Abstract
In mammals, there are four Argonaute (Ago) family proteins that play crucial roles in RNA silencing, a process wherein microRNA (miRNA) mediates inhibition of target mRNA translation. Among the Ago proteins, Argonaute2 (Ago2) uniquely possesses an endoribonuclease (slicer) activity that is critical for the biogenesis of specific miRNAs and mRNA cleavage. This Ago2 slicer activity is required for postnatal development. Despite its important roles, there are still gaps in our understanding of the mechanistic basis of Ago2’s unique functions in vivo due to a limited availability of experimental tools. In order to investigate Ago2’s functions, we generated a new cellular model of Ago2-deficiency in 3T3 mouse embryonic fibroblasts (MEFs). This cell line can be used for investigating general Ago2 functions, but also for further understanding of Ago2’s unique characteristics including the slicer activity, specific amino acid residues, and domains in Ago2 by reconstitution of Ago2 mutants. Here, we describe the methods for establishing Ago2-deficient MEFs and for reconstituting the MEFs with an Ago2 mutant lacking its slicer activity by means of a retrovirus-mediated gene transfer.
Keywords: Ago2, 3T3 protocol, Mouse embryonic fibroblast
1. Introduction
Argonaute (Ago) proteins are essential components of RNA-induced silencing complex (RISC) and play a central role in RNA silencing processes [1]. Among four Ago proteins in mammals, Ago2 is regarded as a special member since only this family member retains endoribonuclease (slicer) activity that generates specific microRNAs (miRNAs) and cleaves target mRNAs [2-6]. Ago2-deficient mice are embryonic lethal and mice lacking Ago2’s slicer activity die shortly after birth with anemia, indicating essential roles of Ago2- and its slicer activity-dependent RNA silencing [2, 6]. In addition to these developmental regulations, it has become clear that Ago2 is involved in the regulation of a variety of cellular and physiological events including tumorigenesis, hypoxia, bone marrow maturation, and pancreatic β cell expansion [7-10]. Previous reports demonstrated that there are several amino acid residues critical for Ago2 functions; slicer activity (D669), RNA recognition (e.g., K533, Q545, K570), epidermal growth factor signaling activity (Y393), and Akt-mediated RNA silencing regulation (S388) [8, 11, 12]. Furthermore, there are several domains critical for Ago2 function including PAZ, an RNA binding module that recognizes the 3′ end of miRNA, or PIWI, a protein domain found in piwi proteins and a large number of related nucleic acid-binding proteins [13]. To investigate the roles of amino acid residues and domains of Ago2 in vivo is of biological interest and critical physiologic importance, as a deeper understanding of their functions could provide us a novel mechanistic basis of RNA silencing and a design of improved therapeutics targeting miRNA-mediated events.
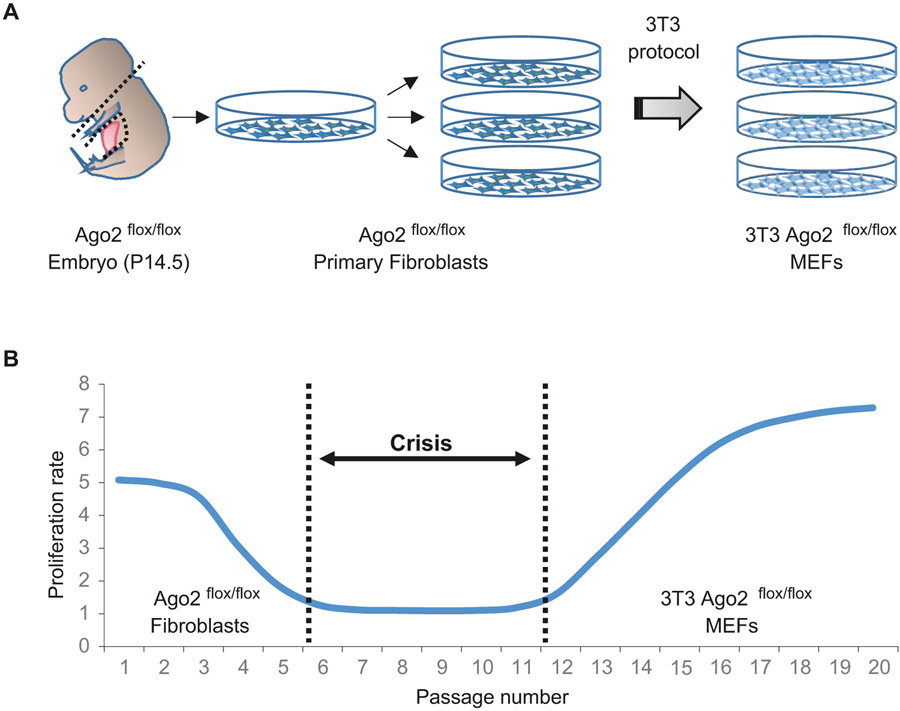
