Abstract
The transcription factor Glioma-Associated Oncogene Homolog 1 (GLI1) is activated by sonic hedgehog (SHH) cascade and is an established driver of pancreatic ductal adenocarcinoma (PDAC). However, therapies targeting upstream hedgehog signaling have shown little to no efficacy in clinical trials. Here, we identify Mixed Lineage Kinase 3 (MLK3) as a druggable regulator of oncogenic GLI1. Earlier, we reported that MLK3 phosphorylated a peptidyl-prolyl isomerase PIN1 on the S138 site, and the PIN1-pS138 translocated to the nucleus. In this report, we identify GLI1 as one of the targets of PIN1-pS138 and demonstrate that PIN1-pS138 is upregulated in human PDAC and strongly associates with the upregulation of GLI1 and MLK3 expression. Moreover, we also identified two new phosphorylation sites on GLI1, T394, and S1089, which are directly phosphorylated by MLK3 to promote GLI1 nuclear translocation, transcriptional activity, and cell proliferation. Additionally, pharmacological inhibition of MLK3 by CEP-1347 promoted apoptosis in PDAC cell lines, reduced tumor burden, extended survival, and reduced GLI1 expression in the Pdx1-Cre x LSL-KRASG12D x LSL-TP53R172H (KPC) mouse model of PDAC. These findings collectively suggest that MLK3 is an important regulator of oncogenic GLI1 and that therapies targeting MLK3 warrant consideration in the management of PDAC patients.
Keywords: Pancreatic Cancer, Cell Biology, Oncogenes, PIN1, GLI1, MLK3
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) has a dismal 5-year survival rate of less than 9% [1]. As most patients present to the clinic with advanced disease, many are not eligible for surgical intervention, and most are refractory to standard of care chemotherapy. As the underlying pathobiology of pancreatic cancer is becoming more apparent, several molecular targets have been proposed to extend survival and reduce disease-associated morbidity [2]. It is well established that the majority of PDAC tumors harbor mutant KRAS and loss of function mutation in tumor-suppressor, p53, yet both of these proteins are ‘undruggable’ [3, 4]. The dismal PDAC patients’ survival highlights the urgency to understand the molecular mechanisms of the disease better and find new druggable targets for clinical management.
To this end, Mixed Lineage Kinase 3 (MLK3) has been proposed as a key regulator of several intersecting MAPK cascades with numerous roles in carcinogenesis [5, 6]. MLK3 has previously been shown to phosphorylate B-RAF directly and is required for ERK-mediated cell proliferation [7, 8], as well as p38 [9] and JNK [10] activation. Beyond the canonical role for MLK3 in MAPK pathway regulation, we have previously demonstrated that in breast cancer, MLK3 directs the activity of Peptidyl-prolyl cis-trans isomerase PIN1 via phosphorylation at S138 [11]. The interaction between MLK3 and PIN1 is important for breast cancer development, driving the cell cycle by promoting cyclin D1 stability and centrosome amplification [11]. Importantly, we also observed that the phosphomimetic PIN1 mutant (PIN1-S138E) was exclusively localized in the nucleus [11]. The protein PIN1 is reported to be overexpressed in almost all cancer; however, its detailed underlying signaling mechanism and cellular targets in cancer are not fully understood [12, 13].
In the present study, GLI1 was identified as a target of phosphorylated PIN1 (i.e., PIN1-S138E) in a non-malignant breast cancer cell line. GLI1 is a classical target of hedgehog signaling, and its activation is required for KRAS-induced tumorigenesis [14] and is particularly pertinent to tumor-associated fibrosis/desmoplasia and chemoresistance in PDAC [15]. GLI1 has been considered a drug target primarily in PDAC [16] and glioblastoma [17, 18], and therefore we planned to define the biology of the MLK3-PIN1-GLI1 axis in PDAC.
Our results established that MLK3-induced phosphorylated PIN1 not only increased the GLI1 transcripts; moreover, the GLI1 was also directly phosphorylated by MLK3 to increase its nuclear translocation and transcriptional activity. The protein stability of phosphorylated GLI1 was relatively higher than phosphodeficient protein, and phosphorylated GLI1 increased the proliferation of pancreatic cancer cell lines. The expressions of p-PIN1 and GLI1 were decreased in the pancreatic tumors of the PDAC mouse model (i.e., KPC) upon treatment with a MLK3 inhibitor, CEP-1347. The desmoplastic content and ductal cell proliferation in mouse pancreatic tissues were similarly reduced, and there was significant cell death in the pancreatic ductal cells by CEP-1347. Moreover, the CEP-1347 significantly extended the survival and loss of lean body mass in the animals. These data suggest that the MLK3/PIN1/GLI1 axis could be an important driver of the neoplastic phenotype in PDAC and that agents targeting MLK3, such as CEP-1347, warrants consideration in the management of pancreatic cancer patients.
2. Materials and Methods
2.1. RT2 Profiler array.
For RT2 Profiler Array, the cell lines expressing either pEGF-PIN1-S138A or pEGF-PIN1-S138E were sorted via FACS sorting on EGFP expression (UIC, RRC Core facility). RNA was isolated/quantified as described previously [19, 20], and a Human Breast Cancer Pathway RT2 Profiler™ PCR Array (PAHS-131Z, Qiagen) was purchased and used per manufacturer specification. The fold changes of gene expression, scatterplot, and heat-map were analyzed by using RT2 PCR array data analysis web portal version 3.5 (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php). Genes with >2-fold change were considered significant and validated by qPCR.
2.2. Kinase assay.
Approximately 5 μg of recombinant GST-GLI1 proteins were incubated with recombinant MLK3 enzyme (E.M.D. Millipore) and kinase assay was carried out following our published report [10, 21, 22].
2.3. Mouse model of PDAC and treatment.
The Pdx1-Cre x LSL-KRASG12D-TP53R172H (KPC) mice were generated as described previously [23]. At 3 months of age, randomly selected animals (N=7/group) were administered daily intraperitoneal injections of either saline (vehicle) or 1.5 mg/kg CEP-1347. The CEP-1347 solution was always made fresh from a stock solution (4 mg/ml in DMSO), supplemented with 35% of PEG-400 (Sigma-Aldrich). After 7 weeks on either vehicle or CEP-1347, animals were sacrificed and tissues were evaluated. All animal procedures were approved by the UIC-IACUC.
2.4. Study approval.
All experiments involving the use of mice were performed following protocols approved by the UIC-IACUC. The de-identified patient slides and information were obtained from the UIC Pathology Core, following IRB approval.
2.5. Statistical analyses.
Unless otherwise indicated, all the experiments presented in the manuscript were repeated at least three times with a minimum of three replicates. A paired two-tail student t-test was used for the comparison between two groups. One-way ANOVA followed by Bonferroni multiple comparison test was done for the multivariate groups using the GraphPad Prism (Version 5.03) software. The significant differences were indicated by *p≤0.05, **p≤0.001, and ***p≤0.0001.
