Abstract
Natural products and secondary metabolites comprise an indispensable resource from living organisms that have transformed areas of medicine, agriculture, and biotechnology. Recent advances in high-throughput DNA sequencing and computational analysis, however, suggests that a vast majority of natural products remain undiscovered. To accelerate the natural product discovery pipeline, cell-free metabolic engineering approaches used to develop robust catalytic networks are being repurposed to access new chemical scaffolds, and new enzymes capable of performing diverse chemistries. Such enzymes may serve as flexible biocatalytic tools to further expand the unique chemical space of natural products and secondary metabolites and provide a more sustainable route to manufacture these molecules in the future. Here, we highlight select examples of natural product biosynthesis in cell-free systems and propose how cell-free technologies could facilitate our ability to access and modify these structures to transform synthetic and chemical biology.
Introduction
Natural products have inspired the development of numerous pharmaceuticals, food preservatives, crop protectants, cosmetics, biological probes, and more. Given their importance to society, the ability to access, manipulate, and engineer natural product chemical space is highly sought after.1 For example, new compounds are constantly needed to combat the ongoing battle against antibiotic resistance.2 Traditional antibiotic discovery approaches have been hampered by rediscovery and have failed to bring new antibiotic scaffolds to market over the last 30 years.3 Despite this challenge, evidence suggests that the majority of natural products remain undiscovered and this critical natural resource has far from dried up.4 In fact, high-throughput DNA sequencing and a growing collection of powerful bioinformatic tools have uncovered unique “cryptic” natural product gene clusters that may serve as next-generation pharmaceuticals or tools.5–11 High-throughput “bottom-up” approaches (e.g., metagenomics) can access the silent genetic potential of microorganisms and lead to a renaissance in the field of natural products.12–19 Unfortunately, methods to express biosynthetic gene clusters lag behind the strategies used to identify them.7 Alleviating this biosynthetic bottleneck would provide two major resources: (i) new scaffolds with unique functions and (ii) new enzymes with biomanufacturing utility. Importantly, these natural product tailoring enzymes may serve as valuable tools to produce natural product-like compounds via combinatorial biosynthesis.20–22
In this minireview, we discuss select advances in cell-free natural product biosynthesis and the exciting opportunity of combining cutting edge cell-free technologies to facilitate natural product pathway assembly, enzyme discovery, and pathway evolution towards combinatorial biosynthesis. First, we highlight recent works as useful blueprints for the assembly of native natural product pathways using cell-free systems. Then, we present high-throughput, cell-free strategies for enzyme characterization and evaluation of their substrate promiscuity, followed by a discussion of exploiting these natural promiscuities to assemble novel pathways and compounds. Finally, we touch on how merging bioorthogonal chemistry with cell-free synthesized natural products can provide chemical diversity not accessible through enzymatic or chemical synthesis alone. Collectively, cell-free systems provide tools with the potential to create and diversify the natural product chemical space at an unprecedented rate, helping to bridge the gap between the pace of biosynthetic gene cluster identification and characterization.
Advantages of cell-free pathway engineering
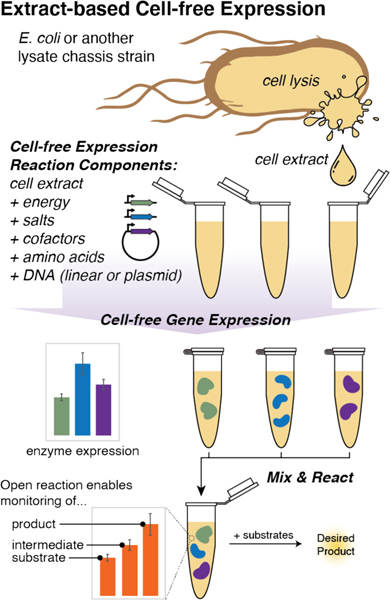
Extract-based cell-free systems offer several unique advantages over complementary in vivo and purified enzyme-based approaches for enzyme characterization and pathway assembly. The foundational principle is that discrete biosynthetic pathways can be constructed through modular assembly of cell-free lysates pre-enriched with enzyme(s) produced either by overexpression in a lysate chassis strain (in vivo production) or by direct cell-free gene expression (Fig. 1).23,24 Cell-free gene expression is not constrained by sustaining life, therefore, much of the ellular resources can be directed towards producing a desired product. High concentrations of desired enzyme homologs or entire pathways can be produced in a few hours from linear DNA templates. Typically performed at small scales (μL), cell-free gene expression (CFE) allows for 100s of enzyme homologs (or variants) to be expressed in parallel without specialized equipment. In addition, cell-free systems offer a controllable and open environment for precise substrate, intermediate, and product manipulation and monitoring (Fig. 1). This enables stepwise reconstruction of complex natural pathways with the ability to easily observe and characterize intermediates and products, rapidly identify biosynthetic bottlenecks, troubleshoot individual steps, and deliver high yields without the need for enzyme purification.25,26 Cell-free systems can bolster in vitro, purified approaches by providing a native-like metabolism and precursor pools. For example, a dehydratase from the nisin biosynthetic pathway eluded reconstitution in vitro for 20 years until the addition of bacterial cell extract allowed the desired dehydration activity.27 These advantages provide a basis for exploring biosynthetic enzymes and engineering natural product pathways with CFE.
