Abstract
Eosinophilic esophagitis (EoE) is often misdiagnosed as GERD; therefore, the goal of the current study is to establish a non-invasive diagnostic and monitoring biomarker that differentiated GERD from EoE. Reports indicates that IL-15 responsive iNKT cells and tissue specific IgE have a critical in EoE pathogenesis, not in GERD. Therefore, we tested the hypothesized that the panel of IL-15-responsive T cell and IgE receptors may be novel non-invasive biomarkers for EoE. Accordingly, the receptors of IL-15 responsive T cells (Vα24, Jα18, γδT, αβT) and IgE (FcεRI & FcεRII) were examined. The data indicates that blood mRNA levels of Vα24, Jα18, γδ T, αβ T and FcεRI are significantly reduced in EoE compared to the GERD patients and normal individuals. The ROC curve analysis indicated FcεRII, Jα18 and δ TCR are the positive predictors that discriminate EoE from GERD. Thus, these molecules will be a novel non-invasive diagnostic biomarker for EoE.
Keywords: EoE, GERD, IL-15Rα, Eosinophils, IgE, FcεRI, FcεRII)
1. INTRODUCTION
The field of eosinophilic esophagitis (EoE) research has expanded greatly in recent years; however, while major advances have occurred in understanding the mechanisms of eosinophil and mast cell (MC) accumulation in the esophagus [1-3] and of esophageal remodeling [4, 5], there has been a significant delay in identifying predictive non-invasive biomarkers. One of the main limitations has been the difficulty to assess response to therapy, given that gastrointestinal endoscopy followed by biopsy is the only reliable method for evaluation. There is an urgent need to define novel biomarkers for EoE; due to a complete lack of alternatives, the diagnosis of EoE can only be performed histologically from esophageal biopsies obtained via upper endoscopy. Some patients with EoE may have <15 eosinophils per high-power field (hpf), and there are clear cases of gastroesophageal reflux disease (GERD) with biopsies that are hard to differentiate from EoE, as these patients may have a proton pump inhibitor (PPI)-responsive EoE [6]. A consensus for diagnostic recommendation was published by the Appraisal of Guidelines for Research and Evaluation (AGREE) in 2018 indicating that esophageal biopsy remains the only reliable diagnostic test for EoE. AGREE recommends that EoE diagnosis should follow PPI-nonresponsive esophageal dysfunction and at least 15 eosinophils/hpf (or approximately 60 eosinophils per mm2) on esophageal biopsy [7]. The panel did not recommend any non-invasive diagnostic markers, and a study of a panel of serum biomarkers found that none correlated with disease, including interleukin (IL) 4, IL-5, IL-6, IL-9, Il-13, TGFα, TGFβ, TNFα, eotaxin-1, eotaxin-2, eotaxin-3, and eosinophil cell-surface molecules like CD101 and CD274 [8-11]. Thus, there remains a need for a novel non-invasive biomarker that differentiates EoE from GERD.
The identification of a reliable EoE biomarker would advance EoE treatment by allowing for more accurate and timely detection of changes resulting from therapy alteration. Avoiding repeated endoscopies would improve patient care; reduce the possibility of complications related to the invasive nature of the procedure, and lower medical costs. The discovery of a biomarker for EoE would change our approach to the diagnosis and management of patients with EoE. EoE was once believed to be a pediatric disease, but it is now well established that it affects a sizable portion of adults [12-14]. Our investigations provide evidence that EoE is caused by a Th2-type allergy to food or other environmental antigens [1, 15-17]. We recently showed that IL-15 expression is induced in esophageal biopsies of pediatric and adult EoE patients [18]. Interestingly, IL-15 induction is correlated with esophageal eosinophilia in human EoE, and IL-15Rα-deficient mice were protected from the induction of experimental EoE [18]. Additionally, we found that high percentages of the T cells that accumulate in the esophagus of EoE patients are iNKT cells [16]. Experimental modeling establishes an essential role of iNKT cells, as CD1d gene-deficient mice are protected from the induction of EoE. [19] Notably, we showed that iNKT stimulation is sufficient to induce EoE in mice [19]. Additionally, induced IgE is reported in EoE but not GERD patients. IgE binds to high-affinity receptors (FcϵRI) on effector cells such as mast cells and basophils. Allergens bind to IgE and initiate an inflammatory cascade resulting in the release of pro-inflammatory mediators that contribute to the acute and chronic symptoms in EoE pathogenesis. [20, 21] We also provide direct evidence that tissue-specific IL-15-induced IgE-mediated responses, not systemic IgE, are critical in promoting EoE pathogenesis. [22] These findings lead us to hypothesize that the receptors of IL-15 and IL-15 responsive IgE (FcεRI and FcεRII), along with iNKT cell (IL-15Rα, Vα24, Jα18) mediators, may be non-invasive biomarkers of EoE, and that the levels of these biomarkers will differentiate EoE from GERD.
Accordingly, we now present data in support of our hypothesis that establishes novel non-invasive biomarkers for EoE. We show that the T cell surface receptors of IL-15Rα, Vα24, Jα18, γδ TCR, and αβ TCR T cells and the receptors of IgE (FcεRI and FcεRII) may be novel non-invasive biomarkers for EoE. To test this hypothesis, blood RNA was isolated from normal individuals, EoE patients, and GERD patients. Real time quantitative PCR analysis was performed for mRNA levels of iNKT cells, and T cell surface receptors were examined and normalized with respective mRNA levels of GADPH.
2. METHODS
2.1. Study Population.
Patients were enrolled for obtaining the blood samples from Cincinnati Center for Eosinophilic Disorder (CCED) of Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, and Tulane Eosinophilic Disorder Center, Tulane University School of Medicine, New Orleans, LA. The data are obtained from the both center and the pooled data were used for the evaluation of EoE biomarker. The samples includes normal individuals, GERD patients, and EoE patients. Eligible patients were aged 2–21 years. Patients with known diagnosis of any systemic or gastrointestinal inflammatory condition other than eosinophilic gastrointestinal disorder were excluded. Patients with symptoms consistent with EoE or GERD were included in this biomarker study following an Institutional Review Board (IRB)-approved protocol.
