Abstract
Tick-borne zoonotic pathogens are increasingly becoming important across the world. In sub-Saharan Africa, tick-borne pathogens identified include viruses, bacteria and protozoa, with Rickettsia being the most frequently reported. This study was conducted to screen and identify Rickettsia species in ticks (Family Ixodidae) infesting livestock in selected districts of southern Zambia. A total of 236 ticks from three different genera (Amblyomma, Hyalomma, and Rhipicephalus) were collected over 14 months (May 2018–July 2019) and were subsequently screened for the presence of Rickettsia pathogens based on PCR amplification targeting the outer membrane protein B (ompB). An overall Rickettsia prevalence of 18.6% (44/236) was recorded. Multi-locus sequencing and phylogenetic characterization based on the ompB, ompA, 16S rRNA and citrate synthase (gltA) genes revealed the presence of Rickettsia africae (R. africae), R. aeschlimannii-like species and unidentified Rickettsia species. While R. aeschlimannii-like species are being reported for the first time in Zambia, R. africae has been reported previously, with our results showing a wider distribution of the bacteria in the country. Our study reveals the potential risk of human infection by zoonotic Rickettsia species and highlights the need for increased awareness of these infections in Zambia's public health systems.
Keywords: Rickettsia, ticks, tick-borne zoonoses, Zambia, febrile illness
Introduction
Vector–borne zoonotic pathogens are of increasing importance worldwide, with many reports of emerging and/or re-emerging pathogens being detected in invertebrate hosts (1). This increase in reports has been attributed to factors such as climate change, land-use changes as well as various anthropogenic activities (2). Amongst the emerging and re-emerging vector-borne pathogens, are those which are transmitted by ticks (3).
Ticks are considered to be amongst the main vectors of zoonotic pathogens (4), and rank only second to mosquitoes in terms of importance as vectors of human pathogens (5, 6). Ticks serve as reservoirs, vectors or amplifying hosts for a variety of pathogens, with a number of these ticks reportedly infesting humans (7–12). Tick-borne zoonoses are emerging across the world (13–15), with their public health impact being on the rise in tropical and subtropical regions (3, 16–18).
Within southern Africa, several human infections by tick-borne zoonotic pathogens have been reported. These include viral (Crimean-Congo hemorrhagic fever), bacterial (Borrelia duttonii, Anaplasma phagocytophilum, Ehrlichia ruminantium, Ehrlichia canis, Rickettsia africae, Rickettsia aeschlimannii, and Rickettsia conorii) and protozoal (Babesia microti) infections (19). The most commonly reported tick-borne pathogen in southern Africa is the obligate intracelluar bacteria of the Rickettsia genus (spotted fever group—SFG), which cause febrile illnesses in humans (20, 21). Across Africa, more than 10 SFG Rickettsiae have been reported in ticks, humans and animals (21). Despite these reports of the pathogens in Africa, the diseases they cause are still neglected (22).
Many human rickettsial infections have been reported within southern Africa, mostly in tourists. Countries within the region which have reported active human infections include South Africa (23–34), Botswana (35), Mozambique (36), and Zimbabwe (37, 38). Although there have been no confirmed clinical cases reported in Zambia, serological evidence of human infection exists (39). Furthermore, infection by rickettsial pathogens has been reported in non-human primates in the country (40). However, information on rickettsial pathogens in ticks is very limited. Presently, to our knowledge, there are only two published reports of Rickettsia in Zambian ticks: one from a tick survey (41), and the other from a tick collected from an exported reptile (42). Considering that ticks play a significant role in the epidemiology of these pathogens, it is essential that they are surveyed to assess the presence of pathogens, which can inform on the potential risk for human infections. Therefore, this study sought to screen and to phylogenetically characterize rickettsial pathogens from ticks collected from cattle in southern Zambia.
Materials and Methods
Tick Collection, Identification, and DNA Extraction
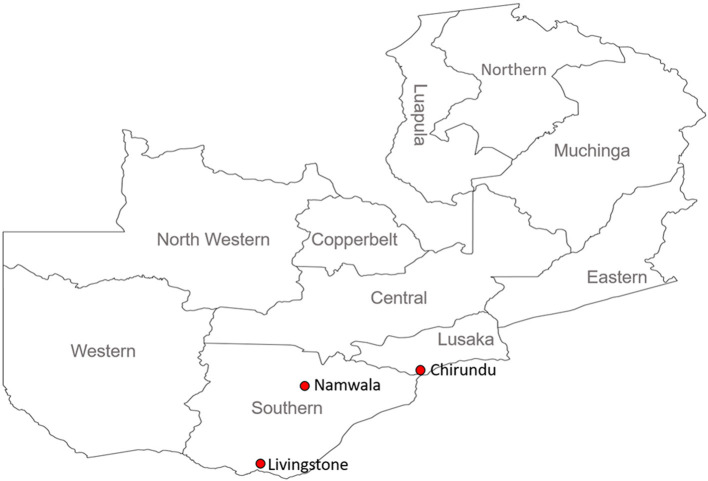
During May 2018 to July 2019, Ixodid ticks were collected from cattle in three districts in the southern part of Zambia (Chirundu, Namwala, and Livingstone; Figure 1). The sampling areas were purposively chosen because the southern region has the highest livestock density in the country, with the majority of the people practising subsistence farming. There is therefore close interaction between the people and the animals. Cattle sampling was chosen as it widely reared and ensured easy collection of the tick samples. The ticks were placed in aerated tubes provided with moisture and transported live to the laboratory for morphological identification using identification keys (43). After morphological identification, the ticks were stored at −80°C awaiting further analysis. For molecular analysis, individual ticks were sterilized by washing briefly in 70% ethanol after which they were washed in phosphate buffered saline (PBS) and transferred into homogenizing tubes containing 200 μL of Dulbecco's modified eagle medium (DMEM) (Sigma–Aldrich®–USA). The ticks were then homogenized in a MicroSmash™ MS-100R homogenizer (TOMY Digital Biology Co., Ltd., Japan), after which the homogenate was centrifuged, with the supernatant separated into a clean micro-centrifuge tube. DNA was extracted from the tick homogenate using the TRI Reagent® protocol (Sigma–Aldrich, USA) according to the manufacturer's recommendation. The extracted DNA was used in subsequent polymerase chain reactions (PCR) to screen for the presence of rickettsial DNA as well as sequencing.
