.
References
- Broughton WJ, Dilworth MJ (1971) Control of leghemoglobin synthesis in snake beans. Biochem J 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Yang G, Sheng Y, Li P, Qiu H, Zhou X, Huang L, Chao Z (2017) Glomus mosseae inoculation improves the root system architecture, photosynthetic efficiency and flavonoids accumulation of liquorice under nutrient stress. Front Plant Sci 8: 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WF, Guan SH, Zhao CT, Yan XR, Man CX, Wang ET, Chen WX (2008) Different Mesorhizobium species associated with Caragana carry similar symbiotic genes and have common host ranges. FEMS Microbiol Lett 283: 203–209 [DOI] [PubMed] [Google Scholar]
- Egamberdieva D, Li L, Lindström K, Räsänen LA (2016) A synergistic interaction between salt-tolerant Pseudomonas and Mesorhizobium strains improves growth and symbiotic performance of liquorice (Glycyrrhiza uralensis Fish.) under salt stress. Appl Microbiol Biotechnol 100: 2829–2841 [DOI] [PubMed] [Google Scholar]
- Hayashi H, Sudo H (2009) Economic importance of licorice. Plant Biotechnol 26: 101–104 [Google Scholar]
- Isah T (2019) Stress and defense responses in plant secondary metabolites production. Biol Res 52: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Yaginuma A, Nakadate S (2018) Characteristic of Stolons of Glycyrrhiza uralensis Cultivated in Tubes. Shouyakugaku Zasshi 72: 87–89 (in Japanese) [Google Scholar]
- Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, et al. (2000) Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res 7: 331–338 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchimi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K, et al. (2002) Complete genome sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res 9: 189–197 [DOI] [PubMed] [Google Scholar]
- Keele BB Jr, Hamilton PB, Elkan GH (1969) Glucose catabolism in Rhizobium japonicum. J Bacteriol 97: 1184–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojoma M, Kohda H, Tani N, Ashida K, Sugino M, Yamamoto A, Horikoshi T (1995) In vitro propagation from axillary buds of Glycyrrhiza glabra L. Plant Tissue Culture Letters 12: 145–149 [Google Scholar]
- Kojoma M, Ohyama K, Seki H, Hiraoka Y, Asazu NS, Sawa S, Sekizaki H, Yoshida S, Muranaka T (2010) In vitro proliferation and triterpenoid characteristics of licorice (Glycyrrhiza uralensis Fischer, Leguminosae) stolons. Plant Biotechnol 27: 59–66 [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL (2004) Versatile and open software for comparing large genomes. Genome Biol 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano G, Shibano M, Watanabe H, Ozaki K (2003) Pharmaceutical botanical studies on some Glycyrrhiza species. Pharmaceut Soc Jpn (Yakugaku Zasshi) 123: 619–631 (in Japanese) [DOI] [PubMed] [Google Scholar]
- Li L, Sinkko H, Montonen L, Wei G, Lindström K, Räsänen LA (2012) Biogeography of symbiotic and other endophytic bacteria isolated from medical Glycyrrhiza species in China. FEMS Microbiol Ecol 79: 46–68 [DOI] [PubMed] [Google Scholar]
- Mylona P, Pawlowski K, Bisseling T (1995) Symbiotic nitrogen fixation. Plant Cell 7: 869–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M (2008) Genome Matcher: A graphical user interface for DNA sequence comparison. BMC Bioinformatics 9: 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama A, Kurokawa K, Enai K, Saitoh H, Kanaya S, Altaf-UI-Amin M, Ogasawara N (2006) Bioinformatics tool for genomic era: A step towards the in silico experiments: Focused on molecular cloning. J Comput Aid Chem 7: 102–115 [Google Scholar]
- Orujei Y, Shabani L, Sharifi-Tehrani M (2013) Induction of glycyrrhizin and total phenolic compound production in licorice by using arbuscular mycorrhizal fungi. Russ J Plant Physiol 60: 855–860 [Google Scholar]