Mouse embryonic fibroblasts (MEFs) have been broadly used as an experimental tool for cellular- and molecular-based biology. Primary fibroblasts isolated from mouse embryos have limited capacity for proliferation. However, the murine fibroblasts can be immortalized by distinct procedures, so that it allows us to easily enable the desired genetic manipulations, reduce culture maintenance time and effort, and facilitate biological replicates in experiments without concern for the limitation of cell proliferation. There are basically two different ways to immortalize the primary mouse fibroblasts. One is to introduce an oncogene(s), such as SV40 large T antigen, into the primary fibroblasts in order to induce malignant transformation. However, the oncogene could disrupt proper cellular processes/events and the MEFs immortalized by the oncogene could manifest biological characteristics as cancer cells. Another method establishing immortalized MEFs is by means of the “3T3” protocol, which means that primary MEFs are transferred (the “T”) every 3 days (the first “3”), and inoculated at the rigid density of 3 × 105 cells per 6 cm dish (the second “3”) continuously (Fig. 1a) [14]. Upon successive passages with the defined 3T3 protocol, MEFs initially undergo decent doublings with each passage, and then go through a senescence crisis where the growth of MEFs virtually ceases (Fig. 1b). By continuing the 3T3 protocol even in the crisis phase, MEFs eventually show a sign of the immortalization with a constant or rising growth rate. There is also a potential issue that the MEFs immortalized by 3T3 protocol (3T3 MEFs) could spontaneously harbor a mutation(s) on endogenous oncogenes/tumor suppressor genes during overcoming the crisis phase. Nonetheless, 3T3 MEFs are regarded as one of the most conventional and useful cellular tools. Fundamental cellular and molecular mechanisms revealed by analysis of the 3T3 MEFs have been widely applied to other cell types in vitro and in vivo.
Fig. 1.
Schemes of establishment of 3T3 MEFs. (a) A scheme of isolating mouse primary fibroblasts to generate immortalized MEFs through the 3T3 protocol. (b) A scheme showing the proliferation rate of MEFs during an immortalization process through 3T3 protocol. The cells go through a senescence crisis once and then become immortalized
In this chapter, we describe an establishment of 3T3 MEFs generated from an embryo carrying a floxed allele that enables conditional inactivation of Ago2 by utilizing adenovirus-mediated cre expression, and reconstitution of Ago2-deficient 3T3 MEFs with an Ago2 mutant by means of retrovirus-mediated gene transfer.
2. Materials
2.1. Immortalization of Ago2flox/flox MEFs by 3T3 Protocol
B6.129P2(129S4)-Ago2tm1.1Tara/J (Ago2flox/flox; Jackson Laboratory).
Culture medium: Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Hyclone) and 1× penicillin-streptomycin solution (Life Technologies).
0.25% trypsin-EDTA (Life Technologies).
Phosphate-buffered saline (PBS) (Life Technologies).
Sterile Fine scissors and Fine forceps.
Cell/Tissue culture incubator with 5% CO2 at 37 °C.
Biological safety cabinet.
Cell/Tissue culture supplies: Plastic tissue culture dishes (10-, 6-cm); sterile plastic serological pipettes; sterile disposable tubes (50 ml) cryovials.
BAMBANKER (WAKO).