3. Results
3.1. MLK3 Phosphorylates PIN1 to Increase GLI1 Expression.
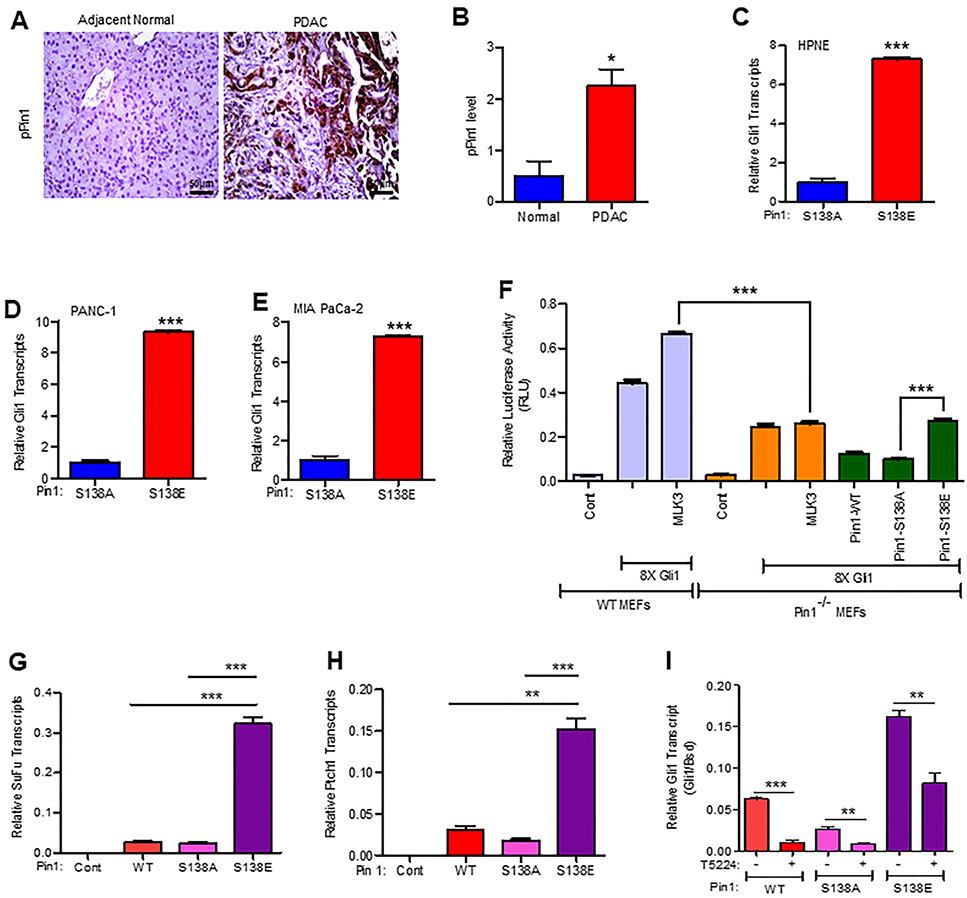
Previously we have demonstrated that one of the primary roles of MLK3 in breast cancer is to direct isomerase activity of PIN1 via phosphorylation, where phosphorylated PIN1 was primarily localized in the nucleus [11]. To identify the targets of phosphorylated-PIN1, we first utilized a non-malignant MCF10A cell line, expressing either a PIN1-S138A variant mimicking a lack of MLK3-induced phosphorylation, or a PIN1-S138E variant mimicking constitutive phosphorylation by MLK3. We performed an RT2 profiler array comparing alterations in known oncogenic pathways and found about 13 folds higher GLI1 mRNA expression in cells expressing PIN1-S138E, compared to PIN1-S138A expressing cells (Supplementary Fig. 1A and B). The GLI1 mRNA expression was further validated by qPCR and observed similar 13 folds increase in presence of the PIN1-S138E (phosphomimetic) variant when compared to the PIN1-S138A (phosphodeficient) variant (Supplementary Fig. 1C). The hedgehog-GLI1 pathway has primarily been reported as a drug target in PDAC [13] and glioblastoma [14, 15], and therefore we next assessed whether PIN1 is similarly phosphorylated at S138 in human PDAC tumors, using antibody raised against PIN1-pS138 site (Supplementary Fig. 1D). We stained patient-derived adjacent normal and PDAC tissues by immunohistochemistry. PIN1-pS138 stained almost exclusively to the neoplastic epithelium and was nearly undetectable in normal tissues (Fig. 1A). The PIN1-pS138 staining was scored and plotted (Fig. 1B). We next determined the levels of p-PIN1 expression in a non-malignant cell line, HPNE, as well as PDAC cancer cell lines PANC-1 and MIA PaCa-2, and AsPC-1. Consistent with data from cancer patients, PDAC cell lines also displayed increased level of p-PIN1 compared to the non-malignant cell line, HPNE (Supplementary Fig. 1E). We next transfected HPNE, PANC-1, and MIA PaCa-2 cell lines either with PIN1-S138A or PIN1-S138E and assessed changes in GLI1 mRNA expression. As observed in breast cancer cells, we also found that all cell lines expressing PIN1-S138E displayed significant upregulation in GLI1 mRNA when compared to PIN1-S138A expressing cells (Figs. 1C-E). To determine the role of MLK3 in PIN1 phosphorylation and its impact on GLI1 gene expression/signaling, we transfected PANC-1 cells with expression vectors for either GLI1 or PIN1, or both together. Besides, we also treated cells transfected with MLK3 and PIN1 together with a pharmacological inhibitor of either PIN1, ATRA (All-Trans-Retinoic Acid) or MLK3 inhibitor, CEP-1347. We then performed a GLI1-luciferase assay using the 8XGLI1 luciferase promoter reporter. Consistent with our previous observations, concomitant overexpression of PIN1 increased GLI1-luciferase expression, which was significantly enhanced upon MLK3 co-expression, and both the inhibitors were able to significantly block the GLI1’s transcriptional activity (Supplementary Figs. 1F-G). To further corroborate that the MLK3-PIN1 axis is necessary for transcriptional activation of GLI1, the murine embryonic fibroblasts (MEFs) derived from PIN1 knockout (PIN1−/−) and PIN1 sufficient (WT) were co-transfected with MLK3 and 8XGLI1 reporter. Interestingly, the significant increased GLI1 transcriptional activation seen in WT MEFs was completely blunted in PIN1−/− MEFs (Fig. 1F). Moreover, the GLI1 transcriptional activity was restored in PIN1−/− MEFs by PIN1-S138E but not by PIN1-S138A expression vectors (Fig. 1F). To determine that transcriptional activation observed in reporter assay is truly translated into increased expression of GLI1 target genes, qPCR was done with mRNA isolated from PANC-1 stable cell lines, expressing either PIN1-WT or -S138A, or -S138E. The transcripts of two well-known GLI1 targets genes, SUFU, and PTCH1 [24] were significantly increased in the stable cell line, expressing PIN1-S138E, compared to either PIN1-WT or -S138A (Figs. 1G and H). Next, we wanted to know how MLK3-induced phosphorylation of PIN1 increases GLI1 transcripts. Earlier, we published that MLK3 acts as a potent activator of JNK [10, 25], and activated JNK is known to phosphorylate transcription factors, FOS and JUN, to form AP-1-complex that finally regulates the transcription of target genes [26]. The FOS and JUN phosphorylated by JNK are not transcriptionally competent. It is reported that enzymatically active PIN1 isomerizes phosphorylated FOS and JUN to form a stable and transcriptionally competent AP-1 complex [27]. We analyzed the promoter of GLI1 to know whether it contains any AP-1 binding site(s). The promoter analysis identified two putative AP-1 binding sites on the GLI1 promoter (Supplementary Table 1). Therefore to know any contribution of AP-1 activation via MLK3/JNK-PIN1 axis in the upregulation of GLI1 transcripts, we utilized stable PANC-1 cell lines, expressing either PIN1-WT or -S138A, or -S138E proteins. These cell lines were treated either with vehicle or AP-1 inhibitor, T5224 [28] and estimated the GLI1 transcript by qPCR. The GLI1 transcripts were reduced by AP-1 inhibitor, even in PANC-1 cells expressing PIN1-S18E (Fig. 1I). We also analyzed the promoters of GLI1 target genes, SuFu, Ptch1, and Cyclin D1. The promoters of all these genes contain more than one AP-1 binding site (Supplementary Table 1), suggesting that GLI1 target genes could also be regulated similarly to GLI1 transcripts. The qPCR data shows that all these GLI1 target genes are similarly regulated via AP-1 activation (Supplementary Figs. 1H-J). These results suggest that phosphorylation of PIN1 by MLK3 is crucial to regulate GLI1 gene expression perhaps via stabilizing AP-1 complex.
Figure 1. MLK3 phosphorylates PIN1 to increase GLI1 expression.
(A) Human adjacent non-malignant (normal) and pancreatic ductal adenocarcinoma (PDAC) tissue sections were stained for the level of PIN1-pS138 by immunohistochemistry. (B) Sections were scored by two investigators, and average values are presented as mean ± SEM. The two groups were compared by the two-tailed Student t-test (N=4/group, *p<0.05). (C-E) The non-malignant pancreas cell line, HPNE and PDAC cell lines, PANC-1 and MIA PaCa-2, were transfected either with a non-phosphorylatable PIN1 mutant (S138A) or PIN1 phospho-mimetic mutant (S138E), and GLI1 mRNA expression was determined by qPCR. Data represent mean ± SD, and comparison between PIN1 -S138A and -S138E was determined by the two-tailed Student t-test (N=3, ***p<0.0001). (F) PIN1 sufficient (WT) or PIN1 knockout (PIN1−/−) MEFs were transfected with an 8XGLI1-Luc reporter vector along with either MLK3 or control empty vector. For a separate transfection, the PIN1−/− MEFs were complemented with PIN1 -WT or - S138A, or -S138E, and GLI1 transcriptional activity was assessed by luciferase assay. Data represent mean ± SD by one-way ANOVA, Bonferroni multiple comparison test (N=4, ***p<0.0001). (G-H) Expressions of SUFU and PTCH1 transcript were determined in the PANC-1 cells stably expressing either PIN1-WT or -S138A, or -S138E by qPCR. I the stable PANC-1 cell lines were treated with vehicle or AP-1 inhibitor, T5224 (5 μM for 24 hrs.) and expression of GLI1 was determined as in G-H. Expression levels were normalized with the blasticidin (Bsd) gene present on the lentiviral expression vector (pLenti6.3/DEST-V5). Data represent mean ± SD by one-way ANOVA, Bonferroni multiple comparison test (N=3, **p<0.001 and ***p<0.0001).
