Abstract
Background
The SARS-CoV-2 pandemic has challenged many of our current routine practices in the treatment and care of patients. Given the critical importance of blood donation and transfusion we analyzed 92 blood samples of individuals infected with SARS-CoV-2 stratified by symptoms.
Study Design and Methods
We therefore tested blood samples for SARS-CoV-2 via RT-PCR targeting the E gene. In addition, we tested each blood sample for anti-SARS-CoV-2 IgG antibodies via ELISA and performed plaque reduction neutralization tests.
Results
SARS-CoV-2 RNA was absent in the blood of mild to asymptomatic patients (57 individuals) and only detectable in individuals with severe COVID-19 who were admitted to the intensive care unit (35 individuals) (n = 6/92 [6.5%]; p = 0.023 Fisher's exact test). Interestingly, anti-spike IgG antibodies were not significantly higher in intensive care unit patients compared to mild patients, but we found that their neutralizing capacity was disproportionately increased (p < 0.001).
Conclusion
Our observations support the hypothesis that there are no potential hazards from blood or plasma transfusion of SARS-CoV-2-positive individuals with mild flu-like symptoms and more importantly of asymptomatic individuals.
Keywords: Blood donation, Blood safety, Plasma, Virus safety
Introduction
As of February 7th, 2021, the COVID-19 pandemic has caused nearly 106 million reported infections and over 2.3 million confirmed deaths [1]. Statistical and serological estimates suggest the true prevalence of SARS-CoV-2 may be between 5 and 50 times the reported case count [2, 3, 4, 5]. The failure to detect infections and in particular those of large numbers of asymptomatic or mild cases raises the question whether SARS-CoV-2 plasma viremia poses a threat to individuals in need of blood donations. As particular individuals with pre-existing conditions are more likely to have a more severe course of COVID-19, knowledge about potential hazards from blood or plasma transfusion is important. SARS-CoV-2, as a novel coronavirus, infects primarily the upper and lower respiratory tract and can cause acute respiratory infections. Similar to rhinovirus or influenza, SARS-CoV-2 can be transmitted from human to human mainly by droplets, aerosols and fomites [6]. The estimated infectiousness starts at 2–3 days before symptom onset and declines rapidly within 7 days [7]. In comparison, transmission of blood-borne viruses like HIV can result from sexual intercourse, maternal-fetal transmission, or exposure-prone procedures in health-care settings like needle stick injuries or blood transfusion [8, 9]. Untreated blood-borne virus infections can have long-lasting viremia and in case of HIV life-long contagiousness [10, 11]. So far, only a few reports from Wuhan detected SARS-CoV-2 RNA in the blood in individuals who were diagnosed with severe symptoms and viral pneumonia [12, 13, 14]. In addition, SARS-CoV-2 RNAemia in the blood has been evaluated in only 3 individuals from Germany with asymptomatic infection [15]. We therefore systematically evaluated the presence of SARS-CoV-2 RNA in the plasma of acutely infected patients stratified by their symptoms.
Materials and Methods
Patient Material Sampling
A subset of acutely SARS-CoV-2 infected individuals were enrolled during a previous study in the hotspot of Gangelt, which is located in the German county of Heinsberg in North Rhine-Westphalia [16]. For this current study, we analyzed in total blood samples from 92 acutely SARS-CoV-2 infected individuals and tested them for SARS-CoV-2 via RT-PCR targeting the E gene. 57 individuals included in this study were tested positive for SARS-CoV-2 via pharyngeal, nasopharyngeal or anal swabs but were not hospitalized. 12 of them were asymptomatic and 45 indicated only mild flu-like symptoms (fever, coughing, loss of taste, loss of smell or dyspnea) [17]. Pharyngeal (55), nasopharyngeal (1) or anal (1) swabs as well as blood were taken at the same time and analyzed via RT-PCR (described in the following). 35 of the 92 individuals had severe symptoms, were admitted to an intensive care unit (ICU) and needed invasive mechanical ventilation because of acute respiratory distress syndrome (ARDS) (Table 1). Pharyngeal (34) or nasopharyngeal (1) swabs and blood were not taken at the same time. Due to treatment and the patient's clinical status blood was drawn shortly after the positive RT-PCR test. These ICU patients were still in critical condition and needed invasive mechanical ventilation.