Figure 1.
General scheme of cell-free gene expression & in vitro reconstitution of a biosynthetic pathway.
High-throughput pathway assembly in the cell-free environment
Using CFE, natural product pathways can be rapidly assembled by combining multiple enzymes at user-defined concentrations. For example, a diketopiperazine was recently produced in the cell-free environment by combining two enzymes in the gramicidin-S biosynthetic gene cluster (Fig. 2).28 The modular and customizable nature of cell-free systems enables combinatorial biosynthesis of complete pathways where individual steps in a given pathway can be replaced with enzyme homologs from different strains, organisms, or even engineered enzymes.25 One early example used pre-enriched extracts to reconstruct a flexible route to aminoglycosides in a stepwise manner.29 These results informed the design of custom aminoglycoside biosyntheses, using enzymes from multiple natural pathways, to produce superior analogs by in vivo production. Similarly, a five enzyme pathway was assembled by CFE for the production of n-butanol.30 This pathway was recently expanded to combinatorially screen over hundreds of pathway designs for the production of 3-hydroxybutyrate and n-butanol.31 Top-performing pathways were then implemented in the non-model organism Clostridium autoethanogenum, thus improving in vivo production by more than 20- and 4-fold, respectively. This is a key example of screening a desired pathway in a high-throughput fashion that would otherwise be slow, or potentially not feasible, in vivo.
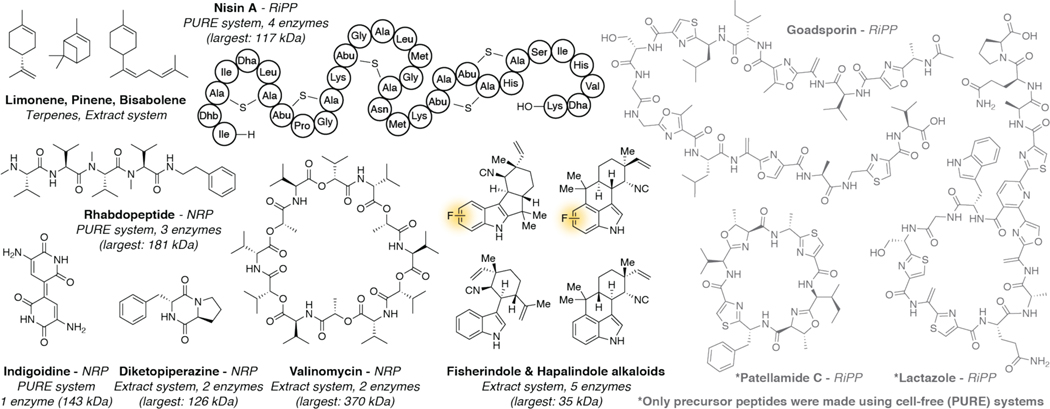
Figure 2.
Natural products made using various cell free systems (PURE or extract-based). The total number of enzymes and the largest enzyme made in each system are noted. Highlighted in yellow are unnatural variants that were made using cell-free technologies. Natural products in grey represent examples where only part of the biosynthesis was reconstituted in a cell-free system. Only the precursor peptides were synthesized using the PURE system and all enzymes were expressed heterologously and purified.
Extract-based cell-free systems have now expanded to produce full natural product pathways spanning many classes of compounds (Fig. 2). Optimal pathways for two monoterpenes (limonene and pinene) and a sesquiterpene (bisabolene) were developed by screening over 150 unique sets of enzymes in 580 discrete pathway conditions.32–34 Three non-ribosomal peptide natural products (NRPs) have been produced using cell free systems. A CFE system was used to express the entire valinomycin gene cluster (>19kb) and produce valinomycin in a one-pot reaction in just a few hours.35 Importantly, the two enzymes within the pathway, Vlm1 (370kDa) and Vlm2 (284kDa), represent two of the largest enzymes ever reported using cell-free protein synthesis. Additionally, lysates pre-enriched with these enzymes produce high titers that rival yields from the native producers in vivo.36 A separate study described similar syntheses of NRPs indigoidine and rhabdopeptide using the commercially available PURExpress system.37 The authors were also able to use CFE to synthesize several other megasynthases from additional NRPs, fatty acid, and polyketide pathways although their native activity was not demonstrated. A series of indole alkaloids was produced using cell-free technology.38 Several unnatural halogenated indole compounds were produced by simply feeding reactions with chemically synthesized precursors. Lastly, the pathway for the prototypical lanthipeptide, nisin, was recently rebuilt using extract-based CFE.39 The authors coupled cell-free nisin biosynthesis with an antibiotic activity assay which allowed them to screen over 3000 analogues and quickly identify 2 variants that were more active than the parent compound. Cell-free systems have been used to synthesize other ribosomally synthesized and post-translationally modified peptide (RiPP) natural products, however only precursor peptides (or analogues) were synthesized this way.40–45 These initial examples of natural product pathways established in cell-free systems mark the starting point of a field that will improve metabolic engineering efforts for a wide variety of natural product classes.
