Abstract
Mixtures risk assessment needs an efficient integration of in vivo, in vitro, and in silico data with epidemiology and human studies data. This involves several approaches, some in current use and others under development. This work extends the Agency for Toxic Substances and Disease Registry physiologically based pharmacokinetic (PBPK) toolkit, available for risk assessors, to include a mixture PBPK model of benzene, toluene, ethylbenzene, and xylenes. The recoded model was evaluated and applied to exposure scenarios to evaluate the validity of dose additivity for mixtures. In the second part of this work, we studied toluene, ethylbenzene, and xylene (TEX)-gene-disease associations using Comparative Toxicogenomics Database, pathway analysis and published microarray data from human gene expression changes in blood samples after short- and long-term exposures. Collectively, this information was used to establish hypotheses on potential linkages between TEX exposures and human health. The results show that 236 genes expressed were common between the short- and long-term exposures. These genes could be central for the interconnecting biological pathways potentially stimulated by TEX exposure, likely related to respiratory and neuro diseases. Using publicly available data we propose a conceptual framework to study pathway perturbations leading to toxicity of chemical mixtures. This proposed methodology lends mechanistic insights of the toxicity of mixtures and when experimentally validated will allow data gaps filling for mixtures’ toxicity assessment. This work proposes an approach using current knowledge, available multiple stream data and applying computational methods to advance mixtures risk assessment.
Keywords: VOCs, PBPK, toxicogenomics, computational systems biology, enrichment analysis
Environmental toxic substances such as volatile organic compounds (VOCs) include a variety of chemicals, some of which have short- and long-term adverse health effects. Volatile organic compounds exposures may increase risks for cancer and exacerbate asthma and other adverse respiratory effects (Bolden et al., 2015; Dehghani et al., 2018; Lim et al., 2014). In fact, numerous studies in humans identified the respiratory tract as a sensitive target of individual VOCs such as benzene, toluene, ethylbenzene, and xylenes and their mixture (BTEX) (ATSDR, 2004, 2007, 2010, 2017). However, the mechanistic understanding of these health effects is lacking.
Studies have suggested that prolonged occupational exposure to VOCs may result in increased levels of biomarkers of oxidative stress, DNA damage and dysregulation of cellular antioxidant defense and trigger processes leading possibly to carcinogenesis (ATSDR, 2004; Lundberg and Milatou-Smith, 1998). Epidemiological studies have reported adverse outcomes to VOCs exposure even if no biological and occupational exposure limits are exceeded (Moro et al., 2010).
Hong and colleagues have identified time-dependent biomarkers and effects of exposure to toluene, ethylbenzene, and xylenes (TEX) using high-throughput methods, such as microarray analysis (Hong et al., 2016). They reported short-term exposure changes in the expression of genes related to the respiratory, kidney, liver, and immune system at short-term exposure. In contrast, long-term exposure was related to genes associated with nervous system and hormone imbalance, and shown to be related to the cell cycle, cell growth such as cancerrelated processes.
Available toxicogenomics databases and computational systems biology tools facilitate the identification of important toxicity pathways and molecules from large data sets. These tasks can be extremely laborious when performed by a classical literature search. Computational systems biology offers more advantages than just providing a high-throughput literature search engine. Experimentally identified genetic targets data of the specific chemical(s) of interest can be uploaded and overlapped into biological networks to gain insights that can be used to formulate hypotheses to link chemical exposure and human diseases. Consequently, this information can also be applied for designing more intelligent animal and cell-based laboratory experiments to test the formulated hypotheses.
Physiologically based pharmacokinetic (PBPK) models have been used to organize pertinent chemical, physical, and pathological information to study characteristics of a chemical including the concentrations of chemicals in various organs/ tissues. Because of their versatility and potential use, numerous PBPK models have been developed and published. However, use of different simulation platforms among various developers can limit model acceptance or use. This limitation also restricts standardized application of models in public health practice. Even experienced PBPK modelers sometimes face problems when trying to apply published PBPK models that lack key information or equations. Apart from educating and training, research is needed to make PBPK models more accessible and in easy-to-use formats. Having models in an easier-to-use, standardized format capable of simulating real life scenarios will greatly increase their value and use, thus enhancing integration of innovative tools and scientific advances into decision-making processes. The Agency for Toxic Substances and Disease Registry (ATSDR) has developed a PBPK model toolkit, these are being recoded into a single language, Berkeley Madonna (Emond et al., 2017; Mumtaz et al., 2012; Ruiz et al., 2010) and are validated and evaluated as part of the methods development.
For mixtures, PBPK is useful to determine the internal dose based on the interaction among the 3 chemicals in the mixture. In addition, the genes expression drives the modification of the biological pathways preceding a disease. Changes in genes expression represent a sensible toxicity endpoint providing information on what can be affecting by the mixture exposure scenarios. There have been several attempts to use genomics and PBPK models to assess network and pathways affected by individual chemical exposure to establish links among chemical-gene-disease interactions and key biological events (Andersen et al., 2017a,b; Maldonado et al., 2017; Sier et al., 2017).
In this work, available mixtures PBPK models, toxicogenomic databases, systems biology tools, and published gene expression data are used to gain insights into the pathways affected by exposure to TEX mixtures.
The aims of this study were to (1) share a new recoded PBPK model for BTEX, part of our toolkit at the ATSDR, with the scientific community, (2) explore the use of individual or combined chemical published data from various sources to generate or hypothesize the potential mechanisms of toxicity of the TEX mixture, and (3) introduce methodology for new testable hypotheses to address some of the chemical mixtures data gaps previously identified by federal agencies.
MATERIALS AND METHODS
We took a 2-pronged approach in this exploratory study. First, we recoded an existing PBPK VOC mixture model for BTEX using Berkeley Madonna software and assessed its reproducibility. In the second step, we used toxicogenomic and computational systems biology tools and available human gene expression data after short- and long-term exposure TEX mixture to gain mechanistic insights of molecular and cellular processes leading to the toxicity of TEX mixture (Figure 1).
Figure 1.
Schematic overview of the physiologically based pharmacokinetic (PBPK) and signaling pathways for potential use in chemical mixtures assessment. A, Representation of the PBPK steps used. Recoding, evaluation, and application of the published (B) toluene, ethylbenzene, and xylene (TEX) PBPK model. B, Toluene, ethylbenzene, and xylene pathways analysis. Representation of the chemical/gene/disease’s interactions component. Use of published high-throughput analysis, toxicogenomic databases, and systems biology platforms to understand pathways impacted by chemical mixtures. C, PBPK/PD and signaling pathways. Representation of exploring the A and B components and future connecting directions.