Table 1.
Molecular detection of SARS-CoV-2 in infected individuals
| Overall | Asymptomatic cases | Mild cases | ICU | p-value | |
|---|---|---|---|---|---|
| Age | 0.0021 | ||||
| Mean ± SD | 53.3±19.2 | 55.5±19.4 | 46.8±19.8 | 61.6±15.2 | |
| Gender | |||||
| Male | 51 | 5 | 21 | 25 | |
| Female | 41 | 7 | 24 | 10 | |
| Swab Ct value | <0.0001 | ||||
| Mean ± SD | 31.9±5.6 | 36.0±2.9 | 33.2±3.8 | 25.7±6.3a | |
| RNAemia (Ct value) | 0.023 | ||||
| Number (mean ± SD) | 6 (35.1±1.8) | 0 | 0 | 6 (35.1±1.8) | |
| Number of symptoms | <0.0001 | ||||
| Mean ± SD | 1.9±1.3 | 0 | 1.9±1.0 | 2.5±1.1b | |
| Patients with ARDS | |||||
| Number (median) | 35 | 0 | 0 | 35 |
ICU, intensive care unit; Ct, threshold cycle; ARDS, acute respiratory distress syndrome. Statistical significance was assessed by Kruskal Wallis test and Fisher's exact test. aCt value from 6 patients were not available due to the fact that RT-PCR was performed elsewhere. bTwo patients were relocated to the University Hospital Bonn with unknown COVID-19 symptoms.
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
The complete virological analyses were performed via RT-PCR with a validated diagnostic protocol [18]. Briefly, swab or plasma samples were homogenized by short vortexing, 140 μL of the media containing sample material were transferred to a sterile 2-mL microcentrifuge tube holding 560 μL AVL buffer (Qiagen). Viral RNA was extracted with the QIAmp Viral RNA Mini kit (Qiagen). The RNA was used as template for two real-time RT-PCR reactions (SuperScriptTMIII One-Step RT-PCR System with PlatinumTM TaqDNA Polymerase, Thermo Fisher) to amplify the sequences of the SARS-CoV-2 E gene (primers E_Sarbeco_F1 and R2, and probe E_Sarbeco_P110) and an internal control for RNA extraction, reverse transcription, and amplification (innuDETECT Internal Control RNA Assay, Analytik Jena #845-ID-0007100), to minimize batch effects and ensure sample stability. Samples were considered positive for SARS-CoV-2 if amplification occurred in the virus-specific reactions and if the internal control's Ct value was within the expected range.
Plasma Isolation
Whole EDTA blood was centrifuged at 600 × g for 10 min (without centrifuge brake) and EDTA plasma was stored until further analysis (−80°C).
ELISA
Frozen plasma was thawed, heat inactivated for 30 min at 56°C, centrifuged for 5 min at 5,000 × g, and the supernatant transferred to a fresh tube. Each sample was tested with the Euroimmun SARS-CoV-2 IgG ELISA kit (EI 2606-9620 G, Euroimmun, Germany) and sample dilution was performed according to the manufacturer's instructions. ELISA of the plasma samples from ICU patients was only possible for 32 individuals due to the lack of enough sample volume.
SARS-CoV-2 Neutralization Assay
A plaque reduction neutralization test was performed with heat-inactivated plasma samples of ICU patients. To this end, the plasma was serially 2-fold diluted starting with 1:2 up to 1:16,384. 120 μL of each plasma dilution was mixed with 80 plaque forming units (PFU) of SARS-CoV-2 in 120 μL OptiPROTMSFM (Thermo Fisher, USA) cell culture medium. After incubation of 1 h at 37°C, 200 μL of each mixture were added to wells of a 24-well plate seeded the day before with 1.5 × 105 Vero E6 cells/well. After incubation for 1 h at 37°C, the inoculum was removed and cells were overlaid with a 1:1 mixture of 1.5% (w/v) carboxymethylcellulose (Sigma-Aldrich, USA) in 2x MEM (Biochrom, Germany) with 4% FBS (Thermo Fisher, USA). After incubation at 37°C and 5% CO2 for 3 days, the overlay was removed and the 24-well plates were fixed using a 6% formaldehyde solution and stained with 1% crystal violet in 20% ethanol.