3.2. The MLK3 and PIN1 are Upregulated in Human PDAC and Positively Associates with GLI1 Expression.
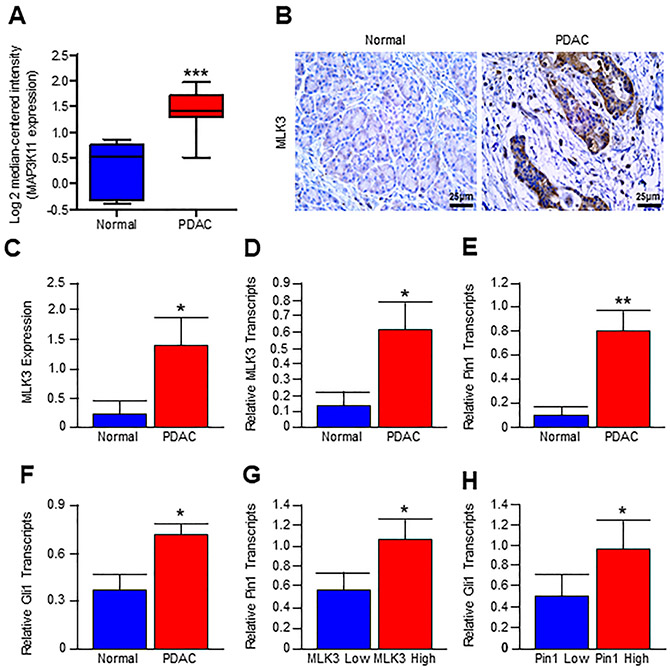
While MLK3 and PIN1 appear to direct GLI1 expression, we next sought to determine whether these events are recapitulated in human pancreatic cancer. We first mined the established Logsdon datasets (Oncomine) and found that PDAC samples have significantly increased MLK3 transcripts compared to non-malignant (normal) tissues (Fig. 2A). To validate the Logsdon datasets, we examined the protein expression of MLK3 in primary PDAC and adjacent normal tissues by immunohistochemistry. The human PDAC tissues displayed significant upregulation of MLK3 protein expression compared to adjacent normal tissue (Figs. 2B and C). Using human pancreatic cancer tissues, we next evaluated the expression of the MLK3 transcript (Fig. 2D) as well as that of its downstream target PIN1 (Fig. 2E), both of which were found to be strongly upregulated in PDAC tissues. We then evaluated patient samples for GLI1 expression, which was also upregulated in tumor tissue (Fig. 2F). Interestingly, patients with high MLK3 expression (above the mean) had significantly more PIN1 expression (Fig. 2G) when compared to patients with low MLK3 expression (below the mean). Similarly, patients with high PIN1 expression had significantly higher GLI1 expression compared to those with low PIN1 expression (Fig. 2H). These data collectively suggest that in human PDAC tissues as well, there is a direct correlation among MLK3, PIN1, and GLI1 expressions.
Figure 2. MLK3/PIN1 axis is upregulated in human PDAC and positively associates with GLI1 expression.
(A) Box plot of MLK3 (i.e., MAP3K11) mRNA expression in normal and pancreatic adenocarcinoma (PDAC) patient using the Logsdon patient datasets (Oncomine, Thermo Fisher, Ann Arbor, MI). Both groups were compared with the two-tailed Student t-test (normal, N=5 and PDAC, N=15, ***p<0.0001). (B) Adjacent Normal and PDAC tumor tissue were stained for MLK3 expression by immunohistochemistry. (C) Sections were scored by two investigators, averaged, and values are presented as mean ± SEM., and two groups were compared by two-tailed Student t-test (N=7/group, *p<0.05). (D-F) Non-malignant (normal) and PDAC tumor tissues were collected, and mRNA expression of MLK3, PIN1, and GLI1 was determined by qPCR. Data represent mean ± SEM, and two groups were compared by two-tailed Student t-test (normal, N=9 and tumor, N=10, *p<0.05 and **p<0.001). (G-H) Using these relative values of mRNA expression in PDAC, samples (N=10) were segregated into MLK3 Low (below the mean) and MLK3 High (above the mean) groups, and Pin1 expression levels were compared and presented as mean ± SEM (*p<0.05). Similarly, mRNA values were segregated into PIN1 Low (below the mean) and PIN1 High (above the mean) groups. The GLI1 expression levels were compared and presented as mean ± SEM (*p<0.05). The comparison between Low and High groups was done by a two-tailed Student t-test (*p<0.05).
3.3. MLK3 Phosphorylates GLI1 at T394 and S1089 Sites.
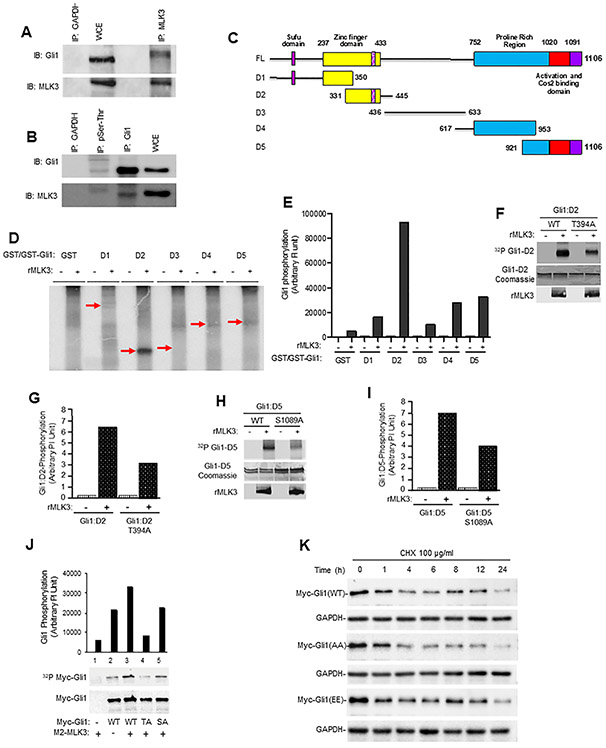
MLK3 is a functional Ser/Thr kinase, and therefore first, we determined any possible functional interaction between MLK3 and GLI1. The endogenous MLK3 from PANC-1 cells was immunoprecipitated and blotted with GLI1 antibody. The MLK3 immunoprecipitation was able to bring down GLI1, which was slightly shifted compared to GLI1 in cell extract (Fig. 3A). The reverse immunoprecipitation of endogenous GLI1 was also able to bring down MLK3 (Supplementary Fig. 2A). To further understand the importance of phosphorylation-dependent interaction between GLI1 and MLK3, the cell extract from PANC-1 was immunoprecipitated with pSer/pThr antibody and blotted with GLI1 and MLK3 antibodies, respectively. Again, the pSer/pThr antibody was able to bring down both GLI1 and MLK3 together from PANC-1 cells (Fig. 3B). Interestingly the protein bands of both GLI1 and MLK3 were shifted to higher molecular weight, suggesting that phosphorylated GLI1 and MLK3 interact with each other (Fig. 3B), perhaps transiently. To further understand the relationship between MLK3 and GLI1, we performed in vitro kinase assay using five bacterially expressed GST-tagged GLI1 protein fragments (Fig. 3C) and recombinant MLK3 (rMLK3) enzyme. Our results demonstrated that MLK3 phosphorylates GLI1, primarily the D2 fragment (AA 331-445) and somewhat to D4 and D5 GLI1 fragments (Figs. 3D and E and Supplementary Fig. 2B). We subsequently repeated this experiment using non-radiolabeled ATP and evaluated all five GST-tagged GLI1 fragments by phosphomass spectrometry, and identified T394 (on D2 fragment) and S1089 (on D5 fragment) as being phosphorylated by MLK3 (Supplementary Figs. 2C and D). To confirm that identified in vitro phosphorylation sites are indeed targeted by MLK3, we created phosphodeficient GLI1-D2 and -D5 expression vectors and expressed in bacteria and made purified proteins. Using these mutant (T394A and S1089A) proteins, we next performed an in vitro kinase assay. The phosphorylation of both T394A and S1089A mutant proteins by rMLK3 enzyme was significantly reduced, suggesting that these two sites on GLI1 could be targeted by MLK3 (Figs. 3F-I). To further confirm that these two p-sites are also targeted by MLK3 in cells, the Myc-tagged mammalian vectors, expressing either wild type (WT) or T394A or S1089A were transfected in HEK-293 cells with the MLK3 expression vector. These cells were radio-labeled with p32-orthophosphate, and GLI1 proteins were pulldown with magnetic beads, coated with anti-Myc antibody. The results clearly show that WT protein is significantly phosphorylated in the presence of MLK3, and phosphorylation of phosphodeficient proteins is less than WT protein (Fig. 3J). Interestingly, the phosphorylation of T394A protein was least, suggesting that this could be the major MLK3 p-site in GLI1 (Fig. 3J). To understand the significance of MLK3-induced phosphorylation on GLI1 protein, we determined the stability of WT, phosphodeficient (AA), and phosphomimetic (EE) GLI1 proteins by cycloheximide chase assay [29, 30]. There was rapid time-dependent degradation of phosphodeficient (AA), compared to phosphomimetic (EE), or WT GLI1 proteins (Fig. 3K and Supplementary Fig. 3A). To know whether MLK3-induced phosphorylation of GLI1 increases its transcriptional activity towards target genes, we transiently transfected PANC-1 cells either with WT or AA or EE GLI1 expression vectors and the expressions of GLI1 target genes, SUFU and PTCH1 were determined by qPCR. The expressions of both SUFU and PTCH1 mRNAs were comparatively higher in PANC-1 cells expressing GLI1 EE than GLI1 AA expression vectors (Supplementary Figs. 3B and C). The expression of SUFU protein was also least in GLI1 AA expressing cells than cells either expressing WT or GLI1 EE proteins (Supplementary Fig. 3D); however, the interaction between SUFU and GLI1 EE proteins was least compared to either WT or GLI1 AA (Supplementary Fig. 3E).