Figure 1.
Map of Zambia showing the study sites (Red dots) in southern Zambia. Sampling sites covered Lusaka (Chirundu) and Southern Provinces (Namwala and Livingstone).
Molecular Screening and Phylogenetic Analysis of Rickettsia
For initial PCR screening, OneTaq® Quick-Load® 2X Master Mix with Standard Buffer (New England BioLabs® Inc., USA) was used to amplify a 429-bp region of the outer membrane protein B (ompB), with cyclic conditions used previously (44). A negative control (using nuclease–free water in place of template DNA) was included in the PCR assays. The PCR primer pairs, expected amplicon size in base pairs (bp) and annealing temperatures of the assays are as shown in Supplementary Table 1. PCR products were electrophoresed on ethidium bromide stained 1.5% agarose gel and then visualized under ultraviolet (UV) light.
For sequencing, all samples positive on the ompB gene were purified using Wizard® SV Gel and Clean-Up System (Promega, Madison, WI, USA). Bidirectional Sanger sequencing was conducted with the purified DNA as a template using Brilliant Dye™ Terminator Cycle Sequencing Kit v3.1 (NimaGen®) according to the manufacturer's protocol. Nucleotide sequences were assembled and edited using GENETYX ATGC software version 7.5.1 (GENETYX Corporation, Tokyo, Japan). For phylogenetic analysis, reference sequences were retrieved from GenBank and aligned along with those determined in this study using ClustalX2. Phylogenetic trees were constructed in MEGA version 6.0 (45) using the Maximum Likelihood method based on the Tamura 3-parameter model and topological support was assessed using the bootstrap method with 1,000 replicates as a confidence interval.
Based on the results from phylogenetic analysis of the ompB, representative samples from each identified species were selected for amplification of a 532-bp fragment of the ompA, a 985-bp fragment of the 16S ribosomal ribonucleic acid (16S rRNA) and a 589-bp fragment of the citrate synthase (gltA) genes for further confimation of the species identity. The amplified segments were subsequently sequenced and phylogenetically analyzed as described for the ompB gene. All the primers used in this study are shown in Supplementary Table 1. The nucleotide sequences obtained in this study have been deposited in the GenBank with accession numbers LC565644–LC565678, LC565679–LC565695, LC565696–LC565701, and LC565702–LC565706 for ompB, ompA, gltA, and 16S rRNA genes, respectively.
Results
Tick Collection and Prevalence of Infection
A total of 236 engorged and semi-engorged adult ticks were collected (Chirundu-36, Namwala-100, and Livingstone-100). On morphological identification, these included Amblyomma spp. (19), Hyalomma spp. (99), and Rhipicephalus spp. (118). The distribution of tick genera in the study areas is shown in Table 1.
Table 1.
Number of ticks collected and the prevalence of infection in the different sampling areas by genus.
| Tick genus | Number of ticks analyzed (Prevalence) | Total number of ticks (Prevalence) | ||
|---|---|---|---|---|
| Chirundu | Namwala | Livingstone | ||
| Amblyomma sp. | 1 (0%) | 14 (28.6%) | 4 (75%) | 19 (36.8%) |
| Hyalomma sp. | 31 (9.7%) | 0 (0%) | 68 (42.6%) | 99 (31.3%) |
| Rhipicephalus sp. | 4 (0%) | 86 (5.8%) | 28 | 118 (5.8%) |
| Total | 36 (8.3%) | 100 (9%) | 100 (32%) | 236 (18.6%) |
Molecular Detection of Rickettsia
Based on the initial screening targeting the ompB gene, the observed overall prevalence of Rickettsia in the ticks was 18.6% (44/236). All the sampled tick genera were found to harbor Rickettsia pathogens, with all sampling areas reporting a different prevalence of infection (Table 1).
Rickettsia Species Identification
Rickettsia specific ompB nucleotide sequences obtained from all the 44 rickettsia-positive tick samples were compared to sequences in GenBank using the basic local alignment search tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast.cgi) (46). The sequences showed high similarity to R. africae (40.9%; 18/44), R. aeschlimannii (52.3%; 23/44), and R. parkeri (6.8%; 3/44), with sequence identity ranging from 98.3 to 99.77% (Supplementary Table 2).