Statistics
Prism (GraphPad Software) was used for statistical analysis. Data are presented as mean ± SD. Differences were estimated with 1-way ANOVA, Fisher's exact test and unpaired t test. p < 0.05 indicates statistical significance.
Results
Patient Characteristics
We analyzed a total of 92 acutely SARS-CoV-2-infected individuals with a median age of 53 years (range: 18–87) of which 55.4% were male. 57 individuals were not hospitalized of which 12 individuals reported no symptoms (termed: asymptomatic). 45 individuals reported mild flu-like symptoms including fever, coughing, loss of taste, loss of smell and dyspnea. We compared those to 35 individuals who had severe COVID-19 and had been hospitalized with ARDS and mechanical ventilation. Individuals with ARDS were significantly older (61.6 ± 15.2) compared to individuals with mild infection (46.8 ± 19.8; p = 0.0015). In addition, there was a significant difference in the Ct values between the patient groups with significantly lower Ct values (25.7 ± 6.3) in the RT-PCR compared to individuals who were asymptomatic (36.0 ± 2.9; p < 0.0001) or with mild symptoms (33.2 ± 3.8; p < 0.0001).
Detectable SARS-CoV-2 RNAemia in Critical Ill Patients
We assessed SARS-CoV-2 RNA by RT-PCR in the plasma from all individuals. However, we were only able to detect SARS-CoV-2 viral load in 6 out of 92 plasma samples (6.5%) (Table 1). All 6 samples were from individuals with severe ARDS and mechanical ventilation, 2 of whom subsequently succumbed to COVID-19. While all 6 individuals were in the ICU with severe symptoms, there were no commonalities between the patients in terms of pre-existing conditions or other medical care. More importantly, however, we were not able to detect any sign of SARS-CoV-2 RNA in individuals with mild or no symptoms. This was statistically significant (p = 0.023, two-tailed Fisher's exact test). Thus, our data indicates that SARS-CoV-2 is only detectable in the blood in rare cases of critical ill patients.
Strong Antibody Neutralizing Titers in ICU Patients' Plasma
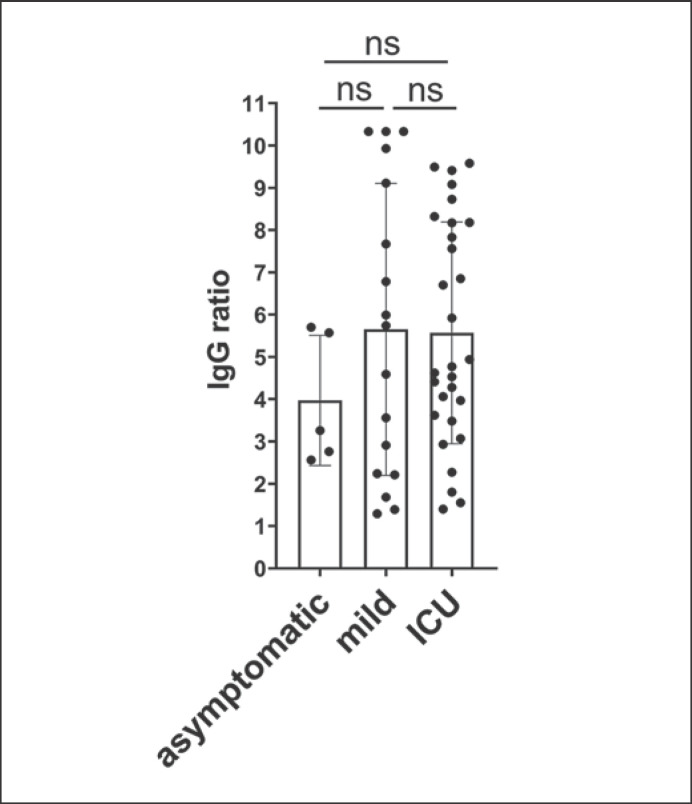
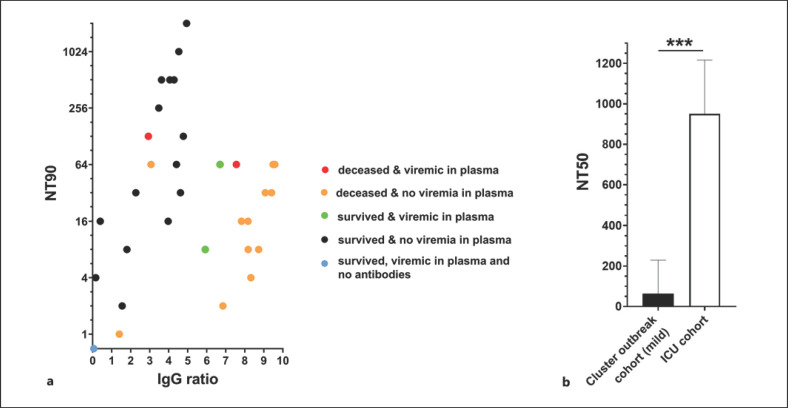