2.2. Genetic Deletion of Ago2 by Adeno-Cre
Pre-packed Cre recombinase adenovirus (Vector Biolabs).
293A cells (Invitrogen).
−80 °C deep freezer.
37 °C water bath.
Anti-Mouse Ago2 Monoclonal Antibody (WAKO).
2.3. Genetic Reconstitution of Human Ago2 by Retrovirus
pMXs-Puro retroviral vector (Cell Biolabs).
PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies).
QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies).
DNA purification kit (Machery-Nagel).
BamHI (NEB), EcoRI (NEB).
DNA ligation kit (Takara).
Platinum-E (Plat-E) retroviral packaging cell line (Cell Biolabs).
Hexadimethrine bromide (Polybrene; Sigma-Aldrich).
Puromycin (Invitrogen).
pFRT/FLAG/HA-DEST EIF2C2 (Addgene).
Top10 competent cells (Invitrogen).
LB medium with/without ampicillin: dissolve 25 g of LB broth powder (Fisher) in about 800 ml of purified water. Heat the solution and frequently agitate until it is completely dissolved. Adjust the pH of the medium to 7.2 using 1 M NaOH and bring volume up to 1 L with purified water. Before use, sterilize by autoclaving for 15 min. If antibiotic is needed, allow the solution to cool to 55 °C or lower, and add ampicillin to the final concentration of 50 μg/ml. Store the solution at room temperature.
LB Agar plates with ampicillin: Suspend 40 g LB agar powder (Fisher) in 1 L of purified water. Heat the solution and frequently agitate until it is completely dissolved. Before use, sterilize by autoclaving for 15 min. Allow the solution to cool to 55 °C, add ampicillin to the final concentration of 50 μg/ml. Mix the solution thoroughly, and pour about 25 ml in each Petri dish. Store the Petri dishes at 4 °C.
Plasmid miniprep kit (Machery-Nagel).
Lipofectamine 2000 (Invitrogen).
Opti-MEM (Invitrogen).
Sterilized 0.45-μm filter.
Hemagglutinin (HA) antibody (Cell Signaling).
3. Method
3.1. Isolate and Immortalize MEF Cells from Ago2flox/flox Mice
Set up breeding pairs with Ago2flox/flox males and females and check females the following day for vaginal plugs to determine if mating has occurred (see Note 1).
At 14.5 days of pregnancy, anesthetize the pregnant Ago2flox/flox female and place it on a dissecting board. Sterilize the fur of the mouse with 70% ethanol. Make a cut on the medioventral line with scissors, and expose the abdominal wall of the mouse.
Cut through the abdominal wall with sterile scissors. Lift up the uterine horns with sterile forceps (see Note 2), and cut off the uterus and place it in tissue culture dish containing PBS for wash.
Carefully dissect the uterus in the PBS, separate the embryos and transfer them to a new dish with PBS. Swirl the dish to remove blood from the embryos.
Draw an embryo and place it on an empty tissue culture dish. Remove the embryonic head, and tear out the red tissue (heart and liver). Place the rest of the embryo in the covered Petri dish containing 3 ml of 0.25% trypsin-EDTA.
Chop up the trimmed embryo with scissors repeatedly to mince the tissue into pieces as small as possible. Pipet the tissue pieces gently several times with a 3 ml syringe loaded with a 18G needle, then place the dish in the 37 °C tissue culture incubator for 5 min.
Remove the dish from the incubator, add 7 ml of DMEM culture medium to the dish in order to inactivate trypsin, and mix the cell suspension gently but thoroughly using a 10 ml serological pipet.
Transfer the cell suspension to a 15 ml conical centrifuge tube. By using an additional 3 ml of culture medium, wash the dish and transfer remaining cells to the tube.
Centrifuge the tube at 200 rcf for 5 min.
Carefully remove and discard approximately 10 ml of supernatant. Add 10 ml of fresh culture medium, and mix the cell suspension gently. Let the cell suspension sit for about 5 min to allow larger embryo fragments to sink to the bottom.
Transfer the supernatant, but not large embryo fragments, as much as possible to a 10 cm dish. Incubate the dish with 5% CO2 at 37 °C overnight and change the culture medium the following morning.
