Abstract
Scope:
Iron deficiency (ID) compromises the health of infants worldwide. Although readily treated with iron, concerns remain about the persistence of some effects. Metabolic and gut microbial consequences of infantile ID were investigated in juvenile monkeys after natural recovery (pID) from iron deficiency or post-treatment with iron dextran and B vitamins (pID+Fe).
Methods and Results:
Metabolomic profiling of urine and plasma was conducted with 1H nuclear magnetic resonance (NMR) spectroscopy. Gut microbiota were characterized from rectal swabs by amplicon sequencing of the 16S rRNA gene. Urinary metabolic profiles of pID monkeys significantly differed from pID+Fe and continuously iron-sufficient controls (IS) with higher maltose and lower amounts of microbial-derived metabolites. Persistent differences in energy metabolism were apparent from the plasma metabolic phenotypes with greater reliance on anaerobic glycolysis in pID monkeys. Microbial profiling indicated higher abundances of Methanobrevibacter, Lachnobacterium and Ruminococcus in pID monkeys and any history of ID resulted in a lower Prevotella abundance compared to the IS controls.
Conclusions:
Lingering metabolic and microbial effects were found after natural recovery from ID. These long-term biochemical derangements were not present in the pID+Fe animals emphasizing the importance of the early detection and treatment of early-life ID to ameliorate its chronic metabolic effects.
Keywords: iron deficiency, anaemia, metabolome, microbiota, microbiome, metabolism, iron supplementation, monkey
Graphical Abstract
LAYMAN’S SUMMARY:
Iron deficiency anemia is a common nutritional and pediatric concern worldwide. In addition to the known effects on red blood cells, iron has diverse effects on oxidative metabolism and synthesis pathways. In this study, lingering metabolic and gut microbial disruptions were observed in juvenile monkeys that had naturally recovery from infantile iron deficiency. These persistent metabolic effects were prevented with the rapid treatment of early-life iron deficiency with iron dextran and B vitamins. These findings highlight the importance of identifying and treating infantile iron deficiency early in life.
1. Introduction
Iron deficiency (ID) is a common nutrient deficiency with the potential for adverse health effects in young children.[1] In addition to iron’s essential role in oxygen transport, heme synthesis, and erythropoiesis, iron is a required cofactor for many enzymes involved in energy and oxidative metabolism.[2] The clinical presentation of ID in infants is anaemia, typically diagnosed by low hemoglobin (Hgb) and by the number and size of red blood cells, although there are many other downstream effects on iron-dependent pathways, including on gluconeogenesis and lipid metabolism. High iron needs have been described in the rapidly growing offspring of many species, including nonhuman primates.[3] This research takes advantage of a well-characterized model of ID in young rhesus monkeys.[4,5]
ID in infant monkeys is similar to the human condition as the primary cause is a depletion of storage iron reserves as growth-related demands for iron exceed the fetal iron endowment and bioavailable iron from breast milk. In this controlled model of nutrition-induced anaemia, between 10–30% of infant monkeys become moderately ID or anemic at 4–6 months of age, which then resolves gradually after nursing with the consumption of dietary iron in solid foods. Alternatively, it can be treated with oral iron ferrous salts or more rapidly remedied by injecting iron dextran. Previous metabolomic studies of serum and cerebrospinal fluid from ID monkeys identified alterations in liver metabolism along with increased bile acid production, and impaired energy metabolism in the CNS.[6,7] These observations indicate a prioritization of iron for red blood cells and the heart over other tissue needs, including the brain.
The capacity to replenish iron reserves naturally from food is influenced by the gut’s ability to absorb dietary iron. In addition to known regulators of iron absorption, especially the hepcidin-ferroportin axis, and intrinsic biochemical processes, such as gut acidity, microorganisms in the gut can also affect iron uptake. This can occur directly through microbial competition for dietary iron and indirectly through the modulation of environmental conditions in the gut and alterations in iron-uptake proteins and transporters in the intestinal epithelium.[8–10] Iron availability can also modify the community structure of the gut microbiota. Previous studies identified differences in the gut microbiome of anaemic infants and demonstrated effects of iron administration on both beneficial taxa, such as Lactobacilli and Bifidobacteria, and pathogenic bacteria, including Escherichia coli, Salmonella typhimurium, and Mycobacterium tuberculosis, and the malarial parasite, Plasmodium.[11,12] However, little is known regarding the enduring effects of anaemia on the intestinal microbiota, especially post-recovery of hematological indices.
To better understand the long-term consequences of early life ID on the global metabolic system, including interactions between the gut microbiota and host, an untargeted metabolic phenotyping approach was used to characterize urinary and plasma metabolic profiles of monkeys who had naturally recovered from ID (pID) or had remained iron sufficient (IS) during the first year of life. Fecal specimens were also acquired and 16S rRNA gene sequencing was used to determine if ID exerted a sustained effect on the community structure of the gut microbiota. In addition, we examined whether administration of iron dextran to anaemic infants (pID+Fe) would lessen long-term effects.
2. Experimental section
2.1. Animals.
A naturalistic animal model of infantile ID was used in this research. Briefly, female monkeys were provided sufficient amounts of iron to meet the recommended nutrient requirements for nonhuman primates (>180 mg/kg[13]). However, it was not sufficient to fulfil the higher needs of gravid females[4]. This prenatal diet results in 20–30% of infant monkeys developing a growth-related ID during the nursing phase, before they recover gradually after weaning by consuming iron in solid foods.[5] Samples were analyzed from 38 young rhesus monkeys (Macaca mulatta), between 7–19 months of age (see Supplementary Materials for more information). They were singleton infants from 38 different mothers, born after unassisted natural deliveries and term pregnancies. Twenty-one served as controls and had remained continuously IS. Based on hematological assessments, 17 were transiently ID between 5–6 months of age. Eleven of these monkeys recovered gradually, a process that takes several months while 6 were administered iron dextran (10 mg) and a prophylactic dose of B vitamin complex (162 mg) IM at weekly intervals for 1–2 months. All other husbandry procedures were standardized, including provision of the same diet to the dams when pregnant and nursing, and then to juveniles at the time of specimen collection (see Supplemental Table 1 for nutrient composition of diet). Husbandry and experimental protocols approved by the institutional Animal Care and Use Committee (L005007, L005009, L005477), University of Wisconsin.