Cell-free platforms facilitate enzyme characterization
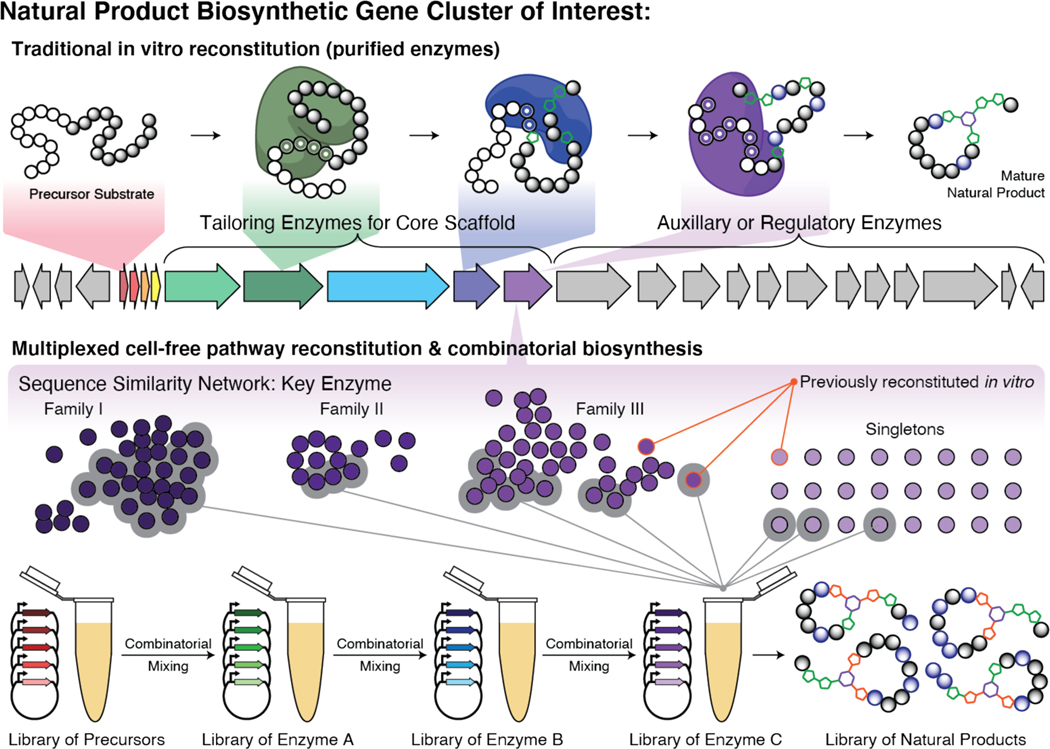
With an ultimate goal of (combinatorial) natural product pathway assembly, an important first step is the evaluation of the constituent biosynthetic enzymes.20 Sequence similarity networks are often used to organize enzyme homologs into families based on protein sequences, and ultimately, their presumed function.46 Typically, the number of homologs tested in a given study is low because traditional biochemical assays rely on heterologous expression and require time-consuming and labor intensive enzyme purification steps. High-throughput enzyme characterization using cell-free systems may enable a large percentage of enzyme families to be examined in parallel (Fig. 3), as has been the case with other metabolic pathways like limonene as described above. This allows for the rapid identification of enzymes with desired qualities, evaluation of substrate promiscuity, and enables one to make broad conclusions about enzymes families rather than single family members.
Figure 3.
Cell-free reconstitution & combinatorial biosynthesis of a natural product. The pathway depicted is loosely based on general RiPP biosynthesis. Large numbers of key enzyme homologs (purple), visualized via sequence similarity networks, can be expressed on small scales in parallel to investigate their function in a high throughput manner. This strategy can be multiplexed to the entire pathway and enable combinatorial biosynthesis of natural product libraries.