Check the cells later in the day. If the cells reach confluency, make cell stocks with BAMBANKER or split the cells. At this time, the passage number of the Ago2flox/flox MEFs is “1.”
To establish 3T3 Ago2flox/flox MEFs, seed the cells at the density of 3 × 105 cells per 6 cm dish, and split the cells with the same density every 3 days. The MEF cells will become immortalized after about 15–20 times of passage. Make cell stocks in the cryovial with an appropriate label including a passage number.
3.2. Genetic Deletion of Ago2 by Adeno-Cre
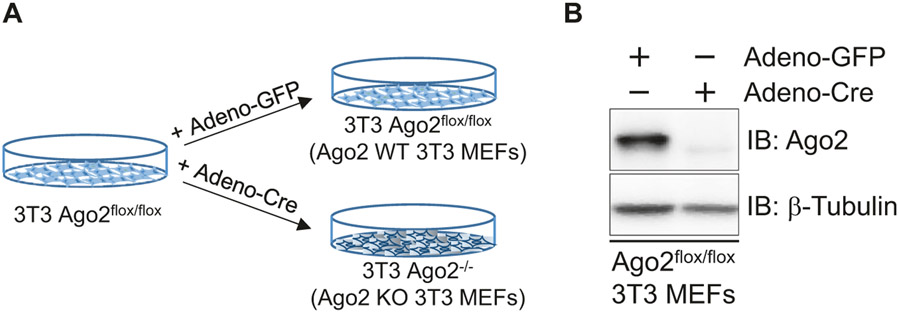
Expression levels of Ago2 in the 3T3 Ago2flox/flox MEFs are intact. Therefore, the 3T3 Ago2flox/flox MEFs can be used as wild-type. In order to generate Ago2-deficient 3T3 MEFs, a cre recombinase needs to be expressed in the cells (Fig. 2a). In this section, the cre gene is transferred into the cells by utilizing an adenovirus vector (see Note 3).
Fig. 2.
Generation of Ago2-deficient 3T3 MEFs. (a) A scheme showing a strategy to delete Ago2 in 3T3 Ago2flox/flox MEFs by adenovirus-mediated cre recombinase (Adeno-Cre) expression. (b) A western blot analysis assessing expression levels Ago2 detected by an anti-Ago2 monoclonal antibody in MEFs treated with Adeno-GFP and Adeno-Cre. β-Tubulin is shown as a control
The 293A cell line, a subclone of the 293 cell line, is used to amplify a replication-incompetent adenovirus. Trypsinize and count the 293A cells, plate them to two 10 cm dishes with 3 × 106 cells per dish. On the following morning, refresh the medium and add a small amount (10–20 μl) of the pre-packed Adeno-Cre virus or a control adenovirus carrying green fluorescent protein (Adeno-GFP) to each dish. Swirl the dish gently to mix.
Keep culturing the cells in the incubator. When about 50% of 293A cells are detaching (about 1 day after transduction), squirt the cells off the dish with a 10 ml pipette. Transfer virus-containing cells and media to 15 ml sterile capped tubes.
Place the 15 ml tube containing harvested cells and media at −80 °C for at least 30 min. Remove tube and place in a 37 °C water bath for no longer than 15 min to thaw. Repeat the freezing and thawing steps twice. Centrifuge the cell lysate at 800 rcf for 10 min. Transfer the supernatant to cryovials in 1 ml aliquots. Store the viral stocks at −80 °C. Titer the virus stock if needed. This virus stock, the 1st amplification of the Adeno-Cre and Adeno-GFP virus, can be used for cell infection to express transgene, further crude amplification, or large amplification and purification for in vivo study.
In order to infect Adeno-Cre and Adeno-GFP virus to 3T3 Ago2flox/flox MEFs, plate the cells to two 6 cm dishes at 5 x 105 cells per dish (Day 0). On the following morning (Day 1), refresh the medium and add approximately 100 μl of the crude viral stocks of Adeno-Cre or Adeno-GFP from step 3 (see Note 4). Refresh the medium on the following morning (Day 2). Continue to culture the cells in the incubator.