2.2. Specimen collection.
Single blood and urine samples and rectal swabs were collected after hematological screening verified Hgb and mean corpuscular volume (MCV) values of the iron-treated monkeys were within the normal range (between 7–19 months of age; Supplemental Table 2). Hematological criteria for designating an earlier history of ID were: Hgb < 110 g/L and MCV < 60 fL. The nutrient and growth-related basis of the ID was confirmed further by elevated levels of the iron-sensitive measure of zinc protophophyrin (ZnPP/H > 150 μM/M), and a normal leukocyte count indicating iron levels were not acutely suppressed due to infection or inflammation. Prior validations of this primate model of ID demonstrated that serum iron was decreased to 15 μM/L as compared to the normal 20 μM/L; transferrin saturation (TSAT) was 18% significantly below the normal 32%; and ferritin was reduced to 10 pmol/L as compared to 25 pmol/L in an IS infant.[4] Serum and urine creatine were also quantified at the time of urine collection to assess for possible confounding differences in kidney function.
2.3. 16S ribosomal RNA gene sequencing.
Total DNA was isolated from rectal swabs with a Mini-Beadbeater-96 (BiospecProducts, Bartlesville, OK) and the PowerSoil DNA Isolation Kit (MoBio, Carlsbad, CA), according to the manufacturer’s protocol. Purified genomic DNA extracts were quantified using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA), and stored at −20 °C in 10 mM Tris buffer until sequenced. PCR amplification of the V4 variable region of the 16S rRNA gene using V4 region specific primers (515F-816R) and amplicon sequencing were performed by Institute for Genomics & Systems Biology at the Argonne National Laboratory (Argonne, IL) on the Illumina MiSeq platform per manufacturer’s guidelines. Microbial profiles were generated from 25 monkeys, representing each condition (IS, 14; pID, 7; pID+Fe, 4).
2.4. Metataxonomic data processing and statistical analyses.
Sequences were analyzed using QIIME 1.9.1 (Quantitative Insights into Microbial Ecology). Sequences were de-multiplexed and quality filtered for high quality sequences (Q > 25). Operational taxonomic units (OTUs) were assigned to the Greengenes (v. 2013_May) database using PyNAST at 97% global identity threshold uclust and the closed-reference OTU picking for non-ambiguous species-level taxonomy matching in QIIME. OTU tables were normalized by dividing each OTU by the known, or predicted, 16S copy number through implementation in PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States). As testing for differential abundance of specific taxa was a primary objective, in addition to testing for differences in microbial community structure, current recommendations against the rarefaction of 16S rRNA data were followed. Instead, data were normalized through the parametric method, metagenomeSeq (implemented in QIIME), to account for unequal sampling. Cases that had less than 1000 sequences per sample were discarded prior to the diversity analyses or taxonomic assignments.
Faith’s Whole Tree index of phylogenetic diversity was generated to examine sample richness, or alpha diversity. Weighted UNIFRAC dissimilarity matrices were used to examine differences in microbial community structure (beta diversity) while accounting for both microbial abundances and phylogenetic relatedness. Both measures were generated using QIIME and principal coordinates analysis (PCoA) obtained through EMPeror. Significance testing was carried out by various implementation in QIIME, and Paleontological Statistics (PAST), Analysis of Similarity (ANOSIM), nonparametric t-tests (Kruskal-Wallis), and graphing. LEfSe (Linear discriminant analysis Effect Size) was used for phylogenetic enrichment analyses and significant testing was carried out at the genus level by Wilcoxon implementation (see Supplementary Information).
2.5. 1H nuclear magnetic resonance spectroscopy-based metabolic profiling.
Urine and plasma samples were analyzed by 1H nuclear magnetic resonance (NMR) spectroscopy. Spectroscopic analysis of all samples was performed on a 700 MHz Bruker NMR spectrometer equipped with a cryoprobe. See Supplemental Materials for detailed methods.
2.6. Pattern recognition and statistical data analysis.
Multivariate modeling was performed in MATLAB with the use of in-house scripts. Initially, principal component analysis (PCA) of the NMR spectra was performed using pareto scaling to visualize patterns and outliers within the data set. This was followed by orthogonal projection to latent structures discriminant analysis (OPLS-DA). Pair-wise comparisons between the study groups were performed to identify discriminatory metabolites between groups. Correlation coefficients plots were generated with the use of back scaling transformation to display the contribution of each metabolite to the sample classification. Color represents the significance of a correlation for each metabolite to the class membership. The predictive performance (Q2Y) of the models was calculated using a 7-fold cross-validation approach and the significance of the OPLS-DA Q2Y values was assessed through permutation testing (1000 permutations).
2.7. Metabolome and bacteriome correlational analysis.
To examine the relationships between the metabolic effects and gut microbial differences, Spearman correlation analyses were run between relative metabolite and OTU abundances (for taxa present over 1%). Hierarchical clustering analysis was used to identify general patterns between the biochemical and microbial variations within the data.