2.2. Patient Characteristics.
Normal (15 nos), GERD (19 nos), and EoE (33 nos) patients were selected without regard to age, atopic status, or gender. Diagnosis was established based on the maximum eosinophil count per high power field (hpf) (400x). “Normal” was defined as having 0 esophageal eosinophils/hpf and no basal layer expansion. Typically, these patients had abdominal pain, some with allergic diseases including asthma or rhinitis, but they had completely normal esophageal endoscopic and microscopic analyses. Patients with EoE were defined as having ≥15 esophageal eosinophils/hpf, as per the recently updated consensus guidelines for diagnosis.[7] Patients with GERD were defined as per the Montreal definition and classification of GERD, a global evidence-based consensus, that reflux of stomach contents causes troublesome symptoms and/or complications with heartburn, and patients are responsive to PPIs [23]. Patients included in this study had EoE or GERD with other allergic diseases such as asthma or atopic dermatitis. The treatment strategies for EoE (including dietary restriction of food allergens or topical fluticasone therapy) and for GERD (including PPI therapy) are listed in Table 1.
Table 1:
Patients Clinical and pathological characteristics
| Patients | Age | Gender | Esophageal disease | Allergic diseases | Current Treatment |
|---|---|---|---|---|---|
| 1 | 9 | M | NL | Asthma | None |
| 2 | 11 | F | NL | Unknown | None |
| 3 | 7 | F | NL | None | H1RA |
| 4 | 9 | F | NL | Unknown | INHGC,LTRA |
| 5 | 4 | M | NL | Rhinitis | LTRA |
| 6 | 12 | M | NL | None | INHGC |
| 7 | 5 | F | NL | Unknown | INHGC |
| 8 | 11 | M | NL | None | PPI, OGC |
| 9 | 9 | M | NL | Asthma | None |
| 10 | 10 | M | NL | None | INHGC, SWGC |
| 11 | 12 | M | NL | None | None |
| 12 | 13 | F | NL | Unknown | PPI |
| 13 | 10 | M | NL | Asthma | INHGC |
| 14 | 12 | M | NL | None | PPI |
| 15 | 8 | F | NL | Rhinitis | None |
| 16 | 14 | F | EoE | Asthma | INHGC |
| 17 | 4 | M | EoE | None | INHGC |
| 18 | 9 | F | EoE | Rhinitis/Asthma/Food allergy | PPI, mesalamine |
| 19 | 8 | M | EoE | Food Allergy | Alimentary diet |
| 20 | 10 | M | EoE | None | None |
| 21 | 8 | F | EoE | Asthma/Eczema | Alimentary diet, INHGC |
| 22 | 9 | F | EoE | Rhinitis/Asthma | Alimentary diet, INHGC |
| 23 | 6 | M | EoE | Unknown | INHGC,B2ARA |
| 24 | 11 | F | EoE | Food Allergy | None |
| 25 | 4 | F | EoE | Asthma | Alimentary diet, INHGC |
| 26 | 5 | M | EoE | None | INHGC |
| 27 | 10 | F | EoE | Asthma/Eczema | None |
| 28 | 6 | M | EoE | None | PPI |
| 29 | 7 | M | EoE | Rhinitis/Asthma | B2ARA |
| 30 | 15 | F | EoE | Food Allergy/anaphylaxis | Alimentary diet, Protonix |
| 31 | 8 | M | EoE | None | Alimentary diet, PPI |
| 32 | 11 | M | EoE | Asthma/Rhinitis | Alimentary diet |
| 33 | 21 | F | EoE | Rhinitis/enzema | Alimentary diet, SWGC |
| 34 | 9 | M | EoE | Unknown | B2ARA |
| 35 | 13 | M | EoE | Food Allergy | SWGC, PPI |
| 36 | 18 | F | EoE | Asthma/Rhinitis | SWGC, H1RA |
| 37 | 8 | M | EoE | Eczema | Alimentary diet, SWGC |
| 38 | 11 | M | EoE | Unknown | PPI |
| 39 | 20 | M | EoE | Rhinitis | Alimentary diet, PPI |
| 40 | 8 | M | EoE | Unknown | Alimentary diet, INHGC |
| 41 | 20 | F | EoE | Asthma | PPI |
| 42 | 11 | M | EoE | None | None |
| 43 | 7 | M | EoE | Asthma/Eczema | INHGC |
| 44 | 11 | F | EoE | Rhinitis | INHGC,PPI |
| 45 | 12 | M | EoE | None | B2ARA |
| 46 | 8 | F | EoE | Food Allergy/anaphylaxis/Asthma | SWGC, Alimentary diet |
| 47 | 11 | M | EoE | Unknown | PPI |
| 48 | EoE | Asthma/Rhinitis | None | ||
| 49 | 20 | M | GERD | Rhinitis | Alimentary diet, |
| 50 | 18 | M | GERD | Unknown | Alimentary diet, INHGC |
| 51 | 20 | F | GERD | Asthma | PPI |
| 52 | 11 | M | GERD | None | None |
| 53 | 7 | M | GERD | Asthma/Eczema | INHGC |
| 54 | 11 | F | GERD | Rhinitis | INHGC,PPI |
| 55 | 12 | M | GERD | None | B2ARA |
| 56 | 8 | F | GERD | Food Allergy/anaphylaxis/Asthma | SWGC, Alimentary diet |
| 57 | 13 | M | GERD | Asthma | OGC |
| 58 | 18 | F | GERD | Unknown | None |
| 59 | 8 | M | GERD | None | H1RA |
| 60 | 11 | M | GERD | Unknown | INHGC,LTRA |
| 61 | 20 | M | GERD | Rhinitis | LTRA, |
| 62 | 8 | M | GERD | None | INHGC, INHGC |
| 63 | 20 | F | GERD | Asthma/ Food allergy | INHGC |
| 64 | 4 | F | GERD | None | PPI |
| 65 | 5 | M | GERD | Rhinitis/enzema | Alimentary diet, SWGC |
| 66 | 10 | F | GERD | Unknown | B2ARA |
| 67 | 6 | M | GERD | Food Allergy | SWGC, PPI |
Abbreviations: NL=normal, M=male, F= female, EoE= eosinophilic esophagitis, gastroesophageal reflux disease GERD. LTRA, leukotriene receptor antagonist; H1RA, H1-receptor antagonist; INHGC, inhaled glucocorticoid; B2ARA, 2 adrenergic receptor antagonist; PPI, proton pump inhibitor; SWGC, swallowed glucocorticoid (fluticasone); OGC, oral glucocorticoid (prednisone).