Recoding, Evaluation, and Application of the BTEX PBPK Model
Model structure and physiological parameters.
We reviewed previously published human VOCs mixture PBPK models (Dennison et al., 2005; Haddad et al., 1999, 2000, 2001; Marchand et al., 2015, 2016; Tardif et al., 1995, 1997). We recoded the BTEX model using Berkeley Madonna software (see Supplementary) version 8.3 for Windows (University of California at Berkeley, California). The recoded PBPK model presented here adapts the original model developed in AcslX (Haddad et al., 1999) using human physiological parameter values given in Table 1 (Marchand et al., 2015; Tardif et al., 1995, 1997). This model was used to predict alveolar air concentration levels in human volunteers exposed for 7h to toluene (17ppm), ethylbenzene (33ppm), and m-xylene (33ppm) alone or in combination.
Table 1.
Physiological Parameter Values for a Standard Adult Male Human
| Parameters | Symbol | Value | ||
|---|---|---|---|---|
| Body weight | BW | 70 | ||
| Cardiac output (l/h) | Qc | 18*BW0.7 | ||
| Alveolar ventilation rate (l/h) | Qalv | 18*BW0.7 | ||
| Compartment volume (% BW) | ||||
| Adipose tissue | Vf | 19 | ||
| Liver | Vl | 2.6 | ||
| Richly perfused tissues | Vr | 5 | ||
| Poorly perfused tissues | Vmp | 62 | ||
| Blood flow to tissue compartment (% Qc) | ||||
| Adipose tissue | Qf | 5 | ||
| Liver | Ql | 26 | ||
| Richly perfused tissues | Qr | 44 | ||
| Poorly perfused tissues | Qmp | 25 | ||
| Parametersa | Symbol | TOLa | EBZa | m-XYLa |
| Blood:air partitioning | Pb:a | 15.6 | 28.0 | 26.4 |
| Liver:air partitioning | Pl:a | 83.6 | 83.8 | 90.9 |
| Adipose tissue:air partitioning | Pf:a | 1021 | 1556 | 1859 |
| Richly perfused tissues:air partitioning | Pr:a | 83.6 | 60.3 | 90.9 |
| Slowly perfused tissues:air partitioning | Ps:a | 27.7 | 26 | 41.9 |
| Maximal rate (mg/h/kg) | Vmax | 4.8 | 7.3 | 5.5 |
| Affinity constant (mg/l) | km | 0.134b | 1.39 | 0.22 |
| Competitive inhibition | ||||
| Inhibitor | Substrate | Inhibition constant Kib | ||
| Toluene | Ethylbenzene | 0.948 | ||
| Toluene | m-Xylene | 0.357 | ||
| Ethylbenzene | Toluene | 0.168 | ||
| Ethylbenzene | m-Xylene | 0.505 | ||
| m-Xylene | Toluene | 0.328 | ||
| m-Xylene | Ethylbenzene | 1.667 | ||
From Tardif et al. (1995).
From Haddad et al. (2000).
Model verification.
The model was assessed by comparing generated simulations to the published model and with human data sets (Tardif et al.,1997). We weighed the performance of the model by calculating the percent median absolute performance error (MAPE%) based on estimates of performance error (PE) between the recoded model and the experimental data (Ruiz et al., 2011), as follows:
| (1) |
Therefore, MAPE% = median (|PE1|, |PE2|, |PE3|……|PEi|…. |PEn|), where PEi is the performance error measure of the model prediction and data at time point i. We assessed the measure for the model bias by considering the median of the true values of PE. This measure is calculated as MPE% = (PE1, PE2, PE3…PEi…. PEn). The accuracy of the prediction was evaluated by percent root median-square performance error (RMSPE%), as follows:
| (2) |
where n is the total number of data points. We also calculated the correlation coefficient between Cmeasured and Cpredicted.
Model Application
Biological hazard indexes.
Traditionally, a hazard index (HI) is calculated using estimated exposures and allowable levels in environmental media. The American Conference of Governmental Industrial Hygienists (ACGIH) Threshold Limit Values (TLVs), and Biological Exposure Indices (BEIs) are intended for use by industrial hygienists in making decisions regarding safe levels of exposure to various chemical substances and physical agents found in the workplace. The TLVs and BEIs represent conditions under which ACGIH believes that nearly all workers may be repeatedly exposed without adverse health effects. BEIs are guidance values for assessing biological monitoring results (https://www.acgih.org/tlv-bei-guidelines/policies-procedures-presentations/overview; last accessed December 2019). The human PBPK models enable calculation of biological hazard indexes (BHIs). A BHI is based on estimated internal dose rather than applied dose.
We used the human PBPK model mixture to calculate BHIs for 8-h exposures to varying simulated mixtures of the 3 chemicals (5–40ppm toluene, 10–50ppm m-xylene, and 10–50ppm ethylbenzene) (Haddad et al., 1999; Tardif et al., 1997). We used the following equation to calculate the BHIs:
| (3) |
where SCi is the simulated venous blood concentration of the component chemical (i) and BEIi is the biological exposure index or blood concentration of the component chemical in a healthy person (ie, at the TLV for toluene [50ppm], m-xylene [100ppm], and ethylbenzene [100ppm]). Simulated venous blood concentration (SCi) values were predicted for the mixture components using the ternary mixture PBPK model. Individual chemical PBPK models were used to calculate the TLV-based BEIs. The BHIs were subsequently compared with exposure concentration-based HI values for each mixture, calculated as follows, where Ei is the exposure level of the chemical:
| (4) |
Exploration of various exposure scenarios: resting and working.
Under Occupational Safety and Health Administration and American Conference of Governmental Industrial Hygienists guidelines, the mixture formula (unity calculation, UC) is used to evaluate exposures to mixtures of chemicals that cause similar toxicities (Dennison et al., 2005). Using this methodology described in the guidelines, the overall exposure is acceptable if exposures are reduced in proportion to the number of chemicals and their respective exposure limits. Most of the occupational exposure limits are derived from studies of humans or animals at rest, often using inhaled concentrations to estimate external exposures. For a variety of exposures to toluene, ethylbenzene, or xylene, UCs were performed, using the equation below (Dennison et al., 2005):
| (5) |
Ui is the exposure level of each chemical and OULi is the occupational expose limit for each chemical. We used the human PBPK model to examine the potential toxicity from coexposure to TEX under resting and working conditions. Instead of using inhaled concentrations to estimate external exposure, we utilized the PBPK model to determine the blood concentration of each chemical and in the UCs. The UCs were based on internal doses of each chemical instead of the traditional external exposure level, thus considering nonlinear PK and the influence of PK interactions on cumulative dose. The chemical effects are assumed to result from systemic absorption and exposure of internal organs to the blood and not from direct toxicity of inhaled concentration at the port of entry.