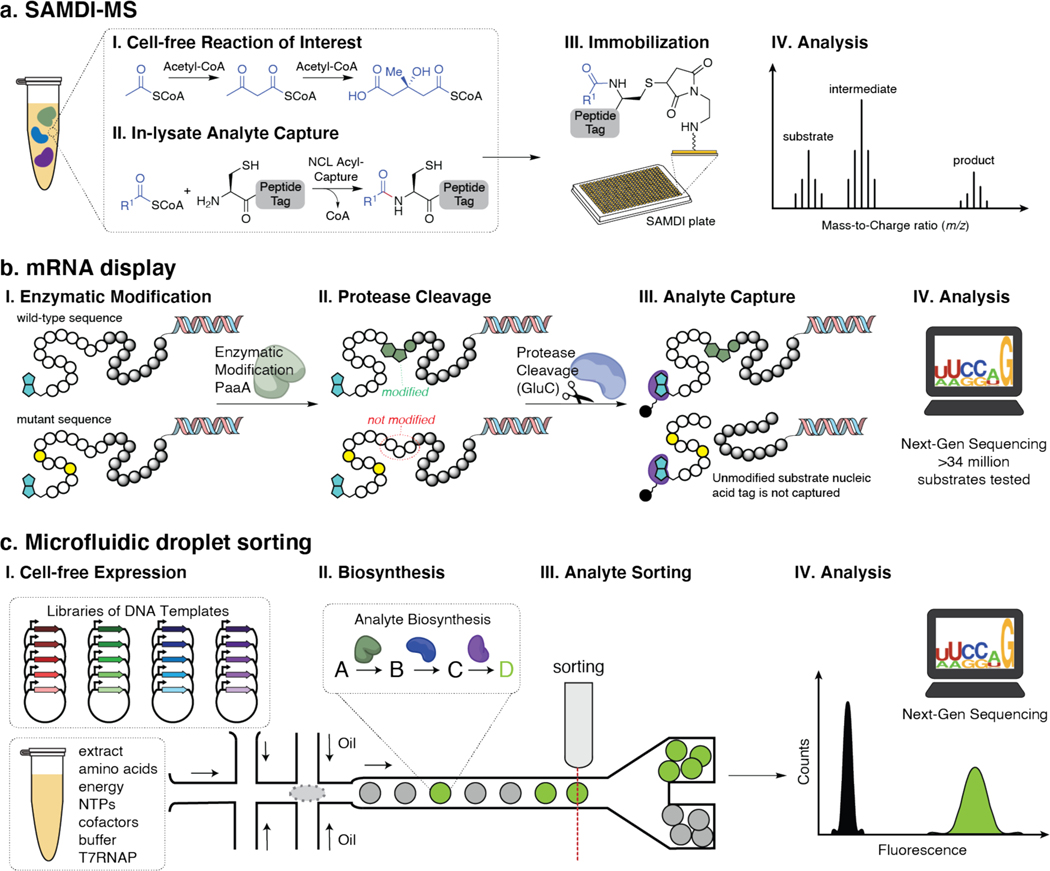
To study enzyme families in depth, the throughput of enzyme expression must be matched with rapid and quantitative biochemical assessment. In this regard, cell-free platforms have already been integrated with high-throughput enzyme screening assays; including, self-assembled monolayers for matrix-assisted desorption/ionization mass spectrometry (SAMDI-MS), mRNA display, in-droplet reaction microfluidics, and next generation sequencing (Fig. 4). By combining cell-free protein synthesis with SAMDI-MS, reactive metabolites are captured on monolayers that can be analyzed by MALDI-TOF MS (Fig. 4a). The approach was used to rapidly screen over 800 unique reaction conditions to optimize the synthesis of hydroxymethylglutaryl-CoA, a biosynthetic precursor of mevalonate and isoprenoid metabolites.47 Using a capture technique similar to native chemical ligation, all acyl intermediates on CoA could be simultaneously analyzed, revealing a full picture of the biocatalytic system. Similarly, SAMDI has also been used to investigate the promiscuity of N-glycosyltransferases (NGTs). More than 3,000 peptide substrates were rapidly screened in 13,903 unique reaction conditions to develop precise peptide acceptor sequences that showed robust glycosylation both in vitro and in vivo.48
Figure 5.
High-throughput methods to rigorously evaluation enzyme function and substrate promiscuity.
mRNA display has been repurposed to intensively investigate the promiscuity of RiPP tailoring enzyme PaaA, in the biosynthesis of Pantocin A (Fig. 4b).49 Over 34 million substrates were screened to reveal PaaA’s broad substrate tolerance. While this current approach was designed specifically around the chemistry performed by PaaA and is not broadly applicable to other RiPP enzymes, it represents a first major step in using massive display libraries to inform broadly on substrate promiscuity of natural product tailoring enzymes. Importantly, it provides another intriguing access point for generating libraries of natural products for drug discovery.