2.3. Patient Blood Collection and Blood RNA Isolation.
Blood samples were obtained from normal individuals and patients with GERD and EoE. RNA from blood was isolated using PAXgene Blood RNA kit from PreAnalytix GmbH, Hombrechtikon, CH.
2.4. Esophageal Eosinophil Analysis.
2.4.1. Hematoxylin and eosin (H&E) analysis:
The eosinophils in esophageal biopsies of normal, GERD, and EoE patients were identified in eosin and hematoxylin (H&E)-stained tissue sections. Esophageal eosinophils were quantified by counting the eosinophils with the assistance of digital morphometry using the Metamorph Imaging System (Universal Imaging Corp, Sunnyvale, MI) and expressed as eosinophils/hpf (400x) per square millimeter as described previously [18].
2.4.2. Immunohistochemical analysis:
Paraffin embedded tissue sections (5μm) were immunostained with antiserum against mouse eosinophil major basic protein (anti-MBP) as described previously. [18, 24, 25]
2.5. Measurement of Biomarkers mRNA Levels by RT-PCR Analysis.
Quantitative PCR was performed on blood RNA (500 ng) and it was subjected to reverse transcription analysis using iScript reverse transcriptase (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. IL-15 receptors, cell surface molecules, and IL-15 responsive T cell and IgE receptors were quantified by real-time PCR using IQ5 (Bio-Rad, Hercules, CA). Results were then normalized to human GAPDH amplified from the same cDNA mix and expressed as relative gene expression. cDNA was amplified using the primers listed in Table 2.
Table 2:
List of human primer sequences
| Genes | Sense and anti-sense primer sequence (5’-3’) |
|---|---|
| h-IL15R-α | F: 5’-CATCACGTGCCCTCCCCCCATG-3′ |
| R: 5′-AGTGGTGTCGCTGTGGCCCTG-3′ | |
| h-Vα24 | F: 5′-GAAAGTTTAGGTTCGTATCTGTTTCA-3′ |
| R: 5′-GATATACAGCAACTCTGGATGCA-3′ | |
| h- Jα18 | F: 5′-TTTTCAACCCTGTACTCCAA-3′ |
| R: 5′-GGATATCAGGCCAGACAGTC -3′ | |
| h-TCR-δ | F: 5′-CTGTGCACTCCACTGACTTTG -3′ |
| R: 5′-GGGTTTATGGCAGCTCTTTG-3′ | |
| h-TCR-γ | F: 5′-AACGGTGCCAGAAAAGCTAC-3′ |
| R: 5′-TGTCTTTGGGATCCATTGTG-3′. | |
| h-TCR-α | F: 5′-GTCCAGCCCTACCCACAG-3′ |
| R: 5′-GACCAGGCAGACCACCAC-3′. | |
| h-TCR-β | F: 5’-TCTCTGTGCCTGGAGTTGG-3’ |
| R: 5’-TCCTCTAGGATGGACAGTCGAG -3’ | |
| h-FcεRI | F: 5’-GGGAACAATTTCTTTGAAGTCAGT-3’ |
| R: 5’-CATTTGTAATGGAGATGTTGTGGT-3’ | |
| h-FcεRII (CD23) | F: 5’-AGCCCCGGTATGCCTGTGAC-3’ |
| R: 5’-GAAGAATCCTGACAGCCTTCA-3’ | |
| h-GAPDH | F: 5′TGGAAATCCCATCACCATCT3′ |
| R: 5′GTCTTCTGGGTGGCAGTGAT3′ |
2.6. Flow cytometry and antibodies.
The total blood cells were stained with the specific cell surface molecule-antibodies for flow cytometer analysis. Total blood cells were isolated as per the protocol described earlier.[8, 26] The following reagents were used for specific antigen analysis: anti-CD3, anti-CD45, anti-Vα24Jα18, hTCR γ/δ, hFcϵRI and their respective isotype controls obtained from eBiosciences San Diego, CA and Bio legend, San Diego, CA. FcR block (anti-CD16/CD32 Ab) was added to all surface staining mixtures. 7AAD was used to exclude dead cells. The cells were incubated for specific antigens with the required combination of antibodies at 4 °C for 45 min followed by two washes. FACS analysis was performed using a FACSCalibur (BD Biosciences, San Jose, CA, USA) and analyzed with FlowJo software version 7.1 (Tree Star).
2.6. Statistical Analysis.
Esophageal eosinophil counts from each patient were calculated by averaging the mean eosinophil counts obtained from the proximal and distal esophagus. The correlation of individual mRNA transcript (biomarker) levels with the overall mean eosinophil counts from the esophagus were evaluated using Spearman’s correlation test. mRNA transcript levels were analyzed with the Mann–Whitney U test for comparisons between the two groups. All statistical tests were compared using t tests or analysis of variance. Values are reported as mean ± standard deviation (SD). p values < 0.05 were considered statistically significant. Statistical tests were performed using GraphPad Prism 5 Software (San Diego, CA).