On BLAST analysis of selected ompA sequences, there was an agreement between the ompA and ompB analysis on all samples which on the latter gene had shown close sequence similarity to R. africae (N383, N384, N385, N386, and N387), with 100% sequence similarity to R. africae KZN26 detected in South Africa (accession no. MH751466). The samples which had shown high sequence similarity to R. parkeri on ompB analysis (CT36, CT40, and CT43), showed 100% sequence similarity to R. africae which was found in India [accession no. MK905242]. Meanwhile, samples which had shown close sequence similarity to R. aeschlimannii on ompB analysis (N320, N323, N330, N345, N349, N356, N358, and N377), showed high sequence similarity (99.81–100%) to a Rickettsia endosymbiont from Turkey (accession no. KT279888), with the exception of sample N381 which showed 100% sequence similarity to R. aeschlimannii from Turkey [accession no. MG920562] (Supplementary Table 3).
BLAST analysis on gltA sequences showed agreement with the ompB sequence identity for R. aeschlimannii (N381), displaying 100% sequence similarity R. aeschlimannii from China [MH267736]. Meanwhile, CT36 which had shown high sequence similarity to R. parkeri on the ompB analysis, showed high sequence similarity to R. africae detected in Egypt [accession no. HQ335126] (99.87% identity) and those that had shown close sequence similarity to R. africae on ompB analysis (N28, N383, and N385), displayed 99.61% sequence similarity to Rickettsia sp. Identified in Slovakia [accession no. HM538186] (Table 2).
Table 2.
Rickettsia identity based on the gltA gene.
| Area | Tick species | DNA ID | Reference strains of Rickettsia species (GenBank Accession Number) | Nucleotide percent identity (%) |
|---|---|---|---|---|
| Chirundu | Hyalomma spp. | CT36 | R. africae Egypt (HQ335126) | 99.87 |
| Namwala | Amblyomma spp. | N28 | Rickettsia sp. Slovakia (HM538186) | 99.74 |
| Namwala | Rhipicephalus spp. | N61 | Rickettsia sp. Slovakia (HM538186) | 99.61 |
| Livingstone | Hyalomma spp. | N381 | R. aeschlimannii China (MH267736) | 100 |
| Livingstone | Amblyomma spp. | N383 | Rickettsia sp. Slovakia (HM538186) | 99.87 |
| Livingstone | Hyalomma spp. | N385 | Rickettsia sp. Slovakia (HM538186) | 99.61 |
The 16S rRNA gene BLAST sequence analysis results revealed agreement with the findings of ompB analysis for samples with close sequence similarity to R. africae (N61, N383) and R. aeschlimannii (N323, N381) with sequence identity ranging from 99.78 to 100% to those detected in Ethiopia [L36098] and China [MH923218], respectively. Meanwhile, the sample that had shown close sequence similarity to R. parkeri on ompB analysis (CT36), showed highest sequence similarity to an uncultured Rickettsia sp. Strain Bel-4109 (MH618379) from Serbia (Table 3).
Table 3.
Rickettsia identity based on the 16S rRNA gene.
| Area | Tick species | DNA ID | Reference strains of Rickettsia species (GenBank accession number) | Nucleotide percent identity (%) |
|---|---|---|---|---|
| Chirundu | Hyalomma spp. | CT36 | Rickettsia sp. Strain Bel–4109 (MH618379) | 100 |
| Namwala | Rhipicephalus spp. | N61 | R. africae Ethiopia (L36098) | 100 |
| Livingstone | Hyalomma spp. | N323 | R. aeschlimannii China (MH923218) | 100 |
| Livingstone | Hyalomma spp. | N381 | R. aeschlimannii China (MH923218) | 100 |
| Livingstone | Amblyomma spp. | N383 | R. africae Ethiopia (L36098) | 99.78 |
Phylogenetic Analysis
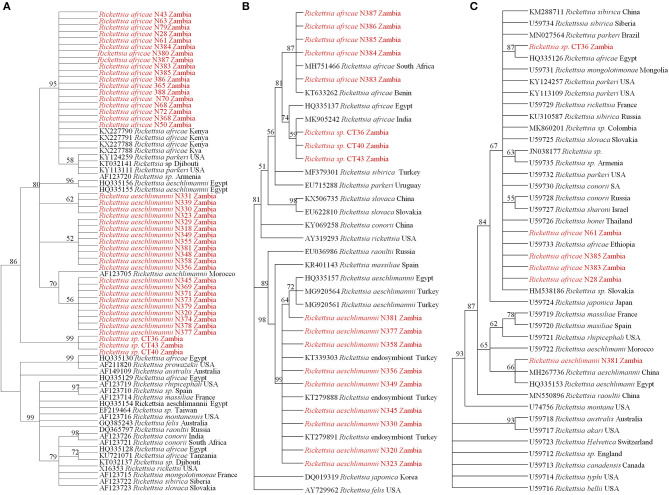
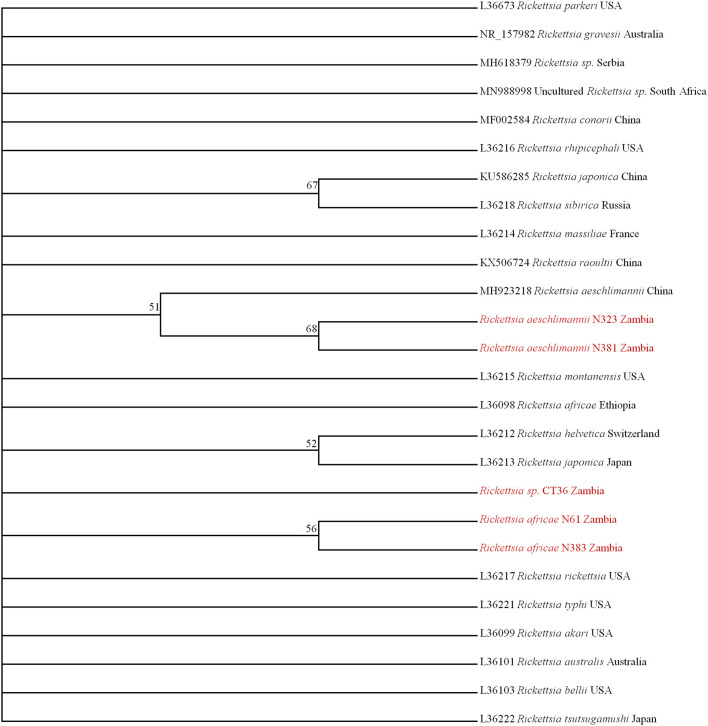