We next compared antibody levels against SARS-CoV-2 in only seroconverted individuals but SARS-CoV-2 RNA positive in the swab. Seropositivity was defined as IgG ELISA (IgG ratio >1.1). While all 92 patients were acutely infected with SARS-CoV-2, anti-spike-IgG was present in 29 out of 32 critically ill patients (90.6%), 5 out of 12 asymptomatic patients (41.7%), and 18 out of 45 patients with mild symptoms (40.0%). Interestingly, we observed similar IgG levels in all individuals regardless of the severity of their symptoms (Fig. 1). In addition, there was no significant difference in the IgG levels of individuals who had RNAemia in the blood compared to those without detectable SARS-CoV-2 RNA (4.6 ± 3.1; 4.3 ± 3.3; p = 0.88, respectively). To further characterize the immune response of critically ill COVID-19 patients, we measured the neutralization capacity of their antibody responses using a plaque reduction neutralization assay. We found that individuals with severe SARS-CoV-2 infection showed very high neutralization capacities (Fig. 2) compared to those with mild or no symptoms [16]. Remarkably, out of 5 patients in intensive care who had detectable viral RNA in the blood, 1 individual had no detectable anti-SARS-CoV-2 IgG, no neutralization capacity of the plasma, but ultimately recovered from COVID-19 (Fig. 2a, blue dot). In contrast, 2 individuals that died of COVID-19 had detectable viral RNA in the blood and SARS-CoV-2-specific IgG with neutralizing capacity (Fig. 2a, red dot). Interestingly, in the other fatal cases, no RNAemia was measurable in the plasma, but specific IgG was detected and neutralizing capacity was high (Fig. 2a, orange dot). To be able to compare neutralization capacity directly between the mild and severe course of COVID-19, we tested an ELISA IgG-level-matched (online suppl. Fig. S1; for all online suppl. material, see www.karger.com/doi/10.1159/000515841) group of volunteers from an earlier study in the neutralization assay [16]. Samples with similar IgG levels as the ICU patients did not even reduce plaques by 90% at the highest serum concentration, already indicating lower neutralization capacities, so we were only able to compare the two cohorts by their NT50 values (Fig. 2b). The neutralization capacity of the ICU patients was significantly higher than those from volunteers of the cluster outbreak cohort at similar SARS-CoV-2-specific IgG levels.
Fig. 1.
Anti-SARS-CoV-2 IgG levels in SARS-CoV-2-positive patients with no symptoms, mild symptoms, or requiring intensive care. IgG levels of only seroconverted patients with positive IgG ELISA (IgG ratio >1.1) were compared according to severity of disease. The category “mild” includes all volunteers reporting at least 1 out of 5 symptoms (fever, coughing, loss of taste, loss of smell or dyspnea).
Fig. 2.