3. RESULTS
3.1. Esophageal eosinophilia is increased in EoE patients compared to GERD or normal patients.
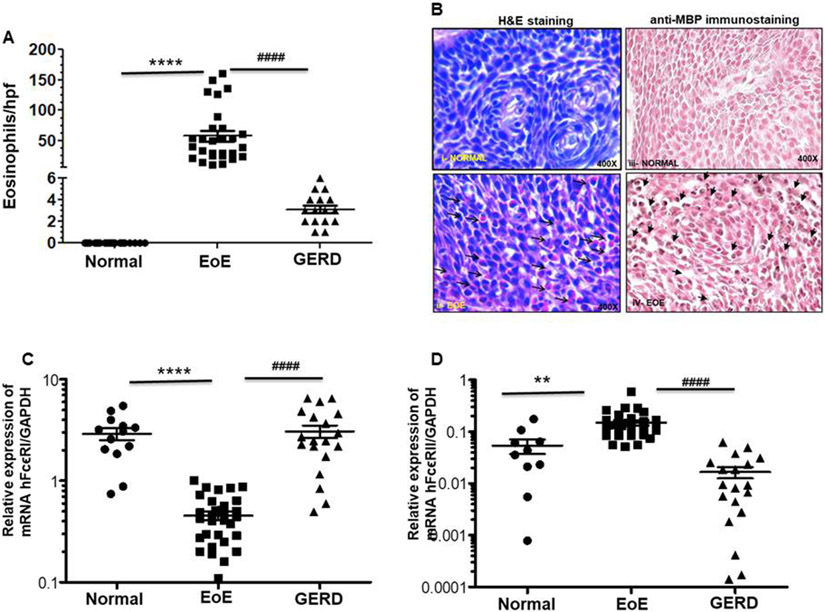
Anti-MBP immunostained accumulated eosinophils in chronic EoE patient biopsies were almost 100-fold higher compared to normal individuals, indicating the chronic nature of the disease (Fig 1A). The mean eosinophil counts in GERD patients were 1 ± 6 eosinophils/hpf, while no eosinophils were observed in esophageal biopsies of normal individuals. Further, the mean eosinophil counts in proximal and distal esophagus of EoE patients were 26 ± 21 and 31 ± 26 eosinophils/hpf, respectively, and we found no significant difference between them (p>0.3) (Suppl. Fig 1A). Further, representative photomicrographs of hematoxylin and eosin (H&E) stained and anti-MBP immunostaining detected induced eosinophils (as shown by arrows) in the muscular mucosa in the EoE patient’s esophagus sections compared to normal individual, (Fig 1B i-iv). Photomicrographs are presented in original magnification 400x.
Figure 1. Analysis of eosinophils and mRNA levels of the IL-15 induced IgE receptors (FcεRI and FcεRII) in EoE patients, GERD patients, and normal individuals.
Eosinophils are significantly detected in the esophageal epithelial mucosa in the EoE patients compared to GERD and normal individuals (A). Each data points indicate individual patient’s eosinophils numbers. Representative photomicrograph showing eosinophils in the esophageal biopsies of normal individuals and EoE patients following H&E and anti-MBP stained tissue sections (original magnification 400X [B (i-vi)]. RT-PCR analysis indicated that mRNA expression levels of FcεRI were decreased and FcεRII increased in blood samples of EoE patients compared to GERD patients and normal individuals (C, D). Each data points indicate individual transcripts expression levels in patients. Data are expressed as mean± SD, n=3; EoE: 33 patients, GERD: 19 patients, normal individual: 15 nos. Some patient data points are missing as they did not show up during PCR analysis. EoE vs Normal **** p<0.0001, ** p<0.006; EoE vs GERD #### p<0.0001; NS, not significant.
3.2. Relative expression of IgE receptor (FCεRI and FCεRII) mRNA levels in the blood of normal, EoE, and GERD patients.
Both animal models and human subject’s research have indicated that allergic sensitization leads to food allergen antigen-specific IgE, and Th2 lymphocytes have been shown to be involved in EoE pathogenesis along with eosinophils, mast cells, basophils, and antigen-presenting cells. Recently, we reported a novel role for IL-15-induced esophageal IgE, and found that systemic IgE induction has no role in in promoting allergen-induced EoE pathogenesis. [22, 27] Here, we tested the hypothesis that EoE patients may have altered mRNA levels of IgE receptors FcεRI and FCεRII compared to normal individuals and GERD patients. Accordingly, mRNA levels of IgE receptors in the blood were examined as novel noninvasive biomarkers for human EoE. Our data indicated that mRNA levels of FCεRI are significantly decreased and FCεRII is increased in EoE patients compared to normal individuals and GERD patients (Fig 1 C, D). FcεR II is a low affinity receptor for IgE, and its increase is consistent with our previous observation that tissue-specific induction of IgE has a role in EoE pathogenesis. [22] Additionally, we also found decreased FCεRI expressing cells in EoE patients compared to normal individuals and GERD patient’s blood samples. The data presented in Suppl. Fig 2 a-e.
3.3. IL-15Rα mRNA levels are decreased in EoE compared to normal individuals and differentiate between EoE and GERD
We previously reported that IL-15 mRNA and protein levels are significantly increased in the esophagus of patients with EoE compared with non-EoE individuals. [18] Therefore, we hypothesized that IL-15Rα mRNA levels in the blood may be novel non-invasive diagnostic biomarkers for EoE and that the levels of these molecules will differentiate EoE from GERD. To study this hypothesis, we examined IL-15Rα mRNA expression in the blood of normal, EoE, and GERD patients. Interestingly, we found highly decreased IL-15Rα mRNA levels in the blood of EoE patients compared to normal individuals and GERD patients (Fig 2A). These findings of reduced IL-15Rα mRNA levels in the blood are distinct from the earlier observed tissue levels of IL-15 Rα mRNA. [18] This reduced IL-15Rα mRNA indicates that most of the T cells expressing IL-15Rα moved to the tissue. We previously reported that induced IL-15Rα-expressing iNKT cells are induced in the esophagus and their trafficking is promoted by induced CXCL16 in EoE patients.[19]
Figure 2. Decreased mRNA levels of the IL-15Rα and IL-15 responsive iNKT cells receptors in blood samples of EoE patients compared to GERD patients and normal individuals.
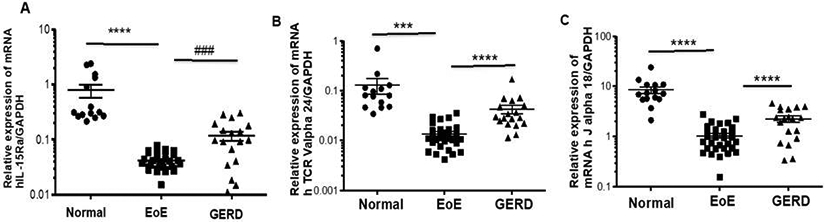
RT-PCR analysis indicated that IL-15Rα (A) and iNKT cell-specific receptor Vα24, Jα18 (B, C) mRNA expression levels are decreased in the blood samples of the EoE patients compared to GERD and normal individuals. Each data points indicate individual transcripts expression levels in patients. Data are expressed as mean± SD, n=3; EoE: 33 patients, GERD: 19 patients, normal individual: 15 nos. Some patient data points are missing as they did not show up during PCR analysis. EoE vs Normal *** p<0.0003, **** p<0.0001; EoE vs GERD #### p<0.0001, ### p<0.001.