On phylogenetic analysis based on the ompB sequence, the Zambian sequences under study formed three distinct clusters, namely R. africae, R. aeschlimannii, and Rickettsia sp. (Figure 2A). It was noteworthy that the Zambian Rickettsia sp. did not cluster with any of the reference sequences from GenBank. Phylogenetic analysis based on the ompA sequence showed Zambian sequences forming two clusters; one grouping with R. africae and another clustering with R. aeschlimannii and Rickettsia endosymbiont (Figure 2B). In addition, phylogenetic analysis of selected samples, based on the gltA sequence showed that Zambian samples clustered with R. africae (CT36, N61, N385, N383, and N28) and R. aeschlimannii (N381) (Figure 2C). Based on 16S rRNA gene sequences, phylogenetic analysis of the Zambian samples clustered with R. africae (N61, N383), R. aeschlimannii (N323, N381) with CT36 not clustering with any reference sequences from GenBank (Figure 3).
Figure 2.
Phylogenetic relationships of the ompB (A), ompA (B), and gltA (C) genes of Rickettsiae detected from ticks in Zambia. Phylogenetic analysis was based on 397, 460, and 596 bp of the ompB, ompA, and gltA genes, respectively. Numbers at branch nodes indicate bootstrap values ≥50%. The reference sequences included in the analyses are shown by their GenBank accession number, species, and country of origin. The Rickettsiae characterized in the present study are in red text.
Figure 3.
Phylogenetic relationships of the 16S rRNA genes of rickettsiae detected in ticks in Zambia. Phylogenetic analysis was based on 795 bp of the 16S rRNA gene. Numbers at branch nodes indicate bootstrap values ≥50%. The reference sequences included in the analyses are shown by their GenBank accession number, species, and country of origin. The Rickettsiae characterized in the present study are in red text.
Taken together, based on nucleotide sequence similarities as determined by BLAST analysis and phylogenetic analysis of four different genes, there were three distinct Rickettsia species identified, namely R. africae, R. aeschlimannii-like species and Rickettsia sp.
Distribution of Rickettsia Species by Sampling Area and Tick Genera
By area of sampling, ticks from Chirundu were infected with Rickettsia sp. (3/3), those from Namwala were infected by R. africae only (9/9), whilst those from Livingstone were infected by R. africae (9/32) and R. aeschlimannii-like species (23/32)]. By tick genera, Amblyomma were infected with R. africae (6/7) and R. aeschlimannii-like species (1/7), Hyalomma were infected with R. aeschlimannii-like species (22/32), R. africae (7/32), and unidentified Rickettsia sp. (3/32), with Rhipicephalus ticks being found only with R. africae (5/5).
Discussion
This study found an overall Rickettsia prevalence of 18.6% in the ticks sampled from the southern part of Zambia. The previous report (41) on Rickettsia prevalence in ticks in Zambia showed a much lower prevalence of 4.6% and the difference in prevalence between the two studies could be attributed to the differences in the proportions of ticks sampled. The study conducted in eastern Zambia by Chitimia-Dobler et al. (41) was predominated by ticks of the Rhipicephalus genus (96% Rhipicephalus, 4% Amblyomma, and Hyalomma), which are considered to always have lower rickettsial infections (41). In contrast, our study had relatively more Amblyomma and Hyalomma species (50% Amblyomma and Hyalomma, 50% Rhipicephalus), tick species considered amongst the principal Rickettsia vectors in the region (21). Indeed, our findings on infection rates in the three tick genera confirm the observations by Parola et al. (21) that Amblyomma and Hyalomma have the highest infection rates of Rickettsia, with Rhipicephalus being amongst those with low infection rates (21). However, since we collected ticks from cattle, our finding of Rickettsia pathogens in the tick species cannot be conclusively used as an indicator of their potential vector role. This is because the identified pathogens could have been picked from cattle during feeding.