Exploration of microarray data.
An upcoming area of research is to input the potential concentrations derived from high-throughput assays such as microarray data, into the existing PBPK model to back-extrapolate external exposure concentrations that would produce equivalent target tissue concentrations research. Through an interactive process both the PBPK modeling and the experimental testing can be improved and/or calibrated. We used the PBPK model to initially simulate the starting points of equivalent external exposure for controls, short- and long-term exposure groups as reported by Hong et al., to estimate potential distribution of blood concentrations (more reflective of internal estimated dose). Initially the simulation was performed for one-third TLVs doses. Following this, different scenarios for incremental changes in TLVs were simulated.
Toluene, Ethylbenzene, Xylene, and Genes Interactions
We examined possible mechanisms by which TEX might interact to cause potential health effects using Comparative Toxicogenomics Database (CTD) and MetaCore.
Chemical-gene-disease interactions using CTD.
The CTD(http://ctdbase.org; last accessed December 2019) contains more than 1 million curated chemical-gene interactions and more than 15 million curated and inferred gene-disease interactions (Davis et al., 2017; Grondin et al., 2018). The National Institute of Environmental Health Sciences funds the database, which can be used to identify genes or environmental chemicals that commonly affect the proteins that they code for. These sets of common genes can be further filtered to only include genes that have been associated with a condition or disease. This approach uses bioinformatics to identify toxicogenomic interactions that are promising targets for further study. The CTD was used to identify (1) top curated genes for each of the 3 VOCs, toluene, xylene, and ethylbenzene, (2) genes in common to them, and (3) top interacting curated diseases in common to all the individually VOCs.
We used the CTD “VennViewer” function to compare the genes associated with each of the VOCs, TEX. Only genes with curated associations with each chemical were included. Genes with an inferred relationship or those with no known effects were excluded. The candidate genes associated with each of 3 chemicals were identified; these genes were then used as an input for the “Set Analyzer,” which identifies common genes that have an association with specific diseases or health conditions. The list of genes that were common amongst the 3 VOCs was imported to MetaCore (version 6.36 build 69400) software to perform an enrichment analysis (https://clarivate.com/cortellis/solutions/early-research-intelligence-solutions/; last accessed December 2019). MetaCore is a tool for functional analysis of different “omics” data, including gene expression data. One of MetaCore’s relevant applications for pathway analysis is the Enrichment Analysis Workflow, which allows the users to understand the biological impact of their data by visualizing the intersection of their data set to curated ontologies which are ranked by significance based on p value (hypergeometric mean calculation).
These curated ontologies (eg, pathway, maps, toxicity biomarkers, networks) are primarily derived from peer-reviewed literature and from the Gene Ontology lexicon.
Enrichment analysis on lung-specific ontologies for TEX microarray data using MetaCore.
Hong and colleagues reported the effects of TEX exposure in humans on gene expression data for short-term (workers exposed to VOCs for less than 10years), long-term (workers exposed to VOCs for more than 10years), and no exposure (control) (Hong et al., 2016). We downloaded their microarray data from the GEO Database (www.ncbi.nlm.nih.gov/geo/; last accessed December 2019) ID GSE68906, which describes the differentially expressed genes (DEGs) in human blood samples collected from industry workers at a factory and chemical production company in Ansan, Korea (Hong et al., 2016). The gene expression data were imported as experiment files into MetaCore, and genes showing statistically significant expression changes within the exposed groups were compared.
MetaCore’s system’s toxicology module, which utilizes organ-specific toxicity biomarkers curated for different ontologies, was used to perform enrichment analysis on lung-specific ontologies. We identified and rank cellular pathways and processes most impacted in this tissue by the gene expression changes in the exposed group.
Our enrichment analysis focused not only on the genes that were common to both long- and short-term exposures, but also on the genes that were unique to the long-term exposure.
RESULTS
Recoding, Evaluation, and Application of the BTEX PBPK Model
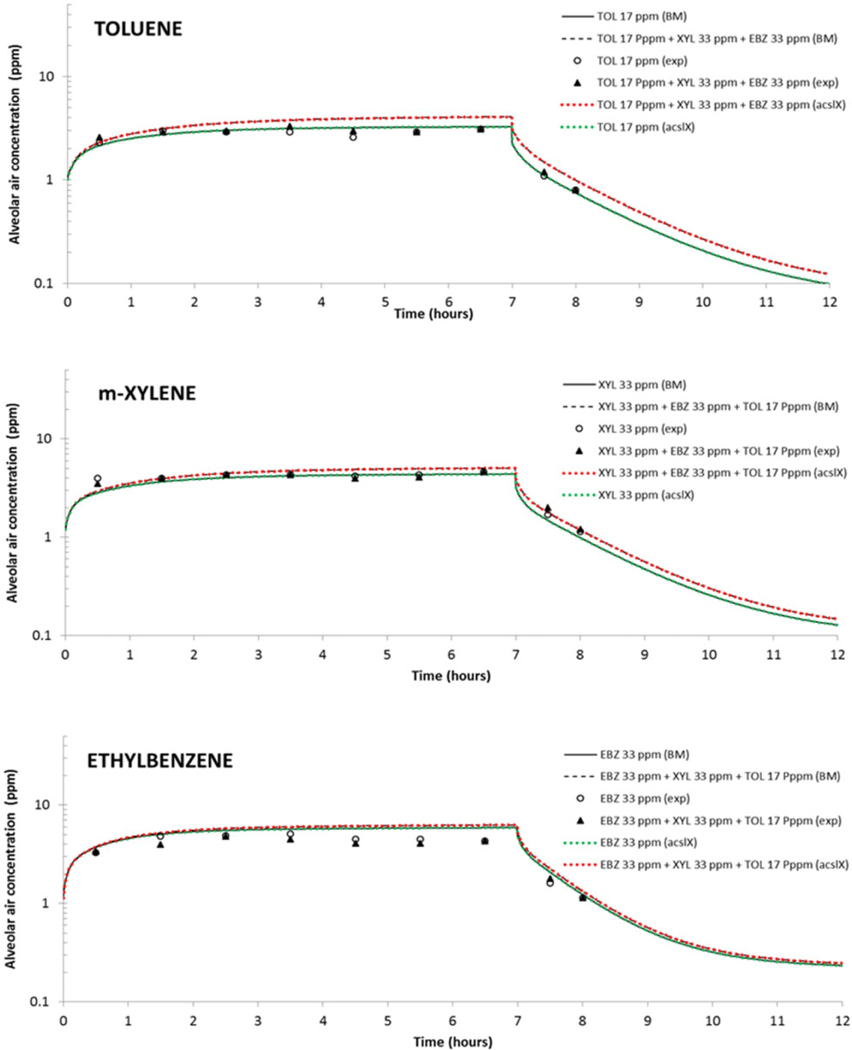
The recoded BTEX PBPK model was assessed for predictability and applicability. By visual comparison, the recoded model simulations were in good agreement with those of the original model as well the experimental data (Figure 2). The simulations were superimposable suggesting a completely matching performance. We also evaluated the model performance by calculating the MAPE%, percent median performance error (MPE%), and RMSPE%, based on estimates of PE (Table 2). The results suggested that the model could simulate and accurately predict the available experimental data and obtained a good fit, as shown. Correlation coefficients (R2) between experimental and predicted data using the recoded model were above 92% for single VOCs and between 89% and 92% for respective VOC mixtures.