2.8. Metabolic reaction network.
MetaboNetworks creates a custom database of chemical reactions that can occur in a supraorganism (host + microbiome).[14] Data were used from the Kyoto Encyclopedia of Genes and Genomes (KEGG) [15] to include reactions mediated by an enzyme encoded by a Macaca mulatta gene or one or more of 3888 bacterial species as well as spontaneous/non-enzymatic reactions. Species-level data were included if the associated phylum had an abundance ≥1% in the 16S rRNA gene sequencing data, comprising the phyla Bacteroidetes (n = 299), Firmicutes (n = 999), Proteobacteria (n = 2509) and Spirochaetes (n = 81). Urinary and plasma metabolites associated with differences between pID and IS or pID and pID+Fe monkeys were mapped in a metabolic reaction network, connecting each pair of identified metabolites via the shortest number of chemical reactions. The involvement of iron in each of the 678 reactions in this network was investigated. All enzymes entries for each metabolite pair were queried for containing iron in different forms: as part of protein (iron-sulfur protein, ferredoxin), as cation (Fe2+, Fe3+, Fe(II), Fe(III), FeII, FeIII) or listed otherwise (e.g. Fe, iron, ferro−, ferri−). Nodes with the text and outer-box coloured in orange are involved in a reaction that requires iron. Edges are colored on the basis of the type of iron involved in the reaction, green for ferredoxin, red for Fe2+, yellow for Fe3+ and blue for reactions involving another form of iron. Supplementary Material details information of all reactions in the network that require iron, including details for compounds involved with KEGG identifiers, and the reactions and enzymes with which they are involved (Supplemental Table 3).
3. Results
Thirty-eight monkeys were IS (n = 21) or ID (n = 17) at the end of the nursing period on the basis of their hematology (i.e., low Hgb <110 g/L) and two other abnormal iron-related measures (see Supplemental Table 2). Eleven of the ID monkeys recovered gradually by consuming bioavailable iron in the diet, while 6 were administered iron dextran and a B vitamin complex weekly for 1–2 months. Fecal, urine and blood specimens were collected later at an older age after weaning and relocation into small social groups of juveniles. We did not detect a systematic difference in urine excretion across the three conditions, nor in the level of serum and urinary creatine that would indicate a long-term effect on renal clearance (Supplementary Table 3).
3.1. Compositional differences in the gut microbiota following ID.
Fecal microbiota were analyzed using MiSeq 16S rRNA gene sequencing. Twenty-five samples passed quality filtering with an average of 2,605 sequences per samples and minimum of 1633 sequences per sample. Significance testing of beta-diversity UniFrac PCoA axis 1 and 2 were performed by planned orthogonal contrasts regression on group membership. Weighted UniFrac measures indicated beta diversity was significantly different in untreated monkeys (pID) compared to IS and pID+Fe monkeys (PCoA 1: F(1,22) = 7.794, p = 0.011; PCoA 2: F(1,22) = 0.111, p = 0.743). No differences were observed between IS monkeys and those that received iron dextran when anaemic (pID+Fe) (PCoA 1: F(1,22) = 0.250, p = 0.622; PCoA 2: F(1,22) = 0.673, p = 0.421). In contrast, unweighted UniFrac values did not significantly differ across the three conditions. The divergent results indicate the effect of untreated ID on the microbial community structure was more related to abundance, as opposed to the presence or absence of specific taxa (Figure 1A and 1B). Significant differences were not observed in phylogenetic richness (alpha diversity) or Faith’s Phylogenetic Diversity: IS vs. untreated pID monkeys (F(1,22) = 3.512, p = 0.074); pID+Fe vs. IS monkeys (F(1,22) = 0.751, p = 0.396) (Figure 1C). However, LEfSe analyses indicated some specific taxa were selectively enriched in each condition. As shown in Table 1, the abundance of Prevotella was higher in IS monkeys compared to monkeys from both ID conditions (LDA = 4.048, p = 0.023). Methanobrevibacter (Archaea), Ruminococcus and Lachnobacterium were more abundant in the untreated pID monkeys as compared to the other two conditions (LDA = 2.965, p = 0.047, LDA = 3.635, p = 0.027 and LDA = 2.947, p = 0.037, respectively). Finally, the pID+Fe group exhibited an enrichment of Rhodoplanes and the CF231 genera and a species of Clostridium that could not be definitively identified but is likely to be Clostridium perfringens (LDA = 3.035, p = 0.004, LDA = 3.009, p = 0.033 and LDA = 2.640, p < 0.001, respectively). Regression of the weighted UniFrac PCoA 1 axis on these taxa indicated that 26.47% and 2.75% of the variance was explained by Prevotella (F(1, 22) = 102.08, p < 0.001, dR-sqr = 0.2647) and Ruminococcus (F(1,22) = 10.62, p = 0.004, dR-sqr = 0.0275, respectively). These two taxa were the primary reason for significant differences in the community structure between the three conditions.