In this study, we report for the first time the presence of R. africae and R. aeschlimannii-like species in Ixodid ticks from the southern part of Zambia. Whilst R. africae has been reported before in ticks from the eastern part of Zambia (41, 42), this study reports for the first time the presence of R. aeschlimannii-like species in the country, adding to the number of SFG Rickettsiae which are reported in Zambia. The previously reported SFG Rickettsiae include R. africae, R. massiliae, R. conorii and R. felis (40–42, 47). To our knowledge, Zambia becomes the third country in southern Africa to report the presence of R. aeschlimannii-like species, with previous reports being in South Africa (48) and Zimbabwe (49). We also report unidentified Rickettsia sp. which showed distinct phylogenetic clustering pattern based on the four genes analyzed in this study, and thus could not be assigned a specific species. This observation further emphasizes the recommendations by Fournier et al. (50) to use multiple genes to ensure proper speciation of Rickettsia species. Alternatively, speciation could be achieved by sequencing the entire full lengths of several diagnostic genes. For example, Moron et al. (51) sequenced the full length ompB gene (~6,000 bp) to phylogenetically compare R. felis with its homologs in spotted fever group (SFG) and typhus group (TG) Rickettsiae.
Our findings add to available data on the prevalence of rickettsial pathogens in ticks within the southern African region. Other countries in the region which have reports of tick surveys for Rickettsia infection include Zimbabwe (49), Botswana (52), Mozambique (53), and South Africa (54–58) with prevalence ranging from as low as 3% to as high as 77%. This study also adds to the growing data on tick-borne (41, 42, 59–61) and tick-associated (62, 63) zoonotic pathogens within Zambia. It further highlights the potential threat posed by tick-borne pathogens to human health and the need to strengthen surveillance of tick-borne diseases in the country.
In conclusion, we have shown the presence of zoonotic SFG Rickettsiae in ticks from the southern part of Zambia, indicating the wider geographical spread of these pathogens within the country and the potential threat they pose to human health. It is therefore important to screen for these pathogens in patients reporting with febrile illnesses of unknown origin. We also report on the presence of R. aeschlimannii-like species in Zambia, an indication of the expanded geographical spread of this pathogen within the southern African region.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by ERES Converge (IRB - 00005948, FWA - 00011697), under reference number 2017-Jul-021. Written informed consent for participation was not obtained from the owners because verbal consent was sought for collection of ticks from the animals.
Author Contributions
SC and ES conceived the study. SC, KChi, KCha, and ES developed the laboratory methodology. KChi and KS conducted most of the laboratory experiments. SC, KN, KCha, and ES supervised the work. SC, KCha, and ES wrote the original draft of the manuscript. SC acquired funding for the study. All authors were involved in data analysis, reviewing, and approving the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge the support from cattle owners and community leaders who facilitated our sample collections in their respective localities.
The content is solely the responsibility of the authors and does not necessarily represent the views of the National Institutes of Health.
Footnotes
Funding. This research work was supported by a grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health Spatial eco-epidemiology of tick-borne rickettsial pathogens under award number R01AI136035 (PI—Gaff H), through a sub-award to the University of Zambia (PI—SC).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.684487/full#supplementary-material
References
- 1.Gubler DJ. Vector-borne diseases. OIE Rev Sci Tech. (2009) 28:583–8. 10.20506/rst.28.2.1904 [DOI] [PubMed] [Google Scholar]
- 2.Jánová E. Emerging and threatening vector-borne zoonoses in the world and in Europe: a brief update. Pathog Glob Health. (2019) 13:49–57. 10.1080/20477724.2019.1598127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estrada-Peña A, Ayllón N, de la Fuente J. Impact of climate trends on tick-borne pathogen transmission. Front Physiol. (2012) 3:64. 10.3389/fphys.2012.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim CM, Yi YH, Yu DH, Lee MJ, Cho MR, Desai AR, et al. Tick-borne rickettsial pathogens in ticks and small mammals in Korea. Appl Environ Microbiol. (2006) 72:5766–76. 10.1128/AEM.00431-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De La Fuente J, Estrada-Pena A, Venzal JM, Kocan KM, Sonenshine DE. Overview: ticks as vectors of pathogens that cause disease in humans and animals. Front Biosci. (2008) 13:6938–46. 10.2741/3200 [DOI] [PubMed] [Google Scholar]