When the infected 3T3 Ago2flox/flox MEFs reach confluency (1–2 days after transduction), plate the infected MEF cells in multiple 6 cm dishes at 5 × 105 cells per dish.
By using the cells on one of the plates, examine expression levels of Ago2 by western blot by detecting with anti-Ago2 antibody (Fig. 2b). If the titer of virus stocks is high enough, expression of Ago2 is completely lost in the 3T3 Ago2flox/flox MEFs infected with Adeno-Cre, but not in cells infected with Adeno-GFP, even after one-time virus infection.
If Ago2 expression remains in the 3T3 Ago2flox/flox MEFs infected with Adeno-Cre, repeat the infection step (step 4). Expression of Ago2 usually disappears within three times of Adeno-Cre infection. Genetic deletion of the Ago2 allele can be confirmed by genotyping using a specific primer set. After confirmation of Ago2 deletion, make stocks of the Ago2-deficient 3T3 MEFs (Ago2 KO 3T3 MEFs) and Adeno-GFP-treated 3T3 Ago2flox/flox MEFs (Ago2 WT 3T3 MEFs) (see Note 5).
3.3. Genetic Reconstitution of Human Ago2 by Retrovirus
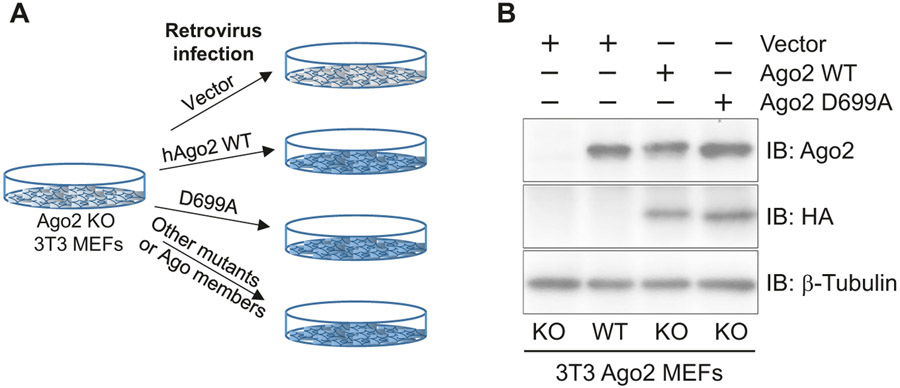
Functional roles of specific amino acids and domains in Ago2 can be investigated in vivo by reintroducing Ago2 mutants in Ago2 KO 3T3 MEFs. In order to reintroduce the Ago2 mutant, one of the most conventional and convenient methods is to utilize a retrovirus-mediated gene transfer that mediates stable genetic modification of treated cells by chromosomal integration of a gene(s) of interest. In this section, Ago2 KO 3T3 MEFs are reconstituted with human Ago2 WT or mutant by means of the retrovirus (Fig. 3a). Through this strategy, we can even replace Ago2 with other Argonaute members in MEFs (Fig. 3a).
Fig. 3.
Reconstitution of human Ago2 in Ago2-deficient 3T3 MEFs. (a) A scheme showing a strategy to reconstitute human Ago2 WT, mutant, or other Ago members by means of retrovirus-mediated gene transfer. (b) A western blot analysis assessing expression levels Ago2 detected by anti-Ago2 that detect both endogenous and reconstituted Ago2 and anti-HA monoclonal antibody that detects exogenous proteins. β-Tubulin is shown as a control
Among the variety of retrovirus vectors, a pMXs-Puro retroviral vector based on the Moloney murine leukemia virus (MMLV) is used in this study. This pMXs-Puro vector can stably integrate and express a gene of interest with a puromycin selection. In order to insert human Ago2 into this vector, human Ago2 coding DNA sequence (CDS) is amplified by PCR with primers that contain a restriction enzyme digestion site, followed by the restriction enzyme digestion and a ligation of the human Ago2 CDS fragment with the pMXs-Puro backbone vector. Retrovirus vector with human Ago2 mutant can also be generated with a mutagenesis kit if necessary. After confirmation of the human Ago2 WT or mutant insertion, the retrovirus vector is ready for transfection to retrovirus packaging cells such as Plat-E cells to produce retrovirus, followed by infection to Ago2 KO 3T3 MEFs for reconstitution.