In a recent study, droplet-based microfluidic sorting together with next-generation sequencing enabled functional screening of a 1 million-membered metagenomic library which revealed previously undiscovered hydrolases and profiled their substrate promiscuity.50 While this study was performed in vivo, this methodology relied on lysing cells within droplets to release enzymes and modify their substrates. Enzyme transformations are sampled and analyzed at the in vitro stage, therefore a cell-free systems screening approach could be easily implemented to accelerate the discovery pipeline (Fig. 4c).
While some of the examples mentioned above describe strategies to determine enzyme specificity/promiscuity outside the context of natural products, we anticipate that these technologies will be useful to investigate natural product tailoring enzymes that modify peptides (RiPPs, NRPs, antimicrobial peptides) and CoA substrates (isoprenoids, polyketides) as well as natural product glycosyltransferases (glycorandomization).
Towards cell-free combinatorial natural product biosynthesis
One of the exciting features of cell-free systems, is the combinatorial assembly strategies that can be used. For example, the ability to exploit the promiscuous nature of natural product tailoring enzymes and assemble hybrid pathways by combining enzymes from different natural product biosynthetic clusters could yield new-to-nature natural products. Screening such pathways in vivo is challenging because troubleshooting variants that produce truncated side-products or non-functional products is time-consuming and can be convoluted. Cell-free extract-based screening methods however, are well positioned to accelerate testing of hybrid pathways and advance the field of natural product synthetic biology towards the long-standing goal of producing custom molecules by rationally designing biosynthetic pathways. Analogous to the natural product workflow, combinatorial biosynthesis of glycan motifs was demonstrated using a small series of glycosyltransferases. In total, 37 putative pathways yielded 23 unique glycans, 18 of which had never been synthesized previously.51
RiPPs represent a suitable entry point for the synthesis of hybrid natural products, as libraries of precursor peptides can be synthesized directly in the cell-free environment and then combined with a library of tailoring enzymes also synthesized via CFE (Fig. 3). RiPPs have been shown to be amenable to hybridization by the successful production of functional thiazoline-lanthipeptide, thiazoline-sactipeptide and thiazoline-lanthionine hybrids in vivo.52 Similarly, enzymes from disparate RiPPs pathways were used to synthesize natural and unnatural thiopeptides based on thiocillin and lactazole scaffolds in vitro with peptide substrates synthesized using the PURE system.44 Furthermore, RiPPs precursor peptides contain discrete recognition sequences that allow tailoring enzymes to recognize and modify their substrates.53 These recognition elements may be engineering to develop superior or more versatile catalysts. For instance, RiPP-tailoring enzymes have been engineered with bound leader peptide fragments, making them constitutively active and able to modify precursors that lack leader peptides.54 This strategy may enhance the already broad substrate promiscuity and enable further substrate sampling.
Integrating biological and chemical synthesis of natural products
In developing new-to-nature pathways, some target transformations may be inaccessible enzymatically or difficult to evolve. In these cases, the open-environment of a cell-free reaction offers the opportunity to incorporate synthetic chemistry through hybrid chemical reactions.55 Chemical transformations may be applied as final tailoring steps or as intermediate steps to allow further enzymatic derivatization. In a recent in vivo study, a hybrid pathway generated several dozen analogs of the natural product violacein, including those that bore aryl-bromides. These unnatural brominated analogues were further derivatized in crude lysates using Suzuki-Miyaura cross couplings to afford an additional 20 analogues that are inaccessible by purely enzymatic means.56 In a separate study, oxidative elimination of Se-phenylselenocysteine using peroxide allowed site selective installation of multiple dehydroalanines (Dhas; a common modification in natural products57) on peptides generated with the PURE system.44 These Dha-bearing peptides served as substrates for pyridine synthases that catalyze unique aza-[4+2] cycloadditions to afford two classes of thiopeptide natural products. Combining both chemical and enzymatic transformations is a useful tactic to generate new-to-nature, natural products that may be challenging to access using either method independently.
Challenges and opportunities for cell-free natural product biosynthesis
Several aspects of the natural product biosynthesis must be considered when applying cell-free technology. For example, polyketides and non-ribosomal peptide natural products are typically synthesized by particularly large enzymes (>150kDa). While such enzymes have been synthesized in CFE systems, there are still relatively few examples.35,37 Several strategies could be used to address this. First, pre-enriched lysates from traditional in vitro expression provide one current alternative strategy to access larger constructs.35 Second, specialized extracts, rich in cold shock proteins or chaperones may provide an attractive solution.58 Third, the expression of each individual enzyme could be separately optimized using lysates from different strains or organisms. Indeed, the idea of organismal amalgams not limited to a single host-derived extract is a tantalizing feature of cell-free systems.