Figure 2.
Comparison between original and recoded physiologically based pharmacokinetic model simulations and experimental data for toluene (17ppm), ethylbenzene (33ppm), and xylene (33ppm) alone and mixed together. All simulations were for 7-h exposures and 5h after exposure. BM: recoded Berkeley Madonna model simulation and AcslX: Original model simulations. Green line shows individual chemical simulations. Red line shows TEX mixture simulations.
Table 2.
Measures of Predictive Performance for Comparing Single TEX and Their Mixtures Experimental Data and the Recoded Model Simulations
| Chemicals | R2 | MPE% | MAPE% | RMSPE% |
|---|---|---|---|---|
| Single chemicals | ||||
| Toluene (Tol) | 0.938 | −4.01 | 8.88 | 12.52 |
| Ethylbenzene (EBz) | 0.937 | −11.07 | 11.34 | 16.15 |
| m-Xylene (Xyl) | 0.928 | 6.57 | 6.57 | 21.33 |
| Mixed chemicals | ||||
| Tol + EBz + Xyl | 0.892 | −13.10 | 13.10 | 17.15 |
| EBz+Xyl + Tol | 0.929 | −23.41 | 23.41 | 23.12 |
| Xyl + Tol + EBz | 0.923 | −3.82 | 17.49 | 16.29 |
Abbreviations: MAPE%, percent median absolute performance error; MPE%, percent median performance error; R2, correlation coefficient; RMSPE%, percent root median-square performance error.
Overall, the recoded PBPK model predictions and experimental data indicated that blood and alveolar air concentrations during and after exposure to the ternary mixture were similar to those measured during and after exposure to each chemical alone. A statistically significant increase was found for blood concentrations of xylene during the simulated mixture exposure but not for toluene and ethylbenzene (Supplementary Table 1). This change was not reflected by the alveolar air concentration data obtained during the same experiment (Figure 2). These results also indicated that the ternary mixture did not significantly modify the metabolism, competitive metabolism inhibition, of the individual components compared with exposure to the individual VOCs alone.
Using our recoded PBPK model, we calculated BHIs for 8-h exposures to varying simulated mixtures: 5–40ppm toluene, 10–50ppm m-xylene, and 10–50ppm ethylbenzene (Supplementary Table 2). Because the PBPK model used to predict the SCi values is interactions-based, the BHI values (based on blood levels) were expected to be the same as the HI values (based on exposure levels) if the toxicokinetic interactions among the mixture components are negligible.
Toluene, Ethylbenzene, Xylene, and Genes Interactions
Common genetic targets identified for TEX using CTD.
Our CTD analysis identified a list of 24 genes common among TEX (Figure 3A). Individually, 826 genes were associated with toluene, 109 genes with ethylbenzene, and 55 genes with xylenes. We have also identified 2 conditions, potentially associated with the overlap of the curated diseases potentially cause by these 3 chemicals that include prenatal exposure delayed effects and micronuclei chromosomal defects (Figure 3B). To understand what pathways are impacted by the list of 24 genes, we performed pathway enrichment analysis using MetaCore. The names, symbols, and functions of the 24 genes are shown in Figure 3A.
Figure 3.
Associated common genes (A) and diseases (B) between ternary volatile organic compounds components from the Comparative Toxicogenomics Database.
Based on our enrichment analysis, a joint mechanism of action was proposed for the combined exposure to TEX mixture. The pathways map shows cross interactions between the downstream targets (Supplementary Figure 1). The analysis shows that the top score enriched pathway map is involved in neural cell death and mitochondrial dysfunction (Supplementary Figure 1). Ubiquitin with dinucleotide deletion acts as a strong inhibitor of the proteasome. As a result, the pool of available proteasomes becomes substantially depleted and not enough for proper degradation of tumor protein (p53), protein kinase C-alpha (PKC-alpha), and mutant Huntingtin. Slow degradation of PKC-alpha results in activation of N-methyl-D-aspartate subclass of ionotropic glutamate receptor (NMDA receptor), followed by mitochondrial dysfunction, alterations in neurotransmitter systems, and neuronal cell death (Fong et al., 2002; Zemskov and Nukina, 2003). In addition, impairing of proteasomal function leads to alterations in neurotransmitter pathways (Wang et al., 2008). These common activation processes can influence or activate each other. Although this computational systems biology-generated global pathway map cannot be considered as a proof of causal linkages without further experimental validation, it provides justification for the mechanistic hypothesis and contributes to new interpretation linking available published toxicology and disease information domains.
Gene expression data and enrichment analysis on lung-specific ontologies using MetaCore.
To understand the toxic impact of VOCs to the target organ lung, we used the system’s toxicology module of MetaCore to compare gene expression data collected from blood samples of individuals exposed for short- and long-term to TEX. The comparison analysis demonstrates that 236 genes are common between the short-term (1) and long-term exposure groups (2) gene expression (Figure 4A). It also shows that 242 genes are unique to short-term exposure (1, orange bar), and 225 genes are unique to long-term exposure (2, blue bar). These findings help explain the overall biological themes represented in each of the studied exposure group networks. The results of the enrichment analysis ranked pathway maps specifically curated for lung toxicity. Ranking was determined by the genes that were common to blood samples of individuals exposed to short- and long-term exposures (Figure 4B, blue stripe bars) and presented from highest log p value to the lowest. Orange and blue bars correspond to log p values that are unique to enrichment of genes from short- and long-term exposures, respectively.
Figure 4.