- 6.Yu Z, Wang H, Wang T, Sun W, Yang X, Liu J. Tick-borne pathogens and the vector potential of ticks in China. Parasit Vectors. (2015) 8:24. 10.1186/s13071-014-0628-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bursali A, Tekin S, Keskin A, Ekici M, Dundar E. Species diversity of Ixodid Ticks feeding on humans in Amasya, Turkey: seasonal abundance and presence of crimean-congo hemorrhagic fever virus. J Med Entomol. (2011) 48:85–93. 10.1603/ME10034 [DOI] [PubMed] [Google Scholar]
- 8.Bursali A, Tekin S, Orhan M, Keskin A, Ozkan M. Ixodid ticks (Acari: Ixodidae) infesting humans in Tokat Province of Turkey: species diversity and seasonal activity. J Vector Ecol. (2010) 35:180–6. 10.1111/j.1948-7134.2010.00075.x [DOI] [PubMed] [Google Scholar]
- 9.Bursali A, Keskin A, Tekin S. Ticks (Acari: Ixodida) infesting humans in the provinces of Kelkit Valley, a Crimean-congo hemorrhagic fever endemic region in Turkey. Exp Appl Acarol. (2013) 59:507–15. 10.1007/s10493-012-9608-z [DOI] [PubMed] [Google Scholar]
- 10.Nava S, Caparrós JA, Mangold AJ, Guglielmone AA. Ticks (Acari: Ixodida: Argasidae, Ixodidae) infesting humans in Northwestern Cordoba Province, Argentina. Medicina (B Aires). (2006) 66:225–8. [PubMed] [Google Scholar]
- 11.Horak IG, Fourie LJ, Heyne H, Walker JB, Needham GR. Ixodid ticks feeding on humans in South Africa: with notes on preferred hosts, geographic distribution, seasonal occurrence and transmission of pathogens. Exp Appl Acarol. (2002) 27:113–36. 10.1023/A:1021587001198 [DOI] [PubMed] [Google Scholar]
- 12.Guglielmone AA, Beati L, Barros-Battesti DM, Labruna MB, Nava S, Venzal JM, et al. Ticks (Ixodidae) on humans in South America. Exp Appl Acarol. (2006) 40:83–100. 10.1007/s10493-006-9027-0 [DOI] [PubMed] [Google Scholar]
- 13.Nicholson WL, Allen KE, McQuiston JH, Breitschwerdt EB, Little SE. The increasing recognition of rickettsial pathogens in dogs and people. Trends Parasitol. (2010) 26:205–12. 10.1016/j.pt.2010.01.007 [DOI] [PubMed] [Google Scholar]
- 14.Piesman J, Eisen L. Prevention of Tick-Borne Diseases. Annu Rev Entomol. (2008) 53:323–43. 10.1146/annurev.ento.53.103106.093429 [DOI] [PubMed] [Google Scholar]
- 15.Vesco U, Knap N, Labruna MB, Avšič-Županc T, Estrada-Peña A, Guglielmone AA, et al. An integrated database on ticks and tick-borne zoonoses in the tropics and subtropics with special reference to developing and emerging countries. Exp Appl Acarol. (2011) 54:65–83. 10.1007/s10493-010-9414-4 [DOI] [PubMed] [Google Scholar]
- 16.Piotrowski M, Rymaszewska A. Expansion of tick-borne rickettsioses in the world. Microorganisms. (2020) 8:1906. 10.3390/microorganisms8121906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parola P, Paddock CD, Raoult D. Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clin Microbiol Rev. (2005) 18:719–56. 10.1128/CMR.18.4.719-756.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parola P, Davoust B, Raoult D. Tick- and flea-borne rickettsial emerging zoonoses. Vet Res. (2005) 36:469–92. 10.1051/vetres:2005004 [DOI] [PubMed] [Google Scholar]
- 19.Chitanga S, Gaff H, Mukaratirwa S. Tick-borne pathogens of potential zoonotic importance in the southern African region. J S Afr Vet Assoc. (2014) 85:1 10.4102/jsava.v85i1.1084 [DOI] [PubMed] [Google Scholar]
- 20.Mediannikov O, Diatta G, Fenollar F, Sokhna C, Trape J-F, Raoult D. Tick-borne rickettsioses, neglected emerging diseases in rural Senegal. PLoS Negl Trop Dis. (2010) 4:9. 10.1371/journal.pntd.0000821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. (2013) 26:657–702. 10.1128/CMR.00032-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chikeka I, Dumler JS. Neglected bacterial zoonoses. Clin Microbiol Infect. (2015) 21:404–15. 10.1016/j.cmi.2015.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Althaus F, Greub G, Raoult D, Genton B. African tick-bite fever: a new entity in the differential diagnosis of multiple eschars in travelers. Description of five cases imported from South Africa to Switzerland. Int J Infect Dis. (2010) 14:274–6. 10.1016/j.ijid.2009.11.021 [DOI] [PubMed] [Google Scholar]
- 24.Caruso G, Zasio C, Guzzo F, Granata C, Mondardini V, Guerra E, et al. Outbreak of African tick-bite fever in six Italian tourists returning from South Africa. Eur J Clin Microbiol Infect Dis. (2002) 21:133–6. 10.1007/s10096-001-0663-3 [DOI] [PubMed] [Google Scholar]
- 25.Consigny PH, Ina S, Fraitag S, Rolain JM, Buffet P. Unusual location of an inoculation lesion in a traveler with african tick-bite fever returning from South Africa. J Travel Med. (2009) 16:439–40. 10.1111/j.1708-8305.2009.00370.x [DOI] [PubMed] [Google Scholar]