Figure 3. MLK3 associates with GLI1 and phosphorylates at Thr394 and Ser1089.
(A) The endogenous MLK3 protein from PANC-1 cell extracts was immunoprecipitated (I.P.) and blotted with an anti-GLI1 antibody. GAPDH antibody was used for control IP, and whole-cell extracts (WCE) were used for input. The membranes were blotted with GLI1 and MLK3 antibodies, respectively. (B) The cell extract from PANC-1 cells was immunoprecipitated either with pSer/pThr or anti-GLI1, or anti-GAPDH (control) and cell extract (30 μg) was used as an input, and membranes were blotted with either GLI1 or MLK3 antibodies. Phosphorylation-dependent interaction between MLK3 and GLI1 was performed using the pSer/pThr antibody. (C) Schema detailing the five GLI1 deletion mutants (i.e., D1-D5) and Full length (FL). (D-E) An in vitro kinase assay was performed using bacterially expressed GLI1 deletion mutant (D1-D5) proteins and recombinant MLK3 enzyme, values presented as an arbitrary unit. (F-I) Bacterially expressed wild type GLI1 (WT), GLI1-T394A, or GLI1-S1089A with and without recombinant MLK3 (rMLK3) enzyme were used in in vitro kinase assays. Phosphorylation of these GLI1 fragments was determined by Phosphoimager and presented as an arbitrary unit (g and i). (J) The confirmation of GLI1 -T394 and -S1089 as the in vivo targets of MLK3 was determined in HEK-293 cells after cell labeling with P32-orthophosphate. (K) The stability of indicated GLI1 protein variants (GLI1 -WT, -AA and -EE) was determined by cycloheximide chase assay, followed by western blotting with an anti-Myc antibody for transfected Myc-tagged GLI1 proteins.
3.4. MLK3-induced GLI1 Phosphorylation Promotes Pancreatic Cancer Cell Proliferation.
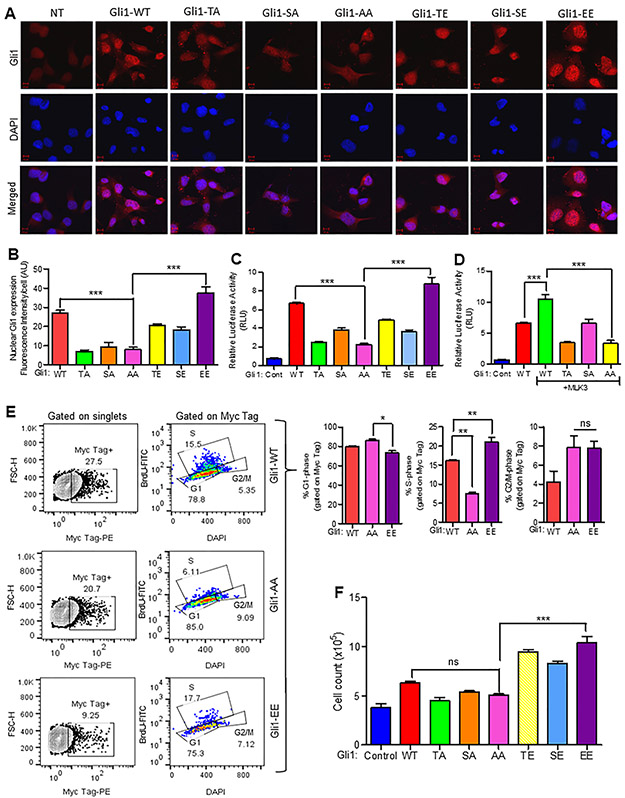
To determine the physiologic relevance of MLK3-induced GLI1 phosphorylation, we first transfected PANC-1 cells with Myc-tagged plasmids expressing, either wild type GLI1 (GLI1-WT), or phosphodeficient GLI1 expression vectors [T394A (TA) or S1089A (SA) or T394A, S1089A (AA)] or phosphomimetic GLI1 expression vectors [T394E (TE) or S1089E (SE) or T394E, S1089E (EE)]. We next assessed GLI1 expression and localization 72 h post-transfection via immunofluorescence for the Myc-tag GLI1 WT or its variants. While GLI1-WT had a diffuse pattern of expression, GLI1 -TA or -SA were primarily in the cytoplasm, whereas GLI1-AA was expressed at comparatively lower levels and found almost exclusively in the cytoplasm. The phosphomimetic GLI1 -TE or -SE were localized in the cytoplasm and slightly in the nuclei; however, GLI1-EE was more strongly expressed and localized in the nuclei compared to the cytoplasm (Figs. 4A and B). Since GLI1 mutant and WT proteins were differentially localized and their transcriptional activities also appeared to be different, therefore we transfected HEK-293 cells with WT or GLI1 phospho-mimetic or phospho-deficient mutant expression vectors to examine their relative expression. Interestingly, the expression of GLI1-EE mutant was the highest, and its phospho-deficient mutant GLI1-AA or GLI1-TA was least (Supplementary Fig. 3F). We, therefore, repeated these transfections and performed a GLI1 luciferase assay using the SUFU promoter (i.e. 8XGLI reporter) as a known transcriptional target of GLI1 [31]. Conceivably, GLI1-WT increased the luciferase activity, while phosphodeficient mutants, either GLI1-T394A (TA) or -S1089A (SA), had reduced activity (Fig. 4C). Moreover, there was a significantly reduced activity in GLI1-T394A, S1089A (AA) compared to GLI1-WT cells (Fig. 4C). Interestingly, while GLI1-T394E (TE) and GLI1-S1089E (SE) single phosphomimetic variants had reduced luciferase activity, the combined GLI1-EE variant had significantly increased activity when compared to the GLI1-AA or any of the phosphodeficient variants (Fig. 4C). To further demonstrate that the transcriptional activity of GLI1 is regulated via MLK3-induced phosphorylation of newly identified p-sites on GLI1, we repeated the luciferase assay, this time expressing GLI1-T394A (TA), -S1089A (SA) and -T394A, S1089A (AA) along with MLK3. Consistent with our previous data, the addition of MLK3 potentiated the transcriptional activity of GLI1-WT; however, the addition of MLK3 failed to enhance the transcriptional activity of GLI1-T394A, whereas GLI1-S1089A had a similar effect like GLI1-WT without MLK3 (Fig. 4D). Interestingly, the transcriptional activity of GLI1-AA was least, even in the presence of MLK3 (Fig. 4D). To understand the physiologic significance of MLK3-induced GLI1 phosphorylation, we transfected PANC-1 cells with WT, AA, and EE variants of GLI1 and determined cell proliferation and cell cycle progression. While there was a small observable differences in G1 phase between cells expressing AA and EE GLI1 (Fig. 4E), however, GLI1-EE alone significantly increased PANC-1 cell proliferation (i.e., S phase), compared to cells expressing GLI1-AA (Fig. 4E). Moreover, there was no difference between AA and EE expressing cells in the G2-M phase (Fig. 4E). Since there was a non-significant difference between GLI1-AA and -EE expressing cells in the G2-M cell cycle transition, therefore, to better understand the impact of MLK3-induced Gli1 phosphorylation on cell proliferation, we repeated the transfection experiment and determined cell proliferation by direct counting the cell numbers. While the addition of GLI1-WT alone had no significant observable effect on cell numbers, however, phosphomimetic versions of GLI1 -EE, -TE, and -SE significantly enhanced PANC-1 cell proliferation, yet failed to do so in the presence of GLI1-AA (Fig. 4F).