A, Hong et al., gene expression comparison for toluene, ethylbenzene, and xylene (TEX) short- and long-term exposures. Gene expression comparison. The gene content is aligned between the 2 uploaded experiments (short- and long-term exposure). The intersection set of experiments is defined as “common” and marked as a blue/white striped bar. The unique genes for the experiments are marked as colored bars 1 and 2, short-term (1): orange bar and long-term (2): blue bar. The genes from the “similar” set are present in all but 1 (any) file. B, Top pathway maps impacted by differentially expressed genes common to both short- and long-term exposures to TEX.
The 2 top-scored lung toxicity pathways (maps with the lowest p values), based on the enrichment distribution sorted by “common” set, showed common themes related to processes involved in respiratory, developmental, and the immune systems (Figure 4B). The 2 top pathway maps are Ephrin signaling and CD28 signaling; these are involved in cell adhesion, cell proliferation, and immune response. Ephrin receptors are major participants in the regulation of cell movement, cell adhesion, and cell survival, whereas CD28 plays a key role in many T-cell processes related to immune signaling pathways.
The first top-scored map (corresponding to the lowest p value) shows Ephrin-mediated signaling (Figure 4B). Short- and long-term exposures upregulate the Ephrin A receptors, as indicated by the red thermometers (Nos 1 and 2, respectively) and downregulate Ephrin B receptors as indicated by the blue thermometers (Nos 1 and 2) as shown in Supplementary Figure 2. Ephrin B proteins have conserved cytoplasmic tyrosine residues that are phosphorylated upon interaction with an EphB receptor and may transduce signals that regulate a cellular response. However, long-term exposures downregulate gene paxillin (see thermometer No. 2, Supplementary Figure 2).
The second top-scored pathway map (second lowest p value), CD28 signaling pathway, is related to induction of immune responses (Supplementary Figure 3). The T-cells receive 2 sets of signals from antigen-presenting cells. The first signal is delivered via T-cell receptor complex (TCR), the second one proceeds via coreceptor CD28. T-cell receptor and CD28 are independent signaling units. However, CD28 amplifies signals triggered by TCR ligation. CD28 is a T-cell surface protein activated by interacting with the B-cell activation antigens CD80 and CD86. The CD28 and CTLA-4 signaling pathways are important participants in a very complex group of regulatory events that maintain immunologic homeostasis. Lung toxicity biomarkers for lung interstitial congestion and lung perforation are major processes in regulation of macrophage migration, suggesting inflammatory response plays a large role in modulation of lung toxicity.
When taking a closer look just at the unique long-term exposure data, the enrichment analysis shows some additional interesting results such as xenobiotic metabolism enzymes are impacted by long-term exposure (Supplementary Figure 4.). The top-scored pathway map (indicating by the lowest p value) for the long-term exposure data is the peroxisome proliferator-activated receptors (PPARs) transcription pathway (Supplementary Figure 4). The PPARs modulate the expression of genes involved in cell proliferation, lipid metabolism, and inflammation.
The top-scored map (corresponding to the lowest p value) for long-term exposure (Supplementary Figure 4) shows that Mitogen-activated protein enzymes (MEK3) are activated in the long-term exposure data set which is an upstream activator of PPARc (Supplementary Figure 4). PPARα activates CYPA13 via transcriptional regulation, as seen in the MetaCore interactions database. As seen in the Supplementary Figure 4, the protein kinase inhibitor of PPARα is downregulated. CYPA13, which is known to metabolize carcinogens in cigarette smoke, is upregulated, whereas CYP4F2 and CYP4F3 are downregulated, both of which are involved in inactivating the inflammatory mediator leukotriene B4. Downregulation of CYP4F2 and CYP4F3 would result in an increased inflammatory response via leukotriene B4.
DISCUSSION
The aims of this study were to (1) share a newly recoded PBPK model for BTEX, part of our ATSDR toolkit, with the scientific community, (2) explore individual or combined chemical published data from various sources to generate or hypothesize about the potential mechanisms of toxicity of the TEX mixture, and (3) introduce methodology for new testable hypotheses to address some of the chemical mixtures data gaps previously identified by federal agencies. Major data gaps for mixtures include linking chemical exposure to cellular pathway disturbance and disease outcome. The application of new technologies and alternative methods, as presented in this study, will not only maximize their use, but will also help gain information into the mechanisms of mixtures toxicity (Figure 1).
Recoding, Evaluation, and Application of the BTEX PBPK Model
We evaluated and applied the recoded model and ascertained its performance to be the same as the original model. The recoded model simulations show a statistically significant increase for blood concentrations of xylene after mixture exposure but not for toluene and ethylbenzene (Supplementary Table 1). This change was not reflected in the alveolar air concentration data obtained during the same experiment (Figure 2).
We demonstrated the applicability of the recoded model using 2 exposure studies. By calculating the BHI and HI, the recoded model results show that metabolic inhibition was not significant during exposure to < 20 to 30ppm, which is about one-third of the current TLV for each chemical. In this range, the additivity assumption held. However, when the exposure increases above this level, metabolic inhibition causes a disproportionately higher tissue dose to occur. In addition, we determined the validity of the additivity assumption by performing UCs for a variety of exposures to TEX. Using internal doses obtained with the recoded model at rest and work, the UCs were 2.9 and 4.6 times, respectively. These results showed that workers with higher activity might experience a significantly higher absorbed dose that could result in 87% higher internal doses. Based on these results, we believe the recoded model can successfully simulate internal doses for varying exposure scenarios (Supplementary Table 3). The BTEX model coded in Berkeley Madonna is now available for use by risk assessors and scientists.
We also used the recoded PBPK model to determine the internal doses to link to the disruptor effects based on the genes expression and mechanism of action implicated. Internal dose, or blood concentration, is a better predictor than external exposure dose (Meek et al., 2013). We applied CTD to identify common genetic targets affected by each VOC in the TEX mixture and to integrate data that relate gene alterations to biological changes. This was followed by an integrative graphical network mapping using MetaCore, which resulted in better understanding of mechanisms of lung toxicity of this mixture. This was a novel approach because it integrated data from multiple sources and justified utilization of PBPK model to predict internal dose, instead of relying on external dose. The following sections discuss each phase of the methodology applied in this work.
Biological Interpretation of Gene Expression Data for Individual and Multiple Chemicals
Common genetic targets identified for TEX using CTD.
This analysis identified 24 genes that are commonly regulated by each of the VOCs and provided insight into potential pathways where in vivo effects have been observed, while recognizing that TEX mixtures would not necessarily act on the same pathways in the same ways to result in additivity effects. The enrichment pathway analysis using the 24 common genes showed that TEX compounds impact the Ubiquitin pathway. This provides evidence that combined exposure of TEX could result in synergistic activation or deactivation of this pathway. The pathway seems relevant to neurological functions and may be relevant in establishing important adverse outcomes for TEX (Supplementary Figure 1).