Enzymes that require special or unknown cofactors, metabolites, or post-translational modifications are ongoing challenges in natural product biosynthesis in general and represent opportunities for cell-free systems. While some known cofactors and metabolites may be added to cell-free premixes, these scenarios may be best approached by using host derived extracts. Indeed, Streptomyces extracts have been prepared and although these systems are less productive than many E. coli counterparts, improvements are being made.59–61 Many interesting natural products are produced by hosts that inhabit anaerobic environments. Creating cell-free systems that function anaerobically to produce oxygen-sensitive enzymes are just starting to be explored.62 Access to these enzymes could open a range of unique and useful chemistries.63,64 Cell-free systems are primed to rapidly explore natural product chemical space and the field is rich with opportunity to expand its access to new biocatalysts and natural products.
Conclusion
The combinatorial biosynthesis of natural products has been a long-standing and evolving goal in the natural product and synthetic biology communities. Key to realizing this goal is generating versatile sets of diverse biocatalysts capable of constructing building blocks, assembling them into core scaffolds, and further diversifying those scaffolds. While tremendous progress has been made using both in vivo and purified in vitro systems, knowledge gaps still exist in how individual biosynthetic enzymes function and how multiple enzymes can be assembled to create novel pathways. In addition, the number of putative biosynthetic gene clusters greatly outnumbers those that have been characterized.
Cell-free technologies have just begun to demonstrate their potential impact on natural product synthesis through production of terpenes, NRPSs, indole alkaloids, saccharides, and RiPPs. The above examples highlight how the user-friendly nature, modularity, and speed at which one can go from gene to natural product can make extract-based cell-free systems an enabling technology to rapidly explore natural product chemical space. Integration with computer-aided pathway design and machine learning, could further accelerate design-build-test cycles.31,65 Although an extract-based cell-free gene expression system has yet, to our knowledge, been used to characterize a previously unknown natural product pathway, all the tools are in place to accomplish this. Taken together, we anticipate that cell-free systems will serve as a powerful, high-throughput engine for enzyme and cryptic pathway characterization, expand our ability to access and manipulate complex chemical space, and establish a new natural product renaissance.
Acknowledgments
M.C.J. acknowledges support from the National Institutes of Health Grant 1U19AI142780–01,the DARPA 1000 Molecules Program HR0011–15-C-0084, the Department of Energy Grant DE-SC0018249, the Office of Energy Efficiency and Renewable Energy Grant DE-EE0008343, the David and Lucile Packard Foundation, and the Camille Dreyfus Teacher-Scholar Program. The U.S. Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright notation thereon. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of DARPA or the U.S. Government. B.V. acknowledges support from the SNSF Early Postdoc.Mobility fellowship P2SKP3_184036. D.A.W. acknowledges support from the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1842165.
M.C.J. has a financial interest in Design Pharmaceuticals Inc. and SwiftScale Biologics. M.C.J.’s interests are reviewed and managed by Northwestern University in accordance with their conflict of interest policies.
Footnotes
Competing interests
All other authors declare no conflicts of interest.
References
- (1).Karageorgis G; Foley DJ; Laraia L; Waldmann H. Nat. Chem 2020, 12 (3), 227–235. [DOI] [PubMed] [Google Scholar]
- (2).Brown ED; Wright GD Nature 2016, 529 (7586), 336–343. [DOI] [PubMed] [Google Scholar]
- (3).Scientific Roadmap for Antibiotic Discovery. Pew Charitable Trusts (11 May 2016); http://www.pewtrusts.org/antibiotic-discovery.