- Ozaki K, Shibano M (2014) Aim for production of Glycyrrhizae Radix in Japan (3): Development of a new licorice cultivar. J Nat Med 68: 358–362 [DOI] [PubMed] [Google Scholar]
- Saeki K, Kouchi H (2000) The lotus symbiont, Mesorhizobium loti: Molecular genetic techniques and application. J Plant Res 113: 457–465 [Google Scholar]
- Safronova V, Belimov A, Sazanova A, Chirak E, Kuznetsova I, Andronov E, Pinaev A, Tsyganova A, Seliverstova E, Kitaeva A, et al. (2019) Two broad host range rhizobial strains isolated from relict legumes have various complementary effects on symbiotic parameters of co-inoculated plants. Front Microbiol 10: 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Ogiwara N, Kaji T (2013) Involvement of autoregulation in the interaction between rhizobial nodulation and AM fungal colonization in soybean roots. Biol Fertil Soils 49: 1141–1152 [Google Scholar]
- Sakamoto K, Ogiwara N, Kaji T, Sugimoto Y, Ueno M, Sonoda M, Matsui A, Ishida J, Tanaka M, Totoki Y, et al. (2019) Transcriptome analysis of soybean (Glycine max) root genes differentially expressed in rhizobial, arbuscular mycorrhizal, and dual symbiosis. J Plant Res 132: 541–568 [DOI] [PubMed] [Google Scholar]
- Sato S, Ikeda H, Furukawa H, Murata Y, Tomoda M (2004) The effects of nutrient solution concentration on inorganic and glycyrrhizin contents of Glycyrrhiza glabra linn. Pharmaceut Soc Jpn (Yakugaku Zasshi) 124: 705–709 (in Japanese) [DOI] [PubMed] [Google Scholar]
- Seki H, Ohyama K, Sawai S, Mizutani M, Ohnishi T, Sudo H, Akashi T, Aoki T, Saito K, Muranaka T (2008) Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc Natl Acad Sci USA 105: 14204–14209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabani L, Ehsanpour AA, Esmaeili A (2010) Assessment of squalene synthase and beta-amyrin synthase gene expression in licorice roots treated with methyl jasmonate and salicylic acid using real-time qPCR. Russ J Plant Physiol 57: 480–484 [Google Scholar]
- Shimoda Y, Mitsui H, Kamimatsuse H, Minamisawa K, Nishiyama E, Ohtsubo Y, Nagata Y, Tsuda M, Shinpo S, Watanabe A, et al. (2008) Construction of signature-tagged mutant library in Mesorhizobium loti as a powerful tool for functional genomics. DNA Res 15: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath P, Sokal R (1973) Numerical Taxonomy. Freeman, San Francisco
- Tang ZH, Li T, Tong YG, Chen XJ, Chen XP, Wang YT, Lu JJ (2015) A systematic review of the anticancer properties of compounds isolated from licorice (Gancao). Planta Med 81: 1670–1687 [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV (2001) The COG database: New development in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29: 22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JM (1970) A Manual for the Practical Study of Root Nodule Bacteria. Blackwell Scientific Publications, Oxford
- Wei GH, Yang XY, Zhang JW, Gao JM, Ma YQ, Fu YY, Wang P (2007) Rhizobialide: A new stearolactone produced by Mesorhizobium sp. CCNWGX022, a rhizobial endophyte from Glycyrrhiza uralensis. Chem Biodivers 4: 893–898 [DOI] [PubMed] [Google Scholar]
- Xie W, Hao Z, Zhou X, Jiang X, Xu L, Wu S, Zhao A, Zhang X, Chen B (2018) Arbuscular mycorrhiza facilitates the accumulation of glycyrrhizin and liquiritin in Glycyrrhiza uralensis under drought stress. Mycorrhiza 28: 285–300 [DOI] [PubMed] [Google Scholar]
- Zheng H, Zhong Z, Lai X, Chen WX, Li S, Zhu J (2006) A LuxR/LuxI-type quorum-sensing system in a plant bacterium, Mesorhizobium tianshanense, controls symbiotic nodulation. J Bacteriol 188: 1943–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]