Competitive metabolic inhibition is the most plausible mechanism of interaction at higher concentrations among the TEX components based on the PBPK studies, as well as in vitro and in vivo metabolism and toxicity studies for some of the binary component mixtures (Tardif et al., 1997). Therefore, due to the apparent lack of competitive metabolic interactions in TEX mixtures below approximately 20ppm of each component (one-third of TLVs), it is plausible that joint neurotoxic actions among the chemicals will be additive at environmental levels of exposure. Exposure to higher concentrations of TEX components (ie, above the threshold for metabolic inhibition) would be expected to lead to greater than additive increases in blood levels of parent compounds and, consequently, increased concern for neurotoxicity. However, it is unclear whether the PBPK model descriptions are adequate for predicting interactions from inhalation of TEX mixtures above approximately 200ppm of each component, because it has not been tested for these high doses (corresponding to the dose reducing the metabolic rate). At a certain dose, the enzymatic saturation is reached, reducing the metabolic rate formation and that may increase the toxicity of the parent compounds.
Studies that directly examined the joint toxic action of TEX chemicals on the nervous system are limited to a few human and animal inhalation studies of some binary mixtures of components. Neurotoxicity studies of the ternary mixtures provide agree with the predictions of the PBPK studies (ie, joint action is expected to be additive at TEX concentrations below approximately 20ppm of each component).
If the concentrations in blood are below the competitive inhibition threshold for each of the components, no change can be expected in the proposed pathways leading to toxicity. However, such changes in constant concentration below the threshold might have effects that will cause some health outcome at long-term exposure.
The CTD results highlighted interconnections, also evident from the independent data analysis from Hong et al. study for the up- and downregulated genes related to neurological diseases, embryonic development, cellular compromise, cellular adhesion, and cellular movement effects.
Gene expression data and enrichment analysis on lung-specific ontologies using MetaCore.
An expressed gene set list (up- and downregulated) at the average short- and long-term exposed group were subdivided into 9 cluster genes using a hierarchical clustering analysis (Hong et al., 2016). In addition, they reported the functional categories and annotations of these genes and created a gene function network that show how key components (consisting of 5 up- and 12 downregulated genes that also exhibit time-dependent changes in methylation) of different pathways interact using DAVID and Ingenuity tools.
By applying enrichment analysis on lung-specific ontologies, this study goes beyond just listing of DEGs to understand the mechanism of the biological impact on the lung tissue. Both approaches, enrichment analysis and gene set analysis are supposed to complement to take gene expression levels and leverage existing knowledge about a given organism to identify the underlying biological processes and mechanisms.
Using the system’s toxicology module of MetaCore, our study used enrichment analysis to understand the toxic impact of TEX to the target organ, lung. As mentioned in the “Materials and Methods” section, the enrichment analysis takes into consideration the number of DEGs that fall on a given curated ontology and uses hypergeometric mean to calculate the significance of this enrichment.
This analysis showed 2 top-scored lung toxicity pathways (maps with the lowest p values), based on the enrichment distribution sorted by “common” set (short- and long-term exposures), were Ephrin signaling and CD28 signaling. These pathways are involved in cell adhesion, cell proliferation, and immune response, suggesting that they may play a crucial role in lung toxicity. Consistent with this hypothesis, Ephrin A receptor stimulation has been previously shown to increase lung vascular permeability (Carpenter et al., 2012; Larson et al., 2008).
In addition, we performed a similar enrichment analysis for the genes that were to unique long-term exposure data, which suggests that xenobiotic metabolism enzymes are impacted by long-term exposures. The top-scored pathway was the PPARs transcription pathway (Supplementary Figure 4). Peroxisome proliferator-activated receptors are nuclear hormone receptors and are ligand-induced transcription factors and, whereas these were not sampled directly in the data set, many upstream interactors (MEK3, AKT, and PKA-reg) of these transcription factors were shown to be upregulated in the long-term exposure group. As shown in Supplementary Figure 4, PPARs play a crucial role in signaling of variety of different biological processes, including inflammation, and cancer. It is therefore not surprising that the PPARs have been explored as candidates for therapeutic intervention to treat variety of lung diseases such as asthma and non-small cell lung cancer (Banno et al., 2018; Li et al., 2011).
Based on our enrichment analyses using the gene lists identified from CTD and Hong et al. data, we hypothesize that exposure to TEX mixture may result in disruption of biological pathways such as Ubiquitin, Ephrins, CD28, and PPARs. Disruption of these proposed pathways can translate into adverse respiratory and neurological outcomes, depending on exposure durations. The different genes mapped to these biological pathways have been related to alterations of the neurotransmitter system including neural cell death, cell proliferation, inflammation, xenobiotic metabolism, signal transduction, and cell processes. These proposed pathways are based on experimental data from multiple laboratories and have not been examined sufficiently to date comprehensively by an integrated research laboratory. Future experimental evaluation of these pathway maps might lead to the development of new predictive markers of TEX effects that could translate into new disease prevention and clinical use strategies. Specific avenues of laboratory research might include, but not limited to, in vitro studies of target cell populations such as lung cells. Complimentary in vivo studies in rodents dosed with TEX mixture could be performed to determine if the observed in vitro study findings are observed after in vivo exposure and can be extrapolated to human exposure scenarios.
PBPK modeling, gene expression data, signaling pathways, and future directions combining PBPK and signaling pathways.
The enrichment analysis performed using CTD and the MetaCore analyses provides mechanistic insights of TEX mixture and could help the PBPK model to elucidate the biological relevant dosimetry. To use this approach a comprehensive characterization of exposure is needed, because humans are exposed to multiple chemicals from multiple sources through their daily activities.
The estimated blood concentrations from the recoded PBPK model represent a gold standard to understand or interpret the gene expression results driven health effects. Using the model, we generated different scenarios of TEX mixture to see the influence of dose and time in the blood concentration (Table 3). In the Hong et al. study, workers were exposed short- or long-term at doses we assumed were close to the TLV values. It is interesting to observe that each chemical from the mixture easily reaches a plateau (Cmax) very quickly (approximatively few days). This suggests that the maximum blood concentrations will not change whether workers are exposed for a month or a year (Table 3). This observation also indicates that the metabolic interaction does not seem important if exposure occurs to the combination of these VOCs and seems to have a limiting impact on the blood concentration. In fact, another parameter that also influence the concentration of chemical in blood is the partition coefficient in tissue. This parameter for each tissue will influence not only the concentration of parent VOCs in blood, but also the availability of the parent VOCs to be metabolized.