Figure 4. MLK3-induced GLI1 phosphorylation promotes its nuclear translocation and cell proliferation.
(A-B) PANC-1 cells were transfected with either a Myc-tagged- GLI1 wild type vector (GLI1-WT), or with phospho-deficient mutants [GLI1 -T394A (TA), -S1089A (SA) and -T394A, S1089A (AA)] or with phospho-mimetic mutants [GLI1 -T394E (TE), -S1089E (SE) and -T394E, S1089E (EE)]. Seventy-two hours post-transfection, the localization of the respective GLI1 proteins was determined by immunofluorescence staining using Myc-tag antibody and DAPI followed by confocal microscopy, Scale bar: 10 μm. The nuclear expression of GLI1 was enumerated by using Image J software (NIH). The data represents mean ± SEM., by one-way ANOVA, Bonferroni multiple comparison test (N=5 cells, ***p<0.0001). (C) PANC-1 cells were transfected either with GLI1 –WT or phospho-deficient mutants [GLI1 -T394A (TA), -S1089A (SA), and -T394A, S1089A (AA)], or phospho-mimetic mutants [GLI1 -T394E (TE), -S1089E (SE), and -T394E, S1089E (EE)]. The relative GLI1 transcriptional activity was assessed after 48 hours post-transfection by luciferase assay using the 8X-GLI1 promoter construct that contains GLI1 binding sites from the SUFU promoter. (D) PANC-1 cells were transfected with GLI1 -WT, -TA, -SA, or -AA along with MLK3, and GLI1 transcriptional activity was measured 48 hours post-transfection by luciferase assay using the SUFU promoter. All Luciferase values are expressed as mean ± SD from three independent experiments, and multivariate comparison was determined by one-way ANOVA, Bonferroni multiple comparison test (***p<0.0001). (E) PANC-1 cells were transfected either with Myc-tagged GLI1 -WT, -AA, or -EE, and cell cycle was determined by BrdU incorporation followed by counterstaining with DAPI. The representative contour plots (left) show Myc-Tag positive cells gated on singlets, and representative pseudo color plots (center) show cell cycle phases, gated on Myc-Tag positive cells. The Bar graphs (right) represent the quantification of cell cycle phases. (F) PANC-1 cells were transfected with the indicated GLI1 expression vectors and cell numbers were counted manually.Data represent mean ± SEM. by one-way ANOVA, Bonferroni multiple comparison test (N=2, *p<0.05 and **p<0.001) ns, non-significant.
3.5. The Pharmacological Inhibitor of MLK3, CEP-1347, Downregulates GLI1 and its Downstream Targets and Induces Cell Death.
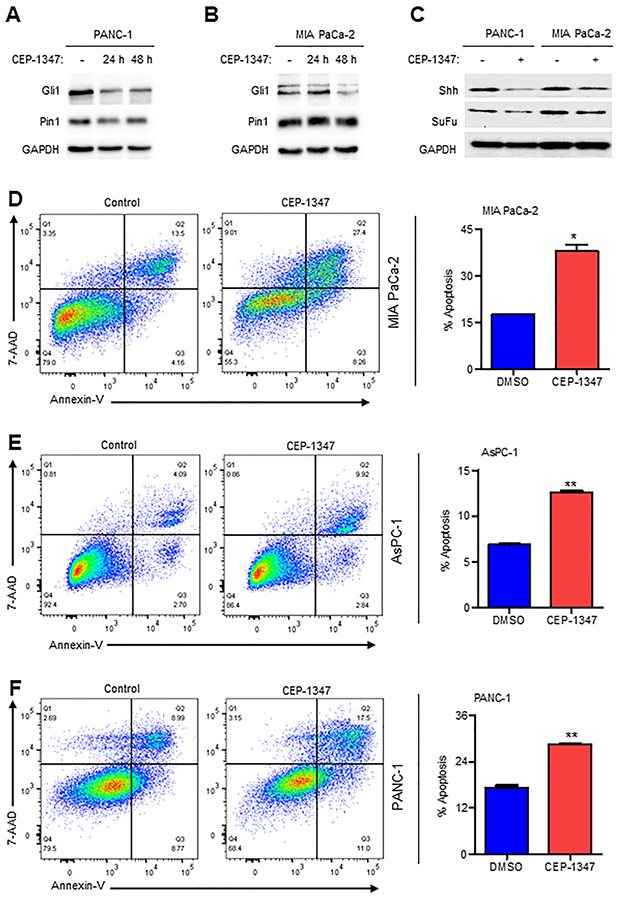
Given the role of the MLK3-PIN1-GLI1 axis in promoting cell proliferation and p-PIN1 upregulating the expression of GLI1 (Fig. 1), we next determined the efficacy of the pharmacological inhibitor of MLK3, CEP-1347, in downregulating protein expression of GLI1 and its downstream targets. Indeed, CEP-1347 was able to downregulate GLI1 protein in PDAC cell lines in a time-dependent manner (Figs. 5A and B) and also the GLI1 downstream target proteins, Sonic Hedgehog (SHH) and Suppressor of Fused (SUFU) (Fig. 5C). Since GLI1 and its target proteins were reduced by CEP-1347 and therefore, we examined the effect of CEP-1347 on pancreatic cancer cell death. The PDAC cell lines, PANC-1, MIA PaCa-2, and AsPC-1, were either treated with vehicle (DMSO) or 500 nM of the CEP-1347, and apoptosis was determined by Annexin V-PI staining. The incubation of PDAC cell lines for 48 h with CEP-1347 promoted significant apoptosis (Figs. 5D-F), suggesting that MLK3 may be a viable target in PDAC.
Figure 5. The pharmacological inhibitor of MLK3, CEP-1347, downregulates GLI1, and its downstream targets and induces cell death.
(A-B) PANC-1 and MIA PaCa-2 cells were treated either with 500 nM of CEP-1347 for 24 or 48 hours or with vehicle (DMSO), and cell extracts were blotted either with GLI1 or PIN1 antibodies. (C) These cell extracts (from a and b) were also blotted with GLI1 downstream target antibodies, SHH and SUFU (D-F) The PANC-1 and MIA PaCa-2 and AsPC-1 cells were treated with 500 nM of CEP-1347 or vehicle control (DMSO) for 24 hours, and apoptosis was assessed with Annexin V-FITC staining. The Pseudo color plots (left) represent 7AAD vs. Annexin V expression, gated on singlets, and bar graphs (right) show quantification of total apoptosis. Data are presented as mean ± SEM, and two groups were compared by two-tailed Student t-test (N=2, *p<0.05 and **p<0.001).