Correlation of neutralizing capacity and anti-SARS-CoV-2 IgG levels in ICU patients' plasma. a SARS-CoV-2 neutralization assays and SARS-CoV-2 IgG ELISAs (Euroimmun) were performed with 32 ICU patients‘ plasma samples. Neutralization assay for the 3 remaining ICU patients were not possible due to the lack of enough sample volume. The neutralization titer 90 (NT90) is the reciprocal value of the plasma dilution at which 90% of plaque formation are suppressed. The IgG ratio is the extinction value measured in ELISA of each sample divided by the extinction of a calibrant provided in the kit. b Plasma samples from a SARS-CoV-2-positive cohort with mild courses of disease (“Cluster outbreak cohort”) were randomly selected in a range of SARS-CoV-2 IgG ratios between 2 and 5. Since plasma samples from the “Cluster outbreak cohort” did not reach NT90 titers, NT50 values were compared between this and the ICU cohort. Note that NT50 = 2,048 was the highest dilution tested, therefore most ICU plasmas reached the maximum NT measurable in that assay. Statistical significance was assessed by an unpaired t test. (***p < 0.001).
Taken together, SARS-CoV-2 RNAemia was only detectable in rare and severe cases of SARS-CoV-2-infected individuals in intensive care but not in individuals with no or mild, flu-like symptoms. In addition, the neutralization capacity of ICU patients' plasma was disproportionately higher than that of mild and asymptomatic COVID-19 cases.
Discussion
In this study, we demonstrated a cohort of 92 acutely SARS-CoV-2-infected patients: 12 patients were asymptomatic, 45 with mild flu-like symptoms, and 35 critically ill patients. Overall, only 6 individuals out of 92 patients had detectable SARS-CoV-2 RNA in their blood. These individuals had a severe course of COVID-19 with ARDS and required mechanical ventilation in the ICU. In individuals who were not hospitalized, asymptomatic or had only mild flu-like symptoms, no RNA was detected in the blood despite a clearly positive result in the RT-PCR from the swabs.
Our data is consistent with the anecdotal reports of individuals, i.e. that SARS-CoV-2 RNA in the blood is only associated with a more severe course of infection, where patients have been hospitalized with COVID-19-induced ARDS requiring mechanical ventilation [15, 16]. Moreover, all 6 patients in our ICU cohort with detectable RNA in the blood differ in their pre-existing conditions, which indicates that most likely the severity of the infection can be associated with SARS-CoV-2 blood RNAemia. Although viral RNA was detected in the blood, it is still unclear whether SARS-CoV-2 can be transmitted due to blood transfusion from critically ill patients. However, only individuals who showed no signs of fever or infection are allowed to donate blood. We found no detectable SARS-CoV-2 RNA in the blood of asymptomatic but acutely SARS-CoV-2-infected individuals, which indicates no potential hazards from blood or plasma transfusion of these individuals. However, further studies should focus on longitudinal analysis with short intervals of blood drawing to ensure no detectable SARS-CoV-2 RNA in the blood throughout the infection.