Figure 1:
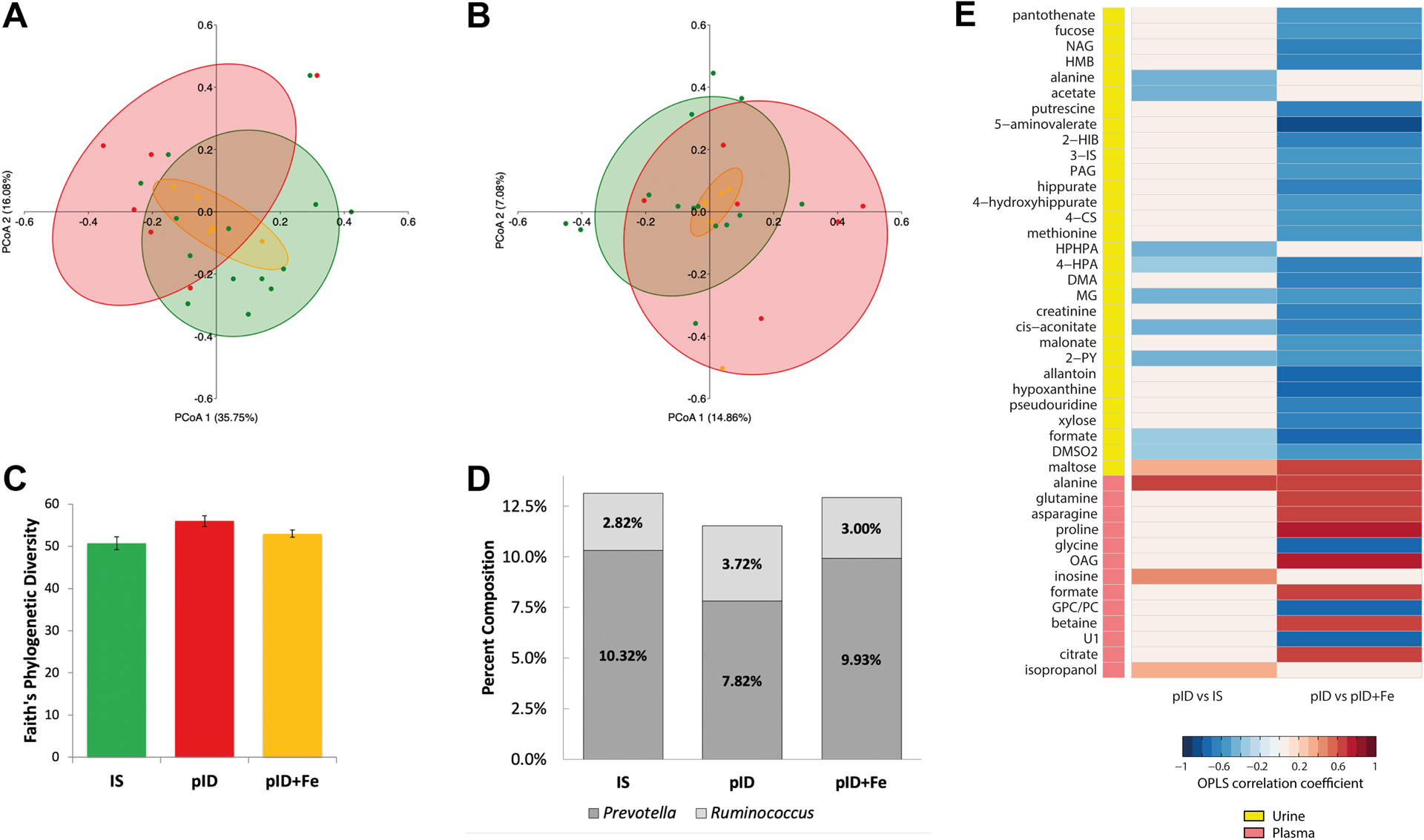
Variation in the community structure of the fecal microbiota measured by 16S rRNA gene sequencing and the urinary and plasma metabolic disruptions associated with gradual recovery from iron deficiency (pID) when compared to continuously iron sufficient (IS) and iron-supplemented monkeys (pID+Fe). (A) Weighted UniFrac analyses revealed significant albeit modest differences in the community structure between young monkeys that had recovered gradually from iron deficiency (pID, red) and those that remained iron sufficient (green, IS) or ones that received iron supplements when anemic (pID+Fe, yellow). Significance testing was performed by regressing PCoA axes 1 and 2 on group membership using planned orthogonal contrasts. (B) Differences were not evident with Unweighted UniFrac analyses indicating variation in community structure was attributable to the abundances, not the presence or absence of taxa. Percentages indicate variance in beta diversity variance accounted for by each PCoA axis. (C) Significant differences in the richness of the community structure were not apparent. (D) Prevotella and Ruminococcus were the two most abundant genera found to differ across the three conditions. Prevotella was highest in IS monkeys, and lowest in the pID animals; conversely, Ruminococus was lowest in IS and highest in the pID group. (E) Colors indicate correlation coefficients extracted from the OPLS-DA models comparing pID and IS urine (Q2Y = 0.10; p = 0.045), and plasma profiles (Q2Y = 0.29; p = 0.029) and the pID and pID+Fe urine (Q2Y = 0.21; p = 0.039) and plasma profiles (Q2Y = 0.45; p = 0.001). Red indicates metabolites that more abundant in the pID samples and blue indicates metabolites less abundant in the pID samples compared to the 2 other groups. The key indicates whether the metabolic change occurred in the urine (yellow) or plasma (pink).
Table 1.
Abundance of enriched taxa in previously iron deficient (pID) and iron-treated (pID+Fe) juvenile rhesus monkeys compared to ones that remained iron sufficient (IS)1.
| IS | pID | pID+Fe | F-Statistic | P-value | |
|---|---|---|---|---|---|
| Methanobrevibacter (Archaea) | 0.04% | 0.09% | 0.02% | 2.205 | 0.134 |
| CF231 | 0.31% | 0.27% | 0.32% | 2.818 | 0.081 |
| Prevotella | 10.32% | 7.82% | 9.93% | 5.832 | 0.009 |
| Lachnobacterium | 0.01% | 0.05% | 0.02% | 2.600 | 0.097 |
| Ruminococcus | 2.82% | 3.72% | 3.00% | 5.889 | 0.009 |
| Rhodoplanes | 0.00% | 0.00% | 0.01% | 9.198 | 0.001 |
Abundance and differences in taxa found to be enriched in LEfSe analyses. Bold font indicates the condition with significant phylogenetic enrichment.