3.6. CEP-1347 Reduces Pancreatic Tumor Burden and Extends Survival in vivo.
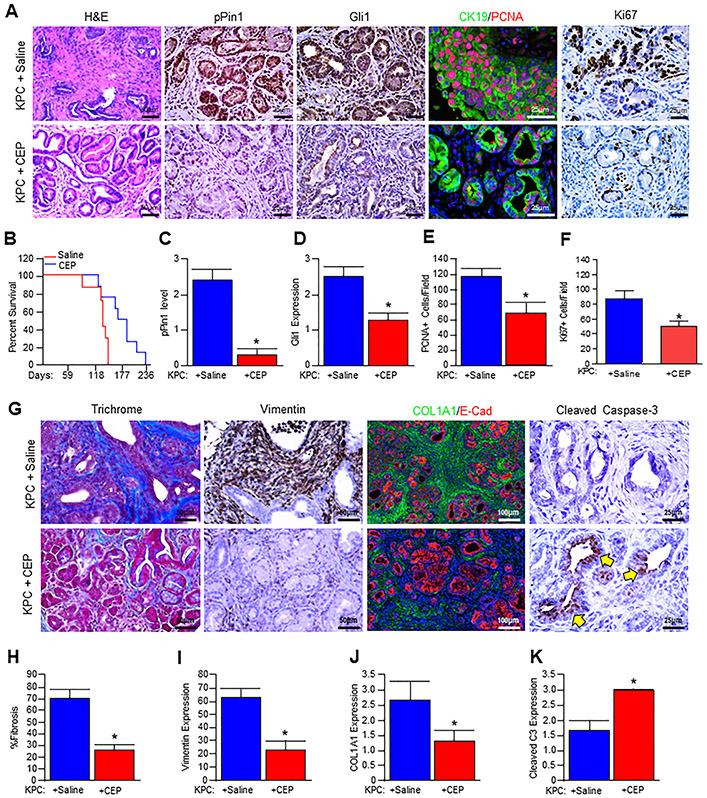
To assess the therapeutic relevance of CEP-1347 in vivo, we first generated cohorts of Pdx1-Cre x LSL-KRASG12D x LSL-TP53R172H (KPC) mice. These animals develop extensive PanIN disease and pancreatic adenocarcinoma, providing a reliable and consistent recapitulation of aggressive human PDAC [23]. Consistent with previous reports [23], saline-treated KPC mice developed invasive carcinoma of the pancreas characterized by a dense tumor stroma (Figs. 6A and G). These animals had high mortality with a mean survival time of 141 days (Fig. 6B), and poor weight gain (Supplementary Fig. 4A) of 1.6 g (8.5%) from the start of injections. Consistent with a reduced tumor burden, CEP-1347-treated mice displayed an overall reduced lesion grade as well as fewer cancerized ducts per high power field (Fig. 6A). This was accompanied by an increased mean survival time of 186 days (Fig. 6B), as well as a significant reduction in cachexia with a mean weight gain of 6.86g (33%) from the start of treatment (Supplementary Fig. 4A). The effective inhibition of MLK3 was determined by evaluating the activity of known MLK3 downstream target, pJNK, which had little to no staining in CEP-1347-treated mice (Supplementary Fig. 4B-C). Similarly, CEP-1347-treated mice also had reduced p-PIN1 expression (Figs. 6A and C), as well as that of GLI1, particularly in the nuclei of tumor tissues (Figs. 6A and D). Since CEP-1347 targets MLK3 and reduced GLI1 protein expression in KPC mouse pancreatic tissues; therefore we determined the effect of CEP-1347 on MLK3 and GLI1 transcripts in KPC mouse pancreatic tissues, treated with either saline or CEP-1347. The mRNA expressions of MLK3 and GLI1 were significantly reduced upon CEP-1347 treatment (Supplementary Fig. 4D-E). CEP-1347 treatment additionally led to a reduced proliferation in neoplastic tissues, determined by dual staining for CK19/PCNA (Figs. 6A and E, and Supplementary Fig. 5A) and Ki67 (Figs. 6A and F). Tissues were next evaluated by Masson’s trichrome staining, which indicated a substantial reduction in the tumor stroma of CEP-1347-treated mice (Figs. 6G and H and Supplementary Fig. 5). Similar results were observed using immunohistochemistry for Vimentin, a mesenchymal marker that localizes predominantly to the tumor stroma, as well as Collagen IA1 (Figs. 6G and I-J, and Supplementary Figs. 5C and D). To determine whether these changes were accompanied by an increase in apoptosis as seen in vitro, we next stained the section with activated/Cleaved Caspase-3, which localized to the neoplastic tissues of CEP-1347-treated animals (Figs. 6G and K). These results clearly suggest that MLK3 inhibitor, is capable of reducing PDAC tumor burden perhaps through MLK3-PIN1-GLI1 axis.
Figure 6. CEP-1347 reduces pancreatic tumor burden and extends survival in vivo.
(A) Pdx1-Cre x LSL-KRASG12D x LSL-TP53R172H (KPC) mice at 3 months of age were treated daily either with saline (vehicle) or 1.5 mg/kg CEP-1347 for seven weeks. At the conclusion of the study, pancreatic tissues from control and treated mice were stained with H&E or via immunohistochemistry or immunofluorescence for PIN1-pS138, GLI1, or CK19/PCNA, or Ki67. (B) Kaplan-Meier curve indicating survival for control (saline) and CEP-1347 treated mice. (C-F) Tissue sections were scored by two investigators and quantified for the indicated proteins. Average values are presented as mean ± SEM, and two groups were compared by the two-tailed Student t-test (N=3, *p<0.05). (G) Pancreatic tissues were next stained with either Masson’s trichrome or via immunohistochemistry or immunofluorescence for Vimentin, COL1A1/E-Cad, or apoptosis surrogate, Cleaved Caspase-3. (H-K) The stained sections were quantified, and average values are presented as mean ± SEM, and two groups were compared by two-tailed Student t-test (N=3, *p<0.05) and presented as mean ± SEM.
4. Discussion
Recent evidence has identified members of the MLK family as being highly relevant to the incidence and progression of several cell processes, including inflammation, metabolism, and a variety of pathologies [32]. The inhibition of MLK proteins by CEP-1347 has already shown early efficacy in neurodegenerative disease [3], and MLK family proteins are now emerging as a novel and druggable oncogenes with important roles in human cancer [6, 33, 34]. Despite these observations, the contributions of MLK family proteins in pancreatic cancer are almost entirely unexplored.
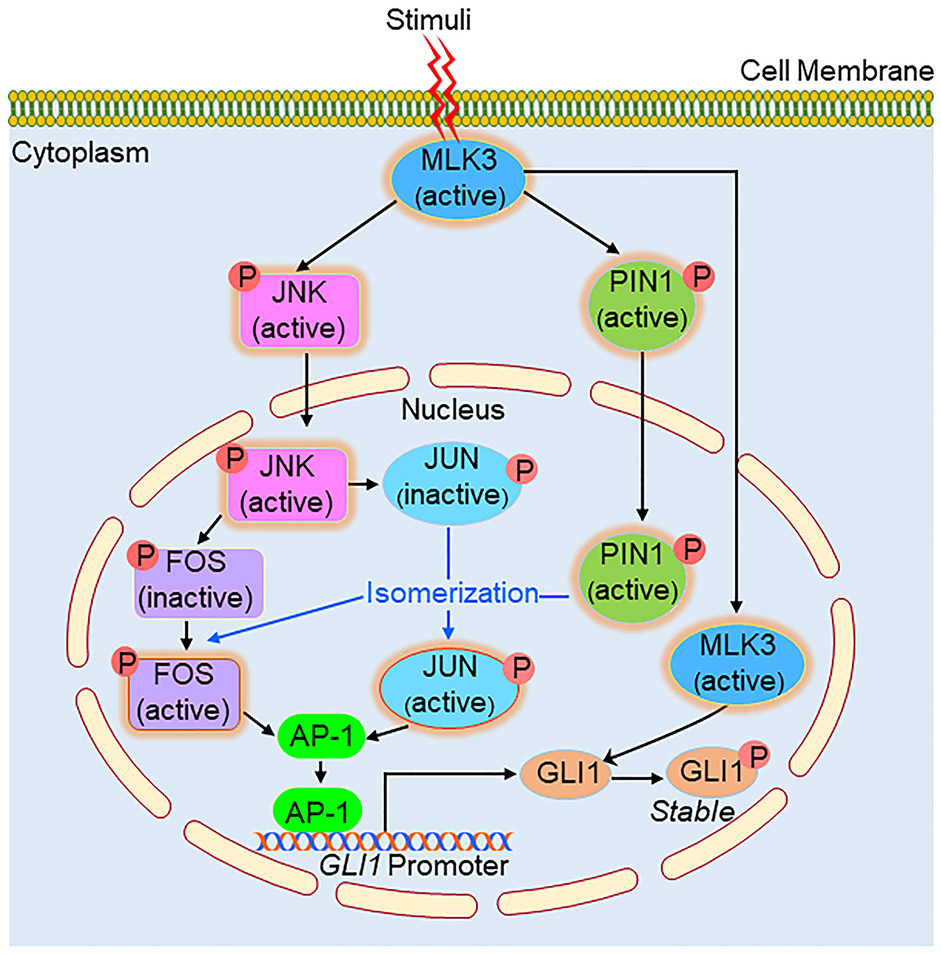
In our previous study, we reported that MLK3 increases the catalytic activity and downstream signaling of PIN1 via direct phosphorylation at the S138 amino acid residue in breast cancer [11]. On further analysis, we determined that this site not only directs cell proliferation in the canonical sense but also is critical for the transcriptional activity of GLI1. Obviously, then the question comes, how phospho-PIN1 upregulates the expression of GLI1 transcripts. Based on our and published results, it appears that MLK3 might regulate GLI1 transcription indirectly via PIN1 phosphorylation (Fig. 7). We have reported that MLK3 potently activates JNK [10, 25], and JNK is known to phosphorylate transcription factors, JUN and FOS, to form the AP-1 complex [26]. The FOS or JUN initially phosphorylated by JNK is reported to be isomerized by catalytically active PIN1 to form a transcriptionally competent AP-1 complex [35]. Since we observed that the AP-1 binding site is present on the GLI1 promoter (Supplementary Table 1) and the AP-1 inhibitor blocked the transcriptional upregulation by constitutive active PIN1-S138E suggests that the MLK3-JNK-AP-1 pathway is one of the mechanisms by which MLK3 upregulates GLI1 transcripts where PIN1 plays an intermediary role (Fig. 7). Alternatively, our data also suggest that GLI1 protein can also be regulated via post-translational modification via MLK3-induced direct phosphorylation, leading to GLI1 protein stability (Fig. 3K and 7). The direct phosphorylation of GLI1 did increase the transcriptional upregulation of the GLI1 target genes as GLI1-EE induced its target proteins SuFu and PTCH1 (Supplementary Fig. 3B-C) and drive the cell cycle (Fig. 4E) and cell proliferation (Fig. 4F).