3.3a. Relative expression of IL-15-responsive iNKT cell receptor (Vα24, Jα18) blood mRNA levels is significantly decreased in EoE patients compared to normal individuals and GERD patients.
Most recently, we reported that iNKT cells are highly expressed in the esophagus, and their levels are reduced in the blood of patients with EoE compared with non-EoE individuals.[19] Therefore, we hypothesized that iNKT cell receptor mRNA expression may be used as a non-invasive biomarker for EoE. To test this, we examined Vα24, Jα18 mRNA expression in the blood of normal individual, EoE, and GERD patients. Interestingly, we found highly decreased Vα24 and Jα18 mRNA levels in the blood of EoE patients compared to non-EoE individuals, including GERD patients (Fig 2 B-C). Additionally, we also found decreased iNKT cells (CD3/anti-hVα24Jα18 antibody) EoE patients compared to normal individuals and GERD patient’s blood samples. Data are presented in Suppl. Fig 2 f-j.
3.3b. Relative expression of IL-15 responsive hTCR α, hTCR β, hTCR γ and hTCR δ T cell receptor blood mRNA levels are significantly decreased in EoE patients compared to normal individuals and GERD patients.
We were also interested in understanding whether TCR receptors such as α and β are also decreased in the blood of EoE patients compared to normal and GERD patients. Therefore, we also tested both α and β TCR mRNA expression in the blood of normal individual, EoE, and GERD patients. Interestingly, we found a highly decreased level of TCRα and TCRβ mRNA in EoE patients compared to non-EoE individuals, including GERD patients (Fig 3 A,B). EoE is a T-cell dependent, Th2 cytokine-mediated allergic disease. We recently established that allergen-induced IL-15 and its responsive and non-responsive T cell subsets have a role in the initiation and progression of EoE pathogenesis. Among T lymphocyte subsets, lymphocytes expressing γδ T cell receptor have been determined to be a key factor for eosinophil accumulation via direct and indirect mechanisms. It has been shown that IL-15 is critical in the initiation, progression, and maintenance of innate immunity of EoE. [16, 18] Therefore, we hypothesized that IL-15 responsive T cell receptor (TCR) mRNA expression levels in the blood may be novel non-invasive diagnostic biomarkers for EoE and that the levels of these molecules will differentiate EoE from GERD. Accordingly, we examined γ and δ TCR mRNA expression in the blood of normal individual, EoE, and GERD patients. Interestingly, we found highly decreased γ and δ TCR mRNA levels in the blood of EoE patients compared to non-EoE individuals, including GERD patients (Fig 3 C,D). Additionally, we also found decreased IL-15 responsive receptor of γ/δ cells (anti-h TCR-γ/δ antibody) in EoE patients compared to normal individuals and GERD patient’s blood samples. Data are presented in Suppl. Fig 2 k-o.
Figure 3. Decreased mRNA levels of IL-15 responsive hTCR α, hTCR β, hTCR γ and hTCR δ T cell receptors in the blood samples of EoE patients compared to GERD patients and normal individuals.

RT-PCR analysis indicated that mRNA expression levels of hTCR α, hTCR β, hTCR γ and hTCR δ T cell receptors are decreased in blood samples of EoE patients compared to GERD and normal individuals (A-D). Each data points indicate individual transcripts expression levels in patients. Data are expressed as mean± SD, n=3; EoE: 33 patients, GERD: 19 patients, normal individual: 15 nos. Some patient data points are missing as they did not show up during PCR analysis. EoE vs Normal ** p<0.005, **** p<0.0001; EoE vs GERD # p<0.01, #### p<0.0001.
3.4. Correlation and receiver operating characteristic (ROC) analysis of IgE receptor (FCεRII), iNKT cell receptor (Jα18), and T cell receptor (TCRδ) to discriminate between patients with EoE compared to GERD.
The mRNA levels of IgE receptor (FCεRII), iNKT cell receptor (Jα18), and T cell receptor (TCRδ) were correlated with peak esophageal eosinophil counts of EoE patients, as shown in Fig 4 A-C. The specificity and sensitivity of these three non-invasive biomarkers (FCεRII, TCRδ and Jα18) were compared between EoE and GERD patients. Cut-off levels were determined by ROC curve analysis to maximize the sensitivity and specificity of all three non-invasive biomarkers that differentiate EoE from GERD. Based on the analysis, the following cut-off points were chosen as mRNA relative expression normalized with GADPH of each molecule: TCRδ (3.01), Jα18 (1.32), and FCεRII (0.75). The ROC curve analysis of D-FCεRII (100%), E-Jα18 (83%) and F-TCRδ (95%) in individual yielded an excellent positive predictive value in support of individual marker for EoE patient that differentiate clearly EoE from GERD (Fig 4 D-F).
Figure 4. Correlation and Receiver operating characteristic (ROC) analysis of IgE receptor (FCεRII), iNKT cell receptor (Jα18) and T Cell receptor (TCRδ) to discriminate between patients with EoE compared to GERD.
A significant statistical correlation between maximum eosinophils number/hpf and FcεRII, Jα18, and δ TCR m RNA expression in human EoE is shown (A-C). p value and r were calculated using Spearman’s correlation test, mRNA vs eosinophils **** p<0.0001. The solid line (no discrimination) indicates values that have no discriminatory value. Note that on the vertical axis, the scale is from no (0%) to complete (100%) sensitivity. The horizontal axis is a reciprocal scale (100% - % specificity). Based on the analysis, the following cut-off points of the individual biomarkers were chosen as an mRNA relative expression normalized with GAPDH of each molecule: A- FcεRII (0.75), B- Jα18 (1.32), and C- δ TCR (3.01). The use of all three biomarkers in individual yielded an excellent positive predictive value for D- FcεRII (100%), E- Jα18 (83%) and F- δ TCR (95%) (D-F). The optimum performance of a diagnostic test is determined either at the highest sum of the sensitivity and specificity or at an acceptable level of sensitivity for the given disease.