3.2. Urinary metabolic disruptions following recovery from infantile ID.
Pairwise OPLS-DA models were constructed to compare the metabolic profiles of urine collected from monkeys in the three conditions. A significant model was obtained comparing the urinary metabolic profiles of IS to pID monkeys (Q2Y = 0.10, p = 0.045; Supplementary Figure 1). Untreated pID monkeys excreted more maltose compared to those continuously IS, and lower amounts of microbial-derived metabolites, including acetate, butyrate, propionate, formate, 3-[3-hydroxyphenyl]-3-hydroxypropanoic acid (HPHPA), and 4-hydroxyphenylacetate (4-HPA). The pID monkeys also excreted less dimethylsulfone (DMSO2), alanine, cis-aconitate, methylguanidine (MG), and N-methyl-2-pyridone-5-carboxamide (2-PY) compared to monkeys that had been continuously IS. Figure 1E shows the correlations between the OPLS predictive component from this pairwise model (pID vs. IS) and these metabolites.
A significant OPLS-DA model was also obtained comparing the urinary metabolic profiles of pID+Fe and pID monkeys (Q2Y = 0.21, p = 0.039; Supplementary Figure 1). Untreated pID monkeys excreted more maltose compared to pID+Fe animals, as well as relatively higher amounts of metabolites associated with inflammatory activity and oxidative stress (i.e., allantoin, pseudouridine, N-acetyl glycoproteins [NAG], fucose), pantothenate (vitamin B5), 5-aminovalerate, methionine, β-hydroxy-β-methylbutyrate (HMB), malonate, cis-aconitate, hypoxanthine, and xylose. In addition, pID monkeys excreted lower amounts of a number of microbial-derived metabolites, dimethylamine (DMA), 3-indoxyl sulfate (3-IS), hippurate, 4-hydroxyhippurate, N-phenylacetylglutamine PAGn, 2-hydroxyisobutyrate (2-HIB), 4-HPA, DMSO2, butyrate, and formate. No differences were observed in urinary metabolites between the pID+Fe treated and IS monkeys suggesting that long-term metabolic effects of early life ID were lessened by iron treatment.
3.3. Plasma metabolic changes subsequent to ID.
Consistent with the urinary biochemical profiles, significant pair-wise OPLS-DA models were obtained comparing plasma from IS and pID monkeys (Q2Y = 0.29, p = 0.029; Supplementary Figure 2) and untreated and treated monkeys (pID vs pID+Fe; Q2Y= 0.45, p = 0.001). Metabolic differences between the three conditions are summarized in Figure 1. The primary metabolic difference was higher circulating amounts of alanine and inosine in the pID monkeys compared to IS monkeys. Compared to the pID+Fe animals, pID monkeys had higher circulating amino acids (alanine, proline, glutamine, and asparagine), formate, betaine, O-acetyl glycoproteins (OAG), and citrate, and lower circulating amounts of glycine and glycerophosphocholine (GPC). No significant differences were found in the plasma metabolites between IS and pID+Fe animals.
3.4. Multi-compartmental metabolic reaction network.
MetaboNetworks was used to construct a multi-compartment metabolic reaction network from the urinary and plasma metabolites identified to differ between pID versus IS or pID versus pID+Fe monkeys (Figure 2; Supplementary Figure 4). This network included reactions that can occur in the rhesus monkey host (solid lines) and/or that can occur in bacteria present in the gut (dashed lines). From this analysis, it is clear that many pathways requiring iron, including those in the host or performed by commensal bacteria, remain perturbed several months after the gradual resolution of ID. Iron is directly involved in the synthesis of several host-derived metabolites that remained altered in monkeys that had previously been ID. These metabolites included citrate, cis-aconitate, alanine, glycine, glutamine, hypoxanthine, and 2PY. Fucose also depends upon iron for its production by the host but is subsequently metabolized by bacteria. Fucose remained lower in the urine of pID animals after the natural resolution of ID compared to the pID+Fe animals. A number of metabolites indirectly dependent upon iron (i.e., their precursors or products depend upon iron) were also identified as different between the three conditions, including betaine, putrescine, creatine, malonate, inosine, and maltose.
Figure 2.