Table 3.
Predicted Blood Concentrations for Different Exposure Scenarios Using the Recoded PBPK Model
| Exposure Concentrations | Exposure Scenarios | |||
|---|---|---|---|---|
| Dose (ppm) | Chemical | 1 Day | 1 Month | 1 Year |
| One-third TLV | Predicted blood concentrations (mg/l) | |||
| 17 | Toluene | 0.20 | 0.21 | 0.21 |
| 33 | Ethylbenzene | 0.64 | 0.70 | 0.72 |
| 33 | Xylene | 0.46 | 0.50 | 0.50 |
| TLV | ||||
| 50 | Toluene | 1.07 | 1.21 | 1.21 |
| 100 | Ethylbenzene | 3.23 | 3.86 | 3.82 |
| 100 | Xylene | 2.78 | 3.23 | 3.30 |
| 2× TLV | ||||
| 100 | Toluene | 2.61 | 2.95 | 3.04 |
| 200 | Ethylbenzene | 8.20 | 9.83 | 9.91 |
| 200 | Xylene | 7.65 | 8.88 | 8.89 |
Exposure time simulations were 8h/day, 5days/week.
Abbreviation: TLV, threshold limit value.
Further research is needed to confirm or validate this proposed approach through experimental testing using internal doses estimated by the PBPK model.
The merit of this approach lies in advancement of mixtures risk assessment methodologies using the available current knowledge of the TEX mixtures toxicity, using contemporary alternative toxicity testing data, and applying computational methods. This work provides a Berkeley Madonna BTEX human PBPK model code to obtain a better dosimetry than external environmental exposures. Second, the enrichment analysis provides a possible pathway for the short- and long-term effects of TEX mixtures.
CONCLUSIONS
Humans are typically exposed to mixtures, but our limited understanding of mixture toxicity presents a major challenge in health risk assessment. Because all chemicals and their combinations cannot be experimentally tested, mixtures risk assessment provides an opportunity to employ data from multiple streams (microarray, HTP, and -omics) and to use multiple techniques (statistical and computational) for screening and quantitative risk assessments. Toward this, we present a conceptual framework for a combination of 3 common VOCs (TEX), using PBPK modeling, toxicogenomics data, systems biology approaches, and microarray data that may link TEX exposures to diseases. Although data gaps primarily need to be filled through experimental laboratory research, scientists can examine potential linkages for combined exposures to these VOCs or others environmental chemicals and cellular pathways, as presented here. Progress can be made through strategic research using a well-conceived framework to pursue future integrated approaches to better assess potential health risks from exposure to mixtures. Toward this, data gaps need to be identified and prioritized. Adequate resources (financial, laboratory, multidisciplinary expert teams, etc.) are necessary to conduct targeted studies to fill such data gaps. Our approach can be applied to other airborne chemicals in the future and may provide support to early risk assessment efforts by unraveling biological pathway responses that may contribute to adverse health outcomes.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to Dr Clem Welsh for his valuable comments, and advice during the preparation of this manuscript. We also appreciate Mr Donald Meadows for his editorial technical support. The findings and conclusions in this manuscript have not been formally disseminated by the Centers for Disease Control and Prevention/the Agency for Toxic Substances and Disease Registry and should not be construed to represent any agency determination or policy. Mention of trade names is not an endorsement of any commercial product.
Footnotes
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
DECLARATION OF CONFLICTING INTERESTS
The author/authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- Andersen ME, Black MB, Campbell JL, Pendse SN, Clewell HJ III, Pottenger LH, Bus JS, Dodd DE, Kemp DC, and McMullen PD (2017a). Combining transcriptomics and PBPK modeling indicates a primary role of hypoxia and altered circadian signaling in dichloromethane carcinogenicity in mouse lung and liver. Toxicol. Appl. Pharmacol 332, 149–158. [DOI] [PubMed] [Google Scholar]
- Andersen ME, Cruzan G, Black MB, Pendse SN, Dodd D, Bus JS, Sarang SS, Banton MI, Waites R, and McMullen PD (2017b). Assessing molecular initiating events (MIEs), key events (KEs) and modulating factors (MFs) for styrene responses in mouse lungs using whole genome gene expression profiling following 1-day and multi-week exposures. Toxicol. Appl. Pharmacol 335, 28–40. [DOI] [PubMed] [Google Scholar]
- Agency for Toxic Substances and Disease Registry (ATSDR). (2004). Toxicological profile for Interaction profile for benzene, toluene, ethylbenzene and xylenes. U.S. Department of Health and Human Services, Public Health Service, Atlanta, GA. [PubMed] [Google Scholar]
- Agency for Toxic Substances and Disease Registry (ATSDR). (2007). Toxicological profile for xylenes. US Department of Health and Human Services. Public Health Service, Atlanta, GA. [Google Scholar]
- Agency for Toxic Substances and Disease Registry (ATSDR). (2010). Toxicological profile for ethylbenzene. US Department of Health and Human Services. Public Health Service, Atlanta, GA. [PubMed] [Google Scholar]
- Agency for Toxic Substances and Disease Registry (ATSDR). (2017). Toxicological profile for toluene. US Department of Health and Human Services. Public Health Service, Atlanta, GA. [Google Scholar]
- Banno A, Reddy AT, Lakshmi SP, and Reddy RC (2018). PPARs: Key regulators of airway inflammation and potential therapeutic targets in asthma. Nucl. Receptor Res 5, pii: 101306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolden AL, Kwiatkowski CF, and Colborn T. (2015). New look at BTEX: Are ambient levels a problem? Environ. Sci. Technol 49, 5261–5276. [DOI] [PubMed] [Google Scholar]
- Carpenter TC, Schroeder W, Stenmark KR, and Schmidt EP (2012). Eph-A2 promotes permeability and inflammatory responses to bleomycin-induced lung injury. Am. J. Respir. Cell Mol. Biol 46, 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, Wiegers J, Wiegers TC, and Mattingly CJ (2017). The comparative toxicogenomics database: Update 2017. Nucleic Acids Res. 45, D972–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghani M, Fazlzadeh M, Sorooshian A, Tabatabaee HR, Miri M, Baghani AN, Delikhoon M, Mahvi AH, and Rashidi M. (2018). Characteristics and health effects of BTEX in a hot spot for urban pollution. Ecotoxicol. Environ. Saf 155, 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison JE, Bigelow PL, Mumtaz MM, Andersen ME, Dobrev ID, and Yang RS (2005). Evaluation of potential toxicity from co-exposure to three CNS depressants (toluene, ethylbenzene, and xylene) under resting and working conditions using PBPK modeling. J. Occup. Environ. Hyg 2, 127–135. [DOI] [PubMed] [Google Scholar]