- 26.Consigny PH, Rolain JM, Mizzi D, Raoult D. African tick-bite fever in French travelers. Emerg. Infect. Dis. (2005) 11:1804–6. 10.3201/eid1111.050852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fournier P, Roux V, Caumes E, Donzel M, Raoult D. Outbreak of Rickettsia africae infections in participants of an adventure race in South Africa. Clin Infect Dis. (1998) 27:316–23. 10.1086/514664 [DOI] [PubMed] [Google Scholar]
- 28.Jensenius M, Fournier P-E, Vene S, Hoel T, Hasle G, Henriksen AZ, et al. African tick bite fever in travelers to rural sub-Equatorial Africa. Clin Infect Dis. (2003) 36:1411–7. 10.1086/375083 [DOI] [PubMed] [Google Scholar]
- 29.McNaught J. A tick-borne fever in the Union of South Africa. J R Army Med Corps. (1911) 16:505. [Google Scholar]
- 30.McQuiston JH, Paddock CD, Singleton JJ, Wheeling JT, Zaki SR, Childs JE. Imported spotted fever rickettsioses in United States travelers returning from Africa: a summary of cases confirmed by laboratory testing at the Centers for Disease Control and Prevention, 1999-2002. Am J Trop Med Hyg. (2004) 70:98–101. 10.4269/ajtmh.2004.70.98 [DOI] [PubMed] [Google Scholar]
- 31.Raoult D, Fournier PE, Fenollar F, Jensenius M, Prioe T, de Pina JJ, et al. Rickettsia africae, a Tick-Borne Pathogen in Travelers to Sub-Saharan Africa. N Engl J Med. (2001) 344:1504–10. 10.1056/NEJM200105173442003 [DOI] [PubMed] [Google Scholar]
- 32.Toutous-Trellu L, Péter O, Chavaz P, Saurat JH. African tick bite fever: not a spotless rickettsiosis! J Am Acad Dermatol. (2003) 48:S18–S9. 10.1067/mjd.2003.122 [DOI] [PubMed] [Google Scholar]
- 33.Tsai KH, Lu HY, Huang JH, Fournier PE, Mediannikov O, Raoult D, et al. African tick bite fever in a Taiwanese traveler returning from South Africa: molecular and serologic studies. Am J Trop Med Hyg. (2009) 81:735–9. 10.4269/ajtmh.2009.09-0101 [DOI] [PubMed] [Google Scholar]
- 34.Wieten RW, Hovius JWR, Groen EJ, Van Der Wal AC, De Vries PJ, Beersma MFC, et al. Molecular diagnostics of Rickettsia africae infection in travelers returning from South Africa to the Netherlands. Vector-Borne Zoonotic Dis. (2011) 11:1541–7. 10.1089/vbz.2011.0653 [DOI] [PubMed] [Google Scholar]
- 35.Smoak BL, McClain JB, Brundage JF, Broadhurst L, Kelly DJ, Dasch GA, et al. An outbreak of spotted fever rickettsiosis in U.S. Army Troops Deployed to Botswana. Emerg Infect Dis. (1996) 2:217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sant'Anna J. On a disease in man following tick-bites and occuring in Lourenco Marques. Parasitology. (1911) 4:87–8. 10.1017/S0031182000002523 [DOI] [Google Scholar]
- 37.Zammarchi L, Farese A, Trotta M, Amantini A, Raoult D, Bartoloni A. Rickettsia africae infection complicated with painful sacral syndrome in an Italian traveller returning from Zimbabwe. Int J Infect Dis. (2014) 29:194–6. 10.1016/j.ijid.2014.10.017 [DOI] [PubMed] [Google Scholar]
- 38.Kelly PJ, Beati L, Matthewman LA, Mason PR, Dasch GA, Raoult D. A new pathogenic spotted fever group rickettsia from Africa. J Trop Med Hyg. (1994) 97:129–37. [PubMed] [Google Scholar]
- 39.Okabayashi T, Hasebe F, Samui KL, Mweene AS, Pandey SG, Yanase T, et al. Short report: prevalence of antibodies against spotted fever, murine typhus, and Q fever rickettsiae in humans living in Zambia. Am J Trop Med Hyg. (1999) 61:70–2. 10.4269/ajtmh.1999.61.70 [DOI] [PubMed] [Google Scholar]
- 40.Nakayima J, Hayashida K, Nakao R, Ishii A, Ogawa H, Nakamura I, et al. Detection and characterization of zoonotic pathogens of free-ranging non-human primates from Zambia. Parasit. Vectors. (2014) 7:490. 10.1186/s13071-014-0490-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chitimia-Dobler L, Dobler G, Schaper S, Küpper T, Kattner S, Wölfel S. First detection of Rickettsia conorii ssp. caspia in Rhipicephalus sanguineus in Zambia. Parasitol Res. (2017) 116:3249–51. 10.1007/s00436-017-5639-z [DOI] [PubMed] [Google Scholar]
- 42.Andoh M, Sakata A, Takano A, Kawabata H, Fujita H, Une Y, et al. Detection of Rickettsia and Ehrlichia spp. in Ticks Associated with Exotic Reptiles and Amphibians Imported into Japan. PLoS One. (2015) 10:e0133700. 10.1371/journal.pone.0133700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker AR, Bouattour A, Camicas JL, Estrada-Pena A, Horak IG, Latif AA, et al. Ticks of Domestic Animals in Africa: A Guide to Identification of Species. Bioscience Reports 42 Comiston Drive, Edinburgh EH10 5QR, Scotland (2003). [Google Scholar]