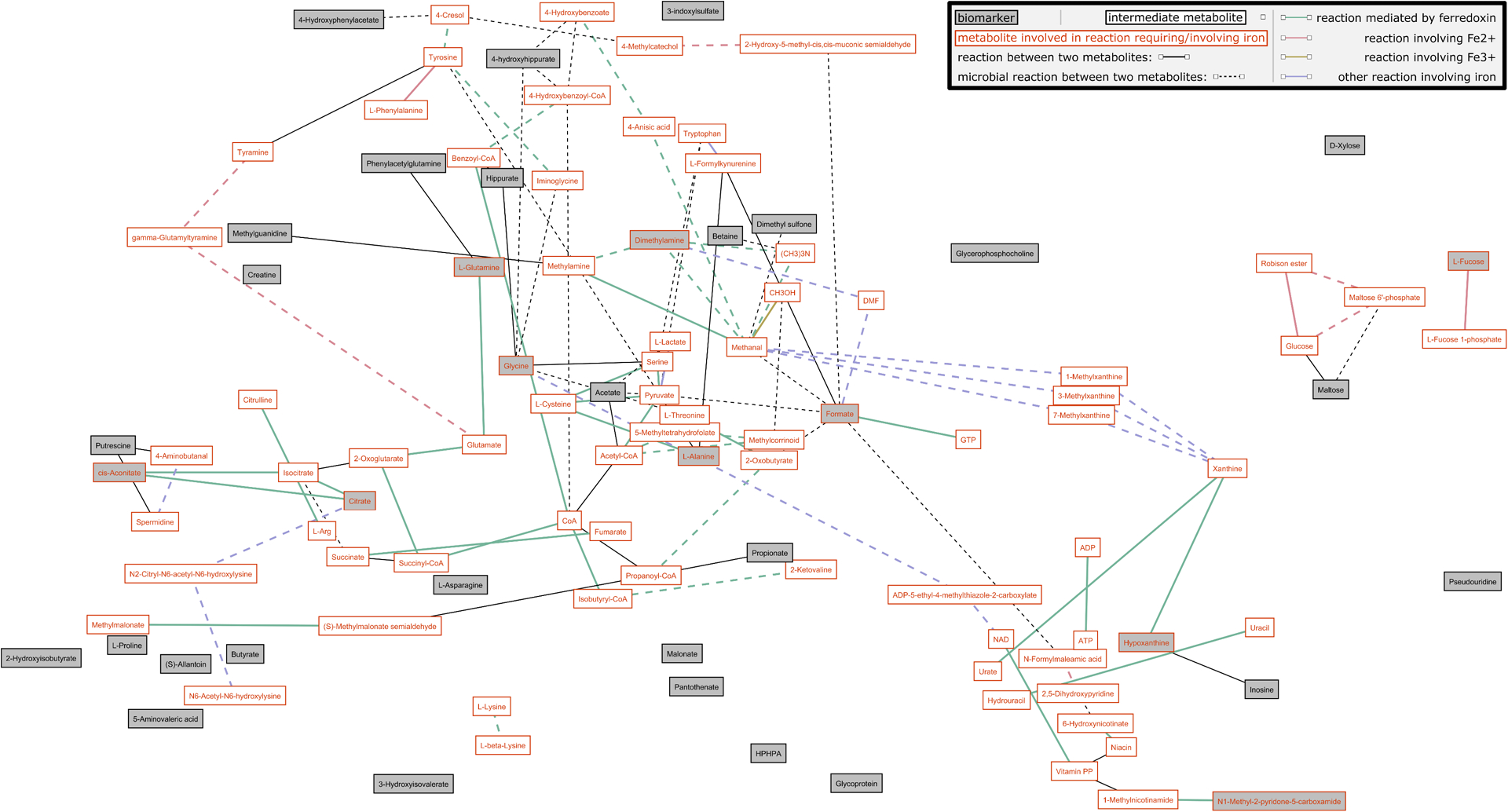
Visualization of the multi-compartmental metabolic reaction network of metabolites identified to differ between monkeys that recovered gradually from ID (pID) when compared to always iron sufficient (IS) or ones supplemented with iron when determined to be ID (pID+Fe). The network includes reactions that can be performed by the monkey host (solid lines) or by bacteria present in the gut (dashed lines). The color of the line indicates reactions that require or involve iron. Grey boxes indicate urinary or plasma metabolites that differed between pID and IS or pID+Fe monkeys. White boxes indicate intermediate metabolites; boxes with an orange border indicate metabolites involved in reactions requiring or involving iron.
Iron is also involved in the bacterial metabolism of formate and DMA, two gut microbial products found to differ between the IS and pID monkeys. Interestingly, correlational analyses identified both DMA and formate to be positively correlated with the genera Prevotella, Flexispira, YRC22 and Anaerovibrio (Supplementary Figure 3). Iron was also found to be indirectly related to the production of several other microbially-derived metabolites that remained different in the pID group. These metabolites included acetate, 4-hydroxyhippurate, 4-HPA, and PAG. Supplementary Table 4 provides information on the enzymes and genes that mediate the reactions involving iron, which are portrayed in the network figure (Figure 2; Supplementary Figure 4 displays intermediate reactions not involving iron).
4. Discussion
These findings indicate that ID in early life can result in a remodeling of maturing metabolic systems, including metabolic processes encoded in the host and microbiome. Consistent with evidence for functional effects, compositional alterations were found in the bacterial community structure. Importantly, these alterations persisted after ID had resolved and the animals were now iron sufficient. A prior history of ID affected a wide range of biochemical pathways directly and indirectly dependent upon iron with important functions related to energy, amino acid, and polyamine metabolism. These results confirm previous reports of effects on energetic pathways in both blood and the CNS, which remained evident months after ID resolved.[6,7] This primate model of microcytic and hypochromic anaemia has been shown to be nutritionally dependent.[4] It typically emerges between 4–6 months of age in nursing infants after a growth-related depletion of prenatally acquired iron stores.[5] Although the ID is transient, more rapid repletion by administering iron dextran lessened the lingering metabolic alterations evident in both blood and urine, even after treatment had ceased and all animals were iron sufficient.
One significant metabolic difference between IS and ID infants involved a dysregulation of the TCA cycle. The TCA cycle includes two enzymes that contain Fe-S clusters, aconitase and succinate dehydrogenase. Aconitase converts citrate to isocitrate via cis-aconitate, while succinate dehydrogenase converts succinate to fumarate. Previous ex vivo analyses of human cells have found reduced aconitase and succinate dehydrogenase activity following iron deprivation.[16] Consistent with reduced aconitase activity, naturally recovered ID monkeys excreted less cis-aconitate and had higher citrate in circulation compared to IS infants. As highlighted in the network analysis, ferredoxin is necessary for NAD+/NADH metabolism, which is essential for driving the TCA cycle. NAD+ has an important role in many metabolic processes and is metabolized via the nicotinamide pathway to 2-PY, which was also excreted in lower amounts by the pID monkeys. Furthermore, alanine, a breakdown product of pyruvate, was higher in the plasma of pID compared to IS monkeys. This may result from pyruvate being unable to enter the impaired TCA cycle, increasing its use in anaerobic glycolysis. Ceyhan et al. have previously reported higher circulating lactate, pyruvate and alanine, end products of anaerobic glycolysis, in anaemic children compared to healthy controls.[17] These effects of ID were not evident when corrected by early administration of iron dextran.