Generate a BamHI-human Ago2-EcoRI nucleotide fragment by PCR using PfuUltra II Fusion HS DNA Polymerase with an appropriate human Ago2 template (e.g., pFRT/FLAG/HA-DEST EIF2C2) (see Note 6). Digest pMXs-Puro retroviral vector and the purified human Ago2 DNA fragments with BamHI and EcoRI. Purify the digested human Ago2 fragments with a DNA purification kit, and the digested backbone vector by gel purification (see Note 7).
Ligate the purified human Ago2 and backbone fragments (follow manufacturer’s protocol in the ligation kit). Transform the ligation product to Top10 competent cells and spread the cells on a LB agar plate containing 50 μg/ml ampicillin. Select 5–10 colonies from the plate after a 37 °C overnight incubation. Culture them overnight in LB medium with 50 μg/ml ampicillin, and isolate plasmid from each colony using a mini-prep kit.
If needed, induce a mutation(s) into human Ago2 by QuikChange Lightning Site-Directed Mutagenesis Kit (follow manufacturer’s protocol). Sequence the plasmids (use the PCR primers in Note 6) to confirm Ago2 insertion.
After confirmation, prepare enough plasmid for transfection (see Note 8), and seed Plat-E cells at 3 × 106 cells in a 6-cm dish (Day 0).
On Day 1, transfect 10 μg of pMXs-Puro retroviral empty vector, pMXs- human Ago2-Puro retroviral vector, or pMXs- human Ago2 mutant-Puro retroviral vector to the Plat-E cells in each 6-cm dish by Lipofectamine 2000.
On Day 2, refresh the culture medium and continue to incubate the cells in the incubator.
After 24 hours, harvest the virus-containing supernatant from the dish of transfected Plat-E cells and filtrate it with a 0.45-μm sterile syringe filter. Aliquot 1 ml of the supernatant into cryovials. This supernatant can be used for gene transfer immediately or stored at −80 °C. Titer the virus stock if needed.
Seed Ago2 KO and Ago2 WT 3T3 MEFs at 3 × 104 cells per well in a 24-well plate.
On the following morning, infect the MEFs with the different dilutions of retroviral supernatant in the presence of 6 μg/ml polybrene for 8–24 h in order to obtain cells expressing human Ago2 WT and mutant at similar levels (see Note 9). Remove and discard the medium from the wells and replace with fresh DMEM culture medium.
After additional 24 h incubation, trypsinize the infected MEFs and plate them in 6-cm dishes with fresh culture medium containing 2 μg/ml puromycin (see Note 10) for selection.
Continue to incubate the cells until the selection is completed (about 3 days). Harvest the selected cells and confirm the expression of human Ago2 WT or mutant protein in Ago2 KO MEF cells with anti-Ago2 antibody by western blot (Fig. 3b). If necessary, compare the expression levels of different groups of cells with Ago2 WT 3T3 MEF cells, and choose the cells expressing similar Ago2 levels as WT cells. Make stocks of the Ago2 KO 3T3 MEFs reconstituted with human Ago2 WT or mutant, and pMXs-Puro retrovirus treated Ago2 KO 3T3 MEFs.
Acknowledgments
We thank Kazutoshi Murakami, Elise Bernhard, and Vishnupriya Borra for discussions. This work was supported by NIH RO1 DK107530, Digestive Disease Research Core Center in Cincinnati (DK078392), and PRESTO from the Japan Science and Technology Agency.
Footnotes
Please refer Information about “how to time mouse pregnancy” described in the website of the Jackson Laboratory.
Do not contaminate the uterus with the fur.
Every procedure in this part contains infectious virus. Please follow the Biological Safety Level 2 work guidelines.
Passaging and transduction can be conducted at the same time. Add the crude virus lysate to medium in the 6 cm dish when seeding the cells on Day 0. Swirl the dish gently to mix. Refresh the medium on the morning of Day 1.