Notably, a number of other analyzed receptors did not correlate with esophageal eosinophilia.
4. Discussion
Eosinophilic esophagitis (EoE) is an immune-mediated, allergen-driven condition characterized by eosinophilic inflammation with resultant tissue remodeling, fibrosis, and esophageal dysfunction. Epidemiologic studies suggest that the prevalence of EoE has increased almost 20-fold over the past 15 years. The diagnosis and monitoring of EoE predominantly rely on symptoms and endoscopy with biopsy eosinophil count (>15 eosinophils/hpf) and at least six weeks of proton pump inhibitor treatment to block gastric acid secretion.[6] The criteria for diagnosis are recommended by an expert panel established as part of the First International Group of EE Researchers (FIGERS). [6] We and other investigators have made significant progress in understanding EoE pathogenesis and developing some diagnostic criteria.[1, 8, 16, 18, 24, 28, 29] Several promising minimally invasive biomarkers for EoE have been previously reported, some of which are able to differentiate EoE from other atopic diseases.[26, 30, 31] However, there remains a need for novel non-invasive biomarkers and potential therapies that differentiate EoE from GERD. An invasive study of a panel of serum biomarkers found that none of the markers correlated with disease, including interleukin (IL) 4, IL-5, IL-6, IL-9, Il-13, TGFα, TGFβ, TNFα, eotaxin-1, eotaxin-2, eotaxin-3, and eosinophil cell surface molecules like CD101 and CD274. [8-11] In addition, multiple studies have shown that EoE is characterized by a significant eosinophilic infiltration and eosinophilic degranulation leading to the release of eosinophilic granule proteins that promote disease pathogenesis. These proteins, including eosinophil cationic protein (ECP), eosinophil-derived neurotoxin (EDN), and eosinophil major basic proteins (MBP), have been evaluated as serologic markers of disease with mixed results.[9, 32, 33] Some studies showed differences in serum eosinophil granule protein levels between EoE subjects and controls, but due to the coexistence of other atopic diseases in EoE patients, there is significant concern regarding the use of these proteins as a biomarker for EoE. [9, 11, 34] Thus, there is a great need to continue with innovative fundamental studies to uncover new possibilities for diagnostic and therapeutic interventions. A non-invasive biomarker is a term often used to refer a molecule measured in blood whose concentration reflects the severity or presence of some disease state. The most attractive biomarker for EoE would be one that is expressed in both esophageal biopsies and in the peripheral blood of EoE patients. We have previously shown that IL-15 and iNKT cells are induced in the esophageal biopsies of EoE patients. [18] In the current study, we evaluated the levels of mRNA expression of several potential molecules related to IL-15 and its responsive iNKT cell surface molecules and receptors in the blood of EoE patients for non-invasive diagnostic markers that differentiate EoE from GERD. GERD is a concern because it is a common condition, and like EoE, it is a frequent indication for upper endoscopy. There is also clinical overlap in the symptoms (heartburn, dysphagia, chest pain) and histology of GERD and EoE. A large number of esophageal biopsies of GERD patients reported greater than 20 eos/hpf. [35] This leads to a potentially complex relationship among esophageal eosinophilia in EoE and GERD patients. Several studies differentiate EoE from GERD based on endoscopic findings and biopsy features; however, no single pathological or genomic marker has been able to distinguish EoE from GERD. Our experimental data identified a combination of potential molecules that are able to predict a diagnosis of EoE in patients before endoscopic evaluation of the esophagus. The data presented in this study show that blood mRNA levels of most IL-15 responsive iNKT cell-related molecules statistically differentiate EoE from normal individuals and GERD patients. The decreased mRNA levels of IL-15Rα, Vα24, Jα18, γTCR and δTCR and cells expressing these cell surface receptors FCεRI, Vα24Jα18 and TCR-γ/δ in the blood cells is due to the receptors expressed NKT and γδTC cells homing into the esophagus. We earlier reported that IL-15 responsive iNKT cells with the help of NKT cells specific chemokine CXCL16 home into the esophagus in EoE [36, 37]. Similarly, the γδTC cells ho induction is observed in EoE; but their role is yet to be established. The mRNA values of Jα18 (R2-0.447) and δ TCR (R2-0.500) in the blood and its correlation with eosinophils further confirm the significance of iNKT cells in EoE. However, the IgE low affinity receptor FcεRII, R2 value (R2-0.238) indicates that its density is more critical compare to the numbers of cells. The presented data of decreased mRNA levels of Vα24, Jα18, γTCR, δ TCR and cells expressing these receptors in the blood of EoE patients strongly suggests that these reduced mRNA levels are indeed a novel biomarker for EoE. Taken together, these correlation data further confirm the significant role of IL-15 responsive NKT cells in the pathogenesis of EoE.
In summary, blood RNA analysis shows that IL-15 and its responsive cell surface molecules including receptors, together with eosinophil correlation and perfect ROC curves, differentiate EoE from GERD. Previously, induced iNKT cells have been reported in experimental and human EoE, and their neutralization protects against an experimental model of EoE. [16] Thus, the presented data suggest that peripheral blood mRNA levels of the reported molecules are relevant biomarkers for monitoring disease activity and suggest a potential role for iNKT cells in EoE pathogenesis.
Supplementary Material
Highlights.
We found that IL-15Rα mRNA levels are decreased in EoE compared to normal individuals and differentiate between EoE and GERD patient samples.
Relative expression of IL-15 responsive γδ T cell receptors blood mRNA levels is significantly decreased in EoE patients compared to normal individuals and GERD patients.
Significantly decreased relative expression of blood mRNA levels of IL-15 responsive iNKT cell receptors (Vα24-Jα18) and IgE receptors were observed in EoE patients compared to normal individuals and GERD patients.
IL-15 responsive iNKT and T cell surface receptors show a strong correlation with esophageal eosinophils and perfect ROC curves that differentiate EoE from GERD.
Acknowledgements
This work was supported by the NIH grant R01 AI080581 (AM). Dr. Mishra is the endowed Schlieder Chair; therefore, we thank the Edward G. Schlieder Educational Foundation for their support. We also thank Eosinophilic Disorder Center of Cincinnati Children’s Hospital Medical Center, Cincinnati OH for providing blood samples from EoE and non-EoE patients and healthy individuals. The authors would like to thank Ms. Loula Burton, Editor for the Office of Research Proposal Development, Tulane University for helping in manuscript editing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Declaration of all sources of funding
All authors have no financial conflict of interest.