- (4).Pye CR; Bertin MJ; Lokey RS; Gerwick WH; Linington RG Proc. Natl. Acad. Sci. U.S.A 2017, 114 (22), 5601–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Donia MS; Cimermancic P; Schulze CJ; Brown LCW; Martin J; Mitreva M; Clardy J; Linington RG; Fischbach MA Cell 2014, 158 (6), 1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Doroghazi JR; Albright JC; Goering AW; Ju K-S; Haines RR; Tchalukov KA; Labeda DP; Kelleher NL; Metcalf WW Nat. Chem. Biol 2014, 10 (11), 963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Navarro-Muñoz JC; Selem-Mojica N; Mullowney MW; Kautsar SA; Tryon JH; Parkinson EI; de los Santos ELC; Yeong M; Cruz-Morales P; Abubucker S; Roeters A; Lokhorst W; Fernandez-Guerra A; Cappelini LTD; Goering AW; Thomson RJ; Metcalf WW; Kelleher NL; Barona-Gomez F; Medema MH Nat. Chem. Biol 2020, 16 (1), 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lok C. Nature News 2015, 522 (7556), 270–273. [DOI] [PubMed] [Google Scholar]
- (9).Blin K; Shaw S; Steinke K; Villebro R; Ziemert N; Lee SY; Medema MH; Weber T. Nucleic Acids Res. 2019, 47 (W1), W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Tietz JI; Schwalen CJ; Patel PS; Maxson T; Blair PM; Tai H-C; Zakai UI; Mitchell DA Nat. Chem. Biol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Merwin NJ; Mousa WK; Dejong CA; Skinnider MA; Cannon MJ; Li H; Dial K; Gunabalasingam M; Johnston C; Magarvey NA Proc. Natl. Acad. Sci. U.S.A 2020, 117 (1), 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kersten RD; Weng J-K Proc. Natl. Acad. Sci. U.S.A 2018, 115 (46), E10961–E10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yamanaka K; Reynolds KA; Kersten RD; Ryan KS; Gonzalez DJ; Nizet V; Dorrestein PC; Moore BS Proc. Natl. Acad. Sci. U.S.A 2014, 111 (5), 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Bachmann BO; Van Lanen SG; Baltz RH J. Ind. Microbiol. Biotechnol 2013, 41 (2), 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Medema MH; Fischbach MA Nat. Chem. Biol 2015, 11 (9), 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wright GD Microb. Biotechnol 2018, 12 (1), 55–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Scott TA; Piel J. Nat. Rev. Chem 2019, 3 (7), 404–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Freeman MF; Gurgui C; Helf MJ; Morinaka BI; Uria AR; Oldham NJ; Sahl H-G; Matsunaga S; Piel J. Science 2012, 338 (6105), 387–390. [DOI] [PubMed] [Google Scholar]
- (19).Milshteyn A; Schneider JS; Brady SF Chem. Biol 2014, 21 (9), 1211–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Kim E; Moore BS; Yoon YJ Nat. Chem. Biol 2015, 11 (9), 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Rodrigues T; Reker D; Schneider P; Schneider G. Nature Chemistry 2016. [DOI] [PubMed] [Google Scholar]
- (22).Walsh CT ChemBioChem 2002, 3 (2–3), 124–134. [Google Scholar]
- (23).Silverman AD; Karim AS; Jewett MC Nat. Rev. Genet 2020, 21 (3), 151–170. [DOI] [PubMed] [Google Scholar]
- (24).Gregorio NE; Levine MZ; Oza JP Methods and Protocols 2019, Vol. 2, Page 24 2019, 2 (1), 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Karim AS; Jewett MC Methods Enzymol. 2018, 608, 31–57. [DOI] [PubMed] [Google Scholar]
- (26).Dudley QM; Karim AS; Jewett MC Biotechnol. J 2015, 10 (1), 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Garg N; Salazar-Ocampo LMA; van der Donk WA Proc. Natl. Acad. Sci. U.S.A 2013, 110 (18), 7258–7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Goering AW; Li J; McClure RA; Thomson RJ; Jewett MC; Kelleher NL ACS Synth. Biol 2017, 6 (1), 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Park JW; Park SR; Nepal KK; Han AR; Ban YH; Yoo YJ; Kim EJ; Kim EM; Kim D; Sohng JK; Yoon YJ Nat. Chem. Biol 2011, 7 (11), 843–852. [DOI] [PubMed] [Google Scholar]
- (30).Karim AS; Jewett MC Metab. Eng 2016, 36, 116–126. [DOI] [PubMed] [Google Scholar]
- (31).Karim AS; Dudley QM; Juminaga A; Yuan Y; Crowe SA; Heggestad JT; Garg S; Abdalla T; Grubbe WS; Rasor BJ; Coar D; Torculas M; Krein M; Liew FE; Quattlebaum A; Jensen RO; Stuart JA; Simpson SD; Köpke M; Jewett MC Nat. Chem. Biol 2019, 10, 319. [DOI] [PubMed] [Google Scholar]
- (32).Dudley QM; Karim AS; Nash CJ; Jewett MC Metab. Eng 2020, doi: 10.1016/j.ymben.2020.05.006. [DOI] [PubMed] [Google Scholar]
- (33).Dudley QM; Nash CJ; Jewett MC Synth. Biol 2019, 4 (1), 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Niu F-X; Huang Y-B; Shen Y-P; Ji L-N; Liu J-ZJ Agric. Food Chem 2020, 68 (7), 2139–2145. [DOI] [PubMed] [Google Scholar]