Certain amino acids can increase iron absorption by chelating iron in the gut lumen, providing a vehicle for absorption. Supplementation with asparagine and glutamine has been associated with an increase in blood iron.[18] The pID monkeys had higher amounts of glutamine, asparagine, and proline in their plasma compared to IS monkeys and those treated with iron. This difference could reflect a compensatory response to the earlier history of ID. Similarly, inosine and its metabolites hypoxanthine and uric acid can increase intestinal iron absorption in the rat and uric acid was found to be lower in anaemic humans.[19,20] Inosine was higher in the plasma of pID monkeys, while two metabolites of inosine (allantoin and hypoxanthine) were excreted in lower amounts. Increased inosine following ID may reflect attempts to compensate for impaired ATP generation following disruptions to the TCA cycle and NAD+ metabolism.
Anaemia has been associated with alterations in the composition and functionality of the gut microbiota.[21] In the current study, modest differences were observed to persist in the gut microbial composition of monkeys with a prior history of ID. Most differences were driven by the relative abundance of two genera, Prevotella and Ruminococcus. Prevotella were lower in the monkeys that naturally recovered from ID. Prevotella are common commensals associated with plant-rich diets, can be associated with better glucoregulation, and are typically reported to be the most predominant taxa in monkeys.[22] Therefore, the lower abundance of Prevotella in the pID monkeys is an important observation for further investigation.
Some selective effects on the gut microbiota of the iron-treated monkeys may also warrant further investigation. Although it cannot be definitively identified with 16S sequencing, it appeared that the abundance of C. perfringens may have been increased in the ID+Fe group many months after the treatment had ended. This particular species has been reported to be an iron-scavenger that can affected by the presence of iron.[23,24] The mechanism through which iron dextran administered intramuscularly could modulate the growth of specific microorganisms in the gut remains to be defined. As systemic iron is not likely to be transferred in large quantities into the gut for excretion, the observed effects are likely to occur through indirect processes such as alterations in the vasculature of the hindgut or modifications to the immune system. Interestingly, C. perfringens and other members of the Clostridium genus have also been found previously to increase after oral iron supplementation in human infants.[11] The benefits of iron supplements therefore have to be weighed against the known effects of elevated iron availability on enteric pathogens.[25,26]
The metabolic profiles also indicated that early life ID resulted in long-term modifications to the functional capacity of commensal microbiota. Gut microbial amino acid degradation appeared to be lower in the pID monkeys, reflected by reduced excretion of several metabolites arising from this activity. These metabolites included 4-hydroxyphenylacetate (4-HPA), 3-(3-hydroxyphenyl)-3-hydroxypropionic acid (HPHPA), 4-cresyl sulfate (4-CS), and PAG. Tyrosine can be converted to 4-HPA, which is then decarboxylated to 4-cresol and sulfated in the liver to form 4-CS. In addition, phenylalanine can be converted to 3-hydroxyphenyproponic acid and undergo beta oxidation to produce HPHPA. Both 4-cresol (the bacterial precursor precursor to 4-cresyl sulfate) and HPHPA can inhibit dopamine β-hydroxylase, the enzyme responsible for converting dopamine to norepinephrine.[27,28] This observation would be consistent with higher norepinephrine and lower dopamine concentrations found previously in the CSF of monkeys that had been ID.[29] It is plausible that the paucity of iron during the maturation and establishment of the infant’s bacteriome may have restricted the expansion of bacteria reliant on protein fermentation for growth. For example, Figure 2 highlights that the bacterial metabolism of tyrosine is dependent on ferredoxin. Some bacterial taxa may remain under-represented even when iron availability increases at an old age.
Indoxyl-sulfate (3-IS), is another microbial-host co-metabolite derived from bacterial protein fermentation that was reduced in monkeys with a history of ID. It is produced by the microbial conversion of tryptophan to indole, which is subsequently absorbed and further metabolized by the host to indoxyl, before being sulfated and excreted in urine as 3-IS. Unless treated with iron, the pID monkeys excreted less 3-IS in their urine, suggesting reduced production of indole by the gut microbiota in a monkey with a history of ID. 3-IS can increase the major iron-regulator, hepcidin in HepG2 cells and increase hepatic hepcidin mRNA expression and plasma hepcidin in mice [30]. Increased hepcidin will reduce iron absorption by the gut and result in the sequestering of iron into tissue. Thus, lower 3-IS following ID could reduce hepcidin and facilitate iron absorption from the gut. In this way, the intestinal microbiota could serve as a nutritional sensor in the gut relaying signals to the host via indole production.
Notwithstanding the novelty of these findings, several limitations should be acknowledged. First, the design was cross-sectional, to compare the three conditions in older juveniles, rather than a prospective, longitudinal design. The ID was initially more severe in the monkeys that received iron treatments, but that initial difference would potentially have worked against our finding that the later metabolism of those previously anaemic monkeys was more comparable to the IS controls. One procedural step should also be mentioned, which is that the overnight urine was acquired with collection pans that would permit some fecal contamination of the urine specimen. Multiple filters were used, and the urine centrifuged, but it is likely that some metabolites, such as butyrate and propionate, may have been partially derived from dissolved fecal material. However, the same collection protocol was used for all animals, so it would not have caused a directional bias in only one condition.
Conclusions and translational implications
Current pediatric practice typically recommends testing for ID when children are one year of age even though consensus reports have proposed earlier screening at 6 months when there are known risk factors for ID. Given the current demonstration of persistent metabolic effects if ID is not corrected by iron treatment, earlier assessment for anaemia is advisable. The observed alterations were associated with energy, monoamine, and polyamine metabolism, and several effects were likely attributable to changes in the gut microbiota. Nevertheless, the benefits of iron treatment still have to be weighed against the possibility of iron-induced perturbations of the gut bacteriome, which highlights the continued importance of optimizing dosing regimens and limiting the duration of treatment.