- Emond C, Ruiz P, and Mumtaz M. (2017). Physiologically based pharmacokinetic toolkit to evaluate environmental exposures: Applications of the dioxin model to study real life exposures. Toxicol. Appl. Pharmacol 315, 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong DK, Rao A, Crump FT, and Craig AM (2002). Rapid synaptic remodeling by protein kinase C: Reciprocal translocation of NMDA receptors and calcium/calmodulindependent kinase II. J. Neurosci 22, 2153–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin CJ, Davis AP, Wiegers TC, Wiegers JA, and Mattingly CJ (2018). Accessing an expanded exposure science module at the comparative toxicogenomics database. Environ. Health Perspect 126, 014501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad S, Beliveau M, Tardif R, and Krishnan K. (2001). A PBPK modeling-based approach to account for interactions in the health risk assessment of chemical mixtures. Toxicol. Sci 63, 125–131. [DOI] [PubMed] [Google Scholar]
- Haddad S, Charest-Tardif G, and Krishnan K. (2000). Physiologically based modeling of the maximal effect of metabolic interactions on the kinetics of components of complex chemical mixtures. J. Toxicol. Environ. Health A 61, 209–223. [DOI] [PubMed] [Google Scholar]
- Haddad S, Tardif R, Viau C, and Krishnan K. (1999). A modeling approach to account for toxicokinetic interactions in the calculation of biological hazard index for chemical mixtures. Toxicol. Lett 108, 303–308. [DOI] [PubMed] [Google Scholar]
- Hong JY, Yu SY, Kim GW, Ahn JJ, Kim Y, Lim S, Son SW, and Hwang SY (2016). Identification of time-dependent biomarkers and effects of exposure to volatile organic compounds using high-throughput analysis. Environ. Toxicol 31, 1563–1570. [DOI] [PubMed] [Google Scholar]
- Larson J, Schomberg S, Schroeder W, and Carpenter TC (2008). Endothelial EphA receptor stimulation increases lung vascular permeability. Am. J. Physiol. Lung Cell. Mol. Physiol 295, L431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Sorenson AL, Poczobutt J, Amin J, Joyal T, Sullivan T, Crossno JT Jr, Weiser-Evans MC, and Nemenoff RA (2011). Activation of PPARgamma in myeloid cells promotes lung cancer progression and metastasis. PLoS One 6, e28133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SK, Shin HS, Yoon KS, Kwack SJ, Um YM, Hyeon JH, Kwak HM, Kim JY, Kim TY, Kim YJ, et al. (2014). Risk assessment of volatile organic compounds benzene, toluene, ethylbenzene, and xylene (BTEX) in consumer products. J. Toxicol. Environ. Health A 77, 1502–1521. [DOI] [PubMed] [Google Scholar]
- Lundberg I, and Milatou-Smith R. (1998). Mortality and cancer incidence among Swedish paint industry workers with long-term exposure to organic solvents. Scand. J. Work Environ. Health 24, 270–275. [DOI] [PubMed] [Google Scholar]
- Maldonado EM, Leoncikas V, Fisher CP, Moore JB, Plant NJ, and Kierzek AM (2017). Integration of genome scale metabolic networks and gene regulation of metabolic enzymes with physiologically based pharmacokinetics. CPT Pharmacometrics Syst. Pharmacol 6, 732–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand A, Aranda-Rodriguez R, Tardif R, Nong A, and Haddad S. (2015). Human inhalation exposures to toluene, ethylbenzene, and m-xylene and physiologically based pharmacokinetic modeling of exposure biomarkers in exhaled air, blood, and urine. Toxicol. Sci 144, 414–424. [DOI] [PubMed] [Google Scholar]
- Marchand A, Aranda-Rodriguez R, Tardif R, Nong A, and Haddad S. (2016). Evaluation and modeling of the impact of coexposures to VOC mixtures on urinary biomarkers. Inhal. Toxicol 28, 260–273. [DOI] [PubMed] [Google Scholar]
- Meek ME, Barton HA, Bessems JG, Lipscomb JC, and Krishnan K. (2013). Case study illustrating the WHO IPCS guidance on characterization and application of physiologically based pharmacokinetic models in risk assessment. Regul. Toxicol. Pharmacol 66, 116–129. [DOI] [PubMed] [Google Scholar]
- Moro AM, Charao M, Brucker N, Bulcao R, Freitas F, Guerreiro G, Baierle M, Nascimento S, Waechter F, Hirakata V, et al. (2010). Effects of low-level exposure to xenobiotics present in paints on oxidative stress in workers. Sci. Total Environ 408, 4461–4467. [DOI] [PubMed] [Google Scholar]
- Mumtaz MM, Ray M, Crowell SR, Keys D, Fisher J, and Ruiz P. (2012). Translational research to develop a human PBPK models tool kit-volatile organic compounds (VOCs). J. Toxicol. Environ. Health A 75, 6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz P, Fowler BA, Osterloh JD, Fisher J, and Mumtaz M. (2010). Physiologically based pharmacokinetic (PBPK) tool kit for environmental pollutants—metals. SAR QSAR Environ. Res 21, 603–618. [DOI] [PubMed] [Google Scholar]
- Ruiz P, Ray M, Fisher J, and Mumtaz M. (2011). Development of a human physiologically based pharmacokinetic (PBPK) toolkit for environmental pollutants. Int. J. Mol. Sci 12, 7469–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sier JH, Thumser AE, and Plant NJ (2017). Linking physiologically-based pharmacokinetic and genome-scale metabolic networks to understand estradiol biology. BMC Syst. Biol 11, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif R, Charest-Tardif G, Brodeur J, and Krishnan K. (1997). Physiologically based pharmacokinetic modeling of a ternary mixture of alkyl benzenes in rats and humans. Toxicol. Appl. Pharmacol 144, 120–134. [DOI] [PubMed] [Google Scholar]
- Tardif R, Lapare S, Charest-Tardif G, Brodeur J, and Krishnan K. (1995). Physiologically-based pharmacokinetic modeling of a mixture of toluene and xylene in humans. Risk Anal. 15, 335–342. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang CE, Orr A, Tydlacka S, Li SH, and Li XJ (2008). Impaired Ubiquitin-proteasome system activity in the synapses of Huntington’s disease mice. J. Cell Biol 180, 1177–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemskov EA, and Nukina N. (2003). Impaired degradation of PKCalpha by proteasome in a cellular model of Huntington’s disease. Neuroreport 14, 1435–1438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.