5. References
- [1].Mishra A, Hogan SP, Brandt EB, Rothenberg ME, An etiological role for aeroallergens and eosinophils in experimental esophagitis, J Clin Invest, 107 (2001) 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, Jameson SC, Kirby C, Konikoff MR, Collins MH, Cohen MB, Akers R, Hogan SP, Assa'ad AH, Putnam PE, Aronow BJ, Rothenberg ME, Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis, J Clin Invest, 116 (2006) 536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Abonia JP, Blanchard C, Butz BB, Rainey HF, Collins MH, Stringer K, Putnam PE, Rothenberg ME, Involvement of mast cells in eosinophilic esophagitis, J Allergy Clin Immunol, 126 (2010) 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Aceves SS, Newbury RO, Dohil R, Bastian JF, Broide DH, Esophageal remodeling in pediatric eosinophilic esophagitis, J Allergy Clin Immunol, 119 (2007) 206–212. [DOI] [PubMed] [Google Scholar]
- [5].Mishra A, Wang M, Pemmaraju VR, Collins MH, Fulkerson PC, Abonia JP, Blanchard C, Putnam PE, Rothenberg ME, Esophageal Remodeling Develops as a Consequence of Tissue Specific IL-5-Induced Eosinophilia, Gastroenterology, 134 (2008) 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, Burks AW, Chehade M, Collins MH, Dellon ES, Dohil R, Falk GW, Gonsalves N, Gupta SK, Katzka DA, Lucendo AJ, Markowitz JE, Noel RJ, Odze RD, Putnam PE, Richter JE, Romero Y, Ruchelli E, Sampson HA, Schoepfer A, Shaheen NJ, Sicherer SH, Spechler S, Spergel JM, Straumann A, Wershil BK, Rothenberg ME, Aceves SS, Eosinophilic esophagitis: updated consensus recommendations for children and adults, J Allergy Clin Immunol, 128 (2011) 3–20 e26; quiz 21-22. [DOI] [PubMed] [Google Scholar]
- [7].Dellon ES, Liacouras CA, Molina-Infante J, Furuta GT, Spergel JM, Zevit N, Spechler SJ, Attwood SE, Straumann A, Aceves SS, Alexander JA, Atkins D, Arva NC, Blanchard C, Bonis PA, Book WM, Capocelli KE, Chehade M, Cheng E, Collins MH, Davis CM, Dias JA, Di Lorenzo C, Dohil R, Dupont C, Falk GW, Ferreira CT, Fox A, Gonsalves NP, Gupta SK, Katzka DA, Kinoshita Y, Menard-Katcher C, Kodroff E, Metz DC, Miehlke S, Muir AB, Mukkada VA, Murch S, Nurko S, Ohtsuka Y, Orel R, Papadopoulou A, Peterson KA, Philpott H, Putnam PE, Richter JE, Rosen R, Rothenberg ME, Schoepfer A, Scott MM, Shah N, Sheikh J, Souza RF, Strobel MJ, Talley NJ, Vaezi MF, Vandenplas Y, Vieira MC, Walker MM, Wechsler JB, Wershil BK, Wen T, Yang GY, Hirano I, Bredenoord AJ, Updated International Consensus Diagnostic Criteria for Eosinophilic Esophagitis: Proceedings of the AGREE Conference, Gastroenterology, 155 (2018) 1022–1033 e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Venkateshaiah SU, Mishra A, Manohar M, Verma AK, Rajavelu P, Niranjan R, Wild LG, Parada NA, Blecker U, Lasky JA, Mishra A, A critical role for IL-18 in transformation and maturation of naive eosinophils to pathogenic eosinophils, J Allergy Clin Immunol, 142 (2018) 301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dellon ES, Rusin S, Gebhart JH, Covey S, Higgins LL, Beitia R, Speck O, Woodward K, Woosley JT, Shaheen NJ, Utility of a Noninvasive Serum Biomarker Panel for Diagnosis and Monitoring of Eosinophilic Esophagitis: A Prospective Study, The American journal of gastroenterology, 110 (2015) 821–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Venkateshaiah SU, Zhu X, Rajavelu P, Niranjan R, Manohar M, Verma AK, Lasky JA, Mishra A, Regulatory effects of IL-15 on allergen-induced airway obstruction, J Allergy Clin Immunol, 141 (2018) 906–917 e906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hines BT, Rank MA, Wright BL, Marks LA, Hagan JB, Straumann A, Greenhawt M, Dellon ES, Minimally invasive biomarker studies in eosinophilic esophagitis: A systematic review, Annals of allergy, asthma & immunology : official publication of the American College of Allergy, Asthma, & Immunology, 121 (2018) 218–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cantu P, Velio P, Prada A, Penagini R, Ringed oesophagus and idiopathic eosinophilic oesophagitis in adults: an association in two cases, Dig Liver Dis, 37 (2005) 129–134. [DOI] [PubMed] [Google Scholar]
- [13].Liacouras CA, Eosinophilic esophagitis in children and adults, J Pediatr Gastroenterol Nutr, 37 Suppl 1 (2003) S23–28. [DOI] [PubMed] [Google Scholar]
- [14].Potter JW, Saeian K, Staff D, Massey BT, Komorowski RA, Shaker R, Hogan WJ, Eosinophilic esophagitis in adults: an emerging problem with unique esophageal features, Gastrointest Endosc, 59 (2004) 355–361. [DOI] [PubMed] [Google Scholar]
- [15].Hogan SP, Mishra A, Brandt EB, Royalty MP, Pope SM, Zimmermann N, Foster PS, Rothenberg ME, A pathological function for eotaxin and eosinophils in eosinophilic gastrointestinal inflammation, Nat Immunol, 2 (2001) 353–360. [DOI] [PubMed] [Google Scholar]