- 44.Choi YJ, Jang WJ, Kim JH, Ryu JS, Lee SH, Park KH, et al. Spotted fever group and typhus group rickettsioses in humans, South Korea. Emerg Infect Dis. (2005) 11:237–44. 10.3201/eid1102.040603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. (2013) 30:2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinform. (2009) 10:421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moonga LC, Hayashida K, Nakao R, Lisulo M, Kaneko C, Nakamura I, et al. Molecular detection of Rickettsia felis in dogs, rodents and cat fleas in Zambia. Parasit Vectors. (2019) 12:168. 10.1186/s13071-019-3435-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pretorius AM, Birtles RJ. Rickettsia aeschlimannii: a new pathogenic spotted fever group Rickettsia, South Africa. Emerg Infect Dis. (2002) 8:874. 10.3201/eid0808.020199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beati L, Kelly PJ, Matthewman LA, Mason PR, Raoult D. Prevalence of rickettsia-like organisms and spotted fever group rickettsiae in ticks (Acari: Ixodidae) from Zimbabwe. J Med Entomol. (1995) 32:787–92. 10.1093/jmedent/32.6.787 [DOI] [PubMed] [Google Scholar]
- 50.Fournier PE, Dumler JS, Greub G, Zhang J, Wu Y, Raoult D. Gene sequence - based criteria for identification of new rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J ClinMicrobiol. (2003) 41:5456–65. 10.1128/JCM.41.12.5456-5465.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moron CG, Bouyer DH, Yu XJ, Foil LD, Crocquet-Valdes P, Walker DH. Phylogenetic analysis of the rompB genes of Rickettsia felis and Rickettsia prowazekii European - human and North American flying - squirrel strains. Am J Trop Med Hyg. (2000) 62:598–603. 10.4269/ajtmh.2000.62.598 [DOI] [PubMed] [Google Scholar]
- 52.Portillo A, Pérez-Martínez L, Santibáñez S, Blanco JR, Ibarra V, Oteo JA. Detection of Rickettsia africae in Rhipicephalus (Boophilus) decoloratus ticks from the Republic of Botswana, South Africa. Am J Trop Med Hyg. (2007) 77:376–7. 10.4269/ajtmh.2007.77.376 [DOI] [PubMed] [Google Scholar]
- 53.Matsimbe AM, Magaia V, Sanches GS, Neves L, Noormahomed E, Antunes S, et al. Molecular detection of pathogens in ticks infesting cattle in Nampula province, Mozambique. Exp Appl Acarol. (2017) 73:91–102. 10.1007/s10493-017-0155-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo H, Adjou Moumouni PF, Thekisoe O, Gao Y, Liu M, Li J, et al. Genetic characterization of tick-borne pathogens in ticks infesting cattle and sheep from three South African provinces. Ticks Tick Borne Dis. (2019) 10:875–82. 10.1016/j.ttbdis.2019.04.008 [DOI] [PubMed] [Google Scholar]
- 55.Halajian A, Palomar AM, Portillo A, Heyne H, Luus-Powell WJ, Oteo JA. Investigation of Rickettsia, Coxiella burnetii and Bartonella in ticks from animals in South Africa. Ticks Tick Borne Dis. (2016) 7:361–6. 10.1016/j.ttbdis.2015.12.008 [DOI] [PubMed] [Google Scholar]
- 56.Halajian A, Palomar AM, Portillo A, Heyne H, Romero L, Oteo JA. Detection of zoonotic agents and a new Rickettsia strain in ticks from donkeys from South Africa: implications for travel medicine. Travel Med Infect Dis. (2018) 26:43–50. 10.1016/j.tmaid.2018.10.007 [DOI] [PubMed] [Google Scholar]
- 57.Kolo AO, Sibeko-Matjila KP, Maina AN, Richards AL, Knobel DL, Matjila PT. Molecular detection of zoonotic Rickettsiae and Anaplasma spp. in domestic dogs and their ectoparasites in Bushbuckridge, South Africa. Vector-Borne Zoonotic Dis. (2016) 16:245–52. 10.1089/vbz.2015.1849 [DOI] [PubMed] [Google Scholar]
- 58.Mtshali K, Khumalo ZTH, Nakao R, Grab DJ, Sugimoto C, Thekisoe OMM. Molecular detection of zoonotic tick-borne pathogens from ticks collected from ruminants in four South African provinces. J Vet Med Sci. (2016) 77:1573–9. 10.1292/jvms.15-0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baba K, Itamoto K, Amimoto A, Kitagawa K, Hiraoka H, Mizuno T, et al. Ehrlichia canis infection in two dogs that emigrated from endemic areas. J Vet Med Sci. (2012) 74:775–8. 10.1292/jvms.11-0401 [DOI] [PubMed] [Google Scholar]
- 60.Qiu Y, Kaneko C, Kajihara M, Ngonda S, Simulundu E, Muleya W, et al. Tick-borne haemoparasites and Anaplasmataceae in domestic dogs in Zambia. Ticks Tick Borne Dis. (2018) 9:988–95. 10.1016/j.ttbdis.2018.03.025 [DOI] [PubMed] [Google Scholar]
- 61.Vlahakis PA, Chitanga S, Simuunza MC, Simulundu E, Qiu Y, Changula K, et al. Molecular detection and characterization of zoonotic Anaplasma species in domestic dogs in Lusaka, Zambia. Ticks Tick Borne Dis. (2018) 9:39–43. 10.1016/j.ttbdis.2017.10.010 [DOI] [PubMed] [Google Scholar]
- 62.Chitanga S, Simulundu E, Simuunza MC, Changula K, Qiu Y, Kajihara M, et al. First molecular detection and genetic characterization of Coxiella burnetii in Zambian dogs and rodents. Parasit Vectors. (2018) 11:40. 10.1186/s13071-018-2629-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiu Y, Nakao R, Namangala B, Sugimoto C. Short report: first genetic detection of coxiella burnetii in zambian livestock. Am J Trop Med Hyg. (2013) 89:518–9. 10.4269/ajtmh.13-0162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.