Figure 7. Schema describing the MLK3-PIN1-GLI1 axis in PDAC.
Once activated, MLK3 directs GLI1 signaling via two distinct pathways. MLK3 is reported to activate JNK and PIN1 via phosphorylation. The activated JNK is known to enter the nucleus and phosphorylates AP-1 transcription components, JUN and FOS, which are transcriptionally incompetent until isomerized by active-PIN1. Once transcriptionally competent AP-1 complex is formed, it binds with GLI1 promoter and increases GLI1 gene expression. MLK3 is reported to be in the cytoplasm and also in the nucleus. The nuclear MLK3 directly phosphorylates GLI1, supporting its retention in the nucleus, protein stability, and enhancing cell proliferation. Combined, both pathways cooperate to enhance pancreatic tumor cell proliferation and survival.
While GLI1 has prognostic relevance to breast cancer [36], it is more often associated with pancreatic cancer development [16]. GLI1 is classically an effector of hedgehog signaling. In this capacity, hedgehog family ligands, i.e., Sonic Hedgehog (SHH) bind the membrane-bound Patched receptor. This event allows for the liberation of Smoothened and activation of GLI transcription factors, which have been implicated in several hallmark features of tumorigenesis [37]. SHH is critical to pancreatic carcinogenesis and cooperates with oncogenic KRAS to drive tumor development at various points in the disease process [38]. Despite these compelling observations, Smoothened inhibitors have failed to show efficacy in clinical trials [39]. Interestingly, it has been reported that oncogenic KRAS directly activates GLI1 via the ERK/MAPK cascade [40]. This offers one likely explanation for the failure of Smoothened inhibitors, as it is well established that Smoothened is not required for the regulation of GLI1 in pancreatic neoplasms [41].
Given the low cost and wide availability of pharmacologic inhibitors of MLK3 such as CEP-1347 and URMC-099 [42, 43], our findings suggest that these agents may impede both receptor-dependent and receptor-independent modulation of GLI1 in PDAC. Indeed, our results indicate that through direct phosphorylation as well as transcriptional enhancement via PIN1, MLK3 is essential for both GLI1 expression and activity in PDAC. Phosphorylation of PIN1 at S138 by MLK3 increases GLI1 transcription in vitro. Similarly, through direct phosphorylation of GLI1, MLK3 directs GLI1 nuclear translocation/activity and cell proliferation. Further, these events appear to be neutralized by the MLK3 inhibitor CEP-1347, which also increased apoptosis in vitro.
We, therefore, administered CEP-1347 to the KPC model of advanced pancreatic cancer, which significantly extended survival and delayed tumor development. CEP-1347-treated animals also displayed dramatic reductions in both p-PIN1 and GLI1 expression, as well as the density of the tumor stroma. A desmoplastic stroma is a main histologic feature of PDAC and establishes a local environment that facilitates tumor growth and invasion [44]. This process is dependent on the activation of pancreatic stellate cells by several signaling pathways, including the SHH/GLI1 axis [39, 45]. Furthermore, tumor cell activation of GLI1 is also believed to play a role in the emergence of drug resistance [46]. As CEP-1347 reduced both epithelial and mesenchymal expression/nuclear localization of GLI1, MLK3 warrants consideration as a target for therapy to overcome chemoresistance in the clinic.
Additionally, cachexia is one of the main comorbidities associated with pancreatic cancer that affects nearly 80% of patients [47] and is the cause of death for 1/3rd of pancreatic cancer patients [48]. Cachexia is diagnosed clinically by a significant weight loss greater/evidence of sarcopenia, along with reduced food intake and systemic inflammation [47]. Consistent with the reduction in tumor burden, animals administered CEP-1347 maintained a healthy weight throughout the study with no evidence of wasting. Should these findings translate to the clinic, in addition to the observed effects on survival and fibrosis, an added benefit to MLK3-inhibition may be the reduction of tumor-associated cachexia. To summarize, these results substantiate MLK3 as a critical pancreatic tumor promoter via its effects on PIN1 and GLI1. Given the availability of compounds targeting MLK3, collectively, our results support that such agents warrant consideration in managing PDAC patients.
Supplementary Material
Highlights.
The transcription factor GLI1 has been implicated in PDAC.
The MLK3-PIN1 axis regulates GLI1 transcriptional activity.
The MLK3 inhibitor, CEP-1347, inhibits GLI1 and reduces the PDAC burden.
Acknowledgements:
This work was supported by the Veterans Affairs Awards [grant numbers: BX004903, BX004855 and BX003296]; the National Institute of Health [grant numbers: CA216410 CA176846, CA178063, CA219764 and CA236031]. We also acknowledge the National Institute of Health shared instrument grant # S100D018445, awarded to U.A.M.S. Proteomics Core Lab, Little Rock, Arkansas, USA
Footnotes
Competing interest statement: The authors attest that we do not have any competing interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Siegel RL, Miller KD, Jemal A, Cancer Statistics, 2017, CA Cancer J Clin, 67 (2017) 7–30. [DOI] [PubMed] [Google Scholar]
- [2].McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS, Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes, World J Gastroenterol, 24 (2018) 4846–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang LH, Besirli CG, Johnson EM Jr., Mixed-lineage kinases: a target for the prevention of neurodegeneration, Annu Rev Pharmacol Toxicol, 44 (2004) 451–474. [DOI] [PubMed] [Google Scholar]
- [4].Weissmueller S, Manchado E, Saborowski M, Morris J.P.t., Wagenblast E, Davis CA, Moon SH, Pfister NT, Tschaharganeh DF, Kitzing T, Aust D, Markert EK, Wu J, Grimmond SM, Pilarsky C, Prives C, Biankin AV, Lowe SW, Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling, Cell, 157 (2014) 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rattanasinchai C, Gallo KA, MLK3 Signaling in Cancer Invasion, Cancers (Basel), 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rana A, Rana B, Mishra R, Sondarva G, Rangasamy V, Das S, Viswakarma N, Kanthasamy A, Mixed Lineage Kinase-c-Jun N-Terminal Kinase Axis: A Potential Therapeutic Target in Cancer, Genes Cancer, 4 (2013) 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chadee DN, Kyriakis JM, MLK3 is required for mitogen activation of B-Raf, ERK and cell proliferation, Nat Cell Biol, 6 (2004) 770–776. [DOI] [PubMed] [Google Scholar]
- [8].Chadee DN, Kyriakis JM, A novel role for mixed lineage kinase 3 (MLK3) in B-Raf activation and cell proliferation, Cell Cycle, 3 (2004) 1227–1229. [DOI] [PubMed] [Google Scholar]
- [9].Kim KY, Kim BC, Xu Z, Kim SJ, Mixed lineage kinase 3 (MLK3)-activated p38 MAP kinase mediates transforming growth factor-beta-induced apoptosis in hepatoma cells, J Biol Chem, 279 (2004) 29478–29484. [DOI] [PubMed] [Google Scholar]
- [10].Rana A, Gallo K, Godowski P, Hirai S, Ohno S, Zon L, Kyriakis JM, Avruch J, The mixed lineage kinase SPRK phosphorylates and activates the stress-activated protein kinase activator, SEK-1, J Biol Chem, 271 (1996) 19025–19028. [DOI] [PubMed] [Google Scholar]
- [11].Rangasamy V, Mishra R, Sondarva G, Das S, Lee TH, Bakowska JC, Tzivion G, Malter JS, Rana B, Lu KP, Kanthasamy A, Rana A, Mixed-lineage kinase 3 phosphorylates prolyl-isomerase Pin1 to regulate its nuclear translocation and cellular function, Proc Natl Acad Sci U S A, 109 (2012) 8149–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen Y, Wu YR, Yang HY, Li XZ, Jie MM, Hu CJ, Wu YY, Yang SM, Yang YB, Prolyl isomerase Pin1: a promoter of cancer and a target for therapy, Cell Death Dis, 9 (2018) 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bao L, Kimzey A, Sauter G, Sowadski JM, Lu KP, Wang DG, Prevalent overexpression of prolyl isomerase Pin1 in human cancers, Am J Pathol, 164 (2004) 1727–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rajurkar M, De Jesus-Monge WE, Driscoll DR, Appleman VA, Huang H, Cotton JL, Klimstra DS, Zhu LJ, Simin K, Xu L, McMahon AP, Lewis BC, Mao J, The activity of Gli transcription factors is essential for Kras-induced pancreatic tumorigenesis, Proc Natl Acad Sci U S A, 109 (2012) E1038–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bailey JM, Swanson BJ, Hamada T, Eggers JP, Singh PK, Caffery T, Ouellette MM, Hollingsworth MA, Sonic hedgehog promotes desmoplasia in pancreatic cancer, Clin Cancer Res, 14 (2008) 5995–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kasai K, GLI1, a master regulator of the hallmark of pancreatic cancer, Pathol Int, 66 (2016) 653–660. [DOI] [PubMed] [Google Scholar]