- [16].Rajavelu P, Rayapudi M, Moffitt M, Mishra A, Mishra A, Significance of para-esophageal lymph nodes in food or aeroallergen-induced iNKT cell-mediated experimental eosinophilic esophagitis, American journal of physiology. Gastrointestinal and liver physiology, 302 (2012) G645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rayapudi M, Mavi P, Zhu X, Pandey AK, Abonia JP, Rothenberg ME, Mishra A, Indoor insect allergens are potent inducers of experimental eosinophilic esophagitis in mice, Journal of leukocyte biology, 88 (2010) 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhu X, Wang M, Mavi P, Rayapudi M, Pandey AK, Kaul A, Putnam PE, Rothenberg ME, Mishra A, Interleukin-15 expression is increased in human eosinophilic esophagitis and mediates pathogenesis in mice, Gastroenterology, 139 (2010) 182–193 e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rayapudi M, Rajavelu P, Zhu X, Kaul A, Niranjan R, Dynda S, Mishra A, Mattner J, Zaidi A, Dutt P, Mishra A, Invariant natural killer T-cell neutralization is a possible novel therapy for human eosinophilic esophagitis, Clinical & translational immunology, 3 (2014) e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].O'Shea KM, Aceves SS, Dellon ES, Gupta SK, Spergel JM, Furuta GT, Rothenberg ME, Pathophysiology of Eosinophilic Esophagitis, Gastroenterology, 154 (2018) 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Caldwell JM, Paul M, Rothenberg ME, Novel immunologic mechanisms in eosinophilic esophagitis, Current opinion in immunology, 48 (2017) 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Venkateshaiah SU, Kandikattu HK, Mishra A, Significance of Interleukin (IL)-15 in IgE associated eosinophilic Esophagitis (EoE), International journal of basic and clinical immunology, 2 (2019) 1–12. [PMC free article] [PubMed] [Google Scholar]
- [23].Vakil N, van Zanten SV, Kahrilas P, Dent J, Jones R, The Montreal definition and classification of gastroesophageal reflux disease: a global evidence-based consensus, The American journal of gastroenterology, 101 (2006) 1900–1920; quiz 1943. [DOI] [PubMed] [Google Scholar]
- [24].Mishra A, Hogan SP, Lee JJ, Foster PS, Rothenberg ME, Fundamental signals that regulate eosinophil homing to the gastrointestinal tract, J Clin Invest, 103 (1999) 1719–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Verma AK, Manohar M, Venkateshaiah SU, Blecker U, Collins MH, Mishra A, Role of Vasoactive Intestinal Peptide in Promoting the Pathogenesis of Eosinophilic Esophagitis (EoE), Cellular and molecular gastroenterology and hepatology, 5 (2018) 99–100 e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Venkateshaiah SU, Manohar M, Verma AK, Blecker U, Mishra A, Possible Noninvasive Biomarker of Eosinophilic Esophagitis: Clinical and Experimental Evidence, Case reports in gastroenterology, 10 (2016) 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mussarat A, Manohar M, Verma AK, Upparahalli Venkateshaiah S, Zaidi A, Sanders NL, Zhu X, Mishra A, Intestinal overexpression of interleukin (IL)-15 promotes tissue eosinophilia and goblet cell hyperplasia, Immunology and cell biology, 96 (2018) 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mishra A, Hogan SP, Brandt EB, Wagner N, Crossman MW, Foster PS, Rothenberg ME, Enterocyte expression of the eotaxin and interleukin-5 transgenes induces compartmentalized dysregulation of eosinophil trafficking, J Biol Chem, 277 (2002) 4406–4412. [DOI] [PubMed] [Google Scholar]
- [29].Mishra A, Rothenberg ME, Intratracheal IL-13 induces eosinophilic esophagitis by an IL-5, eotaxin-1, and STAT6-dependent mechanism, Gastroenterology, 125 (2003) 1419–1427. [DOI] [PubMed] [Google Scholar]
- [30].Straumann A, Greuter T, Lifting the Veil: The Quest for Noninvasive Biomarkers for the Accurate Diagnosis of Eosinophilic Esophagitis, Digestive diseases and sciences, (2020). [DOI] [PubMed] [Google Scholar]
- [31].Wright BL, Kita H, Noninvasive Diagnosis of Eosinophilic Esophagitis: The Nuclear Medicine Option, Mayo Clinic proceedings, 95 (2020) 432–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Min SB, Nylund CM, Baker TP, Ally M, Reinhardt B, Chen YJ, Nazareno L, Moawad FJ, Longitudinal Evaluation of Noninvasive Biomarkers for Eosinophilic Esophagitis, Journal of clinical gastroenterology, 51 (2017) 127–135. [DOI] [PubMed] [Google Scholar]
- [33].Konikoff MR, Blanchard C, Kirby C, Buckmeier BK, Cohen MB, Heubi JE, Putnam PE, Rothenberg ME, Potential of blood eosinophils, eosinophil-derived neurotoxin, and eotaxin-3 as biomarkers of eosinophilic esophagitis, Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association, 4 (2006) 1328–1336. [DOI] [PubMed] [Google Scholar]
- [34].Schlag C, Miehlke S, Heiseke A, Brockow K, Krug A, von Arnim U, Straumann A, Vieth M, Bussmann C, Mueller R, Greinwald R, Bajbouj M, Peripheral blood eosinophils and other non-invasive biomarkers can monitor treatment response in eosinophilic oesophagitis, Alimentary pharmacology & therapeutics, 42 (2015) 1122–1130. [DOI] [PubMed] [Google Scholar]
- [35].Kia L, Hirano I, Distinguishing GERD from eosinophilic oesophagitis: concepts and controversies, Nature reviews. Gastroenterology & hepatology, 12 (2015) 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shimaoka T, Nakayama T, Kume N, Takahashi S, Yamaguchi J, Minami M, Hayashida K, Kita T, Ohsumi J, Yoshie O, Yonehara S, Cutting edge: SR-PSOX/CXC chemokine ligand 16 mediates bacterial phagocytosis by APCs through its chemokine domain, Journal of immunology, 171 (2003) 1647–1651. [DOI] [PubMed] [Google Scholar]
- [37].Germanov E, Veinotte L, Cullen R, Chamberlain E, Butcher EC, Johnston B, Critical role for the chemokine receptor CXCR6 in homeostasis and activation of CD1d-restricted NKT cells, Journal of immunology, 181 (2008) 81–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.