Expression of genes induced by adenovirus is regulated in a temporal manner.
The primers for the PCR are 5′-CGATAGGGA TCC ACCATGTACTCGGGAGCCGGC-3′ (forward) and 5 ′ - CGATAGGAATTCCCTAGGCAGAGTCTGGGACGTCATATGGATAAGCAAAGTACATGGTGCG-3′ (reverse). HA tag is introduced to the sequence as an optional marker.
BamHI and EcoRI locate separately in a multi-cloning site of the pMXs-Puro retroviral vector and there is about 1 kb between these two sites. So after digestion, two fragments with different sizes are observed in the gel. One of them is a fragment around 1 kb and the other one is about 5–6 kb, which is the backbone fragment.
If mini-prep is used, ethanol precipitation is needed for purification of the plasmid to ensure the transfection efficiency.
If it is necessary to adjust the expression level of restored human Ago2 WT similar to the endogenous Ago2 expression level in WT cells, you can transduce the Ago2 KO MEF cells with different titers of the retrovirus, such as 1, 10, 25, 50, or 100 μl of the crude virus to 500 μl of medium.
Ideally, optimized puromycin concentration for Ago2 MEF should be tested before selection by culture the Ago2 KO MEF cells in the presence of different concentration of puromycin, such as 1, 2, 4, 6 μg/ml and so on. Choose the lowest concentration of puromycin that can induce the death of more than 95% of cells, for the following selection experiment. It is always recommended to have Ago2 KO MEF cells (without transduction) as the positive control for selection.
References
- 1.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R (2005) Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123(4):631–640. doi: 10.1016/j.cell.2005.10.022 [DOI] [PubMed] [Google Scholar]
- 2.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ (2010) A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465(7298):584–589. doi: 10.1038/nature09092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D, Lai EC (2010) Conserved vertebrate mir-451 provides a platform for dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A 107(34):15163–15168. doi: 10.1073/pnas.1006432107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A (2007) A slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 21(16):1999–2004. doi: 10.1101/gad.1565607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meister G (2013) Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 14(7):447–459. doi: 10.1038/nrg3462 [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305(5689):1437–1441. doi: 10.1126/science.1102513 [DOI] [PubMed] [Google Scholar]
- 7.Cheng N, Li Y, Han ZG (2013) Argonaute2 promotes tumor metastasis by way of up-regulating focal adhesion kinase expression in hepatocellular carcinoma. Hepatology 57(5):1906–1918. doi: 10.1002/hep.26202 [DOI] [PubMed] [Google Scholar]
- 8.Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, Du Y, Wang Y, Chang WC, Chen CH, Hsu JL, Wu Y, Lam YC, James BP, Liu X, Liu CG, Patel DJ, Hung MC (2013) EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 497(7449):383–387. doi: 10.1038/nature12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ (2010) A novel miRNA processing pathway independent of dicer requires Argonaute2 catalytic activity. Science 328(5986):1694–1698. doi: 10.1126/science.1190809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tattikota SG, Rathjen T, McAnulty SJ, Wessels HH, Akerman I, van de Bunt M, Hausser J, Esguerra JL, Musahl A, Pandey AK, You X, Chen W, Herrera PL, Johnson PR, O’Carroll D, Eliasson L, Zavolan M, Gloyn AL, Ferrer J, Shalom-Feuerstein R, Aberdam D, Poy MN (2014) Argonaute2 mediates compensatory expansion of the pancreatic beta cell. Cell Metab 19(1):122–134. doi: 10.1016/j.cmet.2013.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ (2005) Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434(7033):666–670. doi: 10.1038/nature03514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horman SR, Janas MM, Litterst C, Wang B, MacRae IJ, Sever MJ, Morrissey DV, Graves P, Luo B, Umesalma S, Qi HH, Miraglia LJ, Novina CD, Orth AP (2013) Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Mol Cell 50(3):356–367. doi: 10.1016/j.molcel.2013.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonas S, Izaurralde E (2015) Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16(7):421–433. doi: 10.1038/nrg3965 [DOI] [PubMed] [Google Scholar]
- 14.Todaro GJ, Green H (1963) Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol 17:299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]