- [17].Braun S, Oppermann H, Mueller A, Renner C, Hovhannisyan A, Baran-Schmidt R, Gebhardt R, Hipkiss A, Thiery J, Meixensberger J, Gaunitz F, Hedgehog signaling in glioblastoma multiforme, Cancer Biol Ther, 13 (2012) 487–495. [DOI] [PubMed] [Google Scholar]
- [18].Infante P, Alfonsi R, Botta B, Mori M, Di Marcotullio L, Targeting GLI factors to inhibit the Hedgehog pathway, Trends Pharmacol Sci, 36 (2015) 547–558. [DOI] [PubMed] [Google Scholar]
- [19].Viswakarma N, Jia Y, Bai L, Gao Q, Lin B, Zhang X, Misra P, Rana A, Jain S, Gonzalez FJ, Zhu YJ, Thimmapaya B, Reddy JK, The Med1 subunit of the mediator complex induces liver cell proliferation and is phosphorylated by AMP kinase, J Biol Chem, 288 (2013) 27898–27911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kumar S, Singh SK, Viswakarma N, Sondarva G, Nair RS, Sethupathi P, Sinha SC, Emmadi R, Hoskins K, Danciu O, Thatcher GRJ, Rana B, Rana A, Mixed lineage kinase 3 inhibition induces T cell activation and cytotoxicity, Proc Natl Acad Sci U S A, 117 (2020) 7961–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Barthwal MK, Sathyanarayana P, Kundu CN, Rana B, Pradeep A, Sharma C, Woodgett JR, Rana A, Negative regulation of mixed lineage kinase 3 by protein kinase B/AKT leads to cell survival, J Biol Chem, 278 (2003) 3897–3902. [DOI] [PubMed] [Google Scholar]
- [22].Mishra R, Barthwal MK, Sondarva G, Rana B, Wong L, Chatterjee M, Woodgett JR, Rana A, Glycogen synthase kinase-3beta induces neuronal cell death via direct phosphorylation of mixed lineage kinase 3, J Biol Chem, 282 (2007) 30393–30405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA, Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice, Cancer Cell, 7 (2005) 469–483. [DOI] [PubMed] [Google Scholar]
- [24].Diao Y, Rahman MF, Vyatkin Y, Azatyan A, St Laurent G, Kapranov P, Zaphiropoulos PG, Identification of novel GLI1 target genes and regulatory circuits in human cancer cells, Mol Oncol, 12 (2018) 1718–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sathyanarayana P, Barthwal MK, Kundu CN, Lane ME, Bergmann A, Tzivion G, Rana A, Activation of the Drosophila MLK by Ceramide Reveals TNF-alpha and Ceramide as Agonists of Mammalian MLK3, Mol Cell, 10 (2002) 1527–1533. [DOI] [PubMed] [Google Scholar]
- [26].Bogoyevitch MA, Kobe B, Uses for JNK: the many and varied substrates of the c-Jun N-terminal kinases, Microbiol Mol Biol Rev, 70 (2006) 1061–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hu X, Chen LF, Pinning Down the Transcription: A Role for Peptidyl-Prolyl cis-trans Isomerase Pin1 in Gene Expression, Front Cell Dev Biol, 8 (2020) 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Aikawa Y, Morimoto K, Yamamoto T, Chaki H, Hashiramoto A, Narita H, Hirono S, Shiozawa S, Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1, Nat Biotechnol, 26 (2008) 817–823. [DOI] [PubMed] [Google Scholar]
- [29].Chou TF, Deshaies RJ, Quantitative cell-based protein degradation assays to identify and classify drugs that target the ubiquitin-proteasome system, J Biol Chem, 286 (2011) 16546–16554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kao SH, Wang WL, Chen CY, Chang YL, Wu YY, Wang YT, Wang SP, Nesvizhskii AI, Chen YJ, Hong TM, Yang PC, Analysis of Protein Stability by the Cycloheximide Chase Assay, Bio Protoc, 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R, The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins, Genes Dev, 24 (2010) 670–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Craige SM, Reif MM, Kant S, Mixed - Lineage Protein kinases (MLKs) in inflammation, metabolism, and other disease states, Biochim Biophys Acta, 1862 (2016) 1581–1586. [DOI] [PubMed] [Google Scholar]
- [33].Thylur RP, Senthivinayagam S, Campbell EM, Rangasamy V, Thorenoor N, Sondarva G, Mehrotra S, Mishra P, Zook E, Le PT, Rana A, Rana B, Mixed lineage kinase 3 modulates beta-catenin signaling in cancer cells, J Biol Chem, 286 (2011) 37470–37482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li X, Guo J, Ding AP, Qi WW, Zhang PH, Lv J, Qiu WS, Sun ZQ, Association of Mixed Lineage Kinase Domain-Like Protein Expression With Prognosis in Patients With Colon Cancer, Technol Cancer Res Treat, 16 (2017) 428–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, Lu KP, Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1, EMBO J, 20 (2001) 3459–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].ten Haaf A, Bektas N, von Serenyi S, Losen I, Arweiler EC, Hartmann A, Knuchel R, Dahl E, Expression of the glioma-associated oncogene homolog (GLI) 1 in human breast cancer is associated with unfavourable overall survival, BMC Cancer, 9 (2009) 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Villavicencio EH, Walterhouse DO, Iannaccone PM, The sonic hedgehog-patched-gli pathway in human development and disease, Am J Hum Genet, 67 (2000) 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Morton JP, Mongeau ME, Klimstra DS, Morris JP, Lee YC, Kawaguchi Y, Wright CV, Hebrok M, Lewis BC, Sonic hedgehog acts at multiple stages during pancreatic tumorigenesis, Proc Natl Acad Sci U S A, 104 (2007) 5103–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gu D, Schlotman KE, Xie J, Deciphering the role of hedgehog signaling in pancreatic cancer, J Biomed Res, 30 (2016) 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ji Z, Mei FC, Xie J, Cheng X, Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells, J Biol Chem, 282 (2007) 14048–14055. [DOI] [PubMed] [Google Scholar]
- [41].Nolan-Stevaux O, Lau J, Truitt ML, Chu GC, Hebrok M, Fernandez-Zapico ME, Hanahan D, GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation, Genes Dev, 23 (2009) 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].P.I. Parkinson Study Group, Mixed lineage kinase inhibitor CEP-1347 fails to delay disability in early Parkinson disease, Neurology, 69 (2007) 1480–1490. [DOI] [PubMed] [Google Scholar]
- [43].Marker DF, Tremblay ME, Puccini JM, Barbieri J, Gantz Marker MA, Loweth CJ, Muly EC, Lu SM, Goodfellow VS, Dewhurst S, Gelbard HA, The new small-molecule mixed-lineage kinase 3 inhibitor URMC-099 is neuroprotective and anti-inflammatory in models of human immunodeficiency virus-associated neurocognitive disorders, J Neurosci, 33 (2013) 9998–10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Phillips P, Pancreatic stellate cells and fibrosis, in: Grippo PJ, Munshi HG (Eds.) Pancreatic Cancer and Tumor Microenvironment, Trivandrum (India), 2012. [Google Scholar]
- [45].Li X, Wang Z, Ma Q, Xu Q, Liu H, Duan W, Lei J, Ma J, Wang X, Lv S, Han L, Li W, Guo J, Guo K, Zhang D, Wu E, Xie K, Sonic hedgehog paracrine signaling activates stromal cells to promote perineural invasion in pancreatic cancer, Clin Cancer Res, 20 (2014) 4326–4338. [DOI] [PubMed] [Google Scholar]
- [46].Palle K, Mani C, Tripathi K, Athar M, Aberrant GLI1 Activation in DNA Damage Response, Carcinogenesis and Chemoresistance, Cancers (Basel), 7 (2015) 2330–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Fearon KC, Voss AC, Hustead DS, Cancer Cachexia Study G, Definition of cancer cachexia: effect of weight loss, reduced food intake, and systemic inflammation on functional status and prognosis, Am J Clin Nutr, 83 (2006) 1345–1350. [DOI] [PubMed] [Google Scholar]
- [48].Bachmann J, Ketterer K, Marsch C, Fechtner K, Krakowski-Roosen H, Buchler MW, Friess H, Martignoni ME, Pancreatic cancer related cachexia: influence on metabolism and correlation to weight loss and pulmonary function, BMC Cancer, 9 (2009) 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.