Whether this observed RNAemia in critical patients occurs in the absence or in spite of a SARS-CoV-2-specific immune response can for example be determined by measuring virus-specific antibody titers and their neutralization capacity. We determined in our study that in comparison of antibody levels between groups of patients with different severity of disease, the overall IgG ratios were similar in all individuals regardless of the severity of their symptoms. However, antibody levels do not indicate potential SARS-CoV-2 RNAemia, which is of relevance for transfusion medicine that there is no easy surrogate for potential viremia. Although our data indicate overall similar IgG levels in severe compared to mild courses of the disease, the plasma of most severely ill patients exhibited a strongly potentiated neutralizing capacity over that observed in mild and asymptomatic cases. Yet, similar to a cluster outbreak cohort with mild or no symptoms reported on earlier, this neutralizing capacity in ICU patients could be positively correlated to IgG levels [16]. This situation is different from infections like influenza, where specific antibody titers were higher in the severely ill, but neutralization capacity did not differ significantly between severe and mild cases [19]. Furthermore, in our ICU cohort SARS-CoV-2 RNAemia in the blood could be consistently correlated neither to antibody levels nor to neutralization. Out of 5 ICU patients with RNAemia in the blood 2 deceased patients had anti-SARS-CoV-2 IgG and exhibited strong neutralization, but 1 who was subsequently released from the hospital showed no antibodies and no neutralizing capacity. This indicates that the detection of viral RNA in the blood, IgG levels and the inpatient outcome are not associated with each other. Recent studies already reported evidence that weak seroconversion cannot predict patient outcomes, including death [20, 21]. Our data provide additional evidence that detectable viral RNA in the blood can be a factor to predict the severity of the infection rather than the patient outcomes. It is important to note that differences between individuals in other aspects of the immune response, such as production of inflammatory mediators or T cell responses, are all likely to contribute as well to patient outcomes. However, this phenomenon needs to be analyzed with larger case numbers to reveal whether the presence of neutralizing antibodies against SARS-CoV-2 can be correlated to disease outcome in critical patients. This complex phenomenon therefore requires further studies involving a greater number of severe cases and additional analysis of immune markers like IL-6 to draw clearer conclusions [22]. Interesting, some of the study participants tested negative for SARS-CoV-2-specific antibodies. This might be the case because samples were collected at different time points relative to infection, since depending on the severity of symptoms the participants got tested during different phases of the infection. In COVID-19, this phenomenon is enhanced by the fact that the disease causes a wide range of symptom severity, depending on age, gender, and several other factors. However, our direct comparison of neutralization capacities between mild and severe cases at comparable IgG levels shows that most critically ill COVID-19 patients have superior neutralizing antibodies, which are much more effective than those in the plasma of patients with mild courses of the disease. In spite of this highly potentiated neutralization, be that by antibody affinity maturation or clonal expansion, the presence of these antibodies did not improve the patients' clinical status, throwing into question the efficacy of convalescent serum therapy.
Statement of Ethics
A subset of SARS-CoV-2-infected individuals were enrolled during a previous study in the hotspot of Gangelt, which is located in the German county of Heinsberg in North Rhine-Westphalia [16]. The study was approved by the Ethics Committee of the Medical Faculty of the University of Bonn (approval number 085/20) and has been registered at the German Clinical Trials Register (https://www.drks.de, identification number DRKS00021306). The study was conducted in accordance with good clinical (GCP) and epidemiological practice (GEP) standards and the Declaration of Helsinki. All necessary written patient/participant consents were obtained before and the appropriate institutional forms have been archived.
Conflict of Interest Statement
The authors have declared no competing interest.
Funding Sources
The government of the German Federal State of North Rhine-Westphalia unconditionally provided 65,000 EUR to support the original study where the SARS-CoV-2-infected individuals were previously enrolled. No other financial support by any third parties was received or used for the study.
Author Contributions
E.R., B.S., C.S., and B.M.K. performed, analyzed, and optimized laboratory work. E.R., D.A.A., B.S., C.B., B.M., S.A., R.M.S., and H.S. were involved in the study design, patient sample and data collection. E.R., B.S. and H.S. wrote the first draft of the manuscript. E.R., D.A.A., B.S., C.B., B.M., S.A., C.S., B.M.K., J.O., M.B.F., C.P., R.M.S., G.H., A.-E.H. and H.S. provided substantial contributions to analysis and interpretation of data as well as edits, discussion, and changes of the manuscript.
Supplementary Material
Supplementary data
References
- 1.Coronavirus disease (COVID-19) Situation Report - 190. World Health Organization 2020. [accessed 2020 Aug 28] Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200728-covid-19-sitrep-190.pdf?sfvrsn=fec17314_2.