- (35).Zhuang L; Huang S; Liu W-Q; Karim AS; Jewett MC; Li J. Metab. Eng 2020, 60, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Li J; Jaitzig J; Theuer L; Legala OE; Süssmuth RD; Neubauer PJ Biotechnol. 2015, 193, 16–22. [DOI] [PubMed] [Google Scholar]
- (37).Siebels I; Nowak S; Heil CS; Tufar P; Cortina NS; Bode HB; Grininger M. bioRxiv 2020, 33, 2020.04.04.025353. [DOI] [PubMed] [Google Scholar]
- (38).Khatri Y; Hohlman RM; Mendoza J; Li S; Lowell AN; Asahara H; Sherman DH ACS Synth. Biol 2020, 9 (6), 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Liu R; Zhang Y; Zhai G; Fu S; Xia Y; Ben Hu; Cai X.; Zhang Y; Li Y; Deng Z; Liu T. bioRxiv 2019, 5, 757591. [Google Scholar]
- (40).Goto Y; Ito Y; Kato Y; Tsunoda S; Suga H. Chem. Biol 2014, 21 (6), 766–774. [DOI] [PubMed] [Google Scholar]
- (41).Ozaki T; Yamashita K; Goto Y; Shimomura M; Hayashi S; Asamizu S; Sugai Y; Ikeda H; Suga H; Onaka H. Nat. Commun 2017, 8 (1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Goto Y; Suga H. Bull. Chem. Soc. Jpn 2018, 91 (3), 410–419. [Google Scholar]
- (43).Goto Y; Suga H. ChemBioChem 2020, 21 (1–2), 84–87. [DOI] [PubMed] [Google Scholar]
- (44).Fleming SR; Bartges TE; Vinogradov AA; Kirkpatrick CL; Goto Y; Suga H; Hicks LM; Bowers AA J. Am. Chem. Soc 2019, 141 (2), 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Vinogradov AA; Shimomura M; Goto Y; Ozaki T; Asamizu S; Sugai Y; Suga H; Onaka H. Nat. Commun 2020, 11 (1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Shannon P; Markiel A; Ozier O; Baliga NS; Wang JT; Ramage D; Amin N; Schwikowski B; Ideker T. Genome Res. 2003, 13 (11), 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).O’Kane PT; Dudley QM; McMillan AK; Jewett MC; Mrksich M. Sci. Adv 2019, 5 (6), eaaw9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Kightlinger W; Lin L; Rosztoczy M; Li W; DeLisa MP; Mrksich M; Jewett MC Nat. Chem. Biol 2018, 14 (6), 627–635. [DOI] [PubMed] [Google Scholar]
- (49).Fleming SR; Himes PM; Ghodge SV; Goto Y; Suga H; Bowers AA J. Am. Chem. Soc 2020, 142 (11), 5024–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Colin P-Y; Kintses B; Gielen F; Miton CM; Fischer G; Mohamed MF; Hyvönen M; Morgavi DP; Janssen DB; Hollfelder F. Nat. Commun 2015, 6 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Kightlinger W; Duncker KE; Ramesh A; Thames AH; Natarajan A; Stark JC; Yang A; Lin L; Mrksich M; DeLisa MP; Jewett MC Nat. Commun 2019, 10 (1), 2364–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Burkhart BJ; Kakkar N; Hudson GA; van der Donk WA; Mitchell DA ACS Cent. Sci 2017, 3 (6), 629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Burkhart BJ; Hudson GA; Dunbar KL; Mitchell DA Nat. Chem. Biol 2015, 11 (8), 564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Koehnke J; Mann G; Bent AF; Ludewig H; Shirran S; Botting C; Lebl T; Houssen WE; Jaspars M; Naismith JH Nat. Chem. Biol 2015, 11 (8), 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Swartz JR AIChE J. 2012, 58 (1), 5–13. [Google Scholar]
- (56).Lai H-E; Obled AMC; Chee SM; Morgan RM; Sharma SV; Moore SJ; Polizzi KM; Goss RJM; Freemont PS bioRxiv 2019, 51, 202523. [Google Scholar]
- (57).Bogart JW; Bowers AA Org. Biomol. Chem 2019, 17 (15), 3653–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Higuchi K; Yabuki T; Ito M; Kigawa T. Biotechnol. Bioeng 2020, 117 (6), 1628–1639. [DOI] [PubMed] [Google Scholar]
- (59).Li J; Wang H; Kwon Y-C; Jewett MC Biotechnol. Bioeng 2017, 114 (6), 1343–1353. [DOI] [PubMed] [Google Scholar]
- (60).Li J; Wang H; Jewett MC Biochem. Eng. J 2018, 130, 29–33. [Google Scholar]
- (61).Xu H; Liu W-Q; Li J. ACS Synth. Biol 2020, 9 (5), 1221–1224. [DOI] [PubMed] [Google Scholar]
- (62).Krüger A; Mueller AP; Rybnicky GA; Engle NL; Yang ZK; Tschaplinski TJ; Simpson SD; Köpke M; Jewett MC Metab. Eng 2020. doi: 10.1016/j.ymben.2020.06.004. [DOI] [PubMed] [Google Scholar]
- (63).Buckel W; Golding BT Annu. Rev. Microbiol 2006, 60 (1), 27–49. [DOI] [PubMed] [Google Scholar]
- (64).Nicolet Y. Nat. Catal 2020, 3 (4), 337–350. [Google Scholar]
- (65).Finnigan W; Hepworth LJ; Turner NJ; Flitsch S. ChemRxiv. Preprint 2020, doi: 10.26434/chemrxiv.12571235.v1. [DOI] [Google Scholar]