Supplementary Material
Acknowledgements
Sources of support:
This research was supported by grants from the National Institute of Child Health and Development (HD080201, HD089989), and an award from the Bill and Melinda Gates Foundation.
Footnotes
The authors declare no conflicts of interest.
References
- [1].Camaschella C, Review Series IRON METABOLISM AND ITS DISORDERS Iron Deficiency, 2019.
- [2].Wang CY, Babitt JL, Blood 2019, 133, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Knight LC, Dilger RN, Nutrients 2018, 10, DOI 10.3390/nu10050632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Coe CL, Lubach GR, Busbridge M, Chapman RS, Res. Vet. Sci 2013, 94, 549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lubach GR, Coe CL, J. Nutr 2006, 136, 2345–2349. [DOI] [PubMed] [Google Scholar]
- [6].Sandri BJ, Lubach GR, Lock EF, Georgieff MK, Kling PJ, Coe CL, Rao RB, J. Nutr 2019, DOI 10.1093/jn/nxz274. [DOI] [PMC free article] [PubMed]
- [7].Rao R, Ennis K, Lubach GR, Lock EF, Georgieff MK, Coe CL, Nutr. Neurosci 2018, 21, 40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pagani A, Nai A, Silvestri L, Camaschella C, Front. Physiol 2019, 10, DOI 10.3389/fphys.2019.01294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yilmaz B, Li H, Pharmaceuticals 2018, 11, DOI 10.3390/ph11040098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Deschemin J-C, Noordine M-L, Remot A, Willemetz A, Afif C, Canonne-Hergaux F, Langella P, Karim Z, Vaulont S, Thomas M, Nicolas G, FASEB J 2016, 30, 252–261. [DOI] [PubMed] [Google Scholar]
- [11].Jaeggi T, Kortman GAM, Moretti D, Chassard C, Holding P, Dostal A, Boekhorst J, Timmerman HM, Swinkels DW, Tjalsma H, Njenga J, Mwangi A, Kvalsvig J, Lacroix C, Zimmermann MB, Gut 2015, 64, 731–742. [DOI] [PubMed] [Google Scholar]
- [12].Paganini D, Uyoga MA, Kortman GAM, Cercamondi CI, Moretti D, Barth-Jaeggi T, Schwab C, Boekhorst J, Timmerman HM, Lacroix C, Karanja S, Zimmermann MB, Gut 2017, 66, 1956–1967. [DOI] [PubMed] [Google Scholar]
- [13].Council NR, Nutrient Requirements of Nonhuman Primates: Second Revised Edition, The National Academies Press, Washington, DC: 2003. [Google Scholar]
- [14].Posma JM, Robinette SL, Holmes E, Nicholson JK, Bioinformatics 2014, 30, 893–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kanehisa M, Nucleic Acids Res 2000, 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Oexle H, Gnaiger E, Weiss G, Biochim. Biophys. Acta - Bioenerg 1999, 1413, 99–107. [DOI] [PubMed] [Google Scholar]
- [17].Ceyhan M, Ozalp I, Altay C, Clin. Pediatr. (Phila) 1988, 27, 206–9. [DOI] [PubMed] [Google Scholar]
- [18].Kroe D, Kinney TD, Kaufman N, Klavins JV, Blood 1963, 21, 546–552. [PubMed] [Google Scholar]
- [19].Faelli A, Esposito G, Biochem. Pharmacol 1970, 19, 2551–2554. [DOI] [PubMed] [Google Scholar]
- [20].Oran M, Aydin M, Mete R, Tülübaş F, Avci O, Yilmaz A, Gürel A, Acta Medica Mediterr 2014, 30, 279–283. [Google Scholar]
- [21].Paganini D, Jaeggi T, Cercamondi C, Kujinga P, Moretti D, Zimmermann M, FASEB J 2016.
- [22].Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Björck I, Bäckhed F, Cell Metab 2015, 22, 971–982. [DOI] [PubMed] [Google Scholar]
- [23].Choo JM, Cheung JK, Wisniewski JA, Steer DL, Bulach DM, Hiscox TJ, Chakravorty A, Smith AI, Gell DA, Rood JI, Awad MM, PLoS One 2016, 11, e0162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Awad MM, Cheung JK, Tan JE, McEwan AG, Lyras D, Rood JI, Anaerobe 2016, 41, 10–17. [DOI] [PubMed] [Google Scholar]
- [25].Kortman GAM, Boleij A, Swinkels DW, Tjalsma H, PLoS One 2012, 7, e29968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kortman GAM, Raffatellu M, Swinkels DW, Tjalsma H, FEMS Microbiol. Rev 2014, 38, 1202–1234. [DOI] [PubMed] [Google Scholar]
- [27].Shaw W, Nutr. Neurosci 2010, 13, 135–143. [DOI] [PubMed] [Google Scholar]
- [28].Southan C, DeWolf WE, Kruse LI, Biochim. Biophys. Acta (BBA)/Protein Struct. Mol 1990, 1037, 256–258. [DOI] [PubMed] [Google Scholar]
- [29].Coe CL, Lubach GR, Bianco L, Beard JL, Dev. Psychobiol 2009, 51, 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hamano H, Ikeda Y, Watanabe H, Horinouchi Y, Izawa-Ishizawa Y, Imanishi M, Zamami Y, Takechi K, Miyamoto L, Ishizawa K, Tsuchiya K, Tamaki T, Nephrol. Dial. Transplant 2018, 33, 586–597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