- 2.Sood N, Simon P, Ebner P, Eichner D, Reynolds J, Bendavid E, et al. Seroprevalence of SARS-CoV-2-Specific Antibodies Among Adults in Los Angeles County, California, on April 10-11, 2020. JAMA. 2020;323:2425–7. doi: 10.1001/jama.2020.8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krantz SG, Rao A. Level of underreporting including underdiagnosis before the first peak of COVID-19 in various countries: Preliminary retrospective results based on wavelets and deterministic modeling. Infect Control Hosp Epidemiol. 2020;41:857–9. doi: 10.1017/ice.2020.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell TW, Golding N, Hellewell J, Abbott S, Wright L, Pearson CAB, et al. Reconstructing the early global dynamics of under-ascertained COVID-19 cases and infections. BMC Med. 2020;18:332. doi: 10.1186/s12916-020-01790-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sánchez-Romero M, di Lego V, Prskawetz A, Queiroz BL. An indirect method to monitor the fraction of people ever infected with COVID-19: An application to the United States. PLoS One. 2021;16((1)):e0245845. doi: 10.1371/journal.pone.0245845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.L. Queiroz M, Perera H, Gunawardana B, Manatunge J. Transmission of COVID-19 virus by droplets and aerosols: A critical review on the unresolved dichotomy. Environ Res. 2020;188:109819. doi: 10.1016/j.envres.2020.109819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyerowitz EA, Richterman A, Gandhi RT, Sax PE. Transmission of SARS-CoV-2: A Review of Viral, Host, and Environmental Factors. Ann Intern Med. 2021;174((1)):69–79. doi: 10.7326/M20-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldmann DA. Blood-borne pathogens and nosocomial infections. J Allergy Clin Immunol. 2002;110:S21–6. doi: 10.1067/mai.2002.125337. [DOI] [PubMed] [Google Scholar]
- 9.Deuffic-Burban S, Delarocque-Astagneau E, Abiteboul D, Bouvet E, Yazdanpanah Y. Blood-borne viruses in health care workers: prevention and management. J Clin Virol. 2011;52:4–10. doi: 10.1016/j.jcv.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 10.Vanhamel J, Bruggemans A, Debyser Z. Establishment of latent HIV-1 reservoirs: what do we really know? J Virus Erad. 2019;5:3–9. doi: 10.1016/S2055-6640(20)30275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arts EJ, Hazuda DJ. HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med. 2012;2:a007161. doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang C, Wang Y, Li X, Ren L, Zjao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JY, Ko JH, Kim Y, Kim YJ, Kim JM, Chung YS, et al. Viral Load Kinetics of SARS-CoV-2 Infection in First Two Patients in Korea. J Korean Med Sci. 2020;35:e86. doi: 10.3346/jkms.2020.35.e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang L, Zhao L, Gong H, Wang L, Wang L. Severe Acute Respiratory Syndrome Coronavirus 2 RNA Detected in Blood Donations. Emerg Infect Dis. 2020;26:1631–3. doi: 10.3201/eid2607.200839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corman VM, Rabenau HF, Adams O, Oberle D, Funk MB, Keller-Stanislawski B, et al. SARS-CoV-2 asymptomatic and symptomatic patients and risk for transfusion transmission. Transfusion. 2020;60:1119–22. doi: 10.1111/trf.15841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Streeck H, Schulte B, Kümmerer BM, Richter E, Höller T, Fuhrmann C, et al. Infection fatality rate of SARS-CoV2 in a super-spreading event in Germany. Nat Commun. 2020;11:5829. doi: 10.1038/s41467-020-19509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmithausen RM, Döhla M, Schöβler H, Diegmann C, Schulte B, Richter E, et al. Characteristic Temporary Loss of Taste and Olfactory Senses in SARS-CoV-2-positive-Individuals with Mild Symptoms. Pathog Immun. 2020;5:117–20. doi: 10.20411/pai.v5i1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.To KK, Zhang AJ, Hung IF, Ting X, Ip WCT, Wong RTY, et al. High titer and avidity of nonneutralizing antibodies against influenza vaccine antigen are associated with severe influenza. Clin Vaccine Immunol. 2012;19:1012–8. doi: 10.1128/CVI.00081-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Röltgen K, Powell AE, Wirz OF, Stevens BA, Hogan CA, Najeeb J, et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol. 2020;5:5. doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun B, Feng Y, Mo X, Zheng P, Wang Q, Li P, et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg Microbes Infect. 2020;9:940–8. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi N, Abe R, Hattori N, Matsumura Y, Oshima T, Taniguchi T, et al. Clinical course of a critically ill patient with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J Artif Organs. 2020;23:397–400. doi: 10.1007/s10047-